Abstract
The events regulating human preimplantation development are still largely unknown owing to a scarcity of material, ethical and legal limitations and a lack of reliable techniques to faithfully amplify the transcriptome of a single cell. Nonetheless, human embryology is gathering renewed interest due to its close relationship with both stem cell biology and epigenetic reprogramming to pluripotency and their importance in regenerative medicine. Carefully timed genome-wide transcript analyses of single oocytes and embryos uncovered a series of successive waves of embryonic transcriptional initiation that start as early as the 2-cell stage. In addition, we identified the hierarchical activation of genes involved in the regulation of pluripotency. Finally, we developed HumER, a database of human preimplantation gene expression, to serve the scientific community. Importantly, our work links early transcription in the human embryo with the correct execution of the pluripotency program later in development and paves the way for the identification of factors to improve epigenetic reprogramming.
Keywords: Genome activation, Human embryos, Pluripotency
INTRODUCTION
Preimplantation development, which covers the interval between fertilization and implantation into the uterus, is a highly dynamic cellular and molecular process that lasts ∼6 days in human. In this short period of time, the newly formed embryo must terminate female meiosis, extinguish both gamete programs, avoid apoptosis, initiate mitosis, reorganize its chromatin and start the pluripotency program that, a few days later, will give rise to the first lineage-committed progenitor cell populations. The correct execution of this complex program is the basis of successful development to term (Li et al., 2008; Payer et al., 2003) and is linked to later postnatal development (Katari et al., 2009; Leader et al., 2002; Li et al., 2008). However, very little is known about its molecular and cellular mechanisms owing to the scarcity of human embryos for research, the ethical and legal aspects surrounding their use, and the very limited techniques that can be reliably applied to such small amounts of nucleic acids and proteins.
A few studies have attempted to overcome these limitations by pooling large numbers of embryos (Zhang et al., 2009) or by combining in-house data with published literature (Assou et al., 2010). We have taken advantage of a newly developed whole-transcriptome amplification method (pico-profiling), which has been shown to give highly accurate results from as few as ten somatic cells (Gonzalez-Roca et al., 2010). In contrast to most amplification methods, which deliver high false-positive rates and greatly overestimate overall mRNA expression, pico-profiling has been shown to perform as well as conventional methods when compared against a panel of 878 quantitative PCR assays (Gonzalez-Roca et al., 2010; Shi et al., 2006). Human oocytes and embryos are both scarce and of high intrinsic ethical value. In order to maintain genetic variability among samples, we analyzed the transcriptome of individual metaphase II oocytes, cleavage stage human embryos, blastocysts, and low-passage human embryonic stem cells (ESCs). To make our data easily available, we developed HumER, a searchable database of human preimplantation development.
Importantly, by combining genome-wide transcriptional analysis of single embryos with the disruption of de novo transcription at successive developmental time points, our study identifies for the first time the exact timing of multiple waves of transcriptional activation in human embryos, and, in contrast to the previously accepted time point of embryonic genome activation at the 4- to 8-cell stage (Braude et al., 1988), places the time of initiation of transcriptional activity in the human embryo at the 2-cell stage. Moreover, we followed the network of pluripotency-associated genes, as defined by Boue et al. (Boue et al., 2010), during early development, and found that there are also distinct patterns of activation, initiated around the time of embryonic genome activation, which was unexpected considering their role later in development. Our work links embryonic genome activation with the initiation of the pluripotency program in human embryos and has implications for ESC biology and the epigenetic reprogramming of somatic cells through induced pluripotency.
MATERIALS AND METHODS
Oocyte and embryo collection and culture
Oocytes and embryos were obtained after signed informed consent of the donors or the couples and with the approval of the Ethics Committee of the CMRB and of the Comisión de Seguimiento y Control de la Donación de Células y Tejidos Humanos del Instituto de Salud Carlos III (project number PI052847).
The average oocyte donor age was 28 (range 20 to 33 years). Oocytes were collected by preovulatory follicular puncture after ovarian hyperstimulation, and the cumulus oocyte complexes were allowed to recover for 2 hours in medium G.1 (Vitrolife, Goteborg, Sweden) at 37°C and 6% CO2. Cumulus cells were removed by exposure to hyaluronidase (80 Ui/ml; Hyase, Vitrolife) and mild pipetting. Denuded oocytes were visually evaluated, and those arrested in the metaphase of the second meiotic division (MII) were allocated to this study.
All embryos included in this study have been donated for research according to the Spanish law on Assisted Reproduction Techniques (Act 14/2006). As some of the couples carried a female factor infertility problem and resorted to female oocyte donors to improve their prognosis, the average age of women whose oocytes were fertilized was 24.6 (range 19 to 32 years). Embryos were frozen at the pronuclear stage, 18 to 20 hours after insemination. After thawing, embryos were allowed to develop until the required stage in a sequential culture system (G1/G2, Vitrolife) at 37°C and 6% CO2. Embryo development was monitored every 12 hours. Once the embryos reached the appropriate stage, the zona pellucida (ZP) was removed with pronase (5 mg/ml; Roche, Basel, Switzerland) at 37°C. Dezoned samples were plunged into 45 μl of Lysis Buffer (20 mM DTT, 10 mM Tris-HCl pH 7.4, 0.5% SDS, 1 mg/ml proteinase K) preheated to 65°C. The tube was maintained at 65°C for 15 minutes and then placed at –80°C for later processing.
Human ESC culture
The three human (h) ESC lines used, ES[2], ES[3] and ES[4], were derived as published (Aran et al., 2010; Raya et al., 2008) at the CMR[B]. Low-passage cells of each line were thawed and cultured for one passage over a feeder layer of human foreskin fibroblasts (HFFs; CCD1112Sk ATCC, Manassas, VA, USA) in hES medium at 37°C and 5% CO2. The composition of hES medium is: KO-DMEM (Invitrogen, Carlsbad, CA, USA), 20% KO serum replacement (Invitrogen), Glutamax (2 mM; Invitrogen), penicillin-streptomycin, non-essential amino acids (Lonza, Verviers, Belgium), 2-mercaptoethanol (0.05 mM; Invitrogen) and basic fibroblast growth factor [bFGF (FGF2), 10 ng/ml; Invitrogen]. To avoid contamination of the ESC sample by HFFs, a clump of 50-100 ESCs was mechanically dislodged from a colony and processed as described below.
RNA isolation and whole-transcriptome amplification (WTA)
RNA from single samples was purified using the RNA Clean XP bead suspension (Agencourt Bioscience, Beckman Coulter, Danvers, MA, USA). Genomic DNA is also bound by the beads but is not released as long as it is not sheared (data not shown). Therefore, every effort was made to minimize shearing. RNA was eluted in 20 μl water.
Different proportions of the RNA samples were used for amplification according to the expected RNA content of embryos. For MII, 2-cell, 4-cell, 6-cell and 8- to 10-cell embryos, the entire RNA preparation was used (20 μl); for morulae, 6 μl; for blastocysts, 3 μl; and for ESCs, 4 μl. Library preparation was performed following the distributor's (Sigma-Aldrich, St Louis, MO, USA) recommendations for WTA2. SYBR Green (Sigma-Aldrich) was added to the amplification reaction, which was performed in a CFX real-time instrument (Bio-Rad) to monitor yield. When the SYBR Green signal reached a plateau after 21 cycles, the reaction was stopped. Amplified cDNA was purified and quantified; 10 μg cDNA was fragmented by DNAseI and biotinylated by terminal transferase (Affymetrix, Santa Clara, CA, USA). Hybridization, washing, staining and scanning of Affymetrix Human Gene ST 1.0 arrays were performed following the manufacturer's recommendations.
Real-time PCR
Samples for real-time PCR analysis and validation were obtained as described above. MII oocytes were obtained fresh and processed immediately. The embryos were thawed at the pronuclear stage and cultured in a G1/G2 media system with or without α-amanitin (24 μg/ml; Sigma-Aldrich). After reaching the expected developmental stage, embryos were dezoned and processed as described above. Because α-amanitin treatment eventually leads to cell cycle arrest, α-amanitin-treated embryos were time-matched to their untreated counterparts. However, all the α-amanitin-treated embryos were at the expected developmental stage at collection.
The cDNA libraries were prepared as described above; however, cDNA fragmentation was not performed. Real-time PCR analysis was carried out with 10 ng cDNA and the following TaqMan assays: RBBP6 (Hs00199439_m1), FAM90A1 (Hs00216400_m1), RFPL4B (Hs02518369_s1), TDGF1 (Hs02339499_g1), NANOG (Hs02387400_g1), L1TD1 (Hs01102131_m1), H2AFZ (Hs01888362_g1), ZSCAN4 (Hs00537549_m1), FIGLA (Hs01079386_m1), SOX2 (Hs01053049_s1), KLF4 (Hs00358836_m1), ZP1 (Hs01399328_m1) and POU5F1 (Hs00999634_Gh).
Because α-amanitin treatment might lower the expression of housekeeping genes, thus underestimating the transcriptional level of the genes tested, we spiked an external standard in all cDNA libraries. The standard spiked template is the reverse complement of a 100 bp portion of the Drosophila melanogaster Protein phosphatase 1α (PpalphaA1) mRNA transcript (NM_079760.2), starting from bp 1181. This amplicon matches the target for the TaqMan assay (Dm02151352_g1) used to quantify the spiked-in control. All assays were carried out in triplicate on each biological sample.
Immunocytochemistry
Embryos were cultured as described above. At selected stages of development, their ZP was removed and they were fixed in PBS containing 4% paraformaldehyde for 1 hour at 4°C. Embryos were then washed extensively in TBS (Sigma-Aldrich) and stained with antibodies against SP1 and α-tubulin. DNA content was visualized with DAPI.
Microarray data analysis
cDNA libraries were hybridized on Affymetrix Human Gene 1.0 ST arrays, which interrogate over 28,000 genes with more than 750,000 probes. Scanned images (DAT files) were transformed into intensities (CEL files) by GCOS (Affymetrix). Statistical analysis was performed using the affy package in R (Gautier et al., 2004). Briefly, raw CEL files were imported with gene annotation from NetAffix and data were normalized with GCRMA and summarized at the gene level (32,321 genes). We processed samples in two batches and corrected for batch effect using the ComBat algorithm in R. Microarray data are available at GEO with accession GSE29397
Correlation between samples of the same stage was calculated by linear correlation statistics. Both linear regression and a pairwise comparison by Student's t-test with Benjamini Hochberg correction (P<0.05, with 2-fold changes) were used to analyze differences among stages. Linear correlation coefficients (R2) ranged between 0.988 and 0.937. During development up to 4-cell, correlation coefficients remained rather stable: oocytes, 0.956 to 0.985; 2-cell, 0.984 to 0.988; and 4-cell, 0.984 to 0.986. As development became asynchronous, correlation coefficients diverged among replicates, as indicated by the 6-cell (0.979 to 0.895) and 8- to 10-cell (0.949) groups. Morulae showed R2 between 0.959 and 0.974, blastocysts between 0.968 and 0.974, and for the three ESC lines around 0.98.
K-means clustering was performed with GenePattern (Reich et al., 2006) with the mean of biological replicates per stage. The average log2 intensities after GCRMA normalization and batch correction were filtered for the genes that showed variation during preimplantation development; probesets with a coefficient of variation [100×(standard deviation/mean)] lower than 5% were not considered. K-means clustering divided 16,976 probesets into ten clusters.
For the study of gene expression patterns during early preimplantation development, K-means clustering was performed with the GenePattern software with the mean of MII, 2-cell, 4-cell, 6-cell and 8- to 10-cell samples, with standard Pearson distance metrics, ten clusters and 1000 trials (12,940 probesets). For each of the ten clusters, a heatmap of the cluster was generated with heatmap2 from the gplots library in R (http://CRAN.R-project.org/package=gplots). Gene ontology was investigated using the DAVID analysis wizard (Dennis et al., 2003; Huang da et al., 2009).
To follow the pluripotency network during preimplantation development, we used the UniGene IDs of the genes upregulated in ES and induced pluripotent (iPS) cells and of the genes upregulated only in ESCs from Boue et al. (Boue et al., 2010). Whenever multiple probesets were available, we retained that with the highest average expression value. We selected genes that showed an absolute fold change of at least 2.0 between any two successive stages. We extracted links between proteins using String software (Szklarczyk et al., 2011) with confidence 0.4 and all evidences. We imported the network into Cytoscape (Cline et al., 2007) and overlaid average gene expression values for each stage.
RESULTS
Transcriptome analysis of human embryos reveals substantial changes in expression patterns across developmental stages
Human metaphase II (MII)-arrested oocytes and embryos spanning the preimplantation stages of development were cultured in vitro, staged according to time in culture and cell number (2-cell, 4-cell, 6-cell, 8- to 10-cell, morula, blastocyst), and subjected to genome-wide transcriptional analysis with three biological replicates. The three ESC lines used in the study were derived in the same laboratory using the same method (Aran et al., 2010), had the same karyotype (46XY) and were fully characterized. Moreover, ESCs were derived from embryos that were handled in the same way as those used in the study. In order to compare the transcriptional profiles within and among developmental stages, single oocyte and embryo data were normalized and a principal component analysis (PCA) was carried out to identify transcriptome distances (Fig. 1A). Invariably, samples at the same stage clustered closer together than to any other sample. Directional distances on the three PCA dimensions also indicate a continuum in the direction of changes that is consistent with developmental progression. Next, we employed hierarchical sample classifications of normalized transcriptional profiles. The 24 samples occupied eight clustered groups according to their biological nature; this result was obtained when using as PCA input the genes that were statistically different among stages (Fig. 1B) and also when using all genes expressed from the arrays.
Fig. 1.
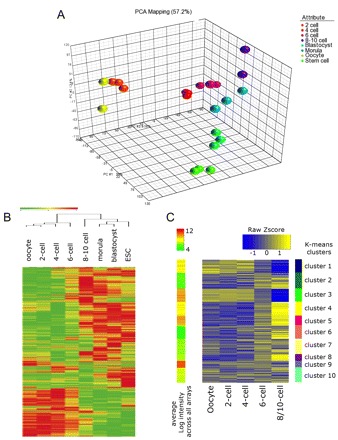
Transcriptome changes during human preimplantation development. (A) Principal component analysis showing that samples at the same stage of development cluster closer together than they do to any other sample. (B) Hierarchical sample classifications of normalized transcriptional profiling of differentially expressed genes. (C) GenePattern K-means analysis identifies ten clusters (1 to 10) of transcripts that change their expression between the MII oocyte and the 10-cell stage. ESC, embryonic stem cell.
Next, we performed a detailed analysis of the qualitative changes across developmental time points (Fig. 2). MII oocytes and 2-cell embryos had similar transcriptional profiles (R2=0.98). Between 2-cell and 4-cell, 606 transcripts were differentially expressed (R2=0.97). Although R2=0.96 paints a more dynamic picture when comparing 4-cell with 6-cell embryos, only 255 transcripts were significantly different, with most of them (n=234) upregulated in 6-cell embryos. This is due to the variability among 6-cell embryos, which increases the threshold for significance in pairwise comparisons.
Fig. 2.
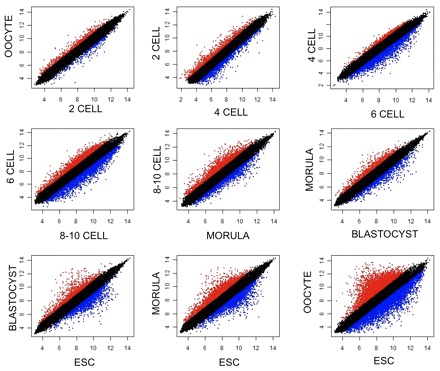
Pairwise changes in the transcriptome during human preimplantation development. Linear regression and pairwise comparison by Student's t-test with Benjamini correction analysis comparing consecutive developmental stages. The overlap between the stage represented on the x-axis (blue) and that represented on the y-axis (red) is in black. OOCYTE, MII oocytes; ESC, embryonic stem cell.
As development proceeds from the 6-cell to the 8- to 10-cell stage, more changes become apparent (R2=0.93). Comparing 8- to 10-cell embryos with morula (R2=0.955), there are 1630 differentially expressed genes, of which 963 are downregulated. Transcriptome changes between morula and blastocyst (R2=0.97) entail 1363 genes. Pairwise comparisons indicate that 1724 genes are differentially expressed between ESC and blastocyst (R2=0.95).
Changes in biological process representation across early human embryo development
We first considered transcripts that were more highly expressed in the 2-cell than 4-cell embryo (n=319) using DAVID (Huang da et al., 2009) software with Ensembl identifiers. PANTHER-based and KEGG-based ontology analyses indicated significant over-representation of pre-mRNA processing and mRNA splicing (see Table S1 in the supplementary material). Conversely, the 287 genes that were upregulated between 2-cell and 4-cell represented processes related to protein production, such as protein biosynthesis and modification (see Table S2 in the supplementary material). The molecular functions most significantly represented involved histones, nucleic acid-binding and ribosomal proteins, suggesting that the first burst of protein production in the embryo is mainly dedicated to building the translation machinery.
Most of the 255 transcripts differentially expressed between 4-cell and 6-cell are upregulated in the 6-cell embryo (n=234), suggesting active transcription in 4-cell stage embryos (see Table S3 in the supplementary material). This hypothesis is supported by the most significant biological processes identified by PANTHER: transcription and transcriptional regulation. This same trends continues as development proceeds from 6-cell to 8- to 10-cell, when the 255 genes that are more highly expressed in the 8- to 10-cell embryo represent protein biosynthesis and modification, as well as transcriptional activity (see Table S4 in the supplementary material).
The passage from cleavage stage to morula is recognizable by the fact that processes related to transcription lose prominence (see Table S5 in the supplementary material), and the 667 genes that are more highly expressed in morula are enriched for metabolic processes involving lipids, proteins, amino acids and carbohydrates (see Table S6 in the supplementary material). Likewise, in the passage from morula to blastocyst (n=1363), processes related to mRNA transcription and transcriptional regulators are downregulated in favor of protein, lipid and hormonal metabolism (see Tables S7 and S8 in the supplementary material).
Maternal mRNA degradation occurs in waves in human embryos
Maternal mRNAs are stored in the cytoplasm of the oocyte and provide the biochemical guide for development until the embryonic genome becomes active (Schultz, 2002). Maternal mRNA degradation patterns provide clues as to the metabolic processes that are in place at any stage of development and indications of its timing (Schultz, 2002). We examined clusters of inherited transcripts in developmental stages from MII to 8-10 cells (Fig. 1C). Clusters 3, 9 and 10 showed patterns indicative of maternal inheritance, with cluster 3 being having the most robust pattern. Cluster 3, or `early maternal', comprises 1594 transcripts with a decrease in expression between MII oocyte and 2-cells, further decreases between 2-cells and 4-cells, and reaching the lowest expression point at 8-10 cells. The 1594 DAVID identifiers in this group show an over-representation of the categories of nucleic acid metabolism, protein trafficking, cell cycle and protein metabolism. An analysis of their promoter binding sites reveals enrichment for the NKX and POU families of transcription factors (NKX3A, NKX6-1, NKX2-5, POU3F2, POU6F1, POU2F1). The nucleic acid metabolism category, in particular, is highly represented (n=237), constituting 14.8% of all genes expressed in this cluster. This analysis revealed a very significant over-representation of macromolecules and protein catabolic processes (see Table S9 in the supplementary material), pointing to active degradation of maternally inherited mRNAs and proteins.
The `late maternal' cluster (cluster 9; n=2243) comprises genes with expression that decreases over time and presents a very sharp downregulation at the 8- to 10-cell stage. The 2243 DAVID identifiers in this cluster are involved in protein metabolism and biosynthesis, as well as mRNA metabolism (see Table S10 in the supplementary material).
Owing to a combination of mRNA degradation, recruitment of maternal transcripts into polysomes, and translation prior to embryonic genome activation (EGA), some transcripts can appear to increase their expression at a given stage (Potireddy et al., 2006). We observed that pairwise comparisons indicate overexpression of some genes at the 2-cell stage and that some genes in cluster 10 show their highest expression level at the 2-cell stage. We attempted to determine whether this apparent increase in transcript abundance could be due to EGA starting at the 2-cell stage using quantitative PCR to analyze gene expression in oocytes and in 2-, 4- and 6- to 10-cell embryos cultured in the presence or absence of the transcription blocker α-amanitin. The results indicated active transcription from the embryonic genome, thus validating the hypothesis of an early wave of EGA in human embryos (Fig. 2A).
Onset of embryonic transcriptional activity at the 2-cell stage in human embryos
Our observation that EGA might occur earlier than at the 4- to 8-cell stage as previously thought in human embryos (Braude et al., 1988), led us to determine its exact timing. We employed real-time PCR in the presence or absence of α-amanitin to determine the time of accumulation of newly synthesized mRNAs and the magnitude of transcriptional activation. We analyzed data from the MII oocyte to the 8- to 10-cell stage with GenePattern K-means clustering. Cluster 1 (n=2072) defines a class of genes with expression that increases significantly at the 2- to 4-cell stage and then decreases as development proceeds. We identify this cluster as `early risers'. Cluster 2 showed a significant increase in expression at the 4-cell stage, and in most cases these genes remained expressed in later development (n=1469). We identified this cluster as `middle risers', as they point to a slightly more advanced wave of gene expression. Finally, genes in cluster 4 (n=1411) increase dramatically in expression at the 6-cell stage and more so still at the 8- to 10-cell stage. Similar patterns, although with different overall intensities, were displayed by clusters 5, 6 and 8. We call this pattern of gene expression, in its entirety, `late risers'. The rise in intensity of late risers – temporally coincident with the reported activation of the embryonic genome (Braude et al., 1988) – and its widespread occurrence across the genome indicate that it is likely to be due mainly to active transcription. However, because of its timing, the rise in intensity of the signal in cluster 1 (early risers) could be due to either active transcription from the newly formed embryonic genome or to a relative (apparent) increase in amount resulting from the degradation of other maternal transcripts.
We analyzed the identities of genes in clusters 1 and 4 by DAVID with Ensembl identifiers (see Tables S11 and S12 in the supplementary material). Analysis of the PANTHER molecular function categories of the early risers (cluster 1) highlighted the presence of transcription factor motifs, such as the zinc-finger and KRAB box. Moreover, PANTHER biological process-based categories highlighted highly significant over-representation of protein biosynthesis and metabolism. Among the late risers (cluster 4), we identified a striking over-representation of the PANTHER biological process category of nucleic acid metabolism, with 216 ID counts ascribed to this category.
EGA in human has been reported to occur by day 3 of preimplantation development (Assou et al., 2010; Dobson et al., 2004), and transcription should not be detectable before this point. Previous studies indicated that waves of transcriptional activation occur during embryogenesis in other species (Flach et al., 1982; Latham et al., 1992). In order to resolve the discrepancy between our observation and the reported literature, we prepared cDNA libraries from MII oocytes, 2-, 4- and 6- to 8-cell stage embryos, cultured from the pronuclear stage in the presence or absence of α-amanitin. A preliminary experiment showed that the dose of α-amanitin used [24 μg/ml, effective in Macaca mulatta embryos (Vassena et al., 2005)] was not toxic for early development (see Table S13 in the supplementary material). Moreover, the dose employed is within the range reported for efficient transcriptional inhibition [11-100 μg/ml (Braude et al., 1988; Flach et al., 1982)]. Alpha-amanitin binds to the free RNA polymerase II core adjacent to the bridge helix (Bushnell et al., 2002), stabilizing the elongation complex in a translocation intermediate that delays both the cycle of nucleotide addition and the translocation of the enzyme along the template DNA (Brueckner and Cramer, 2008), effectively inhibiting transcription. We assayed by quantitative real-time PCR a set of early risers and late risers across the first three cell cycles. We identified a strong inhibitory effect of α-amanitin as early as the 2-cell stage, with the bulk of the effect seen from the 4-cell stage onwards (Fig. 2A).
We selected genes with increased expression between the MII oocyte and 2-cell stage. Real-time PCR showed that the expression of retinoblastoma binding protein 6 (RBBP6), ret finger protein-like 4B (RFPL4B), and the family with sequence similarity 90, member A1 (FAM90A1), increased at the 2-cell stage, and that the increase was α-amanitin sensitive, indicating transcription from the embryonic genome (Fig. 3A). We then asked whether the increase in transcripts at the 4-cell stage could also be due to transcription. We found that, for instance, LINE-1 type transposase domain containing 1 (L1TD1) increased 32-fold at the 4-cell stage compared with the 2-cell stage and that this increase was due to transcription from the embryonic genome (Fig. 3A). Quantitative real-time PCR also confirmed transcription from the embryonic genome of late risers such as ZSCAN4 and H2AFZ (Fig. 3A), which were all highly expressed at the 6- to 8-cell stage and sensitive to α-amanitin. Therefore, in human embryos, transcription from the embryonic genome is underway at the 2-cell stage.
Fig. 3.
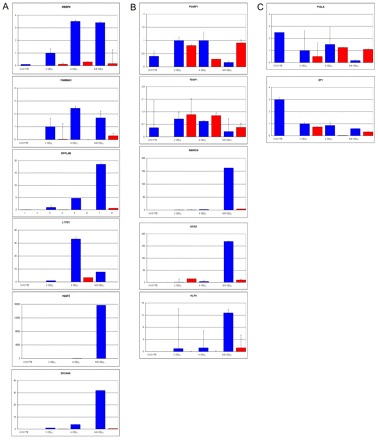
Human embryonic genome activation and pluripotency program initiation. Quantitative real-time PCR analysis of (A) genes that are actively transcribed from the embryonic genome starting at the 2-cell (RBBP6, FAM90A1, RFPL4B), 4-cell (L1TD1) and 6- to 8-cell (ZSCAN4, H2AFZ) stage, (B) genes involved in the initiation of the pluripotency program (POU5F1, TDGF1, NANOG, SOX2 and KLF4) and (C) maternally inherited mRNAs (FIGLA, ZP1). Blue bars, control culture conditions; red bars, α-amanitin-supplemented culture medium. Units on the y-axis indicate fold changes of expression as compared with MII (unit of expression). Error bars indicate s.e.
SP1 expression and localization during human preimplantation development
The major wave of EGA is characterized by a shift from a lack of enhancer function to a dependence on specific enhancers and repressors for the coordinated expression of genes (Lawinger et al., 1999). In mice, SP1-dependent promoters and enhancers are functional from the 2-cell stage, but not in fertilized oocytes (Lawinger et al., 1999); SP1 expression itself increases from the 2-cell stage onwards (Worrad and Schultz, 1997), as does protein localization to the nucleus (Worrad et al., 1994). We found that SP1, although expressed throughout human preimplantation development, peaks at the 2-cell stage (93%). We analyzed the localization of SP1 protein in preimplantation embryos (see Fig. S1 in the supplementary material) and found that it localizes to the nucleus by the 4-cell stage, suggesting an active role for this transcription factor in initiating EGA in human.
Pluripotent program initiation reflects cascades of pluripotency-related gene expression
In order to identify the dynamics of activation of the pluripotency program, we superimposed the pluripotency network recently reported by Boue et al. (Boue et al., 2010) onto the embryo expression data (Fig. 4 and see Fig. S2 in the supplementary material).
Fig. 4.
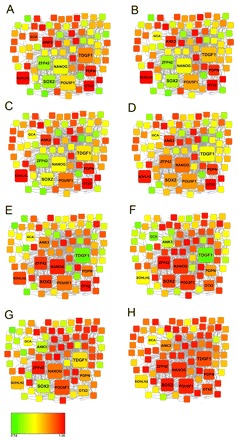
Pluripotency matrix across preimplantation human development. The expression of the main pluripotency-related genes at different stages of development. Red indicates high expression, green indicates low expression. White indicates genes not present on the microarray. The size of the square reflects the relative importance of the gene for the acquisition or maintenance of pluripotency. (A) MII oocytes, (B) 2-cell embryos, (C) 4-cell embryos, (D) 6-cell embryos, (E) 8- to 10-cell embryos, (F) morula stage embryos, (G) blastocyst stage embryos, (H) early passage human ESCs.
Although progression through preimplantation development showed an overall increase in the number of active pluripotency genes that were expressed at a progressively higher level, different, sequential patterns of activation could be identified. A first set of pluripotency-related genes is highly expressed in MII-arrested oocytes, and their expression level decreases during development, reaching the lowest expression level at the morula and blastocyst stages (the adhesion-related genes ANK3, GCA and PDPN; the transcription factors OTX2, SOHLH2 and ZNF462). Conversely, these same genes are very highly expressed in human ESCs, representing a common pluripotency signature between oocyte and ESCs. The core group of pluripotency-related genes [NANOG, POU5F1 (OCT4), ZFP42, SOX2 and TDGF1] was activated progressively from the 2-cell to the 8- to 10-cell stage and remained active until the morula stage. In this general pattern, however, some subpatterns can be discerned. POU5F1 is the first transcript of the core set to become α-amanitin sensitive, at the 4-cell stage, whereas ZFP42 and NANOG started to be highly expressed at the 6-cell and 8- to 10-cell stages, respectively, and remained highly expressed throughout the blastocyst stage. SOX2 expression peaked at the morula stage and decreased in the blastocyst. TDGF1 expression was high in MII oocytes and 2-cell embryos, but decreased thereafter and remained low even at the blastocyst stage.
DISCUSSION
During the first cell cycles, the embryo relies on reserves of mRNA and proteins stored in the oocyte cytoplasm, and it is only later in preimplantation development that EGA occurs, marking the beginning of self-sustained cellular biology.
The translation and degradation of maternally inherited mRNAs stored in the oocyte cytoplasm prior to ovulation is both concomitant with and required for the successful completion of EGA. It has been shown that asynchrony in the maternal to embryonic transition (Braude et al., 1988; Goddard and Pratt, 1983), which is often due to defective cytoplasmic maturation of the growing oocyte (Schramm et al., 2003), frequently results in developmental arrest. We report here the discovery of two waves of maternal mRNA turnover, which are tiled during early development. The first wave, which we term `early maternal', takes place between the MII and 2-cell stage. The genes in this cluster (cluster 3) are characterized by over-representation of the nucleic acid and protein catabolism pathways. During this developmental window, maternal mRNAs are loaded onto polysomes (Potireddy et al., 2006), translocated and degraded, and widespread cytoplasmic catabolism of oocyte-inherited proteins is required for the correct initiation of EGA (Bushati et al., 2008; Lieberfarb et al., 1996; Wang and Latham, 2000). Moreover, the transcripts in this cluster have an over-representation of binding sites for POU transcription factors. POU3F2 (OCT7) expression is restricted to the brain and the embryo in mouse (Scholer et al., 1989), whereas POU2F1 (OCT1) is ubiquitously expressed in both embryonic and adult mouse tissues (Scholer et al., 1989). However, a recent report has highlighted a requirement for POU2F1 in the survival and further differentiation of trophoblast stem cells post-implantation, and has identified Cdx2 as a transcriptional target of POU2F1 in mouse embryos (Sebastiano et al., 2010). The presence of a transcription factor binding site in the promoter sequence of a gene does not mean that the transcription factor binds to the site at all times, or indeed ever. Since the amount of DNA required to evaluate the binding of a protein to chromatin is prohibitive in human embryos, we decided to verify expression of the transcription factors at the time of over-representation of transcripts carrying their binding sites. All three of the POU family transcription factors are expressed during preimplantation development, with POU2F1 having a clearly early maternal expression pattern, thus overlapping with the expression of transcripts carrying its binding site in their promoter. A second wave of maternal mRNA degradation, which we termed `late maternal' (cluster 9), included transcripts that decrease gradually over time without ever recovering their expression levels (n=2234). Unlike the early maternal cluster (cluster 3), which seems more involved in the catabolism of proteins and amino acids, in the late maternal cluster we found significant over-representation of genes involved in the metabolism of mRNA and proteins.
A striking feature of maternal mRNA translation is its dynamism, which results from the interactions between mRNA degradation, unmasking and polyadenylation (Seydoux, 1996), recruitment of maternal transcripts onto polysomes (Potireddy et al., 2006) and translation. In this context, some very stable transcripts can appear to increase in expression at a given stage (Potireddy et al., 2006). Alternatively, their increase might be real but occurs at a time when we do not expect active transcription, as is the case with the 2-cell human embryos. The timing of EGA is species specific; it occurs at the 2-cell stage in mouse (Bensaude et al., 1983; Moore, 1975), at the 4- to 8-cell stage in horse (Brinsko et al., 1995) and at the 8- to 16-cell stage in cow (De Sousa et al., 1998), sheep (Crosby et al., 1988), rabbit (Brunet-Simon et al., 2001) and monkey (Schramm and Bavister, 1999). The timing and the identification of important players in EGA are of great relevance for the field of reproductive medicine and for the reprogramming of somatic nuclei. A seminal work in human provided the first indication of transcription from the embryonic genome between the 4- and 8-cell stages (Braude et al., 1988). Other, more recent reports place it at, or after, day 3 of development (Assou et al., 2010; Dobson et al., 2004). With the exception of the original work by Braude and colleagues, no study so far has addressed the issue of the timing of EGA in human using a transcriptional inhibitor, and none has done so in single embryos and by assaying thousands of transcripts at the same time.
This is the first time that the powerful and newly developed pico-profiling technique has been used to address an outstanding biological question. We coupled pico-profiling with high quality, single-embryo transcriptional arrays to identify patterns of gene expression suggestive of active embryonic transcription, and we then verified them by quantitative PCR after inhibiting transcription (but not short-term embryo development). We identified three waves of transcriptional activation in the human embryo: at the 2-cell stage, the 4-cell stage, and between the 6- and the 8- to 10-cell stages. This last wave is responsible for the transcription of a large number of genes, and we speculate that it should be the human equivalent of the major wave of activation seen in mouse embryos at 26-29 hours post-fertilization (Flach et al., 1982). The genes actively transcribed during the 2-cell and 4-cell waves should represent earlier signs of embryonic genome activity, which might be needed to coordinate the later major activation. We verified some of the genes and confirmed their active transcription as early as the 2-cell stage. Specifically, we confirmed transcription of the genes encoding the DNA-binding proteins RBBP6, which binds to the retinoblastoma (pRB) tumor suppressor protein, FAM90A1, which is a member of the primate-specific family of FAM90A transcripts, and RING finger protein 211 (RFPL4B).
During mouse development, SP1-dependent promoters and enhancers are functional from the 2-cell stage, but not in fertilized oocytes (Lawinger et al., 1999). The over-representation of genes containing binding sites for SP1 among the first transcribed genes supports the proposal that activation of the human embryonic genome occurs earlier than previously thought. We found that SP1 itself, although expressed at high level throughout human preimplantation development, peaks in expression between the 2-cell and the 6-cell stages. When we analyzed the localization of SP1 in developing embryos, we found that although some cytoplasmic vesicles containing SP1 could be seen as early as the 1-cell stage, the transcription factor clearly translocates into the nucleus of 4-cell stage embryos. The nature of human embryo research does not allow for in-depth functional analysis at these early stages, but our results suggest an active role for this transcription factor in initiating the later, major waves of EGA.
One of the key features of the inner cell mass (ICM) is the pluripotency of its blastomeres, i.e. the ability to differentiate into cells of the three germ layers and to give rise to a new individual. One of the questions that remain to be convincingly answered concerning preimplantation development is the timing of initiation of the pluripotency program. During the very first stages of development all cells of an embryo are totipotent, as indicated by the fact that embryo splitting and birth occur following the transfer of 4-cell embryos with a single viable blastomere (Veiga et al., 1987). At the morula stage the first committed cell populations become apparent. At this stage, pluripotency becomes restricted to the cells of the ICM, which express pluripotency-associated genes, such as the transcription factors POU5F1, NANOG, SOX2 and TDGF1, and they exclude markers of trophectodermal lineages such as CDX2. Given this premise, we expected to find sequential activation of most pluripotency-related genes, reaching peaks of expression at the morula and blastocyst stages. However, the superimposition of a pluripotency network (Boue et al., 2010) onto developmental data revealed a more complex picture. First, a portion of the gene network was already active in MII-arrested oocytes, and the expression of these genes tended to decrease during development. This is consistent with a study that found a common signature of expression between oocytes and human ESCs (Assou et al., 2009), as well as with the empirical observation that, although somewhat inefficient, the cytoplasm of the MII oocyte is the best reprogrammer of a somatic nucleus to pluripotency (Latham, 2005). Second, we found that the core regulators of the pluripotency networks are expressed in different patterns; POU5F1 is a master regulator of pluripotency and is the only gene that can, when overexpressed in somatic progenitor cells, reprogram them to a state of pluripotency (Kim et al., 2009a; Kim et al., 2009b). During preimplantation development, POU5F1 appeared to be already expressed in mature MII-arrested oocytes, and increased slightly in expression during development. Real-time PCR analysis revealed that the presence of the POU5F1 transcript during the first two cell cycles is due to a combination of mobilization of stored maternal mRNA, which is mostly responsible for expression to the 2-cell stage, and active transcription from the 2-cell stage onward. Exon probe analysis revealed a mixture of the A and B forms of the gene being expressed during preimplantation development. We therefore analyzed the transcriptional activity of POU5F1 by quantitative real-time PCR and identified a strong presence of the maternal mRNA at the 2-cell stage (low response to α-amanitin). This pattern changed at the 4-cell stage, when most of the expression was due to active transcription, as treatment with α-amanitin decreased the transcript abundance by ∼70% (Fig. 3B). These data indicate the active production of both POU5F1 mRNA and protein from the 4-cell stage, and place POU5F1 among the very first genes to be transcribed in the developing embryo, suggesting a wider role in development than previously thought.
A second expression pattern, exemplified by NANOG, SOX2, KLF4 and ZFP42, is characterized by a very sharp rise in expression at the 6-cell stage from a barely detectable level through the 4-cell stage. Another group of genes, exemplified by PROM1, increased their expression at the morula stage, whereas another group still decreased in expression by the 6-cell stage and resumed expression only in the blastocyst (TDGF1). The sequential activation of these genes might reflect a hierarchy in the cascade of pluripotency-associated transcripts and points to a gradual acquisition of pluripotency in the developing human embryo.
In order to allow the scientific community to benefit from our work, expression data across human preimplantation development have been made available through a free, publicly searchable database called the Human Embryo Resource or HumER (http://intranet.cmrb.eu/Human_embryos/home.html). A box plot shows the percentranks of each gene for each sample. A percentrank is defined as the rank of a value in a dataset as a percentage of the dataset. This function evaluates the relative standing of a value within a dataset. For microarray studies, it means that the probe with the highest intensity will get the rank 100%, whereas the probe with the lowest intensity will get the rank 0%. We estimate that the bottom 40% of ranks reflect noise. This figure is calculated taking into account that not all genes of a genome are expressed in any given cell, at any given time, or under a given condition, and is further based on the fact that, in ESC lines cultured in our institute, the number of presence calls by the mas5calls function is ∼60% when studying gene expression with the Affymetrix HGU-133 plus 2.0 platform. HumER can be interrogated by gene name (or synonym), Ensembl identifier, gene symbol alias or gene description, making it user-friendly for scientists unfamiliar with large dataset analysis. A separate search option, by gene ontology keyword, further widens the scope of this database and allows a more discovery-based approach to gene expression profiles. HuMER complements and expands the data currently available to the scientific community interested in primate preimplantation developmental biology, such as PREGER (www.preger.org), and may contribute to a reduction in the use of precious research material.
In summary, by combining high-fidelity amplification with the genome-wide analysis of gene expression and RNA synthesis inhibition, we have identified waves of activation of the human embryonic genome starting at the 2-cell stage, and we describe how this sequential activation correlates with both the degradation of maternal mRNA transcripts and the initiation of the pluripotency program. Our work links early transcription in the human embryo with the correct execution of the pluripotency program later in development, and paves the way for the identification of factors to improve epigenetic somatic cell reprogramming.
Supplementary Material
Acknowledgments
We thank Ida Paramonov for technical assistance with data analysis and Keith E. Latham and Gustavo Tiscornia for critical reading of the manuscript and helpful discussion. This work was supported by grants from MICINN, Fundacion Cellex, Sanofi-Aventis, CIRM, the Helmsley Foundation and the G. Harold and Leila Y. Mathers Charitable Foundation.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.064741/-/DC1
References
- Aran B., Rodriguez-Piza I., Raya A., Consiglio A., Munoz Y., Barri P. N., Izpisua J. C., Veiga A. (2010). Derivation of human embryonic stem cells at the Center of Regenerative Medicine in Barcelona. In Vitro Cell. Dev. Biol. Anim. 46, 356-366 [DOI] [PubMed] [Google Scholar]
- Assou S., Cerecedo D., Tondeur S., Pantesco V., Hovatta O., Klein B., Hamamah S., De Vos J. (2009). A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics 10, 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S., Boumela I., Haouzi D., Anahory T., Dechaud H., De Vos J., Hamamah S. (2010). Dynamic changes in gene expression during human early embryo development: from fundamental aspects to clinical applications. Hum. Reprod. Update 17, 272-290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaude O., Babinet C., Morange M., Jacob F. (1983). Heat shock proteins, first major products of zygotic gene activity in mouse embryo. Nature 305, 331-333 [DOI] [PubMed] [Google Scholar]
- Boue S., Paramonov I., Barrero M. J., Izpisua Belmonte J. C. (2010). Analysis of human and mouse reprogramming of somatic cells to induced pluripotent stem cells. What is in the plate? PLoS ONE 5, e12664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braude P., Bolton V., Moore S. (1988). Human gene expression first occurs between the four- and eight-cell stages of preimplantation development. Nature 332, 459-461 [DOI] [PubMed] [Google Scholar]
- Brinsko S. P., Ball B. A., Ignotz G. G., Thomas P. G., Currie W. B., Ellington J. E. (1995). Initiation of transcription and nucleologenesis in equine embryos. Mol. Reprod. Dev. 42, 298-302 [DOI] [PubMed] [Google Scholar]
- Brueckner F., Cramer P. (2008). Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat. Struct. Mol. Biol. 15, 811-818 [DOI] [PubMed] [Google Scholar]
- Brunet-Simon A., Henrion G., Renard J. P., Duranthon V. (2001). Onset of zygotic transcription and maternal transcript legacy in the rabbit embryo. Mol. Reprod. Dev. 58, 127-136 [DOI] [PubMed] [Google Scholar]
- Bushati N., Stark A., Brennecke J., Cohen S. M. (2008). Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr. Biol. 18, 501-506 [DOI] [PubMed] [Google Scholar]
- Bushnell D. A., Cramer P., Kornberg R. D. (2002). Structural basis of transcription: alpha-amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc. Natl. Acad. Sci. USA 99, 1218-1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline M. S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., et al. (2007). Integration of biological network and gene expression data using Cytoscape. Nat. Protoc. 2, 2366-2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosby I. M., Gandolfi F., Moor R. M. (1988). Control of protein synthesis during early cleavage of sheep embryos. J. Reprod. Fertil. 82, 769-775 [DOI] [PubMed] [Google Scholar]
- De Sousa P. A., Watson A. J., Schultz R. M. (1998). Transient expression of a translation initiation factor is conservatively associated with embryonic gene activation in murine and bovine embryos. Biol. Reprod. 59, 969-977 [DOI] [PubMed] [Google Scholar]
- Dennis G., Jr, Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003). DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 [PubMed] [Google Scholar]
- Dobson A. T., Raja R., Abeyta M. J., Taylor T., Shen S., Haqq C., Pera R. A. (2004). The unique transcriptome through day 3 of human preimplantation development. Hum. Mol. Genet. 13, 1461-1470 [DOI] [PubMed] [Google Scholar]
- Flach G., Johnson M. H., Braude P. R., Taylor R. A., Bolton V. N. (1982). The transition from maternal to embryonic control in the 2-cell mouse embryo. EMBO J. 1, 681-686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004). affy-analysis of Affimetrix GeneChip data at the probe level. Bioinformatics 20, 307-315 [DOI] [PubMed] [Google Scholar]
- Goddard M. J., Pratt H. P. (1983). Control of events during early cleavage of the mouse embryo: an analysis of the `2-cell block'. J. Embryol. Exp. Morphol. 73, 111-133 [PubMed] [Google Scholar]
- Gonzalez-Roca E., Garcia-Albéniz X., Rodriguez-Mulero S., Gomis R. R., Kornacker K., Auer H. (2010). Accurate expression profiling of very small cell populations. PLoS ONE 5, e14418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57 [DOI] [PubMed] [Google Scholar]
- Katari S., Turan N., Bibikova M., Erinle O., Chalian R., Foster M., Gaughan J. P., Coutifaris C., Sapienza C. (2009). DNA methylation and gene expression differences in children conceived in vitro or in vivo. Hum. Mol. Genet. 18, 3769-3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. B., Greber B., Arauzo-Bravo M. J., Meyer J., Park K. I., Zaehres H., Scholer H. R. (2009a). Direct reprogramming of human neural stem cells by OCT4. Nature 461, 649-643 [DOI] [PubMed] [Google Scholar]
- Kim J. B., Sebastiano V., Wu G., Arauzo-Bravo M. J., Sasse P., Gentile L., Ko K., Ruau D., Ehrich M., van den Boom D., et al. (2009b). Oct4-induced pluripotency in adult neural stem cells. Cell 136, 411-419 [DOI] [PubMed] [Google Scholar]
- Latham K. E. (2005). Early and delayed aspects of nuclear reprogramming during cloning. Biol. Cell 97, 119-132 [DOI] [PubMed] [Google Scholar]
- Latham K. E., Garrels J. I., Chang C., Solter D. (1992). Analysis of embryonic mouse development: construction of a high-resolution, two-dimensional gel protein database. Appl. Theor. Electrophor. 2, 163-170 [PubMed] [Google Scholar]
- Lawinger P., Rastelli L., Zhao Z., Majumder S. (1999). Lack of enhancer function in mammals is unique to oocytes and fertilized eggs. J. Biol. Chem. 274, 8002-8011 [DOI] [PubMed] [Google Scholar]
- Leader B., Lim H., Carabatsos M. J., Harrington A., Ecsedy J., Pellman D., Maas R., Leder P. (2002). Formin-2, polyploidy, hypofertility and positioning of the meiotic spindle in mouse oocytes. Nat. Cell Biol. 4, 921-928 [DOI] [PubMed] [Google Scholar]
- Li X., Ito M., Zhou F., Youngson N., Zuo X., Leder P., Ferguson-Smith A. C. (2008). A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev. Cell 15, 547-557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberfarb M. E., Chu T., Wreden C., Theurkauf W., Gergen J. P., Strickland S. (1996). Mutations that perturb poly(A)-dependent maternal mRNA activation block the initiation of development. Development 122, 579-588 [DOI] [PubMed] [Google Scholar]
- Moore G. P. (1975). The RNA polymerase activity of the preimplantation mouse embryo. J. Embryol. Exp. Morphol. 34, 291-298 [PubMed] [Google Scholar]
- Payer B., Saitou M., Barton S. C., Thresher R., Dixon J. P., Zahn D., Colledge W. H., Carlton M. B., Nakano T., Surani M. A. (2003). Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 13, 2110-2117 [DOI] [PubMed] [Google Scholar]
- Potireddy S., Vassena R., Patel B. G., Latham K. E. (2006). Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev. Biol. 298, 155-166 [DOI] [PubMed] [Google Scholar]
- Raya A., Rodriguez-Piza I., Aran B., Consiglio A., Barri P. N., Veiga A., Izpisua Belmonte J. C. (2008). Generation of cardiomyocytes from new human embryonic stem cell lines derived from poor-quality blastocysts. Cold Spring Harb. Symp. Quant. Biol. 73, 127-135 [DOI] [PubMed] [Google Scholar]
- Reich M., Liefels T., Gould J., Lerner J., Tamayo P., Mesirov J. P. (2006). GenePattern 2.0. Nat. Genet. 38, 500-501 [DOI] [PubMed] [Google Scholar]
- Scholer H. R., Balling R., Hatzopoulos A. K., Suzuki N., Gruss P. (1989). Octamer binding proteins confer transcriptional activity in early mouse embryogenesis. EMBO J. 8, 2551-2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm R. D., Bavister B. D. (1999). Onset of nucleolar and extranucleolar transcription and expression of fibrillarin in macaque embryos developing in vitro. Biol. Reprod. 60, 721-728 [DOI] [PubMed] [Google Scholar]
- Schramm R. D., Paprocki A. M., VandeVoort C. A. (2003). Causes of developmental failure of in-vitro matured rhesus monkey oocytes: impairments in embryonic genome activation. Hum. Reprod. 18, 826-833 [DOI] [PubMed] [Google Scholar]
- Schultz R. M. (2002). The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum. Reprod. Update 8, 323-331 [DOI] [PubMed] [Google Scholar]
- Sebastiano V., Dalvai M., Gentile L., Schubart K., Sutter J., Wu G. M., Tapia N., Esch D., Ju J. Y., Hubner K., et al. (2010). Oct1 regulates trophoblast development during early mouse embryogenesis. Development 137, 3551-3560 [DOI] [PubMed] [Google Scholar]
- Seydoux G. (1996). Mechanisms of translational control in early development. Curr. Opin. Genet. Dev. 6, 555-561 [DOI] [PubMed] [Google Scholar]
- Shi L., Reid L. H., Jones W. D., Shippy R., Warrington J. A., Baker S. C., Collins P. J., de Longueville F., Kawasaki E. S., Lee K. Y., et al. (2006). The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24, 1151-1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Kuhn M., Simonovic M., Roth A., Minguez P., Doerks T., Stark M., Muller J., Bork P., et al. (2011). The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 39, D561-D568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R., Dee Schramm R., Latham K. E. (2005). Species-dependent expression patterns of DNA methyltransferase genes in mammalian oocytes and preimplantation embryos. Mol. Reprod. Dev. 72, 430-436 [DOI] [PubMed] [Google Scholar]
- Veiga A., Calderon G., Barri P. N., Coroleu B. (1987). Pregnancy after the replacement of a frozen-thawed embryo with less than 50% intact blastomeres. Hum. Reprod. 2, 321-323 [DOI] [PubMed] [Google Scholar]
- Wang Q., Latham K. E. (2000). Translation of maternal messenger ribonucleic acids encoding transcription factors during genome activation in early mouse embryos. Biol. Reprod. 62, 969-978 [DOI] [PubMed] [Google Scholar]
- Worrad D. M., Schultz R. M. (1997). Regulation of gene expression in the preimplantation mouse embryo: temporal and spatial patterns of expression of the transcription factor Sp1. Mol. Reprod. Dev. 46, 268-277 [DOI] [PubMed] [Google Scholar]
- Worrad D. M., Ram P. T., Schultz R. M. (1994). Regulation of gene expression in the mouse oocyte and early preimplantation embryo: developmental changes in Sp1 and TATA box-binding protein, TBP. Development 120, 2347-2357 [DOI] [PubMed] [Google Scholar]
- Zhang P., Zucchelli M., Bruce S., Hambiliki F., Stavreus-Evers A., Levkov L., Skottman H., Kerkela E., Kere J., Hovatta O. (2009). Transcriptome profiling of human pre-implantation development. PLoS ONE 4, e7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


