Abstract
Background
Growing evidence suggests that small ubiquitin-like modifier (SUMO) conjugation plays a key role in brain plasticity by modulating activity-dependent synaptic transmission. However, these observations are based largely on cell culture experiments. We hypothesized that episodic and fear memories would be affected by silencing SUMO1–3 expression.
Methods
To investigate the role of SUMO conjugation in neuronal functioning in vivo, we generated a novel Sumo transgenic mouse model in which a Thy1 promoter drives expression of 3 distinct microRNAs to silence Sumo1–3 expression, specifically in neurons. Wild-type and Sumo1–3 knockdown mice were subjected to a battery of behavioural tests to elucidate whether Sumoylation is involved in episodic and emotional memory.
Results
Expression of Sumo1–3 microRNAs and the corresponding silencing of Sumo expression were particularly pronounced in hippocampal, amygdala and layer V cerebral cortex neurons. The Sumo knockdown mice displayed anxiety-like responses and were impaired in episodic memory processes, contextual and cued fear conditioning and fear-potentiated startle.
Limitations
Since expression of Sumo1–3 was silenced in this mouse model, we need to verify in future studies which of the SUMO paralogues play the pivotal role in episodic and emotional memory.
Conclusion
Our results indicate that a functional SUMO conjugation pathway is essential for emotionality and cognition. This novel Sumo knockdown mouse model and the technology used in generating this mutant may help to reveal novel mechanisms that underlie a variety of neuropsychiatric conditions associated with anxiety and impairment of episodic and emotional memory.
Introduction
Small ubiquitin-like modifier (SUMO) conjugation is a post-translational protein modification that targets lysine residues in substrate proteins.1 SUMO1–3 paralogues are widely expressed in mammalian cells. SUMO1 shares about 50% homology with SUMO2 and SUMO3, which are almost identical and therefore usually termed SUMO2/3. SUMO conjugation (SUMOylation) modulates the functions of target proteins in unpredictable ways. For instance, SUMOylation can increase protein stability when SUMO and ubiquitin are competing for the same lysine residue; it can define subcellular localization of proteins; and it can modulate protein–protein and protein–DNA interactions.2 In the brain, SUMO paralogues are highly expressed in neurons, with SUMO1 and SUMO2/3 immunoreactivities prominent in the nucleus and cytoplasm, respectively.3,4 Under physiologic conditions, almost all SUMO2/3 is present as free SUMO2/3. Thermal or metabolic stress increases the levels of SUMO2/3-conjugated proteins, and these accumulate in the nuclei of neurons.3,5
Growing evidence suggests that SUMO conjugation plays a key role in brain plasticity by modulating dendritic spine structure and function, thereby contributing to activity-dependent synaptic transmission.6 Results from various in vitro studies suggest that SUMOylation can regulate the activity of neurons7 and that it is involved in homeostatic synaptic scaling,8 a process that adjusts the strength of synapses to stabilize firing.6,7,9–11 Since alterations in dendritic spine structure and synaptic strength can influence behaviour, especially memory processes, we generated a unique Sumo transgenic mouse model to investigate the role of Sumo conjugation in neuronal functioning associated with cognition in the intact brain. In these Sumo knockdown (Sumo-KD) mice, 3 distinct designed microRNAs (miRNAs) directed against Sumo1–3 are expressed from a single transgene driven by the neuron-specific Thy1 promoter to silence expression of all 3 Sumo paralogues, specifically in neurons.
Methods
Generation of Sumo-KD mice
A fragment containing 3 chained pre-miRNAs that targets SUMO1–3 was previously constructed in our laboratory.12 This fragment, together with an enhanced green fluorescent protein (eGFP) reporter gene, was subcloned into a pThy1 vector to achieve neuron-specific expression of the miRNAs. The linearized vector-free transgene fragment was purified and injected into fertilized FVB/N oocytes at the Duke Transgenic Mouse Facility. Genotyping was performed by polymerase chain reaction (PCR) analysis using mouse tail genomic DNA and Gfp-specific primers (forward primer: 5′-CACATGAAGCAGCACGACTT-3′; reverse primer: 5′-TGCTCAGGTAGTGGTTGTCG-3′). All experiments were conducted according to an approved protocol from the Duke University Institutional Animal Care and Use Committee.
Immunofluorescence and in situ hybridization
Immunofluorescent staining was performed as described previously.5 Briefly, after transcardial perfusion, brains were embedded in paraffin. After deparaffinization, sections were incubated with the primary antibodies at 4°C overnight and then incubated with fluorophore-conjugated secondary antibodies for 1 hour at room temperature. The following antibodies were used: mouse monoclonal anti-Sumo1 (21C7, gift from Dr. Michael J. Matunis, Johns Hopkins University), rabbit polyclonal anti-Sumo2/3 (Covance), rabbit polyclonal anti-Gfp (Invitrogen), mouse monoclonal anti-NeuN (Millipore), mouse monoclonal anti-Gfp (Millipore), Alexa Fluor 488-conjugated goat anti-rabbit (Invitrogen) and Alexa Fluor 594-conjugated goat anti-mouse (Invitrogen). Fluorescent in situ hybridization was performed as described.13 The DNA templates for antisense riboprobe synthesis against Sumo1–3 and Gfp were individually cloned by PCR. The following primers were used: Sumo3 forward primer 5′-AGCGTGACTCGCCCGCTC-3′ and reverse primer 5′-GCGTAATACGACTCACTATAGGGAGACCTGGCCACAGGAGAAG-3′; Sumo2 forward primer 5′-CTTGTGCGCTCCCTCAGT-3′ and reverse primer 5′-GCGTAATACGACTCACTATAGGGAGAAAGGATTTGGTGTTGTTGGG-3′; Sumo1 forward primer 5′-TGTAGAGAAGGGACGGATTTGT-3′ and reverse primer 5′-GCGTAATACGACTCACTATAGGGAGATTTCTTTCTCTCCAGTGAAGCC-3′; eGfp forward primer 5′-TGAGCAAGGGCGAGGAGC-3′ and reverse primer 5′-GCGTAATACGACTCACTATAGGGAGATGTACAGCTCGTCCATGCC-3′. Overviews of whole sections were produced on an Axio Observer Z1 motorized fluorescence microscope (Carl Zeiss MicroImage). Confocal images were captured on a Leica SP5 confocal microscope (Leica Microsystems).
Analysis of dendritic spine density
To determine spine density, dissected brains were placed into Golgi solution for 12 days (FD NeuroTechnologies). Samples were processed as described previously.13 Randomly selected images of dendrites within the area of CA1 stratum radiatum and the dorsolateral amygdala were analyzed using Image J software (National Institutes of Health). The number of spines was counted only on the dendritic region that was clearly in focus and then divided by the length of the dendrite. In total, 40–50 neurons per animal (n = 3 per group) were analyzed.
Behavioural studies
We used male 10–22 week old Sumo-KD and wild-type (WT) littermates that had been backcrossed to C57BL/6 mice for at least 4 generations for behavioural tests. Spontaneous activity was monitored over 30 minutes as distance travelled, vertical activity, thigmotaxis, and centre and corner times, as described previously.14 Activity was assessed as distance travelled (locomotion), vertical activity (rearing), thigmotaxis, centre time, and corner time with Accuscan Instruments equipment. We evaluated episodic short- (STM) and long-term memory (LTM) using the novel object recognition memory test (NORM).15 On day 1, mice were presented with an identical pair of objects. In the STM (20 min) and LTM (24 h) tests, the now familiar object was paired with a novel object. All behaviours were videotaped, and the total time spent with each object was scored by trained observers who were blinded to the mouse genotype using Noldus Observer (Noldus Information Technology). Object recognition scores were calculated by subtracting the total time with the familiar from time spent with the novel object and were then divided by the total amount of time spent with both objects.
Fear conditioning responses were assessed in MedAssociates chambers, as outlined previously.15 After 2 minutes in the conditioning chamber, a 72-dB 12-kHz tone (conditioned stimulus; CS) was presented for 30 seconds and the CS terminated simultaneously with a 2-second 0.4-mA scrambled foot shock (unconditioned stimulus; UCS). The mice were returned to their home cage 30 seconds later. Twenty-four hours later animals were returned to the conditioning chamber for 5 minutes in the absence of the CS and UCS for context testing. The next day mice were placed into a novel environment for 2 minutes, after which the CS was presented for 3 minutes for cued testing. Behaviour was videotaped and scored with the Noldus Observer for freezing behaviours by trained observers blinded to the genotypes of the mice.
Fear-potentiated startle (FPS) was examined over 5 days in MedAssociates chambers, as reported previously.15 On day 1, acoustic startle responses to the 100-, 105- and 110-dB startle stimuli were analyzed. On day 2, mice were re-exposed to each startle stimulus followed by trials where the startle stimulus was preceded with a 30-second 70-dB 12-kHz tone (CS) and by trials where the CS was presented alone. On day 3, animals were given trials where the CS was followed immediately with a 0.25-second 0.4-mA scrambled foot shock (UCS). Day 4 was a hiatus, but on day 5 mice were tested for potentiation of the startle response by the CS using the same procedure described for day 2. Potentiation of the startle response to the CS before and after conditioning was calculated as a ratio of the response to the CS + startle stimuli relative to startle-only responses and expressed as a percentage. Assessment of the sensitivity to foot shock was performed as described previously.16
Statistical analysis
We performed statistical analyses using the SPSS software version 11 (SPSS Inc.). All results are presented as means and standard errors of the mean (SEM) except for spine density data, which are presented as means ± standard deviation (SD). Statistical significance for spine density data was evaluated using a 2-tailed Student t test. We used independent-measures t tests for activity in the open field, while repeated-measures analysis of variance (ANOVA) was used to analyze responses in the NORM, fear conditioning, FPS and shock threshold tests. Bonferroni-corrected pairwise comparisons were used post hoc, and we considered results to be significant at p < 0.05.
Results
Generation and characterization of Sumo-KD mice
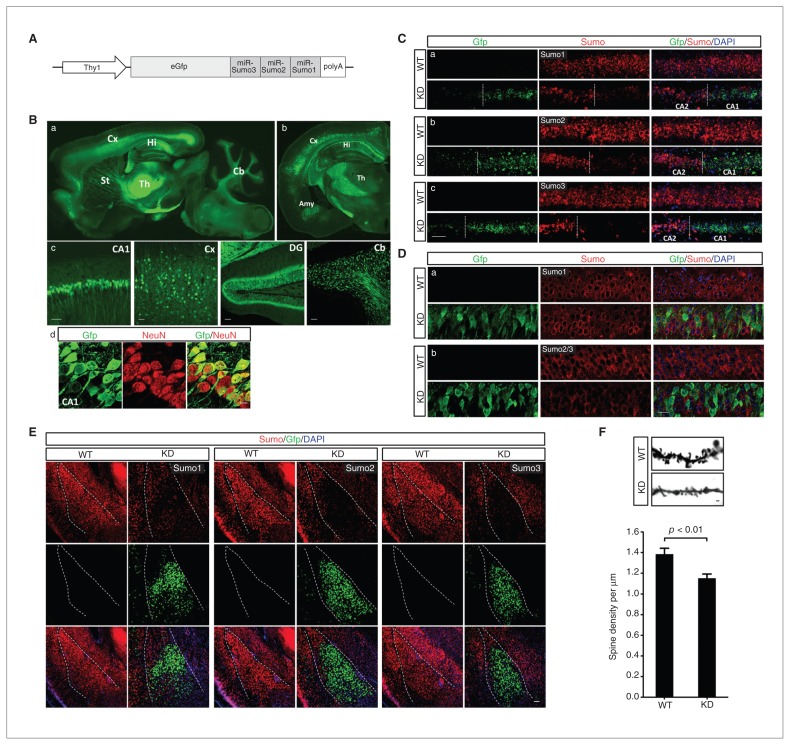
To generate Sumo-KD transgenic mice, we designed and chained 3 pre-miRNAs for silencing Sumo1–3 expression along with Gfp as an indicator of transgene expression. The fragment was cloned into a pThy1 vector to achieve neuron-specific expression (Fig. 1A). We used a 6.5 kb fragment of the Thy1 promoter lacking exon 3 and its flanking introns,17 which are required for expression in non-neuronal cells but not in neurons.18 Six founders were obtained, as verified by PCR genotyping. Consistent with other studies that used the Thy1 promoter to generate transgenic mice,17,19 our 6 lines of Sumo-KD mice showed varying extents of transgene expression from sparse (line 29) to widespread (line 27), as indicated by Gfp expression in the brain. Although sparse lines may be useful for imaging individual neurons, widespread transgene expression is preferred for functional and behavioural studies. Therefore, line 27 was chosen for further characterization in this study (Fig. 1B). The Sumo-KD line 27 mice did not exhibit any developmental or reproductive abnormalities. Sumo-KD mice had the expected litter size of a mean of 6.3 ± an SEM of 0.2,20 with 50.8% and 49.2% male and female animals, respectively. Furthermore, no apparent abnormalities were observed in Nissl-stained brain sections of Sumo-KD mice (see the Appendix, Fig. S1, available at jpn.ca).
Fig. 1.
Generation and characterization of small ubiquitin-like modifier 1–3 knockdown (Sumo-KD) mice. (A) Scheme of the DNA fragment used for generating Sumo-KD mice. (B) Widespread expression of the transgene green fluorescfent protein (Gfp) in KD mouse line 27 (a: sagittal section; b: coronal section; c: different brain areas) and colocalization of Gfp with NeuN (red) in CA1 pyramidal neurons, (d) confirming neuron-specific transgene expression. (C–E) The Gfp-positive cells exhibit (C) a marked decrease in Sumo1–3 mRNA and (D) Sumo2/3 protein levels in the hippocampal CA1 subfield and (E) in Sumo1–3 miRNA levels in the amygdala (outlined). (D) Changes in Sumo1 protein levels were less pronounced than Sumo2/3 levels. (F) Synaptic spine density was significantly reduced in CA1 neurons of Sumo-KD compared with wild type mice. Amy = amygdala; Cb = cerebellum; Cx = cortex; DG = dentate gyrus; Hi = hippocampus; St = striatum; Th = thalamus; WT = wild-type. Scale bars: 50 μm in B-c and C; 10 μm in B-d; 20 μm in D; 100 μm in E; and 1 μm in F. DAPI = 4′,6-diamidino-2-phenylindole.
Green fluorescent protein expression in line 27 mice was found predominantly in cerebral cortex layer V, the hippocampal, amygdala and cerebellar neurons (Fig. 1B), as well as in isolated motor neurons of the spinal cord (data not shown). Neuron-specific expression was confirmed by co-staining with NeuN (Fig. 1B-d). Quantitative analysis revealed that a mean ± SD of 70.9% ± 3.7% and 86.3% ± 2.7% of NeuN-positive cells were also positive for Gfp in the hippocampal CA1 subfield and basolateral amygdala, respectively. As expected, we did not find any Gfp expressing cells that were NeuN-negative. Next, to evaluate the efficacy of Sumo1–3 miRNAs in silencing Sumo1–3 expression, we analyzed Sumo expression at the mRNA and protein levels in the hippocampus by in situ hybridization and immunohistochemistry (Fig. 1C and D). In the hippocampal CA1 subfield, strong Gfp expression was associated with a marked decrease in Sumo1–3 mRNA (Fig. 1C) and Sumo2/3 protein levels, whereas the reduction in Sumo1 protein levels was less pronounced (Fig. 1D). We observed a similar pattern in the amygdala (Fig. 1E) and cerebral cortex (data not shown). In the brain, almost the entire SUMO2/3 pool is present as free SUMO.5 Transient cerebral ischemia and hypothermia activates Sumoylation.3,5 To better illustrate the effects of Sumo1–3 miRNAs expression on Sumoylation, we therefore exposed Sumo-KD mice to transient brain ischemia and compared the intensity and subcellular distribution of Sumo1 and Sumo2/3 immunoreactivity in sham and postischemic brains (See the Appendix, Fig. S2) Together, these data strongly confirm that Sumoylation was effectively reduced in Gfp-positive neurons in mouse line 27. As SUMOylation modulates spinogenesis,21,22 we investigated the effect of Sumo1–3 silencing on synaptic spine density. Silencing Sumo1–3 expression significantly reduced the density of dendritic spines on hippocampal CA1 neurons (Fig. 1F), but not on basolateral amygdala neurons (data not shown).
Behavioural findings
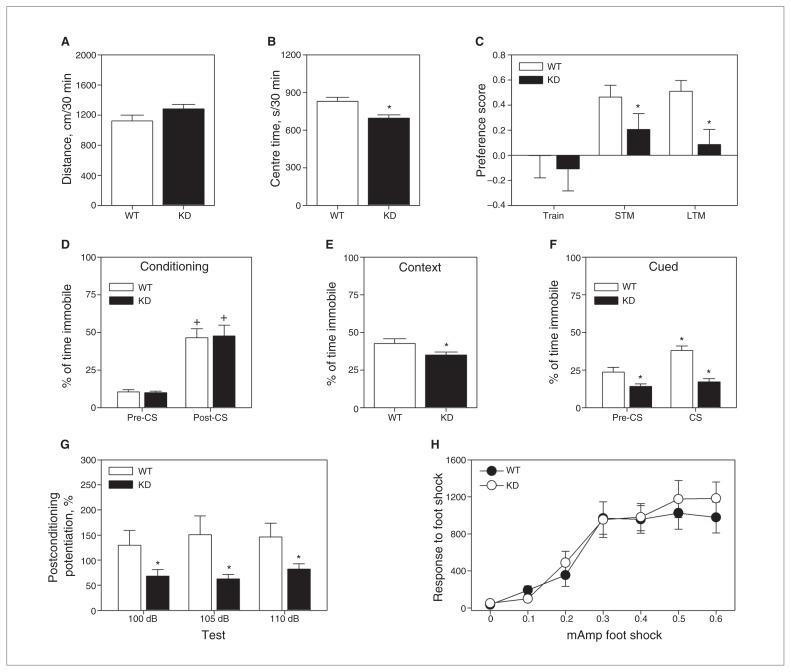
In mouse line 27, Gfp-positive neurons were found mostly in the cerebral cortex, hippocampus and amygdala, brain regions associated with memory processes. We therefore subjected Sumo-KD and WT littermate mice to a battery of behavioural tests to examine the role of Sumoylation in cognition. Spontaneous locomotor (Fig. 2A) and rearing (Table 1) activities were similar between the genotypes. Despite this fact, mutant mice spent less time in the centre zone (t1,30 = 3.226, p = 0.003; Fig. 1B), engaged in more activity in the periphery and spent more time in the corners than the WT animals (Table 1). These data show that while overall activity was similar between genotypes, the Sumo-KD mice appeared to display anxiety-like responses in a novel environment.
Fig. 2.
Behavioural analyses of wild-type (WT) and Sumo1–3 knockdown (Sumo-KD) mice. (A) Spontaneous activity in the open field over minutes depicting locomotion and (B) time in the centre zone. (C) Preference scores for the novel object in the novel object recognition memory (NORM) test, depicting short- (STM) and long-term memory (LTM). (D) Freezing behaviour during the conditioning session and following testing for (E) contextual and (F) cued fear conditioning. (G) Fear-potentiated startle responses of WT and Sumo-KD mice after conditioning. (H) Responses of WT and Sumo-KD mice to different levels of foot shock. There were 16 mice per genotype for all tests except NORM, which had 16 WT and 12 Sumo-KD mice (4 Sumo-KD mice failed to explore the objects). *p < 0.05, WT versus KD mice; +p < 0.05, within genotype pre- versus post-CS (conditioning) or pre-CS versus CS (cued testing). CS = conditioned stimulus.
Table 1.
Results from behavioural assessments of small ubiquitin-like modifier knockdown mice
| Genotype; mean ± SEM | ||||
|---|---|---|---|---|
|
|
||||
| Behavioural test | Wild-type, n = 16 | Sumo-KD, n = 16* | Statistics | p value |
| Open field | ||||
| Vertical activity (beam-breaks) | 451 ± 33.0 | 515 ± 40.1 | t1,30 = 1.237 | 0.23 |
| Thigmotaxis, cm/30 min | 562 ± 50.8 | 691 ± 37.2 | t1,30 = 2.047 | 0.05 |
| Time in corners, s | 262 ± 19.0 | 341 ± 22.4 | t1,30 = 2.686 | 0.012 |
| NORM | ||||
| Preference score at training | +0.002 ± 0.178 | −0.108 ± 0.176 | F1,26 = 1.720 | 0.68 |
| Duration exploring objects, s | F1,26 = 6.419 | 0.018§ | ||
| STM | 40 ± 6.0 | 73 ± 8.0 | 0.002¶ | |
| LTM | 47 ± 8.4 | 73 ± 9.4 | 0.005¶ | |
| Total number of object contacts | F1,26 = 12.263 | 0.002§ | ||
| STM | 51 ± 6.8 | 90 ± 8.4 | — | < 0.001¶ |
| LTM | 50 ± 6.9 | 80 ± 8.5 | — | 0.008¶ |
| FPS | ||||
| Baseline startle response, mAmp† | ||||
| 100 dB | 138 ± 14.4 | 138 ± 14.2 | NS | — |
| 105 dB | 140 ± 10.0 | 131 ± 9.5 | NS | — |
| 110 dB | 151 ± 13.2 | 126 ± 12.6 | NS | — |
| Preconditioning potentiation, %‡ | ||||
| 100 dB | 8.5 ± 8.6 | 14.0 ± 10.5 | NS | — |
| 105 dB | 7.9 ± 8.1 | 10.0 ± 10.2 | NS | — |
| 110 dB | 0.1 ± 8.6 | 12.3 ± 9.4 | NS | — |
ANOVA = analysis of variance; FPS = fear-potentiated startle; LTM = long-term memory; NORM = novel object recognition memory; NS = not significant; SEM = standard error of the mean; STM = short-term memory; Sumo-KD = small ubiquitin-like modifier knockdown.
The NORM test involved 12 Sumo-KD mice because 4 of them failed to explore the objects despite there being no evidence of neophobia.
Repeated-measures ANOVA, startle: F2,60 = 0.071, p = 0.93; startle × genotype interaction: F2,60 = 1.388, p = 0.26.
Repeated-measures ANOVA, preconditioning potentiation: F2,60 = 0.455, p = 0.64; preconditioning potentiation × genotype interaction: F2,60 = 0.62, p = 0.54.
Repeated-measures ANOVA between-subjects tests.
Bonferroni-corrected pairwise comparison.
Episodic memory was assessed in the novel object recognition memory task. No genotype differences in object preferences were observed when 2 identical objects were presented (Table 1). The repeated-measures ANOVA found genotype differences to be significant only in the between-subjects STM and LTM tests (F1,26 = 8.198, all p < 0.008). Bonferroni comparisons revealed that novel object preferences for the STM and LTM tests were reduced for mutants compared with WT animals (all p < 0.047; Fig. 2C). Despite this decreased preference, Sumo-KD mice spent more time exploring and interacting with the objects than the WT controls (Table 1). Hence, their deficiencies in STM and LTM cannot be attributed to neophobia. Together, these results indicate that the Sumo-KD mice are impaired in episodic memory.
Emotional memory was evaluated by fear conditioning. Freezing behaviours were first analyzed before and just after the pairing of the CS with the UCS (Fig. 2D). A repeated-measures ANOVA detected significant within-subject effects only for the test phase (F1,30 = 62.314, p < 0.001). Bonferroni comparisons showed that freezing responses for both genotypes were enhanced in the interval from the pre-CS to the CS–UCS pairing (all p < 0.001). In the test for contextual fear conditioning, all mice showed freezing responses, but these behaviours were decreased in Sumo-KD animals (t1,20 = 2.082, all p < 0.046; Fig. 2E). For cued conditioning, a repeated-measures ANOVA revealed significant within-subject effects for test phase (F1,30 = 17.337, p < 0.001) and a significant test phase × genotype interaction (F1,30 = 7.472, p < 0.010; Fig. 2F). Bonferroni tests showed that freezing during the pre-CS interval in WT mice was increased relative to the Sumo-KD animals (p = 0.017). Interestingly, freezing was not enhanced in the mutants from the pre-CS to the CS phase, whereas freezing was augmented in the WT controls (p < 0.001) and was greater than that in the mutant mice (p < 0.001). Together, these findings demonstrate that the Sumo-KD animals are impaired in both contextual and cued fear conditioning.
Since abnormalities in contextual and cued fear indicate that there may be amygdala dysfunction,23,24 we tested the mice for FPS. On day 1, baseline startle responses to 3 startle stimuli did not differ between the genotypes (Table 1). On day 2, no genotype differences were discerned when the startle stimuli were preceded by the CS (Table 1). On day 3, the CS and UCS were paired. Forty-eight hours later, FPS was assessed. A repeated-measures ANOVA for the between-subject test showed significance for genotype (F1,30 = 5.255, p = 0.029; Fig. 2G). Bonferroni corrections showed that Sumo-KD mice had significant reductions in postconditioning fear-potentiation at all 3 startle intensities compared with WT animals (all p < 0.049). This reduction in fear potentiation supports the fear conditioning results that indicate amygdala dysfunction in Sumo-KD mice. Importantly, the impairments of the mutants in fear conditioning and FPS cannot be explained by genotype differences in their responses to foot shock (Fig. 2H).
Discussion
Here, we present a novel, unique Sumo-KD mouse model, and the first in vivo study to investigate the role of Sumo conjugation in neuronal functioning associated with cognition. To the best of our knowledge, this is the first study reporting generation of transgenic mice in which an RNA polymerase II (Pol II) promoter drives expression of 3 pre-miRNAs simultaneously from 1 transgene to silence expression of 3 distinct genes, in this study Sumo1, Sumo2, and Sumo3. The RNA interference (RNAi) transgenic mice are an inexpensive and rapid alternative to conventional and conditional knockout mice for studying the biological functions of genes of interest in loss-of-function experiments. Several strategies have been developed to generate inducible and reversible RNAi transgenic mice.25,26 In these mice, the tetracycline response element system was used to regulate the expression of short hairpin RNAs (shRNAs). To achieve cell-specific knockdown, these mice need to cross with transgenic mice expressing transactivators only in cells of interest. However, it has not been clearly demonstrated whether this strategy is suitable for knockdown of multiple genes in 1 transgenic animal.
We have used a miRNA-based RNAi approach that exploits the endogenous miRNA processing machinery to produce designed gene-specific shRNAs. Since the Pol II promoter is used in this approach, genes of interest can be knocked down in a tissue- or cell-specific manner.27 Moreover, Pol II promoter–mediated expression of miRNAs is believed to be better tolerated than Pol III promoter–based shRNAs.28,29 Another advantage of this system is the ability to express multiple designed miRNAs from a single construct to silence expression of several genes concurrently, or to increase silencing efficacy, as demonstrated in cell culture studies.27,30,31 In the present experiment, we provided further evidence of the flexibility of this system by advancing from cell cultures to animals and illustrating that expression of multiple genes can be simultaneously silenced in vivo from 1 transgene driven by a Pol II promoter. This application is particularly intriguing for studying a class of similar genes with redundant functions, because using conventional methods to generate a mouse line with several genes deleted is extremely time-consuming and may be practically impossible. In our Sumo-KD mice, expression of Sumo1–3 miRNAs resulted in dramatically lower Sumo1–3 mRNA and Sumo2/3 protein levels, while the effect on Sumo1 protein levels was less pronounced. When we expressed the pre-miRNA sequences targeting Sumo1–3 in cultured cells, the levels of both Sumo1- and Sumo2/3-conjugated proteins were dramatically reduced.12 This suggests that in vivo Sumoylation of proteins by Sumo2/3 is a much more dynamic process than conjugation by Sumo1, as discussed recently.32
To our knowledge, we have provided the first evidence that Sumo conjugation is involved in cognitive processes. The observation that Sumo1–3 miRNA expression results in a more pronounced decrease in levels of Sumo2/3-conjugated proteins than in Sumo1-conjugated proteins implies a pivotal role for Sumo2/3 in this process. To date, studies on the role of SUMOylation on neuronal functions have focused on SUMO1.6 Under physiologic conditions, a large portion of SUMO1, but only a small percentage of SUMO2/3, is conjugated to target proteins in the brain. Metabolic or thermal stress predominantly activates SUMO2/3 conjugation.5,33 Collectively, these observations support a model whereby SUMO2/3 conjugation is a pivotal process associated with neuronal plasticity and protection against various stressful conditions. Despite recent advances, the mechanisms by which global SUMO conjugation contributes to neuronal functions in the brain remains largely unknown. Therefore, the unique Sumo-KD mice will be an invaluable tool for dissecting the roles of Sumoylation in neuronal functions in vivo.
Behavioural analyses revealed that the Sumo-KD mice displayed several phenotypes. In the open field, the mutant mice spent less time in the centre zone, engaged in more thigmotaxis and spent more time in the corners than the WT controls. Since locomotor and rearing activities were not distinguished by genotypes, these findings suggest that the mutant mice were engaging in anxiety-like behaviour.34 Aside from this response, Sumo-KD mice were deficient in STM and LTM in the NORM test. Since Sumo1–3 were knocked down in the hippocampus, it is not surprising that these mice were deficient in episodic memory.35 Besides NORM, the Sumo-KD mice were impaired in both contextual and cued fear conditioning. While deficiency in contextual fear conditioning indicates hippocampal abnormalities, perturbations in both tests are suggestive of amygdala dysfunction.23,24 To confirm this latter possibility, mice were trained and tested for FPS. Although both genotypes showed potentiation to the CS, responses of the Sumo-KD mice were less pronounced than those of the WT controls to all 3 stimulus intensities. Since the Sumo miRNAs were expressed primarily in the basolateral amygdala, our results suggest that Sumo conjugation in these neurons is essential for fear memories.
SUMOylation may regulate emotionality and cognitive processes by a variety of mechanisms. For example, SUMOylation of the protein kinase CASK and the transcription factor MEF2A inhibits or promotes synapse formation, respectively.21,22 MEF2A expression is markedly higher in the hippocampus than in the amygdala,36 brain regions in which the density of dendritic spines was significantly reduced or unchanged in Sumo-KD mice, respectively (Fig. 1F). This suggests that the mechanisms underlying memory processing are not identical in both structures. While long-term potentiation, a form of synaptic plasticity believed to be involved in learning and memory processes, can be elicited in the hippocampus and amygdala, significant differences exist between both brain structures.37 Notably, these differences include changes in spine density following learning processes.38,39
Sumoylation of neurotransmitter receptors and ion channels are believed to regulate the efficiency of synaptic transmission.7,9 Since many transcription and translation factors are targets for SUMO conjugation, the consequent alterations in gene expression and protein synthesis may have unpredictable effects on synaptic plasticity.40 In this way, alterations in SUMOylation may also exert differential effects on various behavioural processes through a variety of mechanisms that may be neuron-, neural circuit– and brain region–specific. The Sumo-KD mouse and the technology used in generating this mutant mouse may help to address these issues, and they may reveal novel mechanisms that underlie a variety of neuropsychiatric conditions.
Limitations
The data presented emphasize a pivotal role for SUMO conjugation in emotionality and cognition. However, since we used a novel mouse model with neuron-specific silencing of all 3 Sumo paralogues, we are not able to distinguish whether the Sumo-KD-induced impairments of neuronal functions reported here resulted from silencing expression of 1 or all 3 Sumo paralogues. Clearly, this important aspect needs to be investigated in future studies using mice with deletion of Sumo1, Sumo2 or Sumo3.
Conclusion
To investigate the role of SUMO conjugation in neuronal functions in vivo, we have generated a novel Sumo-KD mouse model with neuron-specific expression of 3 distinct miRNAs to silence expression of 3 different genes, Sumo1, Sumo2 and Sumo3. Sumo1–3 knockdown mice displayed anxiety-like responses, they were impaired in episodic memory processes, in contextual and cued fear conditioning and in FPS. Our results will help to better understand the molecular mechanisms underlying emotionality and cognition. The novel Sumo-KD mouse model may become an important tool to uncover the role of SUMO conjugation in degenerative diseases and various neuropsychiatric conditions that are associated with anxiety and/or impaired episodic and emotional memory processes.
Acknowledgements
The authors thank Kathy Gage, research development associate, for her excellent editorial contribution in the preparation of this manuscript, Pei Miao for her excellent technical assistance, and Dr. Michael J. Matunis for providing the Sumo1 antibody. Some of the experiments were conducted with equipment and software purchased with a grant from North Carolina Biotechnology Center. This study was supported by funds from the Department of Anesthesiology and the Mouse Behavioural and Neuroendocrine Analysis Core Facility, Duke University Medical Center, by NIH R01 grants HL095552 and NS081299 (to W.P.), and by AHA scientist development grant 12SDG11950003 (to W.Y.).
Footnotes
Competing interests: None declared.
Contributors: W.C. Wetsel, S. Zhao, W. Paschen and W. Yang designed the study. L. Wang, R.M. Rodriguiz, W.C. Wetsel, H. Sheng, X. Liu and W. Paschen acquired the data, which L. Wang, R.M. Rodriguiz, W.C.Wetsel, W. Paschen and W. Yang analyzed. R.M. Rodriguiz, W.C. Wetsel, W. Paschen and W. Yang wrote the article, which all authors reviewed and approved for publication.
References
- 1.Matunis MJ, Wu J, Blobel G. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol. 1998;140:499–509. doi: 10.1083/jcb.140.3.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Yang W, Sheng H, Warner DS, et al. Transient focal cerebral ischemia induces a dramatic activation of small ubiquitin-like modifier conjugation. J Cereb Blood Flow Metab. 2008;28:892–6. doi: 10.1038/sj.jcbfm.9600601. [DOI] [PubMed] [Google Scholar]
- 4.Tirard M, Hsiao HH, Nikolov M, et al. In vivo localization and identification of SUMOylated proteins in the brain of His6-HA-SUMO1 knock-in mice. Proc Natl Acad Sci U S A. 2012;109:21122–7. doi: 10.1073/pnas.1215366110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang L, Ma Q, Yang W, et al. Moderate hypothermia induces marked increase in levels and nuclear accumulation of SUMO2/3-conjugated proteins in neurons. J Neurochem. 2012;123:349–59. doi: 10.1111/j.1471-4159.2012.07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig TJ, Henley JM. Protein SUMOylation in spine structure and function. Curr Opin Neurobiol. 2012;22:480–7. doi: 10.1016/j.conb.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plant LD, Dowdell EJ, Dementieva IS, et al. SUMO modification of cell surface Kv2.1 potassium channels regulates the activity of rat hippocampal neurons. J Gen Physiol. 2011;137:441–54. doi: 10.1085/jgp.201110604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Craig TJ, Jaafari N, Petrovic MM, et al. Homeostatic synaptic scaling is regulated by protein SUMOylation. J Biol Chem. 2012;287:22781–8. doi: 10.1074/jbc.M112.356337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin S, Nishimune A, Mellor JR, et al. SUMOylation regulates kainate-receptor-mediated synaptic transmission. Nature. 2007;447:321–5. doi: 10.1038/nature05736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–35. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loriol C, Khayachi A, Poupon G, et al. Activity-dependent regulation of the sumoylation machinery in rat hippocampal neurons. Biol Cell. 2013;105:30–45. doi: 10.1111/boc.201200016. [DOI] [PubMed] [Google Scholar]
- 12.Yang W, Wang L, Roehn G, et al. Small ubiquitin-like modifier 1–3 is activated in human astrocytic brain tumors and is required for glioblastoma cell survival. Cancer Sci. 2013;104:70–7. doi: 10.1111/cas.12047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Budolfson K, Wang F. Pik3c3 deletion in pyramidal neurons results in loss of synapses, extensive gliosis and progressive neurodegeneration. Neuroscience. 2011;172:427–42. doi: 10.1016/j.neuroscience.2010.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogorelov VM, Rodriguiz RM, Insco ML, et al. Novelty seeking and stereotypic activation of behavior in mice with disruption of the Dat1 gene. Neuropsychopharmacology. 2005;30:1818–31. doi: 10.1038/sj.npp.1300724. [DOI] [PubMed] [Google Scholar]
- 15.Porton B, Rodriguiz RM, Phillips LE, et al. Mice lacking synapsin III show abnormalities in explicit memory and conditioned fear. Genes Brain Behav. 2010;9:257–68. doi: 10.1111/j.1601-183X.2009.00555.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grove M, Demyanenko G, Echarri A, et al. ABI2-deficient mice exhibit defective cell migration, aberrant dendritic spine morphogenesis, and deficits in learning and memory. Mol Cell Biol. 2004;24:10905–22. doi: 10.1128/MCB.24.24.10905-10922.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng G, Mellor RH, Bernstein M, et al. Imaging neuronal subsets in transgenic mice expressing multiple spectral variants of GFP. Neuron. 2000;28:41–51. doi: 10.1016/s0896-6273(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 18.Vidal M, Morris R, Grosveld F, et al. Tissue-specific control elements of the Thy-1 gene. EMBO J. 1990;9:833–40. doi: 10.1002/j.1460-2075.1990.tb08180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Q, Cichon J, Wang W, et al. Imaging neural activity using Thy1-GCaMP transgenic mice. Neuron. 2012;76:297–308. doi: 10.1016/j.neuron.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS ONE. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalizi A, Gaudilliere B, Yuan Z, et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–7. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 22.Chao HW, Hong CJ, Huang TN, et al. SUMOylation of the MAGUK protein CASK regulates dendritic spinogenesis. J Cell Biol. 2008;182:141–55. doi: 10.1083/jcb.200712094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips RG, LeDoux JE. Differential contribution of amygdala and hippocampus to cued and contextual fear conditioning. Behav Neurosci. 1992;106:274–85. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- 24.Fendt M, Fanselow MS. The neuroanatomical and neurochemical basis of conditioned fear. Neurosci Biobehav Rev. 1999;23:743–60. doi: 10.1016/s0149-7634(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 25.Dickins RA, McJunkin K, Hernando E, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat Genet. 2007;39:914–21. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McJunkin K, Mazurek A, Premsrirut PK, et al. Reversible suppression of an essential gene in adult mice using transgenic RNA interference. Proc Natl Acad Sci U S A. 2011;108:7113–8. doi: 10.1073/pnas.1104097108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia XG, Zhou H, Xu Z. Multiple shRNAs expressed by an inducible pol II promoter can knock down the expression of multiple target genes. Biotechniques. 2006;41:64–8. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- 28.Premsrirut PK, Dow LE, Kim SY, et al. A rapid and scalable system for studying gene function in mice using conditional RNA interference. Cell. 2011;145:145–58. doi: 10.1016/j.cell.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mueller C, Tang Q, Gruntman A, et al. Sustained miRNA-mediated knockdown of mutant AAT with simultaneous augmentation of wild-type AAT has minimal effect on global liver miRNA profiles. Mol Ther. 2012;20:590–600. doi: 10.1038/mt.2011.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun D, Melegari M, Sridhar S, et al. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 31.Yang W, Paschen W. Gene expression and cell growth are modified by silencing SUMO2 and SUMO3 expression. Biochem Biophys Res Commun. 2009;382:215–8. doi: 10.1016/j.bbrc.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 32.Demarque MD, Nacerddine K, Neyret-Kahn H, et al. Sumoylation by Ubc9 regulates the stem cell compartment and structure and function of the intestinal epithelium in mice. Gastroenterology. 2011;140:286–96. doi: 10.1053/j.gastro.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 33.Yang W, Sheng H, Warner DS, et al. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metab. 2008;28:269–79. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- 34.Treit D, Fundytus M. Thigmotaxis as a test for anxiolytic activity in rats. Pharmacol Biochem Behav. 1988;31:959–62. doi: 10.1016/0091-3057(88)90413-3. [DOI] [PubMed] [Google Scholar]
- 35.Dere E, Huston JP, De Souza Silva MA. The pharmacology, neuroanatomy and neurogenetics of one-trial object recognition in rodents. Neurosci Biobehav Rev. 2007;31:673–704. doi: 10.1016/j.neubiorev.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Lyons GE, Micales BK, Schwarz J, et al. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–38. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman PF, Ramsay MF, Krezel W, et al. Synaptic plasticity in the amygdala: comparisons with hippocampus. Ann N Y Acad Sci. 2003;985:114–24. doi: 10.1111/j.1749-6632.2003.tb07076.x. [DOI] [PubMed] [Google Scholar]
- 38.Moser MB, Trommald M, Andersen P. An increase in dendritic spine density on hippocampal CA1 pyramidal cells following spatial learning in adult rats suggests the formation of new synapses. Proc Natl Acad Sci U S A. 1994;91:12673–5. doi: 10.1073/pnas.91.26.12673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ostroff LE, Cain CK, Bedont J, et al. Fear and safety learning differentially affect synapse size and dendritic translation in the lateral amygdala. Proc Natl Acad Sci U S A. 2010;107:9418–23. doi: 10.1073/pnas.0913384107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schafe GE, Nadel NV, Sullivan GM, et al. Memory consolidation for contextual and auditory fear conditioning is dependent on protein synthesis, PKA, and MAP kinase. Learn Mem. 1999;6:97–110. [PMC free article] [PubMed] [Google Scholar]