Summary
Continual spermatogenesis relies on the activities of a tissue-specific stem cell population referred to as spermatogonial stem cells (SSCs). Fate decisions of stem cells are influenced by their niche environments, a major component of which is soluble factors secreted by support cells. At present, the factors that constitute the SSC niche are undefined. We explored the role of chemokine (C-X-C motif) ligand 12 (CXCL12) signaling via its receptor C-X-C chemokine receptor type 4 (CXCR4) in regulation of mouse SSC fate decisions. Immunofluorescent staining for CXCL12 protein in cross sections of testes from both pup and adult mice revealed its localization at the basement membrane of seminiferous tubules. Within the undifferentiated spermatogonial population of mouse testes, a fraction of cells were found to express CXCR4 and possess stem cell capacity. Inhibition of CXCR4 signaling in primary cultures of mouse undifferentiated spermatogonia resulted in SSC loss, in part by reducing proliferation and increasing the transition to a progenitor state primed for differentiation upon stimulation by retinoic acid. In addition, CXCL12–CXCR4 signaling in mouse SSCs was found to be important for colonization of recipient testes following transplantation, possibly by influencing homing to establish stem-cell niches. Furthermore, inhibition of CXCR4 signaling in testes of adult mice impaired SSC maintenance, leading to loss of the germline. Collectively, these findings indicate that CXCL12 is an important component of the growth factor milieu of stem cells in mammalian testes and that it signals via the CXCR4 to regulate maintenance of the SSC pool.
Key words: Spermatogonial stem cell, CXCL12–CXCR4, Self-renewal, Migration, Niche
Introduction
The homeostasis of most tissues is dependent on the activities of tissue-specific stem cells. In testes of adult mammals, spermatogonial stem cells (SSCs) provide the foundation for continual spermatogenesis, and functional transplantation revealed that SSCs are present in a subpopulation of undifferentiated spermatogonia (Brinster and Zimmermann, 1994; Kubota et al., 2003). The undifferentiated spermatogonial population is composed of SSCs and non-stem-cell progenitors that periodically commit to terminal differentiation in response to retinoic acid signaling, which is signified by KIT receptor expression (Yoshinaga et al., 1991; de Rooij et al., 1999). The fate decisions of SSCs can be defined as the ability for self-renewal to sustain a foundational stem cell pool and generation of progenitor cells.
Similar to other tissue-specific stem cell populations, fate decisions of SSCs are influenced by extrinsic factors of a cognate niche environment. In general, niche factors influence SSCs by promoting self-renewal divisions while blocking differentiation (Oatley and Brinster, 2008). The components of SSC niches are provided by somatic support cell populations that include Sertoli, Leydig and myoid cells in mammalian testes. In rodents, glial cell line-derived neurotrophic factor (GDNF) promotes the proliferation of undifferentiated spermatogonia in vivo (Meng et al., 2000) and addition of GDNF to media is required for SSC self-renewal in primary cultures of undifferentiated spermatogonia (Kubota et al., 2004). Our previous studies suggest that secretion of colony stimulating factor 1 (CSF-1) from Leydig and myoid cells also plays a crucial role in regulating the self-renewal of SSCs (Oatley et al., 2009). Despite these seminal findings, knowledge of the SSC niche is still rudimentary, and long-term maintenance of SSCs in vitro requires somatic feeder cells (e.g. STO or MEF) that secrete a multitude of soluble factors, even when GDNF is added exogenously to culture media (Kubota et al., 2004). Although primary cultures of mouse undifferentiated spermatogonia can be maintained in vitro without feeders, the number of SSCs declines over time even with GDNF supplementation (Kanatsu-Shinohara et al., 2011). These findings indicate that undiscovered factors produced by feeder cells play crucial roles in maintaining the SSC pool of undifferentiated spermatogonial populations. Furthermore, it is plausible to hypothesize that these same factors are crucial components of niches that influence the fate decisions of SSCs in vivo.
Signaling from chemokine (C-X-C motif) ligand 12 (CXCL12) through C-X-C chemokine receptor type 4 (CXCR4) regulates migration, proliferation and survival of various cell types. In particular, homing of hematopoietic stem cells to bone marrow niches is guided by signaling from CXCL12 (Peled et al., 1999). In the embryonic gonad, CXCL12 guides migration of primordial germ cells (PGC) to the genital ridge (Ara et al., 2003; Molyneaux et al., 2003). At present, the importance of CXCL12–CXCR4 signaling in regulating the activities of SSCs is undefined. However, several lines of indirect evidence suggest that CXCL12–CXCR4 signaling influences postnatal development of the male germline. A recent study showed that formation of the undifferentiated spermatogonial population during neonatal development is disrupted in testes of mice that lack expression of the chromatin modifier SIN3a in Sertoli cells (Payne et al., 2010). Interestingly, CXCL12 expression was found to be significantly reduced in testes of these mice, suggesting an important role in establishing the SSC pool. Also, expression of CXCL12 is reduced in testes of mice deficient for ERM expression, a transcription factor essential for maintenance of the undifferentiated spermatogonial population (Chen et al., 2005). Despite these findings, a direct role of CXCL12–CXCR4 signaling in regulating SSC fate decisions has not been established.
In the current study, we show that in testes of postnatal mice, SSCs express CXCR4 and supporting Sertoli cells secret CXCL12. </emph>CXCL12–CXCR4 signaling was found to promote proliferation and block retinoic acid-induced differentiation of SSCs. In addition, CXCL12–CXCR4 signaling was found to play an important role in regulating migration of SSCs after transplantation into recipient testes. Furthermore, inhibition of CXCL12–CXCR4 signaling in vivo impaired SSC maintenance, resulting in loss of the germline.
Results
CXCL12 is expressed by Sertoli cells and CXCR4 is expressed by undifferentiated spermatogonia in testes of postnatal mice
In mouse testes, prospermatogonia that are derived from PGCs migrate to the basement membrane of seminiferous cords between postnatal days (PD) 0 and 2 and a portion of this population subsequently gives rise to a foundational SSC pool that is fully established around PD 6 (Huckins and Clermont, 1968; Bellvé et al., 1977; Drumond et al., 2011). To locate the expression of CXCL12 in the postnatal mouse testis, we conducted immunofluorescent staining of cross sections from pup (PD 6) and adult (2 months) mouse testes using an antibody that recognizes CXCL12. At both ages, CXCL12 staining was observed within the cytoplasm of Sertoli cells that were identified by co-staining for the marker GATA4 (Fig. 1A). In pup testes, staining appeared to be spread throughout the seminiferous epithelium, whereas, in adult testes staining appeared as distinct foci at the basal membrane of seminiferous tubules (Fig. 1A). Next, we examined expression of CXCR4 in testes of pup and adult mice. Immunofluorescent staining revealed CXCR4 in select germ cells that also stained for the undifferentiated spermatogonial marker PLZF (Fig. 1B). In pup testes, CXCR4 expression was observed on the surface of all PLZF-expressing spermatogonia. In contrast, in adult mice only 46.5% (n = 3 different mice examined) of PLZF-expressing spermatogonia co-stained for CXCR4. Importantly, CXCR4 staining was also present in spermatogonia that expressed ID4 (supplementary material Fig. S1), which is a putative marker of SSCs in testes of mice (Oatley et al., 2011). Collectively, these observations indicate that CXCL12 is secreted by Sertoli cells and CXCR4 is expressed by a subpopulation of undifferentiated spermatogonia including SSCs.
Fig. 1.
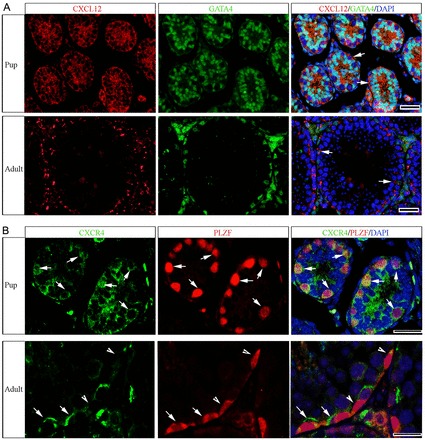
Expression of CXCL12 and CXCR4 within the seminiferous epithelium of testes from postnatal mice. (A) Representative images of immunofluorescent staining for expression of CXCL12 (red) in cross sections of testes from pup (postnatal day 6) and adult (2 months old) mice. Staining is associated with Sertoli cells (arrows) that are labeled by staining for the marker GATA4 (green) and appeared to be concentrated at the basement membrane of seminiferous tubules. (B) Representative images of immunofluorescent staining for expression of CXCR4 (green) in cross sections of testes from pup and adult mice. Staining is observed in cells along the basement membrane that co-stain (red) for expression of the undifferentiated spermatogonial marker PLZF (arrows). Germ cells that stain for PLZF expression but not CXCR4 can also be observed (arrowheads). DAPI was used to stain DNA. Scale bars: 20 µm for all images.
Mouse SSCs express CXCR4
Studying SSCs in vivo is challenging because of the rarity of these cells within the heterogeneous germ cell population. However, the THY1-positive (THY1+) germ cell fraction is enriched for SSCs compared with the unfractionated total cell population of mouse testes (Kubota et al., 2004). Using quantitative (q)RT-PCR analysis, we found that Cxcr4 mRNA abundance is significantly (P = 0.03) enriched by 2.6±0.9-fold in freshly isolated THY1+ fractions (n = 3) compared with corresponding THY1-depleted testicular cell populations of pup mouse testes (Fig. 2A). This characteristic was found to be maintained in the adult testis, with Cxcr4 mRNA abundance being significantly (P = 0.01) greater by 2.7±0.7-fold in the THY1+ fraction (n = 3) compared to corresponding THY1-depleted fractions (Fig. 2B).
Fig. 2.
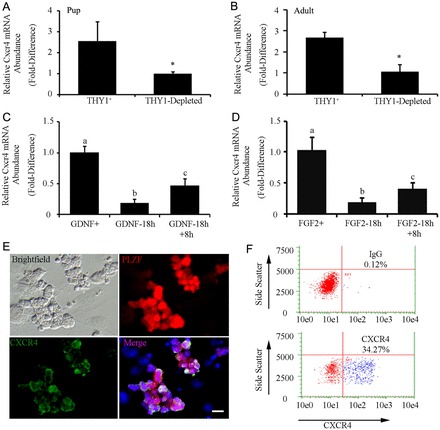
Expression of Cxcr4 mRNA in the undifferentiated spermatogonial population of mouse testes and regulation by the growth factors influencing SSC self-renewal. (A,B) qRT-PCR analysis for relative Cxcr4 transcript abundance in freshly isolated THY1-positive (THY1+) and THY1-depleted cell fractions from pups (postnatal day 6) and adult (2-month old) mice. Data are means ± s.e.m.; *significant difference at P<0.05. (C) qRT-PCR analysis of the relative Cxcr4 transcript abundance in primary cultures of undifferentiated spermatogonia continually supplemented with GDNF (GDNF+), 18 hours after withdrawal of GDNF supplementation (GDNF-18h), and 8 hours after replacement of GDNF following 18 hours withdrawal (GDNF-18h+8h). Data are means ± s.e.m.; different letters denote significant difference at P<0.05. (D) qRT-PCR analysis for relative Cxcr4 transcript abundance in primary cultures of undifferentiated spermatogonia continually supplemented with FGF2 (FGF2+), 18 hours after withdrawal of FGF2 supplementation (FGF2-18h), and 8 hours after replacement of FGF2 following 18 hours withdrawal (FGF2-18h+8h). Data are means ± s.e.m.; different letters denote significant difference at P<0.05. (E) Representative images of immunofluorescent staining of cells expressing CXCR4 (green) and PLZF (red) in primary cultures of undifferentiated spermatogonia. Scale bars: 10 µm. (F) Representative scatter plots from flow cytometric analysis of the percentage of CXCR4+ cells in primary cultures of undifferentiated spermatogonia. The percentage of CXCR4+ cells is indicated in each plot.
Primary cultures of THY1+ undifferentiated spermatogonia provide a valuable model for studying the fate decisions of SSCs (Oatley et al., 2006). When maintained with feeder cell monolayers and culture medium supplemented with GDNF and FGF2 the cells form clumps of SSCs and non-stem cell progenitor spermatogonia (Kubota et al., 2004). The self-renewal of SSCs is supported in these clumps for long periods of time. We found that the abundance of Cxcr4 mRNA in primary cultures of THY1+ undifferentiated spermatogonia is influenced by exposure to GDNF and FGF2. Upon withdrawal of GDNF from the culture medium for 18 hours, Cxcr4 mRNA abundance was significantly (P = 0.001) reduced to only 19.2±6.1% (n = 3 different cultures) of that in cells continually cultured with GDNF (Fig. 2C). Subsequent re-addition of GDNF to the culture medium for 8 hours induced a significant (P = 0.02) increase in Cxcr4 mRNA abundance by 2.8±0.6-fold (n = 3) compared with cells not receiving GDNF following withdrawal (Fig. 2C). Similarly, withdrawal and replacement of FGF2 from the culture medium also influenced Cxcr4 gene expression. After 18 hours without FGF2, the abundance of Cxcr4 mRNA was significantly (P = 0.008) reduced to only 18.7±0.6% (n = 3) of that in cells continually exposed to FGF2 (Fig. 2D). Replacement of FGF2 for 8 hours led to a significant increase (P = 0.05) in Cxcr4 mRNA by 2.5±0.4-fold (n = 3) compared with cells not receiving replacement (Fig. 2D).
Primary cultures of THY1+ undifferentiated spermatogonia form heterogeneous clumps consisting of SSCs and non-stem cell progenitor spermatogonia (Kubota et al., 2004). Immunocytochemical staining of the germ cell clumps suggested that only a portion of this population expresses CXCR4 (Fig. 2E). Flow cytometric analysis of clumps as single cell suspensions revealed that 36.9±4.8% (n = 3 different cultures) of the cells were CXCR4+ (Fig. 2F). Next, we aimed to determine whether the CXCR4+ germ cells possess SSC activity. Thus, we used magnetic-activated cell sorting (MACS) to separate the CXCR4+ fraction from primary cultures of lacZ-expressing undifferentiated spermatogonia, and transplanted the cells into testes of recipient mice. For comparison, we also transplanted the corresponding CXCR4-depleted fraction to assess how much of the SSC population could be captured by separating CXCR4+ cells (Fig. 3A). Examination of recipient testes 2 months after transplantation for colonies of spermatogonia derived from the transplanted cell fractions revealed that SSC activity was significantly (P = 0.05) enriched in the isolated CXCR4+ fraction compared with the CXCR4-depleted fraction (Fig. 3B,C). The CXCR4+ cells generated 178.7±43.1 colonies/105 cells injected (n = 3 different cultures and 12 total recipient testes); whereas, the CXCR4-depleted fraction produced 73.7±24.6 colonies/105 cells injected (n = 3 different cultures and 12 total recipient testes). Thus, the isolated CXCR4+ population was enriched by 2.5-fold compared with the CXCR4-depleted population and contained ∼71% of the entire SSC population.
Fig. 3.
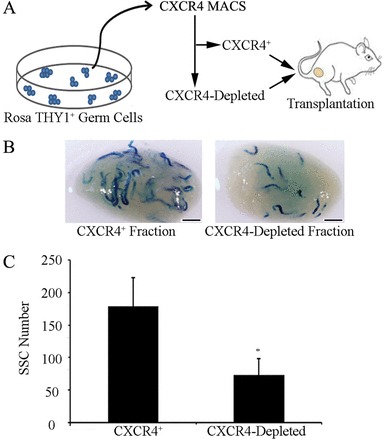
SSC capacity of CXCR4+ undifferentiated spermatogonia. (A) Schematic of the experimental strategy using magnetic-activated cell sorting (MACS) to isolate CXCR4+ cells from primary cultures of THY1+ undifferentiated spermatogonia derived from lacZ-expressing Rosa donor mice. The CXCR4+ and corresponding CXCR4-depleted fractions were transplanted into the seminiferous tubules of recipient mice. (B) Representative images of recipient mouse testes 2 months after transplantation of single-cell suspensions of isolated CXCR4+ or CXCR4-depleted fractions. Each blue segment represents a colony of spermatogonia derived from a transplanted SSC. Scale bars: 2 mm. (C) Comparison of SSC numbers in the CXCR4+ and CXCR4-depleted cell fractions. Data are means ± s.e.m.; n = 3 different cultures and 12 recipient testes. *Significant difference at P<0.05. SSC numbers were derived from X-gal-stained colonies of donor-derived spermatogonia in recipient testes, normalized to the number of cells (105) injected.
CXCR4 signaling is essential for maintenance of SSCs in primary cultures of undifferentiated spermatogonia
The data present above revealed that SSCs express CXCR4, but whether this influences SSC functions remained unknown. First, we evaluated the effects of supplementing the culture medium with recombinant soluble CXCL12 in addition to GDNF and FGF2; however, no differences were observed in growth of the cells or morphology of the clumps compared with cultures maintained with GDNF and FGF2 only (data not shown). These observations led us to investigate whether the STO feeder cells used for co-culture with germ cells secrete CXCL12. Western blot analysis revealed CXCL12 protein in STO cell lysates as well as conditioned media (Fig. 4A), indicating that STO cells are a source of secreted CXCL12 for primary cultures of undifferentiated spermatogonia. Based on this finding, we hypothesized that STO-cell-derived CXCL12 signals through CXCR4 in SSCs to regulate their activities. To explore this possibility, we treated primary cultures of undifferentiated spermatogonia with the CXCR4-specific inhibitor AMD3100 (Donzella et al., 1998) and examined the effects on overall expansion of germ cells and maintenance of SSC content. Because we did not observe staining for CXCR4 expression in STO feeder cells (Fig. 2E), we reasoned that AMD3100 treatment would have a specific effect on the germ cells. We chose to treat cultures with 100 nM AMD3100 because previous studies showed that concentrations of 100 nM to 1 µM effectively inhibited CXCR4 signaling in cancer cells (Cronin et al., 2010). After 1 week of treatment, the total number of germ cells in cultures receiving 100 nM AMD3100 was not significantly different (P = 0.99) from that of vehicle-treated control cultures, suggesting no major effect on overall maintenance of the undifferentiated spermatogonial population (Fig. 4B).
Fig. 4.
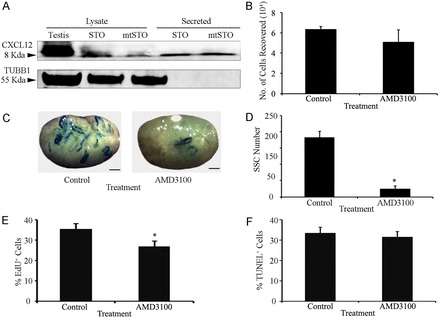
Impact of CXCR4 signaling on maintenance of SSCs in primary cultures of undifferentiated spermatogonia. (A) Representative images of western blot analysis for CXCL12 protein in lysates and conditioned medium from STO and mitotically inactivated (mt) STO feeder cells. TUBB1 was used as a loading control. (B) Quantitative comparison of total germ cell numbers in primary cultures of undifferentiated spermatogonia after 7 days of treatment with the CXCR4-specific inhibitor AMD3100 or vehicle (control). (C) Representative images of recipient mouse testes 2 months after transplantation with lacZ-expressing primary cultures of undifferentiated spermatogonia treated with vehicle or AMD3100 for 7 days. Scale bars: 2 mm. (D) Quantitative comparison of SSC numbers in primary cultures of undifferentiated spermatogonia after 7 days of treatment with AMD3100 or vehicle, using functional germ cell transplantation analysis (12 total recipient testes). (E) Quantitative comparison of the percentage of proliferative cells (EdU+) within the CXCR4+ fraction of primary cultures of undifferentiated spermatogonia treated with the AMD3100 or vehicle (control). (F) Quantitative comparison of the percentage of apoptotic (TUNEL+) cells within the CXCR4+ fraction of undifferentiated primary cultures treated with AMD3100 or vehicle (control) for 3 days. All data are means ± s.e.m. for three different cultures. *Significant difference at P<0.05.
Next, we examined the effects of 100 nM AMD3100 on SSC maintenance using transplantation analysis. In contrast to the effects on total germ cell number, significant (P = 0.003) reduction in the number of SSCs was found after 1 week of AMD3100 treatment compared with treatment with vehicle alone (Fig. 4C,D). Cultures treated with AMD3100 produced 24.9±9.1 colonies/105 cells transplanted (n = 3 different cultures and 12 total recipient testes), which was significantly (P = 0.003) reduced by more than 7-fold compared with the 183.1±18.2 colonies/105 cells transplanted (n = 3 different cultures and 12 recipient testes) produced by vehicle-treated control cultures (Fig. 4D). Loss of SSCs after CXCR4 inhibition could be due to increased apoptosis and/or impaired proliferation. Next, we examined cultures for actively dividing cells by incubation with EdU (5-ethynyl-2’-deoxyuridine), which is incorporated into the DNA of cells in S phase of the cell cycle. After 3 days of treatment, the percentage of cells with EdU incorporation was found to be significantly reduced (P = 0.04) in CXCR4+ fractions of cultures treated with AMD3100 (24.3±2.9%; n = 3 different cultures) compared with vehicle-treated control cultures (34.8±3.0%; Fig. 4E). Examination of the percentages of early and late stage apoptotic cells in the total germ cell population after 1, 3, 5 and 7 days revealed no difference (P>0.05) between AMD3100-treated and vehicle-treated control cultures (supplementary material Fig. S2). Considering that CXCR4+ cells only constitute a sub-fraction of the cultured undifferentiated spermatogonial population, we conducted TUNEL and CXCR4 staining to detect apoptotic cells in the CXCR4+ germ cell fraction, and also found no difference (P>0.05) between control and AMD3100-treated cultures after 3 days of treatment (Fig. 4F). Taken together, these results indicate that CXCL12–CXCR4 signaling plays an important role in regulating the maintenance of SSCs within primary cultures of undifferentiated spermatogonia, in part by influencing self-renewal divisions.
Inhibition of CXCL12–CXCR4 signaling in primary cultures of undifferentiated spermatogonia alters the response to retinoic acid exposure
In the male mouse germline, transition from an undifferentiated to a differentiating state in spermatogonia is regulated by activation of retinoic acid signaling and involves KIT receptor expression (Yoshinaga et al., 1991; de Rooij et al., 1999). At periodic intervals, a majority of the progenitor spermatogonia within the undifferentiated spermatogonial population undergo transition to the differentiating state leaving behind the SSC pool that is responsible for replenishing the lost progenitors (de Rooij et al., 1999). Using flow cytometric analysis, we found that 45.6±0.8% of cells were KIT+ (n = 3 different cultures) in cultures treated for 24 hours with all-trans retinoic acid (AtRA; a bioactive form of retinoic acid), which was an ∼10-fold increase compared with the 4.5±2.1% of cells that were KIT+ (n = 3 different cultures) in vehicle (DMSO)-treated control cultures (Fig. 5A). In cultures pre-treated with AMD3100 for 24 hours, 2.1±0.8% of cells were KIT+ (n = 3 different cultures) 24 hours after treatment with DMSO. In contrast, after AtRA treatment for 24 hours the percentage of KIT+ cells increased to 61.9±3.5% (n = 3 different cultures) in cultures pre-treated with AMD3100, which was significantly (P = 0.001) greater than in the AtRA-treated cultures not receiving AMD3100 (45.6±2.5; Fig. 5B). Also, qRT-PCR analyses revealed that expression of Plzf, a transcription repressor required for SSC maintenance, was significantly reduced, by 31±9% (n = 3 different cultures), in cultures treated with AMD3100 for 7 days compared with control cultures (Fig. 5C). In contrast, expression of Ngn3, a transcription factor involved in formation of progenitor spermatogonia (Yoshida et al., 2004; Kaucher et al., 2012), was significantly increased by 60±19% (n = 3 different cultures) in cultures treated with AMD3100 for 7 days, compared with control cultures (Fig. 5C). Next, we examined the effects of neutralizing STO-cell-secreted CXCL12 on makeup of the cultured undifferentiated spermatogonial population. In cultures treated with a validated CXCL12-blocking antibody for 48 hours, the percentage of KIT+ cells was similar to that of control cultures receiving normal IgG. However, after AtRA treatment for 24 hours, a significantly greater (P<0.05) percentage of the cell population was KIT+ in cultures receiving CXCL12-blocking antibody compare with control cultures (supplementary material Fig. S3), mimicking the effects of AMD3100 treatment for inhibition of CXCR4 signaling. Collectively, these results indicate that inhibition of CXCL12–CXCR4 signaling induces a greater portion of the undifferentiated spermatogonial population to adopt a progenitor cell fate that is primed for transition to a differentiating state upon stimulation by retinoic acid.
Fig. 5.
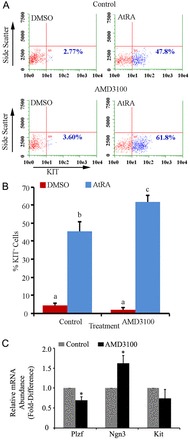
Impact of CXCR4 signaling on the response to retinoic acid of primary cultures of undifferentiated spermatogonia. (A) Representative scatter plots from flow cytometric analysis of the percentage of cells expressing the differentiating spermatogonial marker KIT in primary cultures of undifferentiated spermatogonia 24 hours after treatment with all-trans retinoic acid (AtRA) or DMSO (vehicle control). Cultures were pre-treated with the CXCR4-specific inhibitor AMD3100 or vehicle 24 hours before addition of AtRA or DMSO to the culture media. The percentage KIT+ cells in each plot is provided. (B) Quantitative comparison of the percentage of KIT+ cells 24 hours after DMSO or AtRA treatment in primary cultures of undifferentiated spermatogonia pre-treated with AMD3100 or vehicle (control). Data are means ± s.e.m. for three different cultures and different characters denote significantly different at P<0.001. (C) qRT-PCR analysis of relative transcript abundance of Plzf, Ngn3 and Kit in primary cultures of undifferentiated spermatogonia treated with AMD3100 or vehicle. Data are means ± s.e.m. for three different cultures. *Significant difference at P<0.05.
CXCL12–CXCR4 signaling is crucial for SSC homing and expansion after transplantation
Previous studies showed that CXCL12–CXCR4 signaling plays a critical role in directing the homing of various tissue-specific stem cell populations (Arai et al., 2005; Masztalerz et al., 2007). Transmigration of SSCs is crucial for success of germ cell transplantation because the cells must migrate from the lumen of seminiferous tubules to the basement membrane where they establish SSC-niche units. Within 1 week of transplanting, SSCs penetrate the blood–testis barrier (BTB), complete transmigration to the basement membrane and undergo self-renewal divisions to expand the population (Nagano et al., 1999). To examine whether CXCL12–CXCR4 signaling provides cues for SSC homing to the basement membrane after transplantation, cultured undifferentiated spermatogonia were rendered CXCR4 deficient by stable transduction with lentiviral vectors containing a Cxcr4 shRNA transgene. The abundance of Cxcr4 mRNA in cultures treated with Cxcr4 shRNA lentivirus was significantly (P = 0.005) reduced to only 36.7±13.8% (n = 3 different cultures) of that in control cultures treated with non-targeting shRNA lentivirus (Fig. 6A). Next, cultures of lacZ-expressing cells were stably transduced with Cxcr4 shRNA or control shRNA lentiviral transgenes followed by transplantation into recipient testes (n = 3 different cultures and 27 total recipient testes) to examine effects on colonization ability. Examination of recipient testes at 21 days after transplantation revealed that control shRNA-treated cells generated 73.7±18.8 colonies/105 cells injected, and they contributed to the expanded germ cell layers. In contrast, Cxcr4-shRNA-treated cells produced only 3.3±2.7 colonies/105 cells injected (Fig. 6B,C). To gain a better understanding of the fate of Cxcr4-shRNA-treated cells, we examined the seminiferous tubules of recipient testes for colony formation at earlier time points of 3 and 10 days after transplantation to assess migration to the basement membrane. At 3 days after transplantation, both Cxcr4-shRNA- and control-shRNA-treated SSCs were located in the center of the seminiferous tubules (Fig. 6D). At 10 days after transplantation, donor-derived colonies composed of many donor cells were evident in testes receiving control-shRNA-treated cells, indicative of SSCs that had completed transmigration to the basement membrane and started to expand in number. In contrast, CXCR4-deficient germ cells were observed as only single cells within recipient seminiferous tubules and expanded colonies were not evident (Fig. 6D). These results suggest that CXCR4 signaling is important for SSC migration and expansion following transplantation.
Fig. 6.
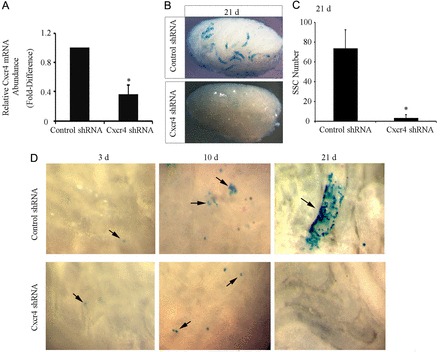
Impact of CXCR4 deficiency on SSC homing and expansion after transplantation. (A) qRT-PCR analysis for relative Cxcr4 transcript abundance in primary cultures of undifferentiated spermatogonia 24 hours after treatment with control shRNA or Cxcr4 shRNA lentivirus. Data are means ± s.e.m. for three different cultures. *Significant difference at P<0.05. (B) Representative images of recipient mouse testes 21 days after transplantation of cultured undifferentiated spermatogonia stably transduced with control or Cxcr4 shRNA lentiviral transgenes. (C) Comparison of SSC numbers in primary cultures of undifferentiated spermatogonia stably transduced with control or Cxcr4 shRNA lentiviral transgenes using functional germ cell transplantation analysis. SSCs were counted in X-gal-stained colonies of donor-derived spermatogonia in recipient testes, normalized to the number of cells (105) injected. Data are means ± s.e.m. for three different cultures and nine total recipient testes for each treatment. *Significant difference at P<0.05. (D) Representative images of seminiferous tubules 3, 10 and 21 days after transplantation with cultured undifferentiated spermatogonia stably transduced with control or Cxcr4 shRNA lentiviral transgenes. Arrows indicate lacZ-expressing donor cells.
Inhibition of CXCL12–CXCR4 signaling results in SSC loss in vivo
The findings presented above indicated that CXCL12–CXCR4 signaling regulates SSC activity in primary cultures of undifferentiated spermatogonia; however, whether a similar role exists in vivo remained undetermined. To address this question, we treated adult mice with AMD3100 to examine the effect of impaired CXCL12–CXCR4 signaling on maintenance of normal spermatogenesis. Males were treated for 7 days with a daily injection of AMD3100 and the testes were examined 28 days later for impaired spermatogenesis, thereby covering a total period of 35 days, which is the time required for a single round of spermatogenesis in mice. In testes of control mice that received vehicle only, spermatogenesis in all seminiferous tubule cross sections examined (n = 3 different mice and 30 total cross sections) appeared normal. In contrast, in testes from mice treated with AMD3100 spermatogenesis appeared abnormal and lacking the germline (Fig. 7A). Staining of these cross sections with an antibody recognizing the germ-cell-specific marker GCNA1 confirmed that tubules with disrupted spermatogenesis lacked the germline and consisted only of Sertoli cells (Fig. 7B). Quantification of this observation revealed that in 8–10% of seminiferous tubule cross sections (n = 3 different mice and 30 total cross sections) spermatogenesis was disrupted. These findings indicate that inhibition of CXCL12–CXCR4 signaling in vivo impairs the ability of SSCs to maintain the germline after one round of terminal germ cell differentiation occurs.
Fig. 7.
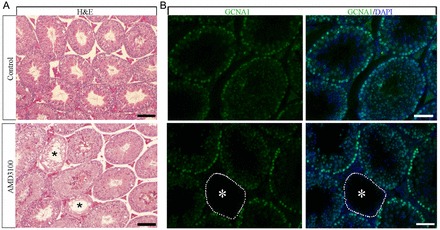
Impact of pharmacological inhibition of CXCR4 signaling on maintenance of spermatogenesis in testes of adult mice. (A) Representative images of Hematoxylin and Eosin (H&E) stained cross sections from testes of adult mice 28 days after receiving daily injections of vehicle or the CXCR4-specific inhibitor AMD3100 for 7 days. Spermatogenesis appeared normal in all seminiferous tubules in cross sections of testes from control animals. In contrast, in 8–10% of seminiferous tubules from males treated with AMD3100 (n = 3 different animals) spermatogenesis was disrupted and all germ cell types were absent (asterisks). Scale bars: 50 µm. (B) Representative images of immunofluorescent staining for the germ-cell-specific marker GCNA1 (green) in cross sections of testes from mice treated with the CXCR4 inhibitor AMD3100. Seminiferous tubules completely lacking germ cells with a Sertoli-cell-only phenotype that is indicative of SSC loss are observed (asterisks). DAPI (blue) was used to stain DNA. Scale bars: 50 µm.
The transcriptome response to CXCL12–CXCR4 signaling in SSCs
To explore the molecular mechanisms by which CXCL12–CXCR4 influences SSC activity, we conducted microarray gene expression profiling on primary cultures of undifferentiated spermatogonia treated with AMD3100 for 24 hours and compared the profile to that of vehicle-treated control cells. These analyses revealed a significant (P<0.05) increase in the abundance of 34 transcripts and decrease in the abundance of 20 transcripts of at least 1.5-fold in cells treated with AMD3100 compared with vehicle-treated controls (supplementary material Table S1). Using Gene Ontology Consortium biological process classification these transcripts were found to fall into seven functional categories including cell adhesion, translational regulation and cell-to-cell communication (Table 1).
Table 1.
Functional classification of genes upregulated or downregulated by at least 1.5-fold in primary cultures of undifferentiated spermatogonia treated with the CXCR4-specific inhibitor AMD3100 compared with vehicle-treated control cultures
| Biological process | Downregulated genes | Upregulated genes |
| Primary metabolic process | Rpl21, Rps27a | Prkg2, Rpl36, Ptpn22 |
| Cell adhesion | Tnc, Mmp13, Tnnt2, Ccbe1 | Hmgb1, Adamts5 |
| Cell cycle | Hmga2, Prkg2 | |
| Signaling transduction | Btc, Prkg2, Ereg, Fgf2, Ptpn22, Pmepa1, Gpr149 | Tax1bp3 |
| Translation | Rpl21, Mtif2, Rps27a | Rpl36, Snord115, U6, Rpl30 |
| Defense response | C3ar1 | |
| Developmental process | Angptl2 | Speer4d |
Of particular interest, the expression of several genes involved in cell adhesion including Mmp13, Tnc, Tnnt2, Ptpn22, Hmgb1 and Ccbe1 was downregulated following CXCR4 inhibition. Maintenance of SSC capacity within primary cultures of undifferentiated spermatogonia requires feeder cells that not only secret growth factors but also provide a monolayer for physical attachment of germ cell clumps that may support SSC activities. We observed that more cells appeared to be detached from the STO feeder cells in cultures treated with AMD3100 than in those treated with vehicle. Quantification of floating cells revealed that 49.1±5.2% of the total cultured germ cell population (n = 3 different cultures) was detached from feeder cells in cultures treated with AMD3100 for 48 hours, which was significantly (P<0.05) greater than the 29.2±3.2% of detached cells in vehicle-treated control cultures (supplementary material Fig. S4). These finding suggest that CXCL12–CXCR4 signaling in SSCs plays an important role in regulating adhesion to an extracellular matrix.
Discussion
Fate decisions of tissue-specific stem cells are greatly influenced by cognate niches and growth factors/cytokines are key components of these microenvironments (de Rooij, 2009; Oatley and Brinster, 2012). Although GDNF was the first identified factor to be essential for SSC maintenance (Meng et al., 2000), other factors, such as CSF-1 (Oatley et al., 2009) and WNT5a (Yeh et al., 2011) have been shown to play important roles in promoting SSC self-renewal. Importantly, maintenance of SSCs within primary cultures of undifferentiated spermatogonia requires co-culture with feeder cell monolayers even when the medium is supplemented with GDNF (Kubota et al., 2004). Therefore, feeders probably secrete undefined factors that are important regulators of SSC functions. In this study, we show that signaling by CXCR4 influences SSC maintenance in primary cultures of mouse undifferentiated spermatogonia. Furthermore, we show that CXCL12, the only known ligand for CXCR4, is expressed by STO feeder cells that support these primary cultures. Although it is possible that undiscovered factors that activate the CXCR4 are expressed by STO cells, our findings indicate that CXCL12 is an important external signal that regulates SSC functions through activation of the CXCR4.
Several lines of indirect evidence from previous studies have suggested that CXCL12–CXCR4 signaling plays an important role in regulating SSC activities (Chen et al., 2005; Payne et al., 2010). In the current study, we provided direct evidence that CXCR4+ undifferentiated spermatogonia possess SSC capacity for regeneration of spermatogenesis. Inhibition of CXCR4 signaling in cultures of undifferentiated spermatogonia was found to decrease SSC proliferation and impair homing, which was evidenced by reduced attachment to feeder cell monolayers and decrease in the number of cells capable of regenerating spermatogenesis following transplantation. Importantly, inhibition of CXCR4 signaling appeared to alter the composition of primary cultures of undifferentiated spermatogonia by increasing the percentage of cells primed to differentiate upon stimulation by retinoic acid. Immunostaining of testis cross sections revealed that CXCL12 is concentrated at the basement membrane of seminiferous tubules in testes of postnatal mice and disruption of CXCL12–CXCR4 signaling resulted in SSC loss in vivo. Together, these findings provide direct evidence that CXCL12–CXCR4 signaling plays an important role in regulating the fate decisions of mouse SSCs.
It is well established that SSCs are a component of the undifferentiated spermatogonial population in mammalian testes, yet specific molecular markers that distinguish SSC from non-stem cell progenitors are currently undefined. Using functional germ cell transplantation analysis, several selection markers for SSC enrichment have been identified including THY1 (Kubota et al., 2004), CDH1 (Tokuda et al., 2007), CD9 (Kanatsu-Shinohara et al., 2004) and ITGB1 (Shinohara et al., 1999). Among these, the THY1+ fraction is the most enriched for SSCs, containing ∼25-fold more SSCs than the unselected total testis cell population of adult mice (Kubota et al., 2004; Oatley et al., 2009). To examine SSC capacity of the CXCR4+ testis cell fraction we chose to fractionate primary cultures of undifferentiated spermatogonia because some testicular somatic cell populations (i.e. Leydig cells) also appeared to express CXCR4 and would therefore complicate assessment of SSC enrichment if isolated directly from testes. Results of transplantation analyses showed that CXCR4+ cells within primary cultures of THY1+ testis cells are capable of re-activating spermatogenesis, fulfilling the definition of SSCs. Comparing colony numbers in recipient testes between the CXCR4+ and CXCR4-depleted fractions suggests that the majority of SSCs within the undifferentiated spermatogonial population express CXCR4. These findings are similar to the hematopoietic (Nie et al., 2008) and neural cell lineages (Carbajal et al., 2010) in which CXCR4 cells represent the stem cell fraction within a heterogonous undifferentiated stem/progenitor pool.
In this study, we found that CXCL12–CXCR4 signaling induces a multifaceted response in SSCs to regulate their fate decisions. Previous studies showed that CXCL12 is a potent regulator of cell migration (Knaut et al., 2003; Molyneaux et al., 2003). Here, we show that inhibition of CXCR4 signaling in primary cultures of undifferentiated spermatogonia results in detachment from feeder cells, indicating impairment of normal cell adhesion properties that may reflect characteristics of SSC homing after transplantation. Also, SSCs deficient for CXCR4 expression failed to produce donor-derived colonies of spermatogonia after transplantation, possibly because fewer cells reached the basement membrane. This suggests that CXCL12–CXCR4 signaling influences the homing ability of SSCs to cognate niches. In support of this, results of microarray analysis revealed that the abundance of transcripts corresponding to protein products that are important for cell adhesion and migration, such as MMP13 (Yu et al., 2011), tenascin-C (Nishio et al., 2005) and Epiregulin (Zhuang et al., 2007) was significantly reduced in cultures with impaired CXCR4 signaling. These findings suggest that CXCL12 controls SSC adhesion and migration by remodeling the extracellular matrix. Previous studies suggest that ITGβ1 (Kanatsu-Shinohara et al., 2008) and RAC regulate homing of SSCs after transplantation into recipient testes (Takashima et al., 2011). However, the specific extrinsic niche factors that stimulate RAC activity and ITGβ1 expression in SSCs are undefined. Signaling from CXCL12 in HSCs was shown to activate RAC1 to regulate cell adhesion and migration (Arai et al., 2005), but whether CXCL12 signaling controls SSC homing via a RAC1-dependent mechanism awaits further investigation.
Growth factors work in concert to sustain the undifferentiated spermatogonial population. Signaling from GDNF is required for maintenance of SSCs (Meng et al., 2000; Kubota et al., 2004). However, testis cell populations expressing GFRα1 or RET, the receptor for GDNF, are not enriched for SSCs (Buageaw et al., 2005; Ebata et al., 2005; Grisanti et al., 2009). These observations suggest that GDNF signaling targets the entire undifferentiated spermatogonial population and not SSCs specifically. The role of FGF2 signaling in the postnatal testis is not clear. Addition of soluble FGF2 to the culture medium in conjunction with GDNF enhances SSC self-renewal in vitro. However, FGF2 alone does not support SSC maintenance, and self-renewal occurs without FGF2, albeit at a reduced rate (Kubota et al., 2004). Interestingly, in the present study we found that both GDNF and FGF2 regulate Cxcr4 mRNA abundance in primary cultures of undifferentiated spermatogonia. Recent studies showed that ETV5, a GDNF-regulated transcription factor in cultured SSCs, influences Cxcr4 transcription (Wu et al., 2011). Also, ETV5 was identified as a downstream target of FGF2 signaling in cultures of SSCs (Ishii et al., 2012). Taken together, these findings indicate that GDNF and FGF2 signaling work in concert to activate ETV5 that then regulates CXCR4 expression in SSCs, thereby controlling the signaling response of CXCL12 to influence self-renewal, proliferation and homing.
The transition between stem and progenitor states is a crucial fate decision for continuity of developmental cell lineages. Maintaining the stem cell state is required for self-renewal to sustain a foundational stem cell pool; whereas, transition to a progenitor state represents priming for commitment to terminal differentiation upon stimulation from extrinsic stimuli. In the present study, we found that CXCL12–CXCR4 signaling is involved in maintaining the stem cell state in primary cultures of undifferentiated spermatogonia. The percentage of cells that initiated differentiation, evidenced by transition to a KIT+ state, was significantly increased after exposure to retinoic acid in cultures subjected to inhibition of CXCR4 signaling. This finding indicates that loss of CXCR4 signaling increased the proportion of progenitor cells within the cultured population that were primed for response to extrinsic inducers of differentiation. Currently, the mechanisms by which retinoic acid signaling controls spermatogonial differentiation are not clear. In the present study, microarray profiling of gene expression revealed that inhibition of CXCR4 signaling led to a decrease of Fgf2 transcript abundance. Previous studies showed that FGF2 signaling antagonizes the response of adipose-derived stem cells to retinoic acid signaling (Quarto et al., 2008). Furthermore, during embryonic development of the gonad, FGF signaling suppresses retinoic acid signaling in male germ cells (Bowles et al., 2010). Based on these observations, it is reasonable to hypothesize that FGF2 secretion from SSCs and/or neighboring progenitors acts in a paracrine or autocrine manner to antagonize retinoic acid signaling in SSCs thereby suppressing transition to a KIT+ differentiating state.
In summary, our findings support a conclusion that CXCL12 is secreted by Sertoli cells to signal via the CXCR4 receptor in SSCs for regulation of self-renewal. The signaling response to CXCL12–CXCR4 activation appears to be multifaceted, involving the regulation of cell cycle progression, prevention of transition to a progenitor state and guiding of SSC homing to cognate niches. Also, our findings suggest that CXCL12, FGF2 and GDNF form a corporative network to influence SSC activities thus adding an important new piece of information to the understanding of the growth factor milieu that constitutes the stem cell niche in mammalian testes. Deciphering the interplay between growth factor signals in regulating fate decisions of mammalian SSCs could impact diagnosis and treatment of certain instances of male infertility. Furthermore, knowledge gained from studying SSCs may be applicable to understanding the stem cell niche of other developmental cell lineages.
Materials and Methods
Animals and antibodies
All animal procedures were approved by the Washington State University or Pennsylvania State University Institutional Animal Care and Use Committees (IACUC). B6;129S-Gt (ROSA)26Sor/J mice (designated Rosa; The Jackson Laboratory, USA) were used as germ cell donors. Recipient mice for germ cell transplantation assays were immunologically compatible F1 progeny of 129SvCP×C57BL/6J mice. Experiments not involving transplantation were conducted with germ cells or tissues from C57BL/6J mice (The Jackson Laboratory, USA).
Primary antibodies were rabbit anti-human PLZF (Santa Cruz Biotechnology, USA), goat anti-mouse GATA4 (Santa Cruz), rabbit anti-human CXCL12 (Cell Signaling Technology, USA), rat anti-mouse CXCR4 (R&D Systems, USA), rat anti-mouse GCNA1 (a generous gift from G. C. Enders, University of Kansas Medical Center, Kansas City, USA), and PE/Cy5-conjugated KIT antibody (Abcam, USA). Secondary antibodies were Alexa-Fluor-546-conjugated donkey anti-rabbit IgG, Alexa-Fluor-488-conjugated donkey anti-goat IgG, Alexa-Fluor-488-conjugated donkey anti-rat IgM, Alexa-Fluor-488-conjugated goat anti-rat IgG and Alexa-Fluor-488-conjugated donkey anti-rabbit IgG (all purchased from Invitrogen, USA).
Histological evaluation and immunofluorescent staining of testis cross sections
Testes were fixed in Bouin's solution and embedded in paraffin. For histological evaluation, sections (5 µm) were stained with Hematoxylin and Eosin. For immunofluorescent staining, after antigen retrieval, sections were incubated with 10% normal donkey or goat serum for 1 hour at room temperature. Primary antibodies were added to sections for overnight incubation at 4°C. Sections were then washed in PBS and incubated with secondary antibodies for 2 hours at room temperature. Slides were mounted with Prolong gold anti-fade reagent containing DAPI (Invitrogen, USA) then viewed by fluorescence microscopy, and digital images were captured with a DD72 microscope camera and CellSens acquisition software (Olympus, USA).
Culture of undifferentiated spermatogonia
Primary cultures of undifferentiated spermatogonia were established from the THY1+ cell fraction of mouse testes as described previously (Kubota et al., 2004; Oatley and Brinster, 2006; Oatley et al., 2011). All cultures were maintained on mitotically inactivated STO feeder cells (ATCC, USA) in mouse serum-free medium (mSFM) (Kubota et al., 2004) that was supplemented with 20 ng/ml GDNF (Peprotech, USA) and 1 ng/ml FGF2 (BD Biosciences, USA).
AMD3100 and CXCL12-blocking-antibody treatments
To examine the effects of CXCR4 inhibition on overall germ cell expansion, AMD3100 (Sigma, USA) diluted in HBSS (Invitrogen, USA) at a concentration of 100 nM was added to mSFM containing GDNF and FGF2 with 5×104 cultured germ cells and replaced every 2 days. After 7 days in culture, germ cell clumps were collected and cell numbers were determined with a hemocytometer. For transplantation analysis, the entire contents of culture wells were collected and single cell suspensions were diluted at 1×106 cells/ml in mSFM. A proportion of cells were used for RNA isolation and gene expression analyses. To examine the effect of CXCR4 inhibition on spermatogonial differentiation, 100 nM AMD3100 or HBSS was added to cultures for 24 hours followed by supplementation with DMSO or 0.5 µM AtRA (Sigma, USA) for 24 hours. To neutralize the function of CXCL12, cells were pre-treated with a CXCL12-blocking antibody (20 µg/ml, R&D Systems, USA) for 48 hours followed by supplementation with DMSO or AtRA for 24 hours. Controls were cells incubated with normal IgG in place of CXCL12-blocking antibody. The percentage of KIT-expressing cells was determined in control and treated cultures using flow cytometric analysis.
Flow cytometric analysis
Flow cytometric analysis was used to determine the percentage of cells expressing CXCR4 or KIT in primary cultures of THY1+ undifferentiated spermatogonia. Briefly, ∼1×105 cells were incubated with the CXCR4 antibody or PE/Cy5-conjugated KIT antibody in DPBS-S (Dulbecco's phosphate-buffered saline with 0.1% Fetal bovine serum, 10 mM HEPES, 10 mM sodium pyruvate, 1 mg/ml glucose and penicillin/streptomycin) for 20 minutes on ice. Cells were then washed with HBSS. For CXCR4 detection, cells were then incubated with Alexa-Fluor-488-conjugated goat anti-Rat IgG secondary antibody on ice for 20 minutes. The percentage of cells staining for CXCR4 or KIT was determined using a Guava PCA 96 personal cytometer (Guava Technologies, USA). Cells incubated with normal rat IgG as the primary antibody were used as the isotype control to set gating parameters.
Isolation of the CXCR4+ fraction from primary cultures of undifferentiated spermatogonia
Magnetic-activated cell sorting (MACS) was used to isolate CXCR4+ germ cells from primary cultures of undifferentiated spermatogonia. Briefly, single cell suspensions were incubated with biotin-conjugated antibody against CXCR4 (BD Biosciences, USA) in DPBS-S for 20 minutes on ice. Cells were then washed in DPBS-S with centrifugation followed by incubation with streptavidin-conjugated magnetic MicroBeads (Miltenyi Biotec, USA) in DPBS-S. The cell suspension was then subjected to separation using MACS separation columns (Miltenyi Biotec). Cells eluted from the columns were considered to be the CXCR4+ fraction and cells passing through the columns were considered the CXCR4-depleted fraction. Fractions were diluted in mSFM at 1×106 cells/ml in preparation for transplantation into recipient testes.
Transduction of cultured undifferentiated spermatogonia with lentivirus
Lentiviral-mediated short hairpin RNA (shRNA) was used to permanently knockdown CXCR4 expression in primary cultures of undifferentiated spermatogonia. To create lentiviral particles, pLKO.1-puro vectors containing Cxcr4 shRNA or scrambled control shRNA sequences (Sigma-Aldrich, USA) were co-transfected with envelope (pMD2.G) and packaging (psPAX2) plasmids into HEK293 cells (ATCC, USA) using Lipofectamine 2000 (Invitrogen, USA). Single cell suspensions of cultured THY1+ undifferentiated spermatogonia were incubated with Cxcr4 shRNA virus or control virus at a multiplicity of infection (MOI) of 5 in the presence of 8 µg/ml polybrene for 18 hours. Cells were then allowed to recover for 24 hours followed by puromycin (1 µg/ml) supplementation for 72 hours to select cells that had stably incorporated the vector. Cells were then collected for RNA extraction to validate knockdown efficiency or for germ cell transplantation to determine SSC capacity.
Germ cell transplantation analysis
Germ cell transplantation assays were conducted as described previously (Brinster and Zimmermann, 1994; Oatley et al., 2006) to determine SSC numbers in CXCR4+ cell fractions and the effects of CXCR4 inhibition on SSC maintenance in culture. Briefly, single cell suspensions of primary cultures established from lacZ-expressing Rosa donors were diluted in mSFM at 1×106 cell/ml, and 10 µl was microinjected into seminiferous tubules of recipient mice that had been pretreated with busulfan (60 mg/kg of body weight) at least 6 weeks previously to deplete endogenous spermatogenesis. For transplantation of Cxcr4-shRNA-treated cells, single cell suspensions were diluted at 3×106 cells/ml. Colonies of donor-derived spermatogonia within recipient testes were examined 2 months after transplantation by X-gal staining.
Immunocytochemistry
Primary cultures of THY1+ undifferentiated spermatogonia were examined for expression of CXCR4 using immunofluorescent staining. Briefly, germ cell clumps were fixed by incubation in 4% paraformaldehyde for 10 minutes followed by incubation in PBS containing 0.1% Triton X-100 (PBS-T). Nonspecific antibody binding was blocked by incubating cells in 5% normal goat and donkey serum diluted in PBS-T for 1 hour at room temperature. Primary antibodies against PLZF and CXCR4 were added to the cells, which were incubated overnight at 4°C. Cells were then washed with PBS followed by incubation with secondary antibodies. DAPI was used to stain DNA, and digital images were captured with a DD72 microscope camera and CellSens acquisition software (Olympus, USA).
Apoptosis analysis
To determine the percentage of apoptotic cells within the CXCR4+ fraction of primary cultures of undifferentiated spermatogonia, ∼1×105 cells were stained for expression of CXCR4 then processed for terminal deoxynucleotidyl transferase-mediated dUPT nick end labeling (TUNEL) using the In Situ Cell Death Detection Kit (Roche Applied Sciences, Germany) following the manufacturer's instructions. The percentage of cells positive for CXCR4 and TUNEL were evaluated by flow cytometric analysis using a Guava PCA 96 personal cytometer. Cells incubated with normal IgG as the primary antibody were used as controls to set gating parameters for determining CXCR4+ cells.
Cell proliferation analysis
To determine the percentage of actively dividing cells within the CXCR4+ fraction of primary cultures of undifferentiated spermatogonia, cells were incubated with 20 nM EdU for 4 hours and then single cell suspensions were collected and stained for CXCR4. Next, the stained cells were processed for detection of EdU that had been incorporated into the DNA of cells that were in S phase of the cycle, using the Click-iT® EdU Alexa Fluor 488 Flow Cytometry Assay Kit (Invitrogen, USA). The percentage of cells positive for CXCR4 and containing EdU was determined using flow cytometric analysis with a Guava PCA 96 personal cytometer. Cells not receiving EdU and incubated with normal IgG as the primary antibody were used as controls to set gating parameters.
Western blot analysis
Protein lysates from STO or mitomycin-C-treated STO (mtSTO) feeder cells were collected in RIPA buffer. For conditioned medium, protein samples were collected by precipitation with ice-cold acetone. For each sample, 25 µg of protein was separated by NuPAGE 4–12% Bis-Tris Gel electrophoresis (Invitrogen, USA), followed by transfer to nitrocellulose membranes. Membranes were incubated with antibody against CXCL12 (1∶1000) overnight at 4°C. Blots were washed in TBS-T and then incubated with HRP-conjugated secondary antibody (1∶5000, Santa Cruz Biotechnology, USA). Signal was detected using PicoWest chemiluminescent substrate (Thermo Scientific, USA) and digital images were captured and analyzed with a luminescent imager (Fuji Medical System). Blots were then stripped with Restore Western Blot stripping buffer (Thermo Scientific, USA) and re-probed with an antibody against TUBB1(1∶3000, Abnova, USA).
RNA isolation and DNA microarray analysis
Total cellular RNA was isolated using Trizol reagent (Invitrogen, USA) and Qiagen RNeasy columns, as described previously (Oatley et al., 2006). Transcriptome analysis was conducted using Affymetrix Mouse Gene 1.0 ST Arrays (Affymetrix, USA). Three different primary cultures of undifferentiated spermatogonia established from C57BL/6 donor mice were treated with AMD3100 or HBSS for 24 hours prior to collection of RNA samples and each was hybridized to a single array. All hybridizations and scanning of arrays were conducted by the Washington State University Genomics Core Facility. Microarray data were analyzed in R using the Limma package (Smyth and Speed, 2003). Quantified signals within arrays were averaged and normalized using the global LOWESS (LOcally WEighted Scatterplot Smoothing) regression algorithm. Contrasts were made to compare control and AMD3100-treated cells. The differentially expressed genes were selected to perform cluster analysis using the CLUSTER/TreeView software, and classified into pathways and biological functions according to the PANTHER (http://www.pantherdb.org/) classification system. A two-class paired test with P-value ≤0.05 was used to identify genes with differential expression at the 1.5-fold level between control and treated groups.
Quantitative RT-PCR
RNA samples were isolated using Trizol reagent and treated with DNase I followed by reverse transcription using oligo(d)T priming and Superscript III reverse transcriptase (Invitrogen, USA). qRT-PCR reactions were performed with validated TaqMan probes and the constitutively expressed gene ribosomal protein S2 (Rps2) using an ABI 7500 Fast Sequence Detection system (Applied Biosystems, USA). CT values for Cxcr4 in each sample were subtracted from corresponding CT values for Rps2 to generate ΔCT values. Relative abundance of Cxcr4 transcript in each sample was determined using the formula: 2−ΔΔCT.
Statistical analysis
All quantitative data are presented as means ± s.e.m. for at least three independent experiments. Differences between means were examined using the general linear model one-way ANOVA function of SAS software (SAS institute, USA). Multiple comparisons analysis was conducted using Tukey's post-hoc test. Differences between means were considered significant at P≤0.05.
Supplementary Material
Footnotes
Funding
This research was supported by the National Institutes of Child Health and Human Development [grant number HD061665 to J.M.O.]; and a Postdoctoral Fellowship from the Lalor Foundation [to Q.E.Y.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.119826/-/DC1
References
- Ara T., Nakamura Y., Egawa T., Sugiyama T., Abe K., Kishimoto T., Matsui Y., Nagasawa T. (2003). Impaired colonization of the gonads by primordial germ cells in mice lacking a chemokine, stromal cell-derived factor-1 (SDF-1). Proc. Natl. Acad. Sci. USA 100, 5319–5323 10.1073/pnas.0730719100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai A., Jin A., Yan W., Mizuchi D., Yamamoto K., Nanki T., Miura O. (2005). SDF-1 synergistically enhances IL-3-induced activation of the Raf-1/MEK/Erk signaling pathway through activation of Rac and its effector Pak kinases to promote hematopoiesis and chemotaxis. Cell. Signal. 17, 497–506 10.1016/j.cellsig.2004.09.007 [DOI] [PubMed] [Google Scholar]
- Bellvé A. R., Cavicchia J. C., Millette C. F., O'Brien D. A., Bhatnagar Y. M., Dym M. (1977). Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J. Cell Biol. 74, 68–85 10.1083/jcb.74.1.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J., Feng C. W., Spiller C., Davidson T. L., Jackson A., Koopman P. (2010). FGF9 suppresses meiosis and promotes male germ cell fate in mice. Dev. Cell 19, 440–449 10.1016/j.devcel.2010.08.010 [DOI] [PubMed] [Google Scholar]
- Brinster R. L., Zimmermann J. W. (1994). Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA 91, 11298–11302 10.1073/pnas.91.24.11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buageaw A., Sukhwani M., Ben–Yehudah A., Ehmcke J., Rawe V. Y., Pholpramool C., Orwig K. E., Schlatt S. (2005). GDNF family receptor alpha1 phenotype of spermatogonial stem cells in immature mouse testes. Biol. Reprod. 73, 1011–1016 10.1095/biolreprod.105.043810 [DOI] [PubMed] [Google Scholar]
- Carbajal K. S., Schaumburg C., Strieter R., Kane J., Lane T. E. (2010). Migration of engrafted neural stem cells is mediated by CXCL12 signaling through CXCR4 in a viral model of multiple sclerosis. Proc. Natl. Acad. Sci. USA 107, 11068–11073 10.1073/pnas.1006375107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Ouyang W., Grigura V., Zhou Q., Carnes K., Lim H., Zhao G. Q., Arber S., Kurpios N., Murphy T. L.et al. (2005). ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 436, 1030–1034 10.1038/nature03894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin P. A., Wang J. H., Redmond H. P. (2010). Hypoxia increases the metastatic ability of breast cancer cells via upregulation of CXCR4. BMC Cancer 10, 225 10.1186/1471-2407-10-225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij D. G. (2009). The spermatogonial stem cell niche. Microsc. Res. Tech. 72, 580–585 10.1002/jemt.20699 [DOI] [PubMed] [Google Scholar]
- de Rooij D. G., Okabe M., Nishimune Y. (1999). Arrest of spermatogonial differentiation in jsd/jsd, Sl17H/Sl17H, and cryptorchid mice. Biol. Reprod. 61, 842–847 10.1095/biolreprod61.3.842 [DOI] [PubMed] [Google Scholar]
- Donzella G. A., Schols D., Lin S. W., Esté J. A., Nagashima K. A., Maddon P. J., Allaway G. P., Sakmar T. P., Henson G., De Clercq E.et al. (1998). AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4, 72–77 10.1038/nm0198-072 [DOI] [PubMed] [Google Scholar]
- Drumond A. L., Meistrich M. L., Chiarini–Garcia H. (2011). Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction 142, 145–155 10.1530/REP-10-0431 [DOI] [PubMed] [Google Scholar]
- Ebata K. T., Zhang X., Nagano M. C. (2005). Expression patterns of cell-surface molecules on male germ line stem cells during postnatal mouse development. Mol. Reprod. Dev. 72, 171–181 10.1002/mrd.20324 [DOI] [PubMed] [Google Scholar]
- Grisanti L., Falciatori I., Grasso M., Dovere L., Fera S., Muciaccia B., Fuso A., Berno V., Boitani C., Stefanini M.et al. (2009). Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells 27, 3043–3052 [DOI] [PubMed] [Google Scholar]
- Huckins C., Clermont Y. (1968). Evolution of gonocytes in the rat testis during late embryonic and early post-natal life. Arch. Anat. Histol. Embryol. 51, 341–354 [PubMed] [Google Scholar]
- Ishii K., Kanatsu–Shinohara M., Toyokuni S., Shinohara T. (2012). FGF2 mediates mouse spermatogonial stem cell self-renewal via upregulation of Etv5 and Bcl6b through MAP2K1 activation. Development 139, 1734–1743 10.1242/dev.076539 [DOI] [PubMed] [Google Scholar]
- Kanatsu–Shinohara M., Toyokuni S., Shinohara T. (2004). CD9 is a surface marker on mouse and rat male germline stem cells. Biol. Reprod. 70, 70–75 10.1095/biolreprod.103.020867 [DOI] [PubMed] [Google Scholar]
- Kanatsu–Shinohara M., Takehashi M., Takashima S., Lee J., Morimoto H., Chuma S., Raducanu A., Nakatsuji N., Fässler R., Shinohara T. (2008). Homing of mouse spermatogonial stem cells to germline niche depends on beta1-integrin. Cell Stem Cell 3, 533–542 10.1016/j.stem.2008.08.002 [DOI] [PubMed] [Google Scholar]
- Kanatsu–Shinohara M., Inoue K., Ogonuki N., Morimoto H., Ogura A., Shinohara T. (2011). Serum- and feeder-free culture of mouse germline stem cells. Biol. Reprod. 84, 97–105 10.1095/biolreprod.110.086462 [DOI] [PubMed] [Google Scholar]
- Kaucher A. V., Oatley M. J., Oatley J. M. (2012). NEUROG3 is a critical downstream effector for STAT3-regulated differentiation of mammalian stem and progenitor spermatogonia. Biol. Reprod. 86, 164 10.1095/biolreprod.111.097386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H., Werz C., Geisler R., Nüsslein–Volhard C., Tübingen 2000 Screen Consortium (2003). A zebrafish homologue of the chemokine receptor Cxcr4 is a germ-cell guidance receptor. Nature 421, 279–282 10.1038/nature01338 [DOI] [PubMed] [Google Scholar]
- Kubota H., Avarbock M. R., Brinster R. L. (2003). Spermatogonial stem cells share some, but not all, phenotypic and functional characteristics with other stem cells. Proc. Natl. Acad. Sci. USA 100, 6487–6492 10.1073/pnas.0631767100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Avarbock M. R., Brinster R. L. (2004). Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 101, 16489–16494 10.1073/pnas.0407063101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masztalerz A., Zeelenberg I. S., Wijnands Y. M., de Bruijn R., Drager A. M., Janssen H., Roos E. (2007). Synaptotagmin 3 deficiency in T cells impairs recycling of the chemokine receptor CXCR4 and thereby inhibits CXCL12 chemokine-induced migration. J. Cell Sci. 120, 219–228 10.1242/jcs.03328 [DOI] [PubMed] [Google Scholar]
- Meng X., Lindahl M., Hyvönen M. E., Parvinen M., de Rooij D. G., Hess M. W., Raatikainen–Ahokas A., Sainio K., Rauvala H., Lakso M.et al. (2000). Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 287, 1489–1493 10.1126/science.287.5457.1489 [DOI] [PubMed] [Google Scholar]
- Molyneaux K. A., Zinszner H., Kunwar P. S., Schaible K., Stebler J., Sunshine M. J., O'Brien W., Raz E., Littman D., Wylie C.et al. (2003). The chemokine SDF1/CXCL12 and its receptor CXCR4 regulate mouse germ cell migration and survival. Development 130, 4279–4286 10.1242/dev.00640 [DOI] [PubMed] [Google Scholar]
- Nagano M., Avarbock M. R., Brinster R. L. (1999). Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol. Reprod. 60, 1429–1436 10.1095/biolreprod60.6.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Y., Han Y. C., Zou Y. R. (2008). CXCR4 is required for the quiescence of primitive hematopoietic cells. J. Exp. Med. 205, 777–783 10.1084/jem.20072513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishio T., Kawaguchi S., Yamamoto M., Iseda T., Kawasaki T., Hase T. (2005). Tenascin-C regulates proliferation and migration of cultured astrocytes in a scratch wound assay. Neuroscience 132, 87–102 10.1016/j.neuroscience.2004.12.028 [DOI] [PubMed] [Google Scholar]
- Oatley J. M., Brinster R. L. (2006). Spermatogonial stem cells. Methods Enzymol. 419, 259–282 10.1016/S0076-6879(06)19011-4 [DOI] [PubMed] [Google Scholar]
- Oatley J. M., Brinster R. L. (2008). Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24, 263–286 10.1146/annurev.cellbio.24.110707.175355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Brinster R. L. (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev. 92, 577–595 10.1152/physrev.00025.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Avarbock M. R., Telaranta A. I., Fearon D. T., Brinster R. L. (2006). Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. USA 103, 9524–9529 10.1073/pnas.0603332103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Oatley M. J., Avarbock M. R., Tobias J. W., Brinster R. L. (2009). Colony stimulating factor 1 is an extrinsic stimulator of mouse spermatogonial stem cell self-renewal. Development 136, 1191–1199 10.1242/dev.032243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley M. J., Kaucher A. V., Racicot K. E., Oatley J. M. (2011). Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol. Reprod. 85, 347–356 10.1095/biolreprod.111.091330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne C. J., Gallagher S. J., Foreman O., Dannenberg J. H., Depinho R. A., Braun R. E. (2010). Sin3a is required by sertoli cells to establish a niche for undifferentiated spermatogonia, germ cell tumors, and spermatid elongation. Stem Cells 28, 1424–1434 10.1002/stem.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peled A., Petit I., Kollet O., Magid M., Ponomaryov T., Byk T., Nagler A., Ben–Hur H., Many A., Shultz L.et al. (1999). Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science 283, 845–848 10.1126/science.283.5403.845 [DOI] [PubMed] [Google Scholar]
- Quarto N., Wan D. C., Longaker M. T. (2008). Molecular mechanisms of FGF-2 inhibitory activity in the osteogenic context of mouse adipose-derived stem cells (mASCs). Bone 42, 1040–1052 10.1016/j.bone.2008.01.026 [DOI] [PubMed] [Google Scholar]
- Shinohara T., Avarbock M. R., Brinster R. L. (1999). beta1- and alpha6-integrin are surface markers on mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 96, 5504–5509 10.1073/pnas.96.10.5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth G. K., Speed T. (2003). Normalization of cDNA microarray data. Methods 31, 265–273 10.1016/S1046-2023(03)00155-5 [DOI] [PubMed] [Google Scholar]
- Takashima S., Kanatsu–Shinohara M., Tanaka T., Takehashi M., Morimoto H., Shinohara T. (2011). Rac mediates mouse spermatogonial stem cell homing to germline niches by regulating transmigration through the blood-testis barrier. Cell Stem Cell 9, 463–475 10.1016/j.stem.2011.08.011 [DOI] [PubMed] [Google Scholar]
- Tokuda M., Kadokawa Y., Kurahashi H., Marunouchi T. (2007). CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol. Reprod. 76, 130–141 10.1095/biolreprod.106.053181 [DOI] [PubMed] [Google Scholar]
- Wu X., Goodyear S. M., Tobias J. W., Avarbock M. R., Brinster R. L. (2011). Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol. Reprod. 85, 1114–1123 10.1095/biolreprod.111.091793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh J. R., Zhang X., Nagano M. C. (2011). Wnt5a is a cell-extrinsic factor that supports self-renewal of mouse spermatogonial stem cells. J. Cell Sci. 124, 2357–2366 10.1242/jcs.080903 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Takakura A., Ohbo K., Abe K., Wakabayashi J., Yamamoto M., Suda T., Nabeshima Y. (2004). Neurogenin3 delineates the earliest stages of spermatogenesis in the mouse testis. Dev. Biol. 269, 447–458 10.1016/j.ydbio.2004.01.036 [DOI] [PubMed] [Google Scholar]
- Yoshinaga K., Nishikawa S., Ogawa M., Hayashi S., Kunisada T., Fujimoto T., Nishikawa S. (1991). Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development 113, 689–699 [DOI] [PubMed] [Google Scholar]
- Yu T., Wu Y., Helman J. I., Wen Y., Wang C., Li L. (2011). CXCR4 promotes oral squamous cell carcinoma migration and invasion through inducing expression of MMP-9 and MMP-13 via the ERK signaling pathway. Mol. Cancer Res. 9, 161–172 10.1158/1541-7786.MCR-10-0386 [DOI] [PubMed] [Google Scholar]
- Zhuang S., Yan Y., Daubert R. A., Schnellmann R. G. (2007). Epiregulin promotes proliferation and migration of renal proximal tubular cells. Am. J. Physiol-Renal. 293, F219–F226 10.1152/ajprenal.00082.2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


