Abstract
In eukaryotes, higher order chromatin structure governs crucial cellular processes including DNA replication, transcription and post-transcriptional gene regulation. Specific chromatin-interacting proteins play vital roles in the maintenance of chromatin structure. We have identified BEND3, a quadruple BEN domain-containing protein that is highly conserved amongst vertebrates. BEND3 colocalizes with HP1 and H3 trimethylated at K9 at heterochromatic regions in mammalian cells. Using an in vivo gene locus, we have been able to demonstrate that BEND3 associates with the locus only when it is heterochromatic and dissociates upon activation of transcription. Furthermore, tethering BEND3 inhibits transcription from the locus, indicating that BEND3 is involved in transcriptional repression through its interaction with histone deacetylases and Sall4, a transcription repressor. We further demonstrate that BEND3 is SUMOylated and that such modifications are essential for its role in transcriptional repression. Finally, overexpression of BEND3 causes premature chromatin condensation and extensive heterochromatinization, resulting in cell cycle arrest. Taken together, our data demonstrate the role of a novel heterochromatin-associated protein in transcriptional repression.
Key words: BEND3, Heterochromatin, Transcriptional repression, SUMO
Introduction
Accurate control of gene expression is crucial for cell survival and is clearly dependent on the chromatin status (Narlikar et al., 2002). Although gene activation is generally associated with euchromatic sites with increased histone acetylation, gene inactivation at heterochromatin is marked with methylation of histone H3 at lysine 9 and hypoacetylation of histones (for reviews, see Dillon and Festenstein, 2002; Hubner and Spector, 2010; Richards and Elgin, 2002). The heterochromatin protein HP1 binds to histone H3 trimethylated at lysine 9 (H3me3K9) and contributes to spreading of heterochromatin, resulting in silencing of gene expression at those sites (Bannister et al., 2001; Jacobs and Khorasanizadeh, 2002; Lachner et al., 2001; Nielsen et al., 2002). Thus, HP1 proteins are implicated in transcriptional repression by establishing specialized, higher-order chromatin structures (Eissenberg and Elgin, 2000; Kellum, 2003a; Kellum, 2003b; Kwon and Workman, 2008; Maison and Almouzni, 2004; Nielsen et al., 2001; Stewart et al., 2005). Recent evidence has also indicated that HP1 proteins participate in transcriptional repression both in heterochromatin and euchromatin (Hediger and Gasser, 2006; Kwon and Workman, 2008). Heterochromatic regions are typically composed of repetitive sequences present at centromeres and telomeres, are usually late replicating and transcriptionally silent (Buhler and Gasser, 2009; Fodor et al., 2010; Schoeftner and Blasco, 2009; Vermaak and Malik, 2009). Our understanding of the molecular network that establishes heterochromatin structure and triggers transcriptional repression remains far from complete.
Transcriptional repression is mediated through several pathways (Cowell, 1994; Johnson, 1995). One pathway involves the recruitment of chromatin regulators, including chromatin remodeling complex, that cause localized histone deacetylation that ultimately results in transcriptional repression of specific genes (Hassig et al., 1997; Kadosh and Struhl, 1998; Nan et al., 1998; Rundlett et al., 1998; Xue et al., 1998; Zhang et al., 1997). Another pathway involves the inactivation of transcription mediated by protein–protein interactions, which prevents the assembly of RNA polymerase or general transcription factors required to establish the preinitiation complex at transcription start sites (Breiling et al., 2001; Inostroza et al., 1992; Meisterernst and Roeder, 1991; Um et al., 1995). Furthermore, small ubiquitin-like modifier (SUMO) protein modification of transcription factors has been associated with repression (Geiss-Friedlander and Melchior, 2007; Gill, 2005; Hay, 2005; Yang and Sharrocks, 2004). SUMOylation is a covalent post-translational modification, whereby SUMO is attached by an iso-peptide linkage to lysine(s) residue in the target protein (Bayer et al., 1998; Johnson, 2004; Mukhopadhyay and Riezman, 2007). Recent studies have shown that SUMOylation of specific chromatin-associated factors regulates gene expression by altering chromatin architecture, which results in localized heterochromatinization and inability of the transcription machinery to interact at specific chromatin sites (Stielow et al., 2008a; Stielow et al., 2008b; Uchimura et al., 2006).
We have identified BEND3 (originally known as KIAA1553), a protein containing four BEN domains, which are thought to be involved in chromatin function and transcription. Localization studies in mammalian cells showed that BEND3 is a heterochromatin-associated protein in which the BEN domain 4 is crucial for heterochromatin association. Tethering BEND3 to a specific gene locus results in transcriptional repression, probably by altering the chromatin structure. Furthermore, BEND3 is SUMOylated and this modification is essential for its repressive function but not for its association with chromatin. Finally, overexpression of BEND3 caused premature chromatin condensation, severe heterochromatinization and cell cycle arrest. A very recent proteomic study has demonstrated that BEND3 is a component of Sall4, a repressive NuRD transcription factor complex implicated in stem cell pluripotency (van den Berg et al., 2010). We provide evidence that BEND3 associates with heterochromatin, interacts with histone deacetylases (HDACs) and Sall4, causes transcriptional repression and can shut down transcription at an actively transcribing genic region. We suggest that BEND3, in conjunction with the NuRD complex, mediates transcriptional repression of specific genes.
Results
BEND3 is highly conserved amongst vertebrates
In a screen to identify novel proteins that associate with heterochromatin and regulate heterochromatin maintenance, we identified BEND3, which contains four BEN domains (Fig. 1A). The BEN domain consists of an all α-fold with four conserved helices with a characteristic signature motif LhxxlFs (l is an aliphatic residue, s is small residue, x is any residue) in helix 2 and an aliphatic residue at the start of helix 3 (Abhiman et al., 2008). BEND3 is highly conserved only amongst vertebrates (Fig. 1B and supplementary material Fig. S1A). It has been suggested that the BEN domain mediates DNA–protein or protein–protein interactions and proteins containing this domain are generally involved in chromatin organization and transcription (Abhiman et al., 2008).
Fig. 1.
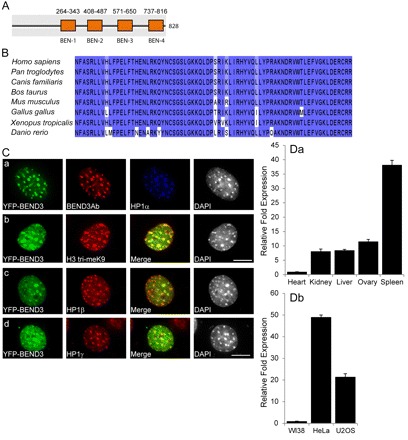
BEND3 associates with heterochromatic regions. (A) Schematic representation of the domain architecture of BEND3 (828aa). Simple modular architecture research tool (SMART)-based domain predictions show four BEN domains (numbers are amino acid positions). (B) Amino acid sequence alignment of the third BEN domain, using the ClustalW2 multiple alignment program, shows very high conservation amongst vertebrates. The color code represents the percentage of conservation, with the darkest shade representing the highest conservation. (C) BEND3 localizes at heterochromatic regions. In NIH3T3 cells, YFP–BEND3 colocalizes with HP1α-, HP1β-, HP1γ- and H3me3K9 (HE tri-meK9)-containing pericentromeric heterochromatic foci. Note that BEND3 antibody detects the transiently transfected YFP–BEND3. DNA was counterstained with DAPI. Scale bars: 10 μm. (D, a) Quantitative RT-PCR of BEND3 expression in various human tissues shows maximum expression in spleen. (b) q-RT-PCR shows higher expression of BEND3 in human cancerous cells than in primary diploid cells and human cell lines. Error bars indicate ±s.d. of three independent experiments.
BEND3 localizes to heterochromatin, which is dependent on the BEN domain 4
To determine the intracellular location of BEND3, we generated various epitope-tagged versions of BEND3, with T7 (an 11 amino acid peptide encoded in the leader sequence of T7 bacteriophage gene10), hemagglutinin (HA) or yellow fluorescent protein (YFP) at either the N-terminus or C-terminus of BEND3. YFP–BEND3 was found to be primarily nuclear with several punctate foci in mouse cells (Fig. 1C) as well as in human cells, including U2OS (supplementary material Fig. S1Ba) and HeLa (supplementary material Fig. S1Bb). Similar observations were made with BEND3–YFP (supplementary material Fig. S1Ca) and T7–BEND3 (supplementary material Fig. S1Cb) demonstrating that the protein bearing different tags on either end has similar localization. Furthermore, in mouse NIH3T3 cells, BEND3 clearly associated with HP1α-, HP1β-, HP1γ- and H3me3K9-containing heterochromatic foci (Fig. 1Ca–d). We generated several antibodies against BEND3 but none of these detected the endogenous BEND3 by immunoblots or immunofluorescence (data not shown). These antibodies, however, could detect exogenously expressed YFP–BEND3 in cells using immunofluorescence, immunoprecipitation, and also immunoblotting (Fig. 1Ca and supplementary material Fig. S1D,E). YFP–BEND3 occurs in several forms, with residues S379, S489 or S503 phosphorylated (supplementary material Fig. S1E), as has been reported for endogenous BEND3 in phospho-proteomic screens (Brill et al., 2009; Olsen et al., 2006).
We used a quantitative real-time PCR approach in order to analyze the expression of endogenous BEND3 in both mouse and human cell lines (Fig. 1D). Analysis of BEND3 expression in human tissues showed maximum levels of expression in spleen and least in heart (Fig. 1Da). Interestingly, BEND3 expression was much higher in transformed or cancerous cell-lines (human U2OS, HeLa; Fig. 1Db and mouse NIH3T3; data not shown) than in primary diploid fibroblasts (WI38; Fig. 1Db).
To determine the region of BEND3 that is essential for its localization to heterochromatin, we generated several mutants of BEND3 spanning each of the four BEN domains (Fig. 2A). YFP-tagged mutants containing the N-terminal fragments, amino acids (aa) 1–357, 1–529 and 1–660 and the C-terminal fragments aa 390–828, 551–828 and 718–828 were transfected into mouse NIH3T3 cells and the localization of these mutants was assessed (Fig. 2B). The mutants expressing YFP–BEND3.718–828 (Fig. 2Be), YFP–BEND3.551–828 (Fig. 2Bf) and YFP–BEND3.390–828 (Fig. 2Bg) localized to pericentric heterochromatic regions similar to that of wild-type BEND3 (BEND3.WT; Fig. 2Ba) suggesting that the BEN domain 4 is vital for heterochromatin localization (Fig. 2A,Be–g). Although the aa 1–357 mutant (Fig. 2Bb) showed few punctate foci, the aa 1–529 and 1–660 mutants showed homogenous nuclear localization and did not associate with heterochromatic regions detected as DAPI-dense foci (Fig. 2Bc,d). Determination of the Pearson coefficient of correlation further corroborated these observations (Fig. 2B, right side of the panel). These results indicate that BEN domain 4 is necessary and sufficient for the localization of BEND3 to heterochromatic regions.
Fig. 2.

BEN domain 4 is crucial for heterochromatin localization of BEND3. (A) Schematic representation of YFP-tagged BEND3 truncation mutants. The lengths of the bars do not truly reflect the length of the mutants. (B) NIH3T3 cells were transfected with YFP–BEND3 wild type or truncation mutants. YFP–BEND3.WT (a) and truncation mutants 718–828 (e), 551–828 (f) and 390–828 (g) localize to the heterochromatic region, indicating that the C-terminus of the protein is necessary for its localization to heterochromatin. Mutant 1–357 (b) shows large nuclear foci, whereas 1–529 (c) and 1–660 (d) are homogenously distributed. DNA was counterstained with DAPI. The Pearson coefficient of correlation is given on the right-hand side of the panel. Scale bar: 10 μm.
To examine the role of BEND3 in heterochromatin organization, we have utilized a modified version of the in vivo cell system devised by David Spector's group (Janicki et al., 2004). A 200 copy transgene array, with each array having 256 copies of the Lac operator, had been integrated into human U2OS cell as a single heterochromatic locus (U2OS-2-6-3) (Janicki et al., 2004). mCherry–LacI (Lac repressor) and rtTa (reverse tetracycline-controlled transcriptional activator) were stably integrated in the 2-6-3 cells, so that the locus containing the transgene array could be readily visualized in living cells by the presence of mCherry–LacI and the heterochromatic locus could be decondensed upon transcriptional activation by the addition of doxycycline (DOX) in the medium (U2OS-2-6-3 CLTon, mCherry–LacI tetracycline activator; Fig. 3A) (Bernard et al., 2010; Prasanth et al., 2010; Shen et al., 2010). YFP–BEND3 localized to the heterochromatic locus in the U2OS-2-6-3 CLTon cells and showed complete colocalization with HP1β (Fig. 3Ba), HP1α (supplementary material Fig. S2B) and H3me3K9 (Fig. 3Bb) at the locus. Transient transfection of BEND3 mutants in U2OS 2-6-3 CLTon cells showed that only the constructs containing BEN domain 4 localize to the heterochromatic locus, similar to our observations of NIH3T3 cells (supplementary material Fig. S2A,C).
Fig. 3.

BEND3 is displaced from decondensed chromatin during activation of transcription. (A) A schematic representation of the gene locus in U2OS 2-6-3 CLTon cells [adapted and modified from (Janicki et al., 2004)]. (B) YFP–BEND3 associates with HP1β- and H3me3K9 (HE tri-meK9)-containing heterochromatic gene locus in CLTon cells. (C, a) YFP–BEND3 (green) associates with the artificially generated heterochromatic gene locus in 2-6-3CLTon cells as shown by its association with mCherry–LacI-containing (red) gene locus. (b) Activation of transcription (DOX+) results in the displacement of BEND3 from the de-condensed gene locus. Note the appearance of CFP–SKL protein in the DOX-treated cells. The coefficient of correlation (right side of the panel) corroborates the overlap of YFP–BEND3 with mCherry–LacI at the condensed locus. (D) Immunoblot analysis of YFP–BEND3-expressing cells, using a GFP antibody, in untreated (0 hours), DOX-treated (4 hours and 8 hours) cells. Note the YFP–BEND3 levels remain constant over time, whereas the CFP–SKL signal increases. (*) denotes non-specific band. Tubulin is shown as a loading control. (E) Live-cell imaging in 2-6-3 CLTon cells demonstrating the displacement of YFP–BEND3.WT from the gene locus during transcriptional activation. The images were taken at every 20 minutes. DOX (1 μg/ml) was infused into the medium at 0.00 minutes. YFP–BEND3, which initially localized to the heterochomatic loci was gradually released upon de-condensation of the gene locus. YFP–BEND3 is still seen localized to the partially heterochromatic spots remaining at the locus. Scale bars: 10 μm.
We then examined whether association of BEND3 with chromatin is specified by the state of the chromatin structure, i.e. whether BEND3 localizes to condensed heterochromatin as well as decondensed chromatin. Significantly, changing the status of the gene locus from heterochromatin to euchromatin by activating DOX-induced transcription as is evident by the decondensation of the locus (Fig. 3Cb, red) as well as the appearance of the reporter protein cyan fluorescent protein (CFP)–SKL (serine-lysine-leucine, peroxisomal targeting signal), resulted in the release of a large fraction of YFP–BEND3 from the gene locus (Fig. 3C). Immunoblots corroborated the increase in CFP–SKL reporter protein in DOX-treated cells (Fig. 3D). Time-lapse imaging of cells expressing YFP–BEND3 clearly demonstrated the removal of YFP–BEND3 from the gene locus during chromatin decondensation (Fig. 3E and supplementary material Fig. S3A, Movies 1 and 2).
BEND3 is SUMOylated at several lysine residues
Recent studies have indicated that SUMOylation modulates the association of proteins to heterochromatic regions (Reo et al., 2010; Shin et al., 2005). Similarly, the SUMO-conjugating enzyme Ubc9/Hus5 has been implicated in heterochromatin organization and transcriptional repression (Shiio and Eisenman, 2003). Using the SILAC (stable isotope labeling with amino acids in cell culture) approach, a previous study compared target protein sets for SUMO1 and SUMO2 and found KIAA1553/BEND3 as a potential target (Vertegaal et al., 2006). Using bioinformatics tools (SUMOplot algorithm) to predict potential SUMOylation sites on BEND3 we identified two sites in BEND3 with the SUMO consensus motif ψKx(E/D) (where ψ is Val, Ile, Leu, Met or Phe, and x is any amino acid) at residues K20 and K512 (Fig. 4Aa). These two consensus sites are highly conserved in mammals (Fig. 4Ab).
Fig. 4.

BEND3 is SUMOylated. (A, a) Schematic representation of SUMOplot prediction of probable SUMOylation sites in BEND3. (b) Alignment of SUMOylation sites of BEND3 in different species showing the potential SUMO target sites are conserved among vertebrates. The SUMOylation consensus sequences (ψKxE) are highlighted. (B) Immunoblot of U2OS cells expressing YFP–BEND3 transfected with or without HA–SUMO1. Arrowheads indicate SUMOylated BEND3. (C) U2OS cells were co-transfected with YFP–BEND3 and HA–SUMO1 or HA–SUMO2 and immunoprecipitation (IP) conducted with anti-GFP antibody. Immunoblots using GFP (a) or HA (b) antibody revealed that both SUMO1 and SUMO2 SUMOylate BEND3. Note the 116 kDa band of YFP–BEND3 and an additional higher molecular mass band representing SUMOylated YFP–BEND3 (*) in the GFP immunoblot (a). The HA antibody detects SUMOylated YFP–BEND3 (*) as well as additional higher molecular mass ones either representing SUMOylated BEND3 or proteins that interact with BEND3 that are also target of SUMOylation (b). (D) SUMO1 SUMOylates BEND3. BEND3 sumo mutants (YFP–BEND3.K20R, YFP–BEND3.K512R or YFP–BEND3.SDM) co-transfected with HA–SUMO1. IP conducted using GFP antibody shows significant loss of SUMOylation in YFP–BEND3.K20R and complete loss of SUMOylation in YFP–BEND3.SDM. SUMOylated YFP–BEND3 (*) in the GFP (a) or HA antibody (b). (E) SUMO3 also SUMOylates BEND3. YFP–BEND3 and YFP–BEND3.mutants co-transfected with HA–SUMO3 show no apparent SUMOylation in YFP–BEND3.K20R and loss of additional higher molecular mass in YFP–BEND3.K512R suggesting K512 SUMOylation follows K20 SUMOylation in GFP or HA immunoblots (a and b). Arrowheads indicate nonspecific bands.
To examine the status of SUMOylation of BEND3 and to determine the lysines that serve as SUMO1 or SUMO2/3 acceptors, we transfected YFP–BEND3 with and without HA–SUMO1 or HA–SUMO2 in human U2OS cells. The SUMOylation status was assessed by immunoblots and immunoprecipitation analysis. Extracts from cells co-transfected with YFP–BEND3 and HA–SUMO1 clearly showed higher molecular mass forms representing SUMOylated BEND3 (Fig. 4B). Immunoblots using GFP antibody showed that YFP–BEND3 yielded an additional, slower migrating form in both SUMO1- and SUMO2-transfected cells (Fig. 4Ca). This form of YFP–BEND3 was also detected by HA antibody indicating that YFP–BEND3 is SUMOylated by SUMO1 or SUMO2 (Fig. 4Cb, asterisk). In addition, HA also detected other higher molecular mass forms that do not correspond to BEND3 and could represent multiple SUMOylation or SUMOylated proteins that interact with BEND3.
To determine which lysine (K) moiety is SUMOylated, we generated mutants in which K20, K512 or both sites (henceforth termed SUMO double mutant, SDM) were mutated to arginines (Fig. 4Aa). Each of the mutants was cotransfected with HA–SUMO1 (Fig. 4D) or HA–SUMO2 (supplementary material Fig. S3B) and assayed as described above. YFP–BEND3 showed two slower-migrating forms when co-transfected with HA–SUMO1 (Fig. 4Da). Mutation of K20, abolished one of the slower-migrating forms of BEND3 completely, whereas the second form was significantly reduced after transfection with either SUMO1 (Fig. 4Da) or SUMO2 (supplementary material Fig. S3Ba). The mutation in K512 also resulted in only one of the slow-migrating forms, corresponding to SUMOylation at K20 residue (Fig. 4Da and supplementary material Fig. S3Ba). It is worth noting that in the K20 mutant, a weak, slower-migrating band of YFP–BEND3 was present, suggesting that the SUMOylation at K512 predominantly depends on initial SUMOylation at K20 (Fig. 4Da and supplementary material Fig. S3Ba). When both K20 and K512 were mutated simultaneously (SDM), the YFP–BEND3 mutant showed reduced mobility in SDS-PAGE and loss of slower-migrating forms of BEND3. It is also possible that SUMO3 contributes to BEND3 SUMOylation. To address this possibility, we co-transfected HA–SUMO3 and YFP–BEND3 and observed the presence of slower migrating forms of YFP–BEND3 that were abolished in the YFP–BEND3.K20R, whereas only one of the two slower migrating bands was abolished in the YFP–BEND3.K512R mutant (Fig. 4E). The significant reduction of SUMO markers in the K20R mutant suggests that SUMOylation at K20 may be needed for efficient SUMOylation at the K512 site. Thus, SUMOylation of BEND3 probably occurs sequentially, with K20 preceding K512. Quantification of the immunoblots revealed that in YFP–BEND3.K20R mutant, SUMOylation is lost at both the sites, whereas in cells expressing the YFP–BEND3.K512R mutant, SUMOylation continues to occur at the K20 residue, with increased intensity (Fig. 4E). Our data also suggest that there is redundancy in terms of which SUMO is accepted by BEND3 at K20. Interestingly, localization studies in cells transfected with YFP–BEND3.K20R, YFP–BEND3.K512R, YFP–BEND3.K20R;K512R (SDM) revealed that these mutants are efficiently recruited to the heterochromatin in NIH3T3 cells as well as at the CLTon heterochromatic locus (supplementary material Fig. S3C and Fig. S4A and data not shown). These data suggest that SUMOylation is not crucial for the association of BEND3 to heterochromatic regions, indicating that localization of BEND3 to heterochromatin is independent of its post-translational modification by SUMOylation.
BEND3 can efficiently repress transcription
Post-translational modifications, especially SUMO modification, have broad impacts on biological processes. SUMOylation of transcription factors, chromatin-modifiers and chromatin-binding proteins has frequently been linked to transcriptional repression (Gill, 2005). To address the functional relevance of BEND3 SUMOylation and its preferential association with heterochromatic regions, we generated a triple fusion protein expressing YFP–LacI–BEND3 so that BEND3 was directly targeted to the stably integrated gene locus in 2-6-3 CLTon cells through a Lac operator–repressor interaction. As described previously, we can activate transcription of the reporter gene at the 2-6-3 locus by addition of doxycycline, which results in decondensation of the chromatin locus, activation of transcription and finally translation of the reporter protein CFP–SKL that is targeted to the cytoplasmic peroxisomes (Janicki et al., 2004; Prasanth et al., 2010). In cells that expressed YFP–LacI constructs, addition of DOX resulted in chromatin decondensation and transcriptional activation of a reporter gene locus (Fig. 5Aa). These cells also showed the presence of CFP–SKL in the peroxisomes, indicating efficient translation of the DOX-induced reporter RNA. By contrast, tethering of YFP–LacI–BEND3 did not allow DOX-induced decondensation of the chromatin locus and no visible CFP–SKL protein product was produced, suggesting that tethering BEND3 to the gene locus inhibits transcriptional activation (Fig. 5Ab). Note that a large fraction of YFP–LacI–BEND3 is predominantly localized at the locus. However, upon overexposure of the image, the punctate nuclear distribution of BEND3 becomes evident (supplementary material Fig. S4D). Similarly, in YFP–LacI–BEND3-transfected cells treated with DOX, immunoblot analysis revealed that the CFP–SKL protein levels were reduced to 69% of the level in cells transfected with YFP–LacI alone (supplementary material Fig. S4E). We believe that the CFP–SKL in the YFP–LacI–BEND3-transfected sample, came from the CLTon cells that were not transfected with YFP–LacI–BEND3. Tethering YFP–LacI–BEND3.K20R or –BEND3.K512R to the locus also did not allow activation of transcription in +DOX (DOX treated) cells (Fig. 5Ac,d). Interestingly, targeting YFP–LacI–BEND3.SDM, in the presence of DOX partially restored chromatin decondensation, with visible levels of CFP–SKL reporter protein in the cytoplasm, suggesting that SUMOylation of BEND3 is essential for its role in repression of transcription (Fig. 5Ae). The quantification of the open (decondensed) and closed (condensed) chromatin locus upon targeting of either YFP–LacI–BEND3 or its mutants to the locus in the absence or presence of DOX further corroborated that BEND3 prevents decondensation, whereas the double SUMO mutant of BEND3 restored the DOX-induced chromatin opening of the gene locus (Fig. 5B).
Fig. 5.

BEND3 represses transcription and SUMOylation of BEND3 plays crucial roles in transcriptional repression. (A, a) 2-6-3 CLTon cells show decondensation of the gene locus [YFP–LacI (green) and mCherry–LacI (red)] upon transcriptional activation (+DOX). CFP–SKL at peroxisomes is a readout for efficient translation of the reporter mRNA. Transient transfection of YFP–LacI–BEND3 (b), YFP–LacI–BEND3.K20R (c) and YFP–LacI–BEND3.K512R (d) prevents the DOX-induced decondensation of the gene locus. Note that these cells also lack CFP–SKL signal. However, transfection of YFP–LacI–BEND3.SDM (e) relieved repression. Scale bar: 10 μm. (B) Statistical analysis of condensed (closed) gene locus of 2-6-3 CLTon cells (with and without DOX activation). Error bars are ± s.d. from three independent experiments (~150 cells in each experiment). (C) BEND3 represses transcription of the reporter gene in a conventional luciferase assay. Gal4-BEND3.WT or GAL4-BEND3.SDM co-transfected with LacZ and luciferase expression vectors into U2OS cells. Relative activations of the firefly luciferase are presented after normalization against the co-transfected LacZ. BEND3 represses transcription >20 fold relative to the Gal4 backbone whereas the BEND3.SDM relieves the repression by ~50%. MAD protein was used as transcriptional repressor control. The error bars represent the means and standard deviation from three independent triplicate reactions.
To determine whether BEND3 can repress transcription, we fused BEND3 or the double SUMO mutant (SDM) of BEND3 to the C-terminus of the GAL4 DNA-binding domain and tested their role in transcriptional activity in human U2OS cells. GAL4–BEND3 expression constructs were transfected together with the GAL4–luciferase reporter, and 48 hours post-transfection, luciferase activity was measured and normalized. Although GAL4–BEND3 caused more than a 20-fold repression, the double mutant relieved the repression significantly (P=0.027, t-test; Fig. 5C). Results obtained from CLTon cells and from luciferase assays indicate that BEND3 can efficiently repress transcription, qualitatively and quantitatively and that its SUMOylation is crucial for its repressive ability.
BEND3 can shut down transcription
Using CLTon cells, we demonstrated that BEND3 at the gene locus represses induction of transcription. Next we wanted to determine whether targeting BEND3 to the transcriptionally active gene locus has any effect on continuous transcription. We activated transcription in the CLTon cells by addition of doxycycline. After 6 hours, the cells were transfected with YFP–LacI, YFP–LacI–BEND3 or YFP–LacI–BEND3.SDM and the decondensation at the locus and the cellular levels of CFP–SKL reporter protein were assessed 24 hours post-transfection (Fig. 6A). In YFP–LacI-expressing control cells the locus remained decondensed with efficient CFP–SKL reporter translation (Fig. 6Ba). However, in a large fraction of YFP–LacI–BEND3-expressing cells the locus was condensed (>70%, n=285) and there were reduced levels of CFP–SKL reporter protein (Fig. 6Bb). In the BEND3.SDM-expressing cells the chromatin was decondensed (similar to that observed for control YFP–LacI-transfected cells; Fig. 6Bc,C) and CFP–SKL protein was present. These data indicate that BEND3 can cause recondensation of a highly transcribing gene locus and efficient repression of transcription.
Fig. 6.

Tethering BEND3 to a transcriptionally active gene locus results in inhibition of transcription. (A) Schematic representation of the experiment. (B) Introduction of YFP–LacI, YFP–LacI–BEND3.WT and YFP–LacI–BEND3.SDM in actively transcribing cells shows a decondensed gene locus and CFP–SKL signal in YFP–LacI-expressing cells (a; n=275); a condensed locus and absence of CFP–SKL in YFP–LacI–BEND3.WT (b; n=285) and a decondensation of the locus and apparent CFP–SKL in YFP–BEND3.SDM-expressing cells (c; n=311). Scale bar: 10 μm. (C) Statistical analysis of the condensed locus in the above experiment. Error bars represent the means and standard deviation from three independent experiments. (D) Co-transfection of HA–BEND3 and FLAG–HDAC1, –HDAC2 or –HDAC3, followed by immunoprecipitation with HA antibody shows interaction of BEND3 with HDAC2 and HDAC3. (E) Co-transfection of HA–BEND3 and FLAG–Sall4, followed by immunoprecipitation with either HA or FLAG antibody shows interaction between BEND3 and Sall4.
To determine whether the transcriptional repression induced by BEND3 is exerted through its interaction with histone deacetylases, we co-transfected HA–BEND3 with FLAG–HDAC1, 2 or 3 and carried out immunoprecipitation using HA antibody, and immunoblots with HA and FLAG antibodies. Our data showed interaction of BEND3 with HDAC2 and 3 (Fig. 6D). Recent proteomic analysis has identified that BEND3 is a component of Sall4, a transcription repressor that also associates with the NuRD complex (van den Berg et al., 2010). We carried out co-immunoprecipitation of BEND3 and Sall4 in cells co-transfected with HA–BEND3 and FLAG–Sall4. BEND3 and Sall4 could efficiently interact with each other (Fig. 6E), corroborating the previously reported proteomic analysis. The interaction of BEND3 with HDAC and Sall4 suggests that the ability of BEND3 to repress transcription is exerted through its interaction with histone deacetylases as well as Sall4.
BEND3-tethered gene locus fails to recruit RNA polymerase II
To determine how BEND3 represses transcription, we examined the recruitment of various transcription factors to the gene locus in the presence of BEND3. In CFP–LacI-expressing control cells, DOX treatment resulted in the production of reporter mRNA, as observed by the accumulation of MS2BP–YFP (MS2BP–YFP binds to the bacteriophage MS2-repeat-containing reporter RNA) at the gene locus in ~76% of cells (Fig. 7Aa and supplementary material Fig. S4Ca). CFP–LacI–BEND3-expressing cells did not show accumulation of MS2BP–YFP at the locus, demonstrating the absence of active transcription (Fig. 7Ab and supplementary material Fig. S4Ca). However, CFP–LacI–BEND3.SDM-expressing cells continued to respond to DOX and activate transcription (Fig. 7Ac and supplementary material Fig. S4Ca). We then examined the association of RNA polymerase using an antibody that recognizes the initiation competent form of RNA polymerase (pol) II [H14 antibody recognizes Ser5 phosphorylated CTD (C-terminal domain)] (Bregman et al., 1995) (Fig. 7B) as well as an antibody that recognizes the unphosphorylated form of RNA pol II (8WG16, supplementary material Fig. S4B). Whereas RNA polymerase was clearly present at the gene locus in 76% of CFP–LacI-expressing cells (Fig. 7Ba and supplementary material Fig. S4Cb), only 40% of CFP–LacI–BEND3-expressing cells showed recruitment of RNA polymerase II (Fig. 7Bb and supplementary material Fig. S4Cb). The CFP–LacI–BEND3.SDM-expressing cells, however, efficiently recruited RNA pol II to the locus (Fig. 7Bc and supplementary material Fig. S4Cb). Similar results were obtained with the 8WG16 antibody, with RNA pol II associated with the CLTon locus in 61% of CFP–LacI-expressing cells, 25% of CFP–LacI–BEND3-expressing cells and 60% of CFP–LacI–BEND3.SDM-expressing cells (supplementary material Fig. S4B). These results were further corroborated by the association of Cdk9, a component of the phosphorylated TEF-B kinase complex that phosphorylates the Ser2 of RNA pol II CTD, which is primarily the elongation-competent form of RNA pol II. Cdk9 was not enriched in the DOX-induced gene locus in ~74% of the cells expressing CFP–LacI–BEND3 (Fig. 7Cb and supplementary material Fig. S4Cc) but continued to associate with the locus in cells expressing CFP–LacI or CFP–LacI–BEND3.SDM (Fig. 7Ca,c and supplementary material Fig. S4Cc). These data indicate that the BEND3-tethered gene locus failed to assemble the transcription pre-initiation complex possibly by altering the chromatin structure at that site. Finally, we analyzed the recruitment of the rtTa transcription activator at the gene locus in the presence of BEND3. Interestingly, both CFP–LacI-as well as CFP–LacI–BEND3-expressing cells showed significant accumulation of YFP–rtTA at the locus (Fig. 8a,b) suggesting that BEND3 does not influence the recruitment of transcription activators to the transcription site.
Fig. 7.

BEND3 inhibits pre-initiation complex assembly at the reporter gene locus in CLTon cells. (A) Recruitment of MS2BP–YFP as an indicator of active transcription at the gene locus (+DOX) in cells expressing CFP–LacI (a), CFP–LacI–BEND3 (b) or CFP–LacI–BEND3.SDM (c). (B) Immunofluorescence localization of the transcription-initiation-competent form of RNA pol II using H14 antibody in cells expressing CFP–LacI (a) CFP–LacI–BEND3 (b) or CFP–LacI–BEND3.SDM (c). (C) Recruitment of YFP–CDK9 at the gene locus (+DOX) in cells expressing CFP–LacI (a), CFP–LacI–BEND3 (b) and CFP–LacI–BEND3.SDM (c). Note the absence of RNA pol II and CDK9 at the gene locus in CFP–LacI–BEND3-expressing cells. Scale bar: 10 μm. Note the loss of CFP–SKL signal in cells where H14 staining has been conducted following a pre-extraction procedure using detergent.
Fig. 8.

The rtTa activator is recruited to the locus irrespective of BEND3 localization. Recruitment of YFP–rtTa (activator) at the gene locus (+DOX) in cells expressing CFP–LacI (a) and CFP–LacI–BEND3 (b).
Over-expression of BEND3 induces hyper heterochromatinization
Our data demonstrated that BEND3 localizes to heterochromatin and is involved in transcriptional repression. To address the functional relevance of BEND3 association with heterochromatin, we carried out BEND3 over-expression studies in mouse and human cells. Strikingly, BEND3-overexpressing cells showed premature chromatin condensation as evident by a ‘worm-like’ appearance of DAPI that overlapped with YFP–BEND3 in human U2OS cells (~11% of cells transfected with 500 ng YFP–BEND3 and ~32% with 1 μg YFP–BEND3; Fig. 9Ac, supplementary material Fig. S5A), HeLa (Fig. 9Bb), MCF7 (data not shown) and mouse embryonic fibroblasts (Fig. 9Cb). The over-heterochromatinized regions were surprisingly not labeled by H3me3K9-containing heterochromatin (supplementary material Fig. S5Be) but were decorated by the polycomb-mediated heterochromatin proteins detected by EZH2 and H3K27 staining (compare supplementary material Fig. S5Ba,c with S5Bd,f). The overall distribution of H3me3K9 was also significantly altered, suggesting that there was change in global chromatin organization in BEND3-overexpressed cells (compare, supplementary material Figure S5Bb and e). Overexpression of YFP–C1 vector alone did not alter chromatin structure (Fig. 9Aa) indicating that overexpression of BEND3 influences chromatin structure. Note that the cells expressing lower levels of BEND3 showed punctate nuclear staining of BEND3 (Fig. 9Ab,Ba,Ca and supplementary material Fig. S1B). Interestingly, in generating the YFP–BEND3-expressing stable cell line, transfection of 1–5 μg (supplementary material Fig. S1D) of YFP–BEND3 into human U2OS cells followed by drug selection resulted in complete cell death within 5 days, indicating that overexpression of BEND3 is detrimental to cell survival. Finally, the stable cell line was generated using 100 ng of YFP–BEND3 (supplementary material Fig. S5C and Fig. S1E) suggesting that the levels of BEND3 are very carefully maintained in the cell. Furthermore, exogenous overexpression of BEND3 (>1 μg DNA) in human cells resulted in cell cycle arrest, which was determined by immunofluorescence analyses using mini-chromosome maintenance (MCM) and proliferating cell nuclear antigen (PCNA) antibodies (supplementary material Fig. S5D). The wild-type BEND3-expressing cell population consisted of 46.6% MCM-positive (+)/PCNA-negative (−; G1 phase), 49% MCM+/PCNA+ (early S), 1.5% MCM−/PCNA+ (late S) and 2.9% MCM−/PCNA− (G2) in the entire population (Prasanth et al., 2004a). By contrast, the control cell population consisted of 46.4% MCM+/PCNA−, 30% MCM+/PCNA+, 3.6% MCM−/PCNA+ and 20% MCM−/PCNA−. These results suggested that BEND3 overexpression halts cell cycle progression with cells arresting in early S phase (supplementary material Fig. S5D).
Fig. 9.

Overexpression of BEND3 causes hyper heterochromatinization. (A-C) Transient overexpression of YFP–BEND3 (but not YFP-C1 vector alone Aa) (1–2 μg) in U2OS cells (Ac), HeLa (Bb) and MEF (Cb) causes premature chromosome condensation and extensive heterochromatinization. Note the worm-like appearance of DNA as visualized by DAPI. Low levels of BEND3 expression with nuclear punctate localization are evident in Ab, Ba and Ca. DNA was counterstained with DAPI. Scale bar: 10 μm.
Our data demonstrate that BEND3 localizes to heterochromatic regions and its overexpression causes extensive heterochromatinization in mammalian cells. Furthermore, BEND3 is SUMOylated, and this modification is crucial for its function in repressing transcription.
Discussion
In eukaryotic cells, repression of transcription is a key mode of gene regulation and is modulated by several transcriptional repressors that have been classified as passive or active (Thiel et al., 2004). Passive repressors compete with activators for DNA binding, whereas active repressors influence chromatin organization through histone deacetylation or histone methylation and heterochromatin formation (Thiel et al., 2004). The methylation on histone H3 at K9 serves as a binding site for the heterochromatin protein HP1 that through homo- and heteromerization results in spreading of heterochromatin status in the adjoining areas (Brasher et al., 2000; Cowieson et al., 2000; Li et al., 2002; Maison and Almouzni, 2004; Ye et al., 1997). Thus the interplay of histone methyltransferases and HP1 is crucial in the establishment and maintenance of heterochromatic sites that ensure gene silencing at those sites.
Protein domains are independent units within a given protein that can function and exist as stable units. The presence of protein domains is often linked to specific functions; for example, basic leucine zipper domains are found in several DNA-binding proteins (Busch and Sassone-Corsi, 1990), cadherin repeats mediate cell–cell adhesion (Hatzfeld, 1999), zinc-finger DNA-binding domains bind DNA (Berg, 1990), WD domains mediate protein–protein interactions (Smith, 2008). Prediction based on secondary structure alignment has identified a novel domain BEN, an all α-fold with four conserved helices. The BEN domain appears to be lost from nematodes and urochordates, suggesting that it is an early lineage-specific advancement in animals (Abhiman et al., 2008). BEN domain proteins have been suggested to be involved in protein–protein or protein–DNA interactions that mediate chromatin organization and/or transcription (Abhiman et al., 2008). NAC1, a BEN domain-containing protein has been shown to bind to HDACs (Korutla et al., 2007), whereas SMAR1 is a transcriptional repressor by virtue of its interaction with the SIN3 complex (Rampalli et al., 2005). We have identified BEND3, a quadruple BEN-domain-containing protein that can efficiently repress transcription. We have also demonstrated that BEND3 associates with heterochromatin and upon overexpression causes premature chromatin condensation (PCC), extensive heterochromatinization and cell cycle arrest. This is similar to overexpression of nimA in fission yeast. Accumulation of NIMA, a protein kinase required for G2–M transition, has been shown to induce extensive chromatin condensation (Krien et al., 1998; O'Connell et al., 1994). In a subset of microcephaly patient cells PCC has been observed and mutations in MCPH1 or condensins have been linked to such hyper-condensed DNA (Griffith et al., 2008; Wood et al., 2008). Previous studies have demonstrated that distinct heterochromatic structures are formed upon oncogene-induced cellular senescence, which efficiently result in silencing of E2F target genes (Narita et al., 2003). Although several factors can lead to PCC, we suggest that overexpression of BEND3 results in global repression of transcription by altering chromatin structure. The heterochromatinized sites are enriched in the polycomb protein EZH2, a member of the PRC2 complex that is known to bring about repression through a process of heterochromatinization (Cao et al., 2002; Margueron et al., 2008).
A recent proteomic study to identify Oct4-interacting proteins in embryonic stem cells has led to the identification of several transcription factors, including Sall4, Tcfcp2l1, Esrrb and Dax1 (van den Berg et al., 2010). Sall4, Tcfcp2l1 and Esrrb were found to associate with BEND3 and Nac1, in addition to their interaction with the NuRD and SWI–SNF complex (van den Berg et al., 2010). Nac1, also a BEN-domain-containing protein has previously been reported to associate with HDACs (Korutla et al., 2007). Proteomic analysis of Sall4, Tcfcp2lI and Esrrb has revealed that BEND3 and Nac1 could be part of a complex that associates with NuRD and mediates stem cell pluripotency (van den Berg et al., 2010). The NuRD complex is known to have chromatin remodeling and deacetylase activity, which together regulate gene expression (Denslow and Wade, 2007). It is also well established that NuRD, through its interaction with specific transcription factors, regulates the expression of specific genes. We provide evidence that BEND3 associates with Sall4 and HDACs. There is also recent evidence that Sall4 associates with the NuRD and represses transcription of genes (PTEN and SALL1) involved in embryonic stem cell leukemogenesis and kidney development (Lu et al., 2009). Sall4, a pluripotency gene, is present in adult tissues in the hematopoietic stem cells (HSC) as well as leukemic stem cells and plays a key role in self-renewal of stem cells, possibly by recruiting epigenetic modulators to specific gene targets (Lu et al., 2009). The interaction of BEND3 with Sall4, its significant enrichment in spleen, a niche for HSC, tempts us to propose that BEND3 is an important molecule in stem cell pluripotency. Strikingly, BEND3 also associates with HDAC2, a component of the NuRD complex that represses genes involved in cell signaling pathways (Wang et al., 2009).
SUMOylation of BEND3 is required for its ability to repress transcription. SUMO modification has previously been linked to transcriptional repression (Gill, 2005), however, the mechanism by which SUMOylation represses transcription remain to be elucidated. It has been suggested that SUMOylated proteins are involved in recruitment of HDACs to chromatin sites, which in turn results in histone deacetylation. Recent work has also shown that SUMOylation is crucial for proper heterochromatin organization in Drosophila through the modification of SU(VAR)3-7 at K839 (Reo et al., 2010) as well as in MBD1 and MCAF1 (Uchimura et al., 2006). Similarly, SUMO-modified Sp3 has been shown to repress transcription by promoting locus-specific heterochromatic gene silencing (Stielow et al., 2008b). SUMO modification has also been implicated in the maintenance of heterochromatin stability in fission yeast (Shin et al., 2005). Interestingly, the SUMO-deficient mutant of BEND3 cannot repress transcription, but continues to be associated with heterochromatin in mammalian cells, suggesting that these two events are independent.
Utilizing an artificial in vivo gene locus we have demonstrated that BEND3 inhibits transcription. Similarly, BEND3 can shutdown transcription of an actively transcribing gene locus when introduced exogenously. We further provide evidence that a BEND3-tethered gene locus fails to recruit RNA pol II and associated factors. This might be similar to a group of repressors, including NC2 and Eve, that inhibit transcription by ‘quenching’ through interaction with activators or general transcription factors thereby inhibiting the interaction between TBP and TFIIA and TFIIB (Kamada et al., 2001; Kim et al., 2003; Li and Manley, 1998; Um et al., 1995). Similarly, ZNF76 represses transcription through its interaction with TBP and further SUMOylation modulates this repression (Zheng and Yang, 2004).
BEND3 is located at 6q21 in humans, a region frequently deleted in leukemias and lymphomas (Zhang et al., 2000), and also associated with several other human malignancies including carcinomas of breast, ovary and prostate (Hyytinen et al., 2002; Morelli et al., 2002; Orphanos et al., 1995a; Orphanos et al., 1995b). A recent study reported a possible tumor suppressor HACE1 at that locus (Zhang et al., 2007). Interestingly, the overexpression of BEND3 causes excessive heterochromatinization and transcriptional shut-down, and therefore in the endogenous context its fine-tuned balance in the cell is crucial for the maintenance of cellular homeostasis.
Materials and Methods
Cell culture and plasmids
HeLa, MCF7, U2OS and wild-type mouse embryonic fibroblast cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing high glucose, supplemented with penicillin–streptomycin and 10% fetal bovine serum (FBS). U2OS-2-6-3 CLTon cells were grown in DMEM with 10% Tet system approved FBS (Clontech). NIH3T3 cells were gown in DMEM supplemented with 10% bovine calf serum (Hyclone).
The coding region of human BEND3 was PCR amplified and cloned into pEYFP-C1 (Clontech), pEYFP-N3, pEYFP–LacI vector and pECFP-LacI vector (modified from pEGFP-LacI; kindly provided by Miroslav Dundr) (Kaiser et al., 2008). BEND3 truncation mutants were created by PCR and inserted into pEYFP-C1 vector. NLS signal sequence was added to the C-terminal mutants. Site-directed mutagenesis was used to make SUMO mutants in pEYFP–BEND3 or pEYFP-LacI-BEND3 as template as per the manufacturer's instructions (Stratagene). The full-length SUMO1 and SUMO2 genes were PCR amplified from cDNA and cloned into pCGN vector and HA–SUMO3 was a gift from Jie Chen (University of Illinois at Urbana-Champaign, Urbana, IL, USA). FLAG–Sall4 was a gift from Hitoshi Niwa and Ryuichi Nishinakamura (Niwa et al., 1991; Yuri et al., 2009). All clones were confirmed by sequencing.
The NCBI accession numbers for BEND3 sequences are: Homo sapiens, NP_001073919.1; Pan troglodytes, XP_527466.2; Bos taurus, XP_603724.2; Mus musculus, NP_950193.1; Canis familiaris, XP_539070.2; Gallus gallus, XP_419805.2; Danio rerio, XP_001923103.1.
qPCR analysis for BEND3
The expression level of BEND3 was quantified against a standard curve by real-time RT-PCR using the SYBR Green I fluorogenic dye in the StepOnePlus Real-Time PCR System (Applied Biosystems) and data were analyzed using the StepOne system software. The primer sets for human BEND3 were: forward 5′-GCAGGACTCCAGCAAACGAAAG-3′, reverse 5′-GCTGTTCTCACGGTTCCGCATG-3′. Using Q-Gene, a Microsoft Excel script package (Muller et al., 2002), the qPCR values of BEND3 were first normalized against the levels of GAPDH of the same samples and then adjusted such that the samples with lowest expression of BEND3 [heart (see Fig. 1Da) and WI38 cells (see Fig. 1Db)] had a value of 1. The data is presented as mean normalized expression.
Transfection
For transient transfection, Lipofectamine 2000 (Invitrogen) was used according to the manufacturer's protocols. For transfection in U2OS 2-6-3 CLTon cells, 100 ng CFP or YFP–LacI constructs were used. For co-transfection, 100 ng LacI constructs and 500 ng plasmid DNA of the desired proteins were used. Cells were fixed in 2% (w/v) formaldehyde, 16–24 hours post-transfection, and then DAPI stained and mounted in p-phenylenediamine (PPD). In order to induce transcription from the locus in U2OS 2-6-3 CLTon cells, 1 μg/ml DOX was added 16–24 hours post-transfection for 4 hours.
Immunofluorescence analysis
Immunofluorescence analysis was carried out as described previously (Prasanth et al., 2004b). Briefly, cells were either fixed in 2% formaldehyde for 15 minutes at room temperature and then permeabilized in 0.5% Triton X-100 in PBS for 7 minutes on ice, or pre-extracted in CSK buffer (10 mM PIPES pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2) + 0.5% Triton X-100 for 5 minutes on ice and then fixed with 2% formaldehyde. Chilled methanol was applied (for 5 minutes) afterwards if needed, according to the specific requirement for the antibodies. The 2-6-3 CLTon cells were pre-extracted in CSK buffer + 0.5% Triton X-100 for 3 minutes and fixed with 1% formaldehyde for 5 minutes. To block the cells PBS + 1% normal goat serum (NGS) was used. It was incubated with primary antibodies in a humidified chamber for 1 hour and then secondary antibody was added for 45 minutes. All washes between each step were done in PBS + 1% NGS. DNA was stained with DAPI. Cells were mounted in either PPD or Vectashield (Vector Laboratories Inc.). Cells were examined using a Zeiss Axioimager z1 fluorescence microscope (Carl Zeiss Inc.) equipped with Chroma filters (Chroma Technology). Axiovision software (Zeiss) was used to collect digital images from a Hamamatsu ORCA-cooled CCD camera. Images were also acquired using a Delta Vision optical sectioning deconvolution instrument (Applied Precision) on an Olympus microscope.
The antibodies used for immunofluorescence were: anti-MCM3 (1:400), anti-PCNA PC10 (1:150), anti-HP1α (1:100; Chemicon), anti-HP1β (1:100; Chemicon), anti-H3–Tri-MeK9 (1:300; Upstate), anti-EZH2 (1:200; BD Biosciences), anti-H3K27 (1:400; Upstate), anti-RNA POLII (H14, 1:50; 8WG16, 1:40).
Live-cell microscopy
U2OS 2-6-3 CLTon cells transiently transfected with 100 ng pEYFP-BEND3 were used for live-cell imaging. 24 hours after transfection, the cells were transferred to a FCS2 live-cell chamber (Bioptechs Inc.) mounted onto the stage of a Delta Vision optical sectioning deconvolution instrument (Applied Precision) on an Olympus microscope and kept at 37°C in L-15 medium (minus Phenol Red) containing 30% FBS. After 20 minutes 1 μg/ml DOX was infused and time-lapse images acquired with a 633 1.42 NA objective lens were captured using a Coolsnap CCD camera.
Immunoprecipitation and immunoblotting
U2OS cells were co-transfected with 500 ng YFP–BEND3 and 1 μg HA–SUMO1, HA–SUMO2 or HA–SUMO3. Cells were lysed, 24 hours post-transfection, in RIPA buffer containing 25 mM Tris (pH 8), 125 mM NaCl, 0.1% NP-40, 1 mM EDTA, with or without 25 mM N-ethylmaleimide, and protease and phosphatase inhibitors for 30 minutes. Lysate was pre-cleared with Gammabind Sepharose beads for 1 hour and incubated with appropriate antibodies overnight. The immune complex was recovered with Gammabind Sepharose beads by incubating for 1.5 hours. The beads were washed three times in RIPA buffer and 60 μl Laemmli buffer was added. The complex was heat denatured at 95°C for 5 minutes. For immunoblots, anti-GFP (1:500; Covance), anti-HA (1:1000), anti-FLAG (M2 FLAG 1:500; Sigma) and anti-α-tubulin (1:10,000; Sigma-Aldrich) antibodies were used.
Luciferase assay
BEND3.WT and BEND3.SDM were PCR amplified and cloned into the pGAL4 vector under the SV40 promoter. Gal4–BEND3 (200 ng) or GAL4–BEND3.SDM (200 ng) co-transfected with β-galactosidase expression plasmid CMVβ (100 ng) and luciferase expression plasmid (100 ng) into U2OS cells using Lipofectamine 2000 reagent. Cells were lysed 48 hours post-transfection using Bright-Glo lysis buffer (Promega) and luciferase activity was measured using a Bright-Glo luciferase assay kit (Promega). Galactosidase activity was measured using chlorophenol red–β-D-galactosidase (CPRG) as substrate and used for transfection normalization. Relative activations of the firefly luciferase are presented after normalization against the co-transfected LacZ MAD-DBD, which was used as a transcriptional repressor control.
Supplementary Material
Acknowledgments
We thank members of the Prasanth laboratory for discussions and suggestions. We thank J. Chen, M. Dundr, B. Freeman, T. Nakamira, R. Nishinakamura, H. Niwa, E. Seto, D. Spector, B. Stillman, E. T. Yeh and R. Zheng for providing reagents and suggestions. We thank P. Newmark and D. Rivier for critical reading of the manuscript. This work was supported by the start-up funds, UIUC and NSF award (0843604) to S.G.P., and ACS award RSG-11-174-01RMC to K.V.P.
Footnotes
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.086603/-/DC1
References
- Abhiman S., Iyer L. M., Aravind L. (2008). BEN: a novel domain in chromatin factors and DNA viral proteins. Bioinformatics 24, 458-461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A. J., Zegerman P., Partridge J. F., Miska E. A., Thomas J. O., Allshire R. C., Kouzarides T. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120-124 [DOI] [PubMed] [Google Scholar]
- Bayer P., Arndt A., Metzger S., Mahajan R., Melchior F., Jaenicke R., Becker J. (1998). Structure determination of the small ubiquitin-related modifier SUMO-1. J. Mol. Biol. 280, 275-286 [DOI] [PubMed] [Google Scholar]
- Berg J. M. (1990). Zinc finger domains: hypotheses and current knowledge. Annu. Rev. Biophys. Biophys. Chem. 19, 405-421 [DOI] [PubMed] [Google Scholar]
- Bernard D., Prasanth K. V., Tripathi V., Colasse S., Nakamura T., Xuan Z., Zhang M. Q., Sedel F., Jourdren L., Coulpier F., et al. (2010). A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 29, 3082-3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasher S. V., Smith B. O., Fogh R. H., Nietlispach D., Thiru A., Nielsen P. R., Broadhurst R. W., Ball L. J., Murzina N. V., Laue E. D. (2000). The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 19, 1587-1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman D. B., Du L., van der Zee S., Warren S. L. (1995). Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J. Cell Biol. 129, 287-298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiling A., Turner B. M., Bianchi M. E., Orlando V. (2001). General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412, 651-655 [DOI] [PubMed] [Google Scholar]
- Brill L. M., Xiong W., Lee K. B., Ficarro S. B., Crain A., Xu Y., Terskikh A., Snyder E. Y., Ding S. (2009). Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5, 204-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler M., Gasser S. M. (2009). Silent chromatin at the middle and ends: lessons from yeasts. EMBO J. 28, 2149-2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch S. J., Sassone-Corsi P. (1990). Dimers, leucine zippers and DNA-binding domains. Trends Genet. 6, 36-40 [DOI] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R. S., Zhang Y. (2002). Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298, 1039-1043 [DOI] [PubMed] [Google Scholar]
- Cowell I. G. (1994). Repression versus activation in the control of gene transcription. Trends Biochem. Sci. 19, 38-42 [DOI] [PubMed] [Google Scholar]
- Cowieson N. P., Partridge J. F., Allshire R. C., McLaughlin P. J. (2000). Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 10, 517-525 [DOI] [PubMed] [Google Scholar]
- Denslow S. A., Wade P. A. (2007). The human Mi-2/NuRD complex and gene regulation. Oncogene 26, 5433-5438 [DOI] [PubMed] [Google Scholar]
- Dillon N., Festenstein R. (2002). Unravelling heterochromatin: competition between positive and negative factors regulates accessibility. Trends Genet. 18, 252-258 [DOI] [PubMed] [Google Scholar]
- Eissenberg J. C., Elgin S. C. (2000). The HP1 protein family: getting a grip on chromatin. Curr. Opin. Genet. Dev. 10, 204-210 [DOI] [PubMed] [Google Scholar]
- Fodor B. D., Shukeir N., Reuter G., Jenuwein T. (2010). Mammalian Su(var) genes in chromatin control. Annu. Rev. Cell Dev. Biol. 26, 471-501 [DOI] [PubMed] [Google Scholar]
- Geiss-Friedlander R., Melchior F. (2007). Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947-956 [DOI] [PubMed] [Google Scholar]
- Gill G. (2005). Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15, 536-541 [DOI] [PubMed] [Google Scholar]
- Griffith E., Walker S., Martin C. A., Vagnarelli P., Stiff T., Vernay B., Al Sanna N., Saggar A., Hamel B., Earnshaw W. C., et al. (2008). Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat Genet. 40, 232-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassig C. A., Fleischer T. C., Billin A. N., Schreiber S. L., Ayer D. E. (1997). Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89, 341-347 [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. (1999). The armadillo family of structural proteins. Int. Rev. Cytol. 186, 179-224 [DOI] [PubMed] [Google Scholar]
- Hay R. T. (2005). SUMO: a history of modification. Mol. Cell 18, 1-12 [DOI] [PubMed] [Google Scholar]
- Hediger F., Gasser S. M. (2006). Heterochromatin protein 1, don't judge the book by its cover! Curr. Opin. Genet. Dev. 16, 143-150 [DOI] [PubMed] [Google Scholar]
- Hubner M. R., Spector D. L. (2010). Chromatin dynamics. Annu. Rev. Biophys. 39, 471-489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyytinen E. R., Saadut R., Chen C., Paull L., Koivisto P. A., Vessella R. L., Frierson H. F., Jr, Dong J. T. (2002). Defining the region(s) of deletion at 6q16-q22 in human prostate cancer. Genes Chromosomes Cancer 34, 306-312 [DOI] [PubMed] [Google Scholar]
- Inostroza J. A., Mermelstein F. H., Ha I., Lane W. S., Reinberg D. (1992). Dr1, a TATA-binding protein-associated phosphoprotein and inhibitor of class II gene transcription. Cell 70, 477-489 [DOI] [PubMed] [Google Scholar]
- Jacobs S. A., Khorasanizadeh S. (2002). Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295, 2080-2083 [DOI] [PubMed] [Google Scholar]
- Janicki S. M., Tsukamoto T., Salghetti S. E., Tansey W. P., Sachidanandam R., Prasanth K. V., Ried T., Shav-Tal Y., Bertrand E., Singer R. H., et al. (2004). From silencing to gene expression: real-time analysis in single cells. Cell 116, 683-698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. D. (1995). The price of repression. Cell 81, 655-658 [DOI] [PubMed] [Google Scholar]
- Johnson E. S. (2004). Protein modification by SUMO. Annu. Rev. Biochem. 73, 355-382 [DOI] [PubMed] [Google Scholar]
- Kadosh D., Struhl K. (1998). Histone deacetylase activity of Rpd3 is important for transcriptional repression in vivo. Genes Dev. 12, 797-805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser T. E., Intine R. V., Dundr M. (2008). De novo formation of a subnuclear body. Science 322, 1713-1717 [DOI] [PubMed] [Google Scholar]
- Kamada K., Shu F., Chen H., Malik S., Stelzer G., Roeder R. G., Meisterernst M., Burley S. K. (2001). Crystal structure of negative cofactor 2 recognizing the TBP-DNA transcription complex. Cell 106, 71-81 [DOI] [PubMed] [Google Scholar]
- Kellum R. (2003a). HP1 complexes and heterochromatin assembly. Curr. Top Microbiol. Immunol. 274, 53-77 [DOI] [PubMed] [Google Scholar]
- Kellum R. (2003b). Is HP1 an RNA detector that functions both in repression and activation? J. Cell Biol. 161, 671-672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Park C. H., Lee M. S., Carlson B. A., Hatfield D. L., Lee B. J. (2003). A novel TBP-interacting zinc finger protein represses transcription by inhibiting the recruitment of TFIIA and TFIIB. Biochem. Biophys. Res. Commun. 306, 231-238 [DOI] [PubMed] [Google Scholar]
- Korutla L., Degnan R., Wang P., Mackler S. A. (2007). NAC1, a cocaine-regulated POZ/BTB protein interacts with CoREST. J. Neurochem. 101, 611-618 [DOI] [PubMed] [Google Scholar]
- Krien M. J., Bugg S. J., Palatsides M., Asouline G., Morimyo M., O'Connell M. J. (1998). A NIMA homologue promotes chromatin condensation in fission yeast. J. Cell Sci. 111, 967-976 [DOI] [PubMed] [Google Scholar]
- Kwon S. H., Workman J. L. (2008). The heterochromatin protein 1 (HP1) family: put away a bias toward HP1. Mol. Cells 26, 217-227 [PubMed] [Google Scholar]
- Lachner M., O'Carroll D., Rea S., Mechtler K., Jenuwein T. (2001). Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410, 116-120 [DOI] [PubMed] [Google Scholar]
- Li C., Manley J. L. (1998). Even-skipped represses transcription by binding TATA binding protein and blocking the TFIID-TATA box interaction. Mol. Cell. Biol. 18, 3771-3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kirschmann D. A., Wallrath L. L. (2002). Does heterochromatin protein 1 always follow code? Proc. Natl. Acad. Sci. USA 99 Suppl. 4, 16462-16469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Jeong H. W., Kong N., Yang Y., Carroll J., Luo H. R., Silberstein L. E., Yupoma, Chai L. (2009). Stem cell factor SALL4 represses the transcriptions of PTEN and SALL1 through an epigenetic repressor complex. PLoS ONE 4, e5577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maison C., Almouzni G. (2004). HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5, 296-304 [DOI] [PubMed] [Google Scholar]
- Margueron R., Li G., Sarma K., Blais A., Zavadil J., Woodcock C. L., Dynlacht B. D., Reinberg D. (2008). Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol. Cell 32, 503-518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisterernst M., Roeder R. G. (1991). Family of proteins that interact with TFIID and regulate promoter activity. Cell 67, 557-567 [DOI] [PubMed] [Google Scholar]
- Morelli C., Karayianni E., Magnanini C., Mungall A. J., Thorland E., Negrini M., Smith D. I., Barbanti-Brodano G. (2002). Cloning and characterization of the common fragile site FRA6F harboring a replicative senescence gene and frequently deleted in human tumors. Oncogene 21, 7266-7276 [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay D., Riezman H. (2007). Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201-205 [DOI] [PubMed] [Google Scholar]
- Muller P. Y., Janovjak H., Miserez A. R., Dobbie Z. (2002). Processing of gene expression data generated by quantitative real-time RT-PCR. Biotechniques 32, 1372-1374, 1376, 1378-1379 [PubMed] [Google Scholar]
- Nan X., Ng H. H., Johnson C. A., Laherty C. D., Turner B. M., Eisenman R. N., Bird A. (1998). Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386-389 [DOI] [PubMed] [Google Scholar]
- Narita M., Nunez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003). Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703-716 [DOI] [PubMed] [Google Scholar]
- Narlikar G. J., Fan H. Y., Kingston R. E. (2002). Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475-487 [DOI] [PubMed] [Google Scholar]
- Nielsen A. L., Oulad-Abdelghani M., Ortiz J. A., Remboutsika E., Chambon P., Losson R. (2001). Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7, 729-739 [DOI] [PubMed] [Google Scholar]
- Nielsen P. R., Nietlispach D., Mott H. R., Callaghan J., Bannister A., Kouzarides T., Murzin A. G., Murzina N. V., Laue E. D. (2002). Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416, 103-107 [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193-199 [DOI] [PubMed] [Google Scholar]
- O'Connell M. J., Norbury C., Nurse P. (1994). Premature chromatin condensation upon accumulation of NIMA. EMBO J. 13, 4926-4937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006). Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635-648 [DOI] [PubMed] [Google Scholar]
- Orphanos V., McGown G., Hey Y., Boyle J. M., Santibanez-Koref M. (1995a). Proximal 6q, a region showing allele loss in primary breast cancer. Br. J. Cancer 71, 290-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanos V., McGown G., Hey Y., Thorncroft M., Santibanez-Koref M., Russell S. E., Hickey I., Atkinson R. J., Boyle J. M. (1995b). Allelic imbalance of chromosome 6q in ovarian tumours. Br. J. Cancer 71, 666-669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth K. V., Camiolo M., Chan G., Tripathi V., Denis L., Nakamura T., Hubner M. R., Spector D. L. (2010). Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol. Biol. Cell 21, 4184-4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S. G., Mendez J., Prasanth K. V., Stillman B. (2004a). Dynamics of pre-replication complex proteins during the cell division cycle. Philos. Trans. R. Soc. Lond. B 359, 7-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth S. G., Prasanth K. V., Siddiqui K., Spector D. L., Stillman B. (2004b). Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance. EMBO J. 23, 2651-2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S., Pavithra L., Bhatt A., Kundu T. K., Chattopadhyay S. (2005). Tumor suppressor SMAR1 mediates cyclin D1 repression by recruitment of the SIN3/histone deacetylase 1 complex. Mol. Cell. Biol. 25, 8415-8429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reo E., Seum C., Spierer P., Bontron S. (2010). Sumoylation of Drosophila SU(VAR)3-7 is required for its heterochromatic function. Nucleic Acids Res. 38, 4254-4262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards E. J., Elgin S. C. (2002). Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108, 489-500 [DOI] [PubMed] [Google Scholar]
- Rundlett S. E., Carmen A. A., Suka N., Turner B. M., Grunstein M. (1998). Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature 392, 831-835 [DOI] [PubMed] [Google Scholar]
- Schoeftner S., Blasco M. A. (2009). A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 28, 2323-2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Sathyan K. M., Geng Y., Zheng R., Chakraborty A., Freeman B., Wang F., Prasanth K. V., Prasanth S. G. (2010). A WD-repeat protein stabilizes ORC binding to chromatin. Mol. Cell 40, 99-111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiio Y., Eisenman R. N. (2003). Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. USA 100, 13225-13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. A., Choi E. S., Kim H. S., Ho J. C., Watts F. Z., Park S. D., Jang Y. K. (2005). SUMO modification is involved in the maintenance of heterochromatin stability in fission yeast. Mol. Cell 19, 817-828 [DOI] [PubMed] [Google Scholar]
- Smith T. F. (2008). Diversity of WD-repeat proteins. Subcell. Biochem. 48, 20-30 [DOI] [PubMed] [Google Scholar]
- Stewart M. D., Li J., Wong J. (2005). Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment. Mol. Cell. Biol. 25, 2525-2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stielow B., Sapetschnig A., Kruger I., Kunert N., Brehm A., Boutros M., Suske G. (2008a). Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell 29, 742-754 [DOI] [PubMed] [Google Scholar]
- Stielow B., Sapetschnig A., Wink C., Kruger I., Suske G. (2008b). SUMO-modified Sp3 represses transcription by provoking local heterochromatic gene silencing. EMBO Rep. 9, 899-906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiel G., Lietz M., Hohl M. (2004). How mammalian transcriptional repressors work. Eur. J. Biochem. 271, 2855-2862 [DOI] [PubMed] [Google Scholar]
- Uchimura Y., Ichimura T., Uwada J., Tachibana T., Sugahara S., Nakao M., Saitoh H. (2006). Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J. Biol. Chem. 281, 23180-23190 [DOI] [PubMed] [Google Scholar]
- Um M., Li C., Manley J. L. (1995). The transcriptional repressor even-skipped interacts directly with TATA-binding protein. Mol. Cell. Biol. 15, 5007-5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010). An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369-381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D., Malik H. S. (2009). Multiple roles for heterochromatin protein 1 genes in Drosophila. Annu. Rev. Genet. 43, 467-492 [DOI] [PubMed] [Google Scholar]
- Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., Lamond A. I. (2006). Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics 5, 2298-2310 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhang H., Chen Y., Sun Y., Yang F., Yu W., Liang J., Sun L., Yang X., Shi L., et al. (2009). LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer. Cell 138, 660-672 [DOI] [PubMed] [Google Scholar]
- Wood J. L., Liang Y., Li K., Chen J. (2008). Microcephalin/MCPH1 associates with the Condensin II complex to function in homologous recombination repair. J. Biol. Chem. 283, 29586-29592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Wong J., Moreno G. T., Young M. K., Cote J., Wang W. (1998). NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 2, 851-861 [DOI] [PubMed] [Google Scholar]
- Yang S. H., Sharrocks A. D. (2004). SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13, 611-617 [DOI] [PubMed] [Google Scholar]
- Ye Q., Callebaut I., Pezhman A., Courvalin J. C., Worman H. J. (1997). Domain-specific interactions of human HP1-type chromodomain proteins and inner nuclear membrane protein LBR. J. Biol. Chem. 272, 14983-14989 [DOI] [PubMed] [Google Scholar]
- Yuri S., Fujimura S., Nimura K., Takeda N., Toyooka Y., Fujimura Y., Aburatani H., Ura K., Koseki H., Niwa H., et al. (2009). Sall4 is essential for stabilization, but not for pluripotency, of embryonic stem cells by repressing aberrant trophectoderm gene expression. Stem Cells 27, 796-805 [DOI] [PubMed] [Google Scholar]
- Zhang L., Anglesio M. S., O'Sullivan M., Zhang F., Yang G., Sarao R., Mai P. N., Cronin S., Hara H., Melnyk N., et al. (2007). The E3 ligase HACE1 is a critical chromosome 6q21 tumor suppressor involved in multiple cancers. Nat. Med. 13, 1060-1069 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Iratni R., Erdjument-Bromage H., Tempst P., Reinberg D. (1997). Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89, 357-364 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Matthiesen P., Harder S., Siebert R., Castoldi G., Calasanz M. J., Wong K. F., Rosenwald A., Ott G., Atkin N. B., et al. (2000). A 3-cM commonly deleted region in 6q21 in leukemias and lymphomas delineated by fluorescence in situ hybridization. Genes Chromosomes Cancer 27, 52-58 [DOI] [PubMed] [Google Scholar]
- Zheng G., Yang Y. C. (2004). ZNF76, a novel transcriptional repressor targeting TATA-binding protein, is modulated by sumoylation. J. Biol. Chem. 279, 42410-42421 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


