Abstract
Previously, we have shown that human DNA polymerase (Pol) η has two functional PCNA binding motifs, PIP1 and PIP2, and that a C-terminal deletion of Polη that lacks the ubiquitin-binding UBZ domain and the PIP2 domain but retains the PIP1 domain promotes normal levels of translesion synthesis (TLS) opposite a cis-syn TT dimer in human cells. Here, we identify two PIP domains in Polκ, and show that TLS occurs normally in human fibroblast cells in which the pip1 or pip2 mutant Polκ is expressed, but mutational inactivation of both PIP domains renders Polκ non-functional in TLS opposite the thymine glycol lesion. Thus, the two PIP domains of Polκ function redundantly in TLS opposite this DNA lesion in human cells. However, and surprisingly, whereas mutational inactivation of the PIP1 domain completely inhibits the stimulation of DNA synthesis by Polκ in the presence of PCNA, RFC, and RPA, mutations in PIP2 have no adverse effect on PCNA-dependent DNA synthesis. This raises the possibility that activation of Polκ PIP2 as a PCNA binding domain occurs during TLS in human cells, and that protein-protein interactions and post transcriptional modifications are involved in such activation.
Keywords: PCNA binding, DNA polymerase κ, translesion synthesis, thymine glycol
Introduction
Humans have four Y-family DNA polymerases (Pols) - η, ι, Rev1, and κ. Structure and function studies with these Pols have indicated that they promote replication through DNA lesions in a highly specialized manner. For example, Polη has the unique ability to replicate through UV induced cyclobutane pyrimidine dimers (CPDs) because it can accommodate the CPD in its active site (Biertumpfel et al. 2010; Johnson et al. 1999a; Johnson et al. 1999b; Masutani et al. 1999; Silverstein et al. 2010). Polι synthesizes DNA opposite template purines using Hoogsteen base pairing (Nair et al. 2005a; Nair et al. 2004; Nair et al. 2006b). The ability of Polι to push the template purine into the syn conformation provides a mechanism for incorporating the correct nucleotide (nt) opposite adducts that impair the Watson-Crick edge but not the Hoogsteen edge of the template purine (Nair et al. 2006a). Rev1 DNA pol is highly specialized for C incorporation opposite template G for which it uses an arginine residue for pairing with C (Nair et al. 2005b; Swan et al. 2009). Because of its ability to evict the N2-adducted template guanine from the DNA helix into a large solvent-filled cavity, Rev1 could incorporate a C opposite bulky N2-adducted guanines such as N2-dG BPDE. Although Polκ can function at the nt insertion step opposite certain DNA lesions (Yoon et al. 2010), it is particularly well-adapted to performing the extension step of TLS opposite minor groove DNA lesions such as N2-dG BPDE (Lone et al. 2007).
Pols η, ι, and κ have been shown to interact physically and functionally with PCNA (Haracska et al. 2001a; Haracska et al. 2001b; Haracska et al. 2002). PCNA, which has been loaded onto DNA by RFC in the presence RPA, stimulates DNA synthesis by these Pols on undamaged and damaged DNAs. PCNA binding does not increase their processivity for DNA synthesis but enhances the catalytic efficiency (kcat/Km) of nt incorporation. Rad6-Rad18 mediated PCNA ubiquitylation at its lysine 164 residue plays a crucial role in the targeting of translesion synthesis (TLS) Pols to PCNA (Haracska et al. 2004; Hoege et al. 2002; Stelter & Ulrich 2003), but how this PCNA modification regulates the TLS process has remained unclear.
PCNA binding PIP domains have been previously identified in yeast and human Polη, and both harbor a PIP domain in the C-terminus (Haracska et al. 2001a; Haracska et al. 2001c). However, whereas mutational inactivation of the PIP domain in yeast Polη causes a complete loss of its ability to physically and functionally interact with PCNA (Haracska et al. 2001c), mutational inactivation of the human Polη (hPolη) C-terminal PIP domain does not confer a complete defect in its ability to physically and functionally interact with PCNA (Acharya et al. 2008). The residual PCNA binding in this hPolη derives from an additional PIP domain present between residues 437 and 444 just C-terminal to the PAD region of its polymerase domain. We have designated this Polη PIP as PIP1, and the C-terminal PIP as PIP2 (Acharya et al. 2008). Biochemical and cellular studies have indicated a redundant role of these PIP domains, as mutational inactivation of both PIP domains completely abrogates the PCNA-dependent stimulation of DNA synthesis by hPolη and increases the UV sensitivity of cells similar to that seen in XPV cells which lack functional Polη (Acharya et al. 2008). And, hPolη harboring mutations in both the PIP1 and PIP2 domains fail to co-localize with PCNA in UV irradiated human fibroblast cells (Acharya et al. 2008).
Here, we identify two PIP domains in human Polκ and show that these PIP domains function redundantly in promoting TLS by Polκ in human cells. However, the two PIP domains differ in their effects on stimulation of PCNA-dependent DNA synthesis by Polκ in vitro. We discuss the possible implications of these two PIP domains in Polκ for TLS, and raise the possibility that the two PIP domains that have been previously identified in human Polι could also be functionally relevant for its role in TLS in human cells (Haracska et al. 2005; Vidal et al. 2004).
Results
PCNA binding motifs in DNA Pols η, ι, and κ
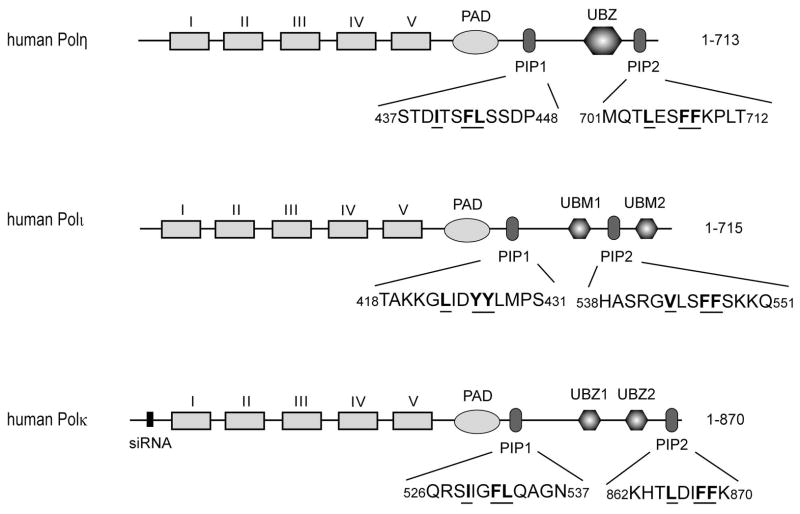
Although human Pols η, ι, and κ, each harbor two potential PCNA binding PIP motifs (Fig. 1), a role for both the PIP motifs has been demonstrated only for hPolη. Mutational inactivation of both PIP1 and PIP2 renders Polη completely defective for PCNA binding in vitro as well as in complementing the UV sensitivity of XPV cells (Acharya et al. 2008). Furthermore, our observation that human Polη (1-475) protein that lacks the C-terminus containing the ubiquitin-binding UBZ domain and the PIP2 domain carries out proficient TLS opposite a cis-syn TT dimer in vivo has provided further evidence that PCNA binding by PIP1 is sufficient for Polη’s ability to function in TLS opposite CPDs in human cells (Acharya et al. 2010). Although two potential PCNA binding motifs are present in Polι, biochemical studies have indicated that only PIP1 is required for PCNA binding and that Polι harboring mutations in PIP1 lacks the ability to carry out PCNA-dependent DNA synthesis in vitro (Haracska et al. 2005; Vidal et al. 2004). However, since the role of PIP1 and PIP2 in Polι has not been examined in TLS in human cells, it is not known whether only PIP1 or both PIP1 and PIP2 can function in TLS in human cells. Two potential PCNA binding motifs, PIP1 and PIP2, are present in Polκ, and they resemble the two PIP motifs in Polη in their location relative to the PAD and UBZ domains, and in harboring the same conserved hydrophobic residues (Fig. 1).
Figure 1.
PCNA binding motifs in human Pols η, ι, and κ. Pols η, ι, and κ each contain two PCNA binding PIP domains. The highly conserved hydrophobic residues in the PIP domains are indicated in bold and underlined. Regions I – V and PAD indicate the positions of these polymerase domains, and the ubiquitin binding UBZ or UBM domains are indicated. The N-terminal region where the sequence of Polκ was changed to siRNA resistant form is indicated by a narrow black rectangle.
Redundant roles of PIP1 and PIP2 in Polκ-mediated TLS opposite thymine glycol (Tg) in human cells
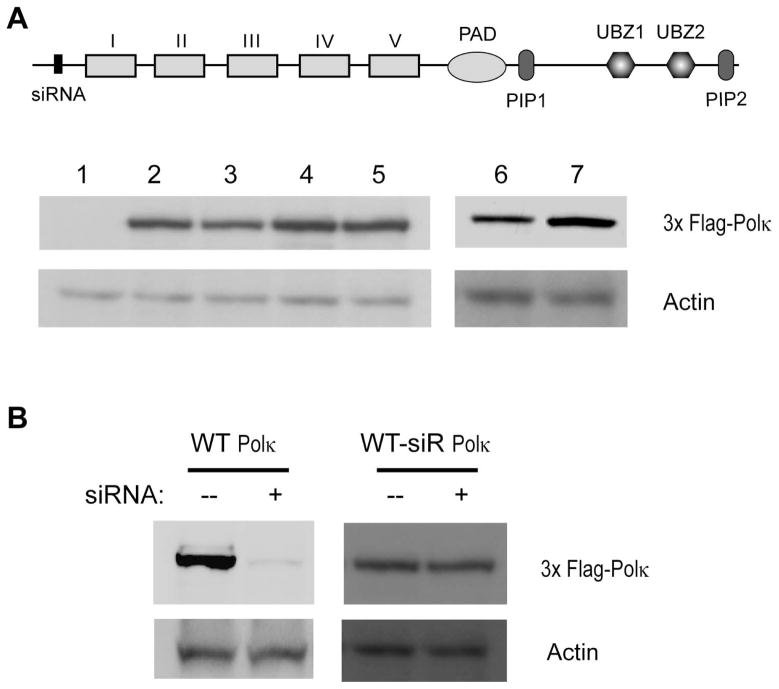
To determine the role of PIP1 and PIP2 in Polκ mediated TLS, we generated a C-terminal deletion of Polκ (1-856) which lacks the last 14 amino acid residues that harbor the PIP2 domain. Additionally, we constructed alanine mutations in the conserved hydrophobic residues F868 and F869 of PIP2 and F532 and L533 in PIP1 (Fig. 1). The siRNA resistant full-length wild type Polκ protein (1-870), C-terminally truncated Polκ (1-856) lacking PIP2, and Polκ PIP mutant proteins, harboring the F868A, F869A mutations in PIP2, F532A, L533A mutations in PIP1, or the F532A, L533A and F868A, F869A mutations that would inactivate both PIP domains, were expressed in human fibroblast (HF) cells (Fig. 2) and their effects on TLS opposite the thymine glycol (Tg) lesion carried on the leading strand template of SV40-based plasmid were examined. We have previously shown that TLS mediated by the consecutive action of Polκ and Polζ at the nucleotide insertion and the subsequent extension step, respectively, promotes an error-free mode of replication through the Tg lesion (Yoon et al. 2010). As shown in Table 1, in human fibroblast (HF) cells treated with control (NC) siRNA, TLS opposite the Tg lesion occurs with a frequency of ~25%, and in cells treated with Polκ siRNA, the TLS frequency is reduced to ~14%. We next examined the effects of Polκ pip mutant proteins expressed in cells from which genomic Polκ had been depleted by siRNA treatment (Fig. 2). As expected, the frequency of TLS is reduced to ~15% in cells from which genomic Polκ has been depleted and which carry the vector plasmid with no Polκ, whereas wild type levels of TLS are restored in cells carrying the plasmid which expresses full-length wild type Polκ. Interestingly, in cells expressing C-terminally truncated (1-856) Polκ, or pip1 or pip2 mutant Polκ, TLS occurs at normal levels. Thus, the inactivation of either PIP domain does not adversely affect the ability of Polκ to function in TLS in human cells; however, in cells expressing Polκ protein in which both PIP1 and PIP2 have been mutationally altered, or which express the C-terminally deleted (1-856) Polκ protein lacking PIP2 and mutationally altered PIP1, TLS is reduced to the same level as that seen upon Polκ depletion. These observations indicate that Polκ can use PIP1 or PIP2 for binding PCNA and that either of these PIP domains is sufficient for TLS opposite the Tg lesion in human cells.
Figure 2.
Expression of wild type and mutant Polκ in normal HF
(A) Western blot analysis of Polκ expression in normal HF cells. Stably expressing 3X Flag-human Polκ was detected by anti-Flag antibody. Actin is used as a loading control. Lane 1, vector control; lane 2, wild type Polκ with siRNA resistant sequence; lane 3, Polκ containing mutations in the PIP1 domain and siRNA resistant sequence; lane 4, Polκ containing mutations in the PIP2 domain and siRNA resistant sequence; lane 5, Polκ containing mutations in the PIP1 and PIP2 domains and siRNA resistant sequence; lane 6, wild type Polκ with siRNA resistant sequence; lane 7, C-terminal deletion of Polκ (1-856) with siRNA resistant sequence. The siRNA target site (siRNA), the polymerase domains, PIP1, PIP2, and UBZ domains are shown in full length (1-870) Polκ. (B) siRNA depletion of 3X Flag-human Polκ in human cells. Cells expressing 3X Flag-wild type or 3X Flag-siRNA resistant mutant Polκ are treated with Polκ siRNA for 48 h. siRNA depletion is determined by Western blotting with anti-Flag antibody. Actin is used as a loading control. siR, siRNA resistant.
Table 1.
Effects of mutations in PCNA binding domains PIP1 and PIP2 in Polκ on TLS opposite the 5R,6S Tg lesion carried on the leading strand template of SV-40 based plasmid in human fibroblast (HF) cells
| siRNA | Vector expressing | # of Kan+ colonies | # of blue colonies among Kan+ | TLS (%) |
|---|---|---|---|---|
| NC | --- | 462 | 114 | 24.7 |
| Polκ | --- | 384 | 52 | 13.5 |
| Polκ | No Polκ | 276 | 42 | 15.2 |
| Polκ | WT Polκ (1-870) | 168 | 47 | 28.0 |
| Polκ | C-terminal Δ of Polκ (1-856) | 154 | 42 | 27.3 |
| Polκ | Polκ pip1 mutation (F532A, L533A) | 136 | 35 | 25.7 |
| Polκ | Polκ pip2 mutation (F868A, F869A) | 112 | 30 | 26.8 |
| Polκ | Polκ pip1 and pip2 mutations (F532A, L533A, F868A, F869A) | 206 | 25 | 12.1 |
| Polκ | Polκ pip1 mutation (F532A, L533A) with C-terminal Δ (1-856) | 148 | 23 | 15.5 |
Requirement of PIP1, but not PIP2, for PCNA stimulated DNA synthesis by Polκ
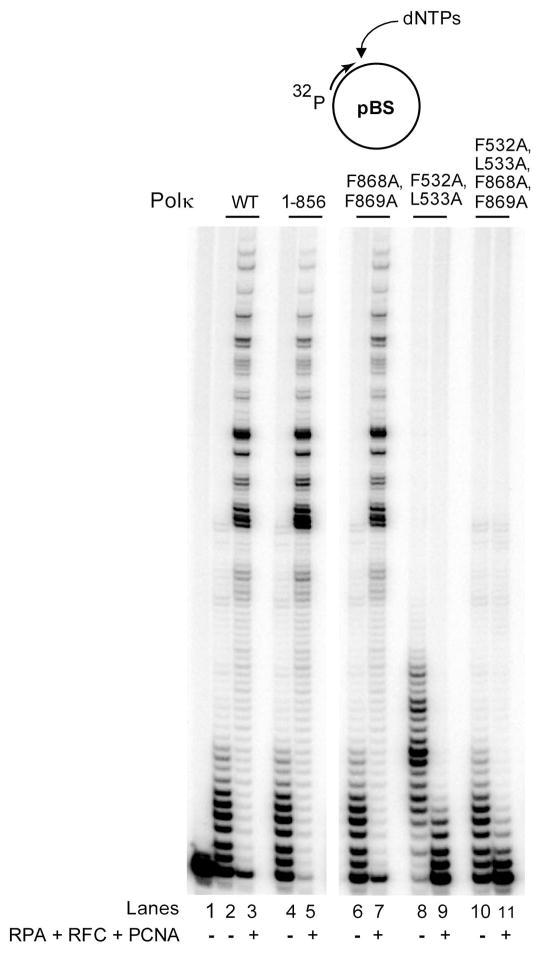
Next, we examined the effects of mutations in the PIP1 and PIP2 domains on PCNA stimulated DNA synthesis by Polκ in vitro. For this purpose, we examined DNA synthesis by Polκ in the presence of PCNA, RFC, and RPA on single stranded pBS circular DNA primed with a 5′-32P labeled oligomer at a unique site. As we have shown previously (Haracska et al. 2002), the DNA synthetic activity of Polκ is enhanced in the presence of PCNA, RFC, and RPA (Fig. 3, compare lanes 2 and 3). An enhancement of PCNA dependent DNA synthesis also occurs with C-terminally deleted Polκ (1-856) protein in which the entire PIP2 domain is absent, and a similar enhancement of DNA synthesis with PCNA is observed with the Polκ protein harboring the F868A, F869A mutations in PIP2 (Fig. 3, lanes 5 and 7). But, PCNA stimulated DNA synthesis by Polκ is completely inhibited by the F532A, L533A mutations in PIP1, and as expected by mutations in both the PIP1 and PIP2 domains (Fig. 3, lanes 9 and 11).
Figure 3.
Requirement of PIP1 domain of Polκ for stimulation of its DNA synthetic activity by PCNA. The complete standard reaction mixture contained 1 nM of Polκ, with PCNA (100 ng), RFC (50 ng), RPA (200 ng), a mix of all four dNTPs (each at 100 μM), 500 μM ATP and DNA substrate (10 nM). PCNA, RFC, and RPA were added (+) or not added (−) to the reaction mixture as indicated.
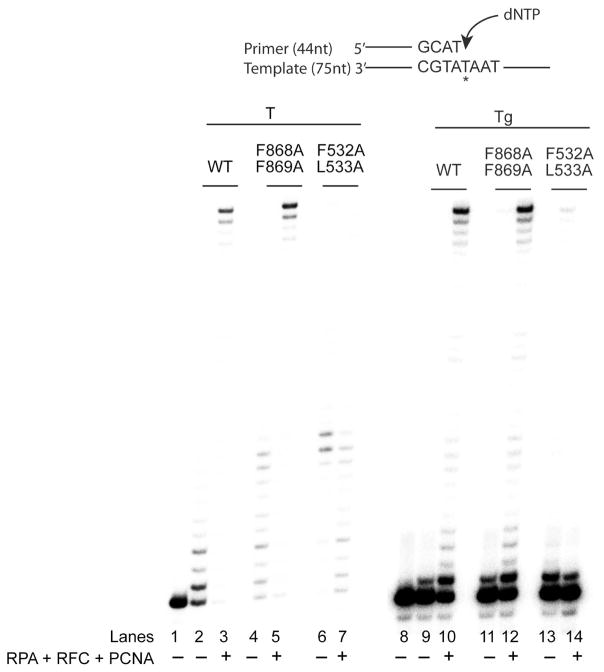
To determine if both PIP domains become functional opposite a Tg lesion, we examined the effects of mutations in PIP1 and PIP2 on PCNA stimulated DNA synthesis by Polκ on Tg containing DNA. For this purpose, a 75 nucleotide template containing an undamaged T or a Tg lesion was hybridized with a 44 nucleotide primer, and DNA synthesis by Polκ carrying mutations in PIP1 or PIP2 was examined in the presence of PCNA, RFC, and RPA. As shown in Fig. 4, on undamaged DNA, mutations in the PIP2 domain had no adverse effect on PCNA-stimulated DNA synthesis (lanes 4 and 5), whereas mutations in the PIP1 domain were inhibitory to PCNA-stimulated DNA synthesis (lanes 6 and 7). Compared to DNA synthesis on undamaged DNA, a Tg lesion presented a strong block to DNA synthesis by wild type Polκ. In the absence of PCNA, Polκ inserted a nucleotide (nt) opposite the Tg lesion but was inhibited at further extension (Fig. 4, lane 9). In the presence of PCNA, RFC, and RPA, Polκ shows an enhanced proficiency for inserting a nt opposite the Tg lesion; moreover, it could extend from the inserted nt, but the extension step was very considerably blocked. (Fig. 4, lane 10). Mutations in the PIP2 domain did not affect PCNA-stimulated DNA synthesis by Polκ (Fig. 4, compare lanes 11 and 12 with lanes 9 and 10); by contrast, PCNA-stimulated DNA synthesis opposite the Tg lesion was inhibited by mutations in the PIP1 domain (Fig. 4, lanes 13 and 14).
Figure 4.
Requirement of PIP1 for PCNA-stimulated DNA synthesis by Polκ opposite a Tg lesion. DNA synthesis by Polκ was examined on linear DNA substrates containing an undamaged T (lanes 1–7) or a Tg lesion in the template strand (lanes 8–14) in the presence (+) or absence (−) of PCNA, RFC, and RPA. DNA synthesis reactions were carried out as described in the Figure 3 legend and in Experimental Procedures. Because a Tg lesion presents a strong block to synthesis by Polκ at the extension step, the contrast in lanes 8–14 has been enhanced to clearly show that mutations in the PIP1 domain inactivate PCNA-stimulated DNA synthesis by Polκ opposite the Tg lesion. T with an asterisk indicates the position of an undamaged T or a Tg lesion in the template strand.
Although both on undamaged and Tg-containing DNAs, the mutational inactivation of PIP1 but not PIP2, is inhibitory to PCNA-stimulated DNA synthesis by Polκ, the extent of PCNA-dependent stimulation of synthesis by Polκ is much greater on undamaged DNA than on damaged DNA. That is because following the insertion of A opposite the Tg lesion, Polκ is strongly inhibited at extending from it (Fischhaber et al. 2002), and our genetic analyses have indicated that relication through the Tg lesion in human cells occurs via the sequential action of Polκ and Polζ, in which following nt insertion by Polκ, Polζ would extend (Yoon et al. 2010)
Discussion
Even though human TLS Pols η, κ, and ι each harbor two PCNA binding PIP motifs, they differ in their use of these two motifs for stimulation of PCNA-dependent DNA synthesis and for promoting lesion bypass in human cells. Polη uses PIP1 or PIP2 for binding PCNA and for replicating through UV induced DNA lesions (Acharya et al. 2008). Particularly striking in this regard is the observation that Polη (1-475) protein, where the C-terminal portion containing the ubiquitin binding UBZ domain and the PIP2 domain has been deleted, but in which the PIP1 domain is retained, functions normally in promoting replication through a cis-syn TT dimer in human cells (Acharya et al. 2010). By contrast, even though Polκ has two PIP domains which resemble the PIP1 and PIP2 domains of Polη, the Polκ PIP domains affect TLS opposite the Tg lesion in human cells differently than that seen for the effects of the Polη PIP domains opposite the UV lesion. Since inactivation of neither PIP domain impairs the ability of Polκ to carry out TLS opposite Tg in human cells, either of the Polκ PIP domains, PIP1 or PIP2, is sufficient for Polκ’s ability to function in TLS opposite this DNA lesion. However, and rather quite surprisingly, only the inactivation of the PIP1 domain affects DNA synthesis by Polκ in the presence of PCNA, RFC, and RPA, whereas mutations in PIP2 have no adverse effect on PCNA-stimulated DNA synthesis by Polκ.
How does one reconcile the different effects of PIP1 and PIP2 domains on Polκ function in TLS in humans cells vs. that inferred from their effects on PCNA-dependent DNA synthesis in vitro? Although the observation that only PIP1 is required for PCNA-dependent DNA synthesis in vitro may imply that only PIP1 affects the ability to Polκ to function with PCNA, the observation that the ability of Polκ to function in TLS opposite the Tg lesion is impaired only when both the PIP domains have been mutationally altered, implies that both PIP1 and PIP2 can function in TLS opposite Tg in human cells equally well. That raises the possibility that the activation of PIP2 as a functional PCNA binding domain occurs in human cells only when Polκ function is required for TLS opposite DNA lesions.
As determined by surface plasmon resonance (SPR), the Polη 20-mer (694-713) C-terminal peptide containing the PIP2 domain binds PCNA with an estimated Kd of 0.40 μM (Hishiki et al. 2009). However, even though the Polκ PIP2 domain shares considerable sequence similarity in the conserved hydrophobic residues with Polη PIP2, the C-terminal 20-mer Polκ (851-870) peptide or the 15-mer Polκ (856-870) peptide containing the PIP2 domain showed no signal for PCNA binding by SPR, unless the four C-terminal residues PLTH of Polη were added to the Polκ C-terminus. The Polκ (856-870) peptide with the added PLTH residues bound PCNA with a Kd of ~ 4.9 μM (Hishiki et al. 2009). Because deletion of the C-terminal PLTH residues from the Polη PIP2 domain greatly reduces its affinity for PCNA, this sequence is important for Polη’s ability to bind PCNA via PIP2 (Hishiki et al. 2009). But, since the Polκ PIP2 lacks the PLTH residues present in Polη PIP2, how does Polκ PIP2 become functional in PCNA binding? One possibility is that post-translational modifications, such as phosphorylations, enhance the ability of Polκ PIP2 to bind PCNA; in addition, protein-protein interactions that promote Polκ function in TLS in human cells could also affect the proficiency of Polκ PIP2 for PCNA binding. The possibility that Polκ PIP2 is a weak PCNA binding domain and that its potential for PCNA binding is enhanced by the binding of UBZ1 and/or UBZ2 domains of Polκ to the lysine 164-linked ubiquitin moiety on PCNA, seems unlikely, as our studies with yeast and human TLS polymerases have yielded no evidence for such a role of ubiquitin binding domains in TLS Pols (Acharya et al. 2007; Acharya et al. 2008; Acharya et al. 2010; Haracska et al. 2006). Another possibility that Polκ PIP2 has a PCNA-independent function in vivo is also rendered unlikely from the observation that PIP1 and PIP2 play redundant roles in TLS opposite the Tg lesion in human cells. Since either the PIP1 and PIP2 domain in Polκ can be used for TLS opposite the Tg lesion in human cells, and since TLS does not occur upon mutational inactivation of both PIP domains, we surmise that both the PIP domains provide functional PCNA binding sites in Polκ.
Similar to that in Polη and Polκ, Polι also has two potential PCNA binding domains; however, biochemical studies have indicated that only PIP1 functions in PCNA-stimulated DNA synthesis by Polι, as mutational inactivation of only PIP1 and not of PIP2 confers impaired DNA synthesis with PCNA, RFC, and RPA (Haracska et al. 2005; Vidal et al. 2004). However, the roles of Polι PIP1 and PIP2 domains have not been examined for TLS in human cells, and it may turn out that similar to that for Polκ PIP domains, the two PIP domains of Polι are also functional for TLS in human cells.
Experimental procedures
Generation of Polκ mutations
All the Polκ constructs were made siRNA resistant by changing the wild type Polκ sequence, 5′-GCTCAAATCACCAGCCAAC-3′ to an siRNA resistant sequence, 5′-GCACAGATAACTAGTCAAC-3′. The C-terminal truncation that contained residues (1-856) was generated by amplifying this segment of the Polκ ORF by PCR. Mutations in the PIP1 and PIP2 domains of Polκ were generated by the site directed mutagenesis kit (Invitrogen) with primers containing the F532A, L533A mutations in PIP1 or the F868A, F869A mutations in PIP2. All the constructs were verified by automated DNA sequencing.
Stable expression of wild type and mutant Polκ in normal HF cells
Wild type and mutant Polκ genes were subcloned into pCMV7-3xFlag-zeo vector (Sigma). The vectors were transfected into normal HF (MRC5) cells by Lipofectamine 2000 reagent (Invitrogen). After 24h incubation, 0.5 μg/ml of Zeocin (Invitrogen) was added to the culture media. After 3 days of incubation, cells were washed with PBS buffer and were continuously cultured with the media containing 250 ng/ml of Zeocin for ~ 2 weeks. For checking protein expression by Western analysis, cells were washed with PBS buffer and lysed with RIPA buffer (1x PBS, 1% IP-40, 0.5% sodium deoxycholate, 0.1% SDS). After 1h incubation on ice, the cellular mixture was centrifuged and the supernatant was collected. Equivalent amounts (approximately 30 μg) of prepared cellular extracts were separated on a 10% SDS-polyacrylamide gel and transferred to a PVDF membrane (Bio-rad). The membranes were probed with affinity purified rabbit polyclonal antibodies against Polκ followed by secondary antibodies conjugated with horseradish peroxidase. The signals were detected using ECL-Plus (Amersham).
In vivo translesion synthesis assays in normal HF cells
The duplex plasmid that carries thymine glycol on the leading strand template has been described previously (Yoon et al. 2010). Stably transfected HF cells were plated in 6-well plate with 80% confluence. After 24h incubation, the duplex plasmid DNA (2 μg) carrying the Tg lesion on the leading strand template was transfected with Lipofectamine 2000 (Invitrogen). After 30 h incubation, plasmid DNA was rescued from cells by the alkaline lysis method and digested with DpnI to remove unreplicated plasmid DNA. The plasmid DNA was then transformed into E. coli XL1-Blue super competent cells (Stratagene). Transformed bacterial cells were diluted in 1mL SOC media and plated on both LB/amp (50 μg/mL ampicillin, Sigma) and LB/kan (25 μg/mL kanamycin, Sigma) plates containing 1 μM isopropyl-1-thio-β-D-galactopyranoside (IPTG) (Roche) and 100 μg/mL of X-Gal (Roche). After 16 h incubation at 37°C, blue and white colonies were counted from kanamycin plates. The actual TLS frequency was determined from the number of blue colonies out of total colonies growing on LB/kan plates. Details of these methods have been described previously (Yoon et al. 2009).
Proteins
WT and mutant Polκ proteins fused to GST were purified as described, and the GST portion was removed by treatment with PreScission protease (Johnson et al. 2006).
DNA Polymerase assays
The single stranded circular DNA (pBluescript, 3KB) annealed to a 41 nt 5′-P32 labelled primer, which is complementary to the F1 origin of plasmid (5′-CCC CCG ATT TAG AGC TTG ACG GGG AAA CCG GCG AAC GTG GC-3′), was used as a substrate for the DNA polymerase assay. The standard DNA Pol reaction mixture contained 1 nM Polκ, 10 nM DNA substrate, 40 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 150 mM NaCl, 1 mM DTT, 100 μg of BSA/ml, 500 μM ATP, and 100 μM each dGTP, dATP, dTTP, and dCTP. The reaction was carried out in the absence or presence of PCNA (100 ng), RFC (50 ng), and RPA (200 ng) at 37 °C for 10 min after the addition of WT or mutant Polκ (1 nM) protein to the reaction mixture. The reaction was stopped by the addition of loading buffer (40 μL) containing EDTA (20 nM), 95% formamide, 0.3% bromophenol blue, and 0.3% cyanol blue. The reaction products were resolved on 10% polyacrylamide gels containing 8 M urea. The reaction products were visualized with a Molecular Dynamics STORM PhosphorImager.
For examining the effects of mutations in the PIP domains on the proficiency of DNA synthesis by Polκ opposite a Tg lesion, linear DNA substrates were generated in which a 75 nt template 5′-AGCAAGTCACCAATGTCTAAGAGTTCGTATXATGCCTACACTGGAGTACCGGAGCATCGTCGTGACTGGGAAAAC-3′, containing a T or a Tg lesion at the position indicated by X, was bound to biotin at both ends, and after annealing to the 44 nt 5′-32P labelled oligonucleotide 5′-GTTTTCCCAGTCACGACGATGCTCCGGTACTCCAGTGTAGGCAT-3′ primer, both of these biotins were coupled to streptavidin. The DNA synthesis reactions using undamaged and Tg-containing linear DNA substrates were carried out as described above for assays with circular DNA.
Acknowledgments
This work was supported by National Institutes of Health Grants ES012411 and ES02833.
References
- Acharya N, Brahma A, Haracska L, Prakash L, Prakash S. Mutations in the ubiquitin binding UBZ motif of DNA polymeraseη do not impair its function in translesion synthesis during replication. Mol Cell Biol. 2007;27:7266–7272. doi: 10.1128/MCB.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Yoon JH, Gali H, et al. Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase η in translesion DNA synthesis. Proc Natl Acad Sci USA. 2008;105:17724–17729. doi: 10.1073/pnas.0809844105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya N, Yoon JH, Hurwitz J, Prakash L, Prakash S. DNA polymerase η lacking the ubiquitin-binding domain promotes replicative lesion bypass in human cells. Proc Natl Acad Sci USA. 2010;107:10401–10405. doi: 10.1073/pnas.1005492107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biertumpfel C, Zhao Y, Kondo Y, et al. Structure and mechanism of human DNA polymerase η. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. Human DNA polymerase κ bypasses and extends beyond thymine glycols during translesion synthesis in vitro, preferentially incorporating correct nucleotides. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- Haracska L, Acharya N, Unk I, et al. A single domain in human DNA polymerase ι mediates interaction with PCNA: implications for translesion DNA synthesis. Mol Cell Biol. 2005;25:1183–1190. doi: 10.1128/MCB.25.3.1183-1190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, et al. Physical and functional interactions of human DNA polymerase η with PCNA. Mol Cell Biol. 2001a;21:7199–7206. doi: 10.1128/MCB.21.21.7199-7206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Johnson RE, Unk I, et al. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc Natl Acad Sci USA. 2001b;98:14256–14261. doi: 10.1073/pnas.261560798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Kondratick CM, Unk I, Prakash S, Prakash L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol Cell. 2001c;8:407–415. doi: 10.1016/s1097-2765(01)00319-7. [DOI] [PubMed] [Google Scholar]
- Haracska L, Torres-Ramos CA, Johnson RE, Prakash S, Prakash L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in. Saccharomyces cerevisiae Mol Cell Biol. 2004;24:4267–4274. doi: 10.1128/MCB.24.10.4267-4274.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Unk I, Johnson RE, et al. Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol Cell Biol. 2002;22:784–791. doi: 10.1128/MCB.22.3.784-791.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Unk I, Prakash L, Prakash S. Ubiquitylation of yeast proliferating cell nuclear antigen and its implications for translesion DNA synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:6477–6482. doi: 10.1073/pnas.0510924103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishiki A, Hashimoto H, Hanafusa T, et al. Structural basis for novel interactions between human translesion synthesis polymerases and proliferating cell nuclear antigen. J Biol Chem. 2009;284:10552–10560. doi: 10.1074/jbc.M809745200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999a;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. Yeast and human translesion DNA synthesis polymerases: expression, purification, and biochemical characterization. Methods Enzymol. 2006;408:390–407. doi: 10.1016/S0076-6879(06)08024-4. [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science. 1999b;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- Lone S, Townson SA, Uljon SN, et al. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol Cell. 2007;25:601–614. doi: 10.1016/j.molcel.2007.01.018. [DOI] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase ι incorporates dCTP opposite template G via a G.C+ Hoogsteen base pair. Structure. 2005a;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science. 2005b;309:2219–2222. doi: 10.1126/science.1116336. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase iota. Nat Struct Mol Biol. 2006a;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. Replication by human DNA polymerase ι occurs via Hoogsteen base-pairing. Nature. 2004;430:377–380. doi: 10.1038/nature02692. [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash S, Prakash L, Aggarwal AK. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-ι active site. Structure. 2006b;14:749–755. doi: 10.1016/j.str.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase eta. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structure of the human REV1-DNA-dNTP ternary complex. J Mol Biol. 2009;390:699–709. doi: 10.1016/j.jmb.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal AE, Kannouche P, Podust VN, Yang W, Lehmann AR, Woodgate R. Proliferating cell nuclear antigen-dependent coordination of the biological functions of human DNA polymerase ι. J Biol Chem. 2004;279:48360–48368. doi: 10.1074/jbc.M406511200. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Bhatia G, Prakash S, Prakash L. Error-free replicative bypass of thymine glycol by the combined action of DNA polymerases κ and ζ in human cells. Proc Natl Acad Sci USA. 2010;107:14116–14122. doi: 10.1073/pnas.1007795107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Prakash L, Prakash S. Highly error-free role of DNA polymerase η in the replicative bypass of UV induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci USA. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]