Abstract
Central to the ABCE model of flower development is the antagonistic interaction between class A and class C genes. The molecular mechanisms underlying the A-C antagonism are not completely understood. In Arabidopsis thaliana, miR172 is expressed in the inner floral whorls where it downregulates the class A gene APETALA 2 (AP2). However, what controls this predominantly inner whorl-specific expression of miR172 is not known. We show that the LEUNIG (LUG) and SEUSS (SEU) co-repressors repress miR172 expression in the outer whorls of A. thaliana flowers. The recruitment of LUG/SEU to the miR172 promoters is dependent on AP2, suggesting that AP2 represses the expression of its cognate microRNA. Our study provides new insights into the molecular mechanisms underlying the A-C antagonism and shed light on the transcriptional regulation of miR172 during flower development.
Keywords: APETALA 2, Co-repressors, Flower development, LEUNIG, SEUSS, miR172, Arabidopsis
INTRODUCTION
The ABCE model of flower development (Coen and Meyerowitz, 1991; Krizek and Fletcher, 2005; Theissen and Saedler, 2001) successfully explains how floral organ identity is established through the combinatorial action of A, B, C and E classes of genes, all of which, except APETALA 2 (AP2), encode MADS box transcription factors. Central to the model is the antagonistic interaction between the A and C class genes. Whereas class A activity is repressed by class C genes in the inner two whorls, class C activity is repressed by class A genes in the outer two whorls (Bowman et al., 1991; Coen and Meyerowitz, 1991; Drews et al., 1991).
The class C gene AGAMOUS (AG) encodes a transcriptional repressor of the class A gene APETALA 1 (AP1) in the inner two whorls (Gustafson-Brown et al., 1994; Mandel et al., 1992). Two interacting transcriptional co-repressors, LEUNIG (LUG) and SEUSS (SEU), have been shown to repress AG transcription in the outer two whorls (Conner and Liu, 2000; Franks et al., 2002; Liu and Meyerowitz, 1995; Sridhar et al., 2004). Neither LUG nor SEU contains any known DNA-binding domains; their binding to the AG cis-regulatory elements is dependent on a direct physical interaction between SEU and the two MADS box proteins AP1 and SEPALLATA 3 (SEP3) (Gregis et al., 2006; Gregis et al., 2009; Sridhar et al., 2006).
AP2 is another transcriptional repressor of AG (Bowman et al., 1991; Drews et al., 1991). Using chromatin immunoprecipitation, AP2 was shown to directly bind within the AG second intron (Yant et al., 2010). Ectopic AP2 activity via the pAP3:AP2r transgene resulted in the repression of AG mRNA, supporting a direct repression of AG by AP2 (Wollmann et al., 2010). It is not yet known whether the repressive activity of AP2 depends on its recruitment of transcriptional co-repressors such as LUG/SEU.
The outer whorl-specific activity of AP2 has been largely attributed to a microRNA (miRNA), miR172, that specifically blocks AP2 mRNA translation as well as causing AP2 mRNA cleavage in the inner two whorls (Aukerman and Sakai, 2003; Chen, 2004; Schwab et al., 2005). In situ hybridization has revealed that miR172 is expressed most strongly in the inner whorls (Chen, 2004). A recent re-examination by in situ hybridization revealed that the spatial expression patterns of AP2 mRNA and miR172 are largely complementary, although there is a transient overlap in the second and possibly third floral whorls (Wollmann et al., 2010). Nevertheless, what determines the inner whorl-specific expression of miR172 remains unknown. A genome-wide analysis of AP2 binding sites using ChIP-seq indicated that miR172b is a target of AP2 (Yant et al., 2010), suggesting a possible role of AP2 in miR172 regulation.
Here, we show that LUG directly and negatively regulates miR172c and miR172e expression in sepals. This direct repression of miR172 by LUG also requires SEU and AP2, suggesting that AP2 might recruit the SEU-LUG co-repressor complex to repress miR172 transcription in the outer floral whorls. Our study provides important insights into miRNA regulation and suggests a positive-feedback loop, in which AP2 maintains its own outer whorl-specific activity by repressing the expression of its cognate miRNA.
MATERIALS AND METHODS
Plant materials
lug-3, seu-1 and ap2-2 mutants and 35S:GFP-LUG; lug-16, ProSEU:GFP-SEU; seu-1, ProSEU:GFP-SEU; ap2-2 and 35S:AP2m3 transgenic lines, all in the Landsberg erecta (Ler) background, were grown at 20°C under long-day conditions (16 hours light/8 hours dark). 35S:GFP-LUG in pAVA393 (Conner and Liu, 2000) was excised with HindIII and SacI and cloned into pCAMBIA2300 (Cambia, Brisbane, Australia), which was used to transform and rescue lug-16. ProSEU:GFP-SEU, previously shown to rescue seu-1 (Azhakanandam et al., 2008), was crossed into ap2-2+/+ant-9 to yield ProSEU:GFP-SEU (WT) and ProSEU:GFP-SEU (ap2-2). 35S:AP2m3 was described previously (Chen, 2004; Zhao et al., 2007).
RNA expression
RNA was isolated from inflorescences using TRI reagent (Sigma-Aldrich, St Louis, MO, USA). For small RNA blots, 30 μg total RNA was separated on a 15% acrylamide gel, transferred, and cross-linked onto Hybond-N nylon membrane using EDC [N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride; Sigma-Aldrich] (Pall and Hamilton, 2008). [γ32P]ATP and the mirVana Probe & Marker Kit (Ambion, Austin, TX, USA) were used to label the miR172 oligo (see Table S1 in the supplementary material). The small RNA blot was hybridized and washed as previously described (Lau et al., 2001; Lee and Ambros, 2001).
For northern blots, PCR-amplified AP2 first exon or 5S RNA was labeled with [α32P]dCTP (Perkin-Elmer, Waltham, MA, USA) with Ready-To-Go DNA Labeling Beads (Amersham, Piscataway, NJ, USA). Then 15 μg total RNA was separated on a 1% agarose gel, transferred onto BrightStar-Plus membrane (Ambion), hybridized and washed using the Northern Max-Gly Kit (Ambion). The signal intensity was measured using Image J (NIH).
For AP2 quantitative (q) RT-PCR (see Table S1 in the supplementary material), the QuantiSure First-Strand cDNA Kit (Accugen Bioscience, Rockville, MD, USA) was used, followed by qRT-PCR with SsoFast EvaGreen Supermix on a CFX96 real-time PCR machine (BioRad, Hercules, CA, USA). The CtAP2 was subtracted from the CtGAPC to yield the ΔCtWT and ΔCtlug3. The Pfaffl formula 2−ΔΔCt (Livak and Schmittgen, 2001), where −ΔΔCt=ΔCtWT–ΔCtlug3, gave the expression difference. Error bars represent the s.d. of two biological replicates (three technical repeats each).
For Fig. S1 in the supplementary material, RNAs isolated from wild-type (Ler) and lug-3 inflorescence tissues were enriched for small RNAs, labeled with Cy5, and hybridized to a custom-designed miRNA chip as described (Pang et al., 2009). Average expression values were first normalized against internal positive controls (tRNAs) before they were compared between wild type and lug-3.
miR172 in situ hybridization
A direct tandem oligonucleotide concatamer (4×) of the sense miR172 strand sequence 5′-AGAATCTTGATGATGCTGCAG-3′, with a T7 RNA polymerase-binding site (5′-CCCTATAGTGAGTCGTATTA-3′) at the 3′ end, served as the template for T7 transcription with the digoxygenin (DIG) RNA Labeling Mix (Roche, Basel, Switzerland). The miR172 sense probe was made by a similar strategy. In situ hybridization was carried out based on Carr and Irish (Carr and Irish, 1997); slides were hybridized at 42°C overnight and were washed three times at 42°C for 30 minutes with 0.2× SSC.
Chromatin immunoprecipitation (ChIP)
Two to three grams of inflorescences with stage 1-11 flowers were used in ChIP based on Sridhar et al. (Sridhar et al., 2006). Protein A Dynabeads (Dynal/Invitrogen, Carlsbad, CA, USA) and a polyclonal anti-GFP antibody (AB290-50, Abcam, Cambridge, MA, USA) were used. qPCR with the QuantiTect SYBR Green PCR Kit (Qiagen, Valencia, CA, USA) was performed on the CFX96 qPCR machine. The percent efficiency for each primer is shown in Table S4 in the supplementary material.
Based on Mukhopadhyay et al. (Mukhopadhyay et al., 2008), the Ct value for the plus antibody sample (+AB) and for −AB was independently subtracted from the Ct value of the input to yield ΔCT. Then ΔCT+AB was subtracted from ΔCT−AB to yield the ΔΔCT for each sample. The Pfaffl formula was used to calculate the fold enrichment.
Yeast two- and three-hybrid assays
The Matchmaker System (Clontech, Mountain View, CA, USA) was used. ANT-pGAD424 (Krizek and Sulli, 2006) and AP2delta-pGAD424 were gifts of B. A. Krizek. SEU (1-563)-pGBT9 and LUH-pGAD424 were reported previously (Sridhar et al., 2004; Sitaraman et al., 2008). To make AP2-pGADT7, AP2 was excised from pGG30 with NcoI and EcoRI and cloned into pGADT7. For P426-GAPD, the GAPD promoter was excised with BamHI and EcoRI, cloned into pCR2.1-TOPO, excised again with SacI and EcoRI and cloned into p426 GAL, replacing GAL. For P426-SEU, SEU was excised from pCRII-TOPO-SEU (Franks et al., 2002) with HindIII and XhoI and cloned into p426 GAPD at HindIII-SalI. Constructs were introduced one at a time into Saccharomyces cerevisiae strain PJ694A according to the Yeast Protocols Handbook (Clontech).
Bimolecular fluorescence complementation (BiFC)
AP2, SEU and LUH were cloned into pUC-SPYNE and pUC-SPYCE (Walter et al., 2004). Specifically, full-length AP2, SEU or LUH was amplified (for primers, see Table S1 in the supplementary material), cloned into pGEM-T Easy (Promega, Fitchburg, WI, USA) for AP2 and SEU or into pCR8/GW/TOPO (Invitrogen) for LUH, excised with SalI and XmaI, and then cloned into pUC-SPYNE and pUC-SPYCE. Helios Gene Gun (BioRad) was used according to a published procedure (Hollender and Liu, 2010). Results were imaged under a Zeiss Axio Observer.Z1 inverted microscope. The experiment was performed twice.
RESULTS AND DISCUSSION
miR172 expression is altered in lug-3 flowers
lug mutant flowers exhibit partially homeotic transformation of sepals to carpelloid sepals, indicating an expansion of C function into the first whorl (Liu and Meyerowitz, 1995). To test whether miR172, which is expressed in the inner floral whorls (Chen, 2004; Wollmann et al., 2010), expands into lug outer whorls, small RNA blots and in situ hybridization were performed. Increased miR172 was detected in lug-3 flowers (Fig. 1A), consistent with preliminary microarray data showing increased expression of miR172a-e (see Fig. S1 in the supplementary material). This increased miR172 in lug-3 correlates with a decrease in AP2 mRNA (Fig. 1B).
Fig. 1.
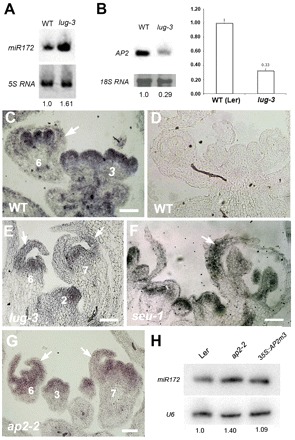
Increased and ectopic miR172 expression in lug-3, seu-1 and ap2-2. (A) A small RNA blot showing miR172 RNA levels in lug-3 and wild-type (WT) Arabidopsis thaliana inflorescences. 5S RNA provides the loading control, with the relative signal intensity shown beneath each lane. (B) Northern and qRT-PCR showing AP2 mRNA levels. 18S RNA provides the loading control. Error bars indicate s.d. (C) In situ hybridization showing miR172 RNA expression in wild-type flowers. Numbers indicate floral stages. (D) miR172 sense probe in wild type. (E) miR172 expression in lug-3 flowers. Arrows indicate sepals with ectopic miR172 expression. (F) miR172 RNA expression in seu-1 flowers. (G) miR172 expression in ap2-2. (H) A small RNA blot showing miR172 expression in ap2-2 and 35S:AP2m3 flowers. U6 provides a loading control. Scale bars: 50 μm.
In situ hybridization was used to examine miR172 expression in lug-3 as well as in seu-1 because SEU is a known partner of LUG (Sridhar et al., 2006). The miR172 antisense probe was made by T7 transcription from a direct tandem oligonucleotide concatamer (4×) of the sense miR172 strand. Using a similar probe, miR172 was previously shown to be expressed in the entire floral meristem, but the expression was abated from sepals in stage 7 and older flowers (Chen, 2004). We showed that miR172 RNA is no longer present in the sepals of stage 6 and older wild-type flowers (Fig. 1C and see Table S2 in the supplementary material). By contrast, in lug-3 and seu-1 mutant flowers, miR172 expression was detected in the sepals of stage 6 or older flowers (Fig. 1E,F). This ectopic miR172 expression was, however, sometimes found in only one of the two sepals (Fig. 1E,F). Further quantification revealed that 82% of the lug-3 (14 of 17) or seu-1 (18 of 22) sepals at stages 6 to 11 showed ectopic miR172 expression (see Table S2 in the supplementary material). This correlates well with the incomplete sepal-to-carpel transformations in lug-3 and seu-1 mutants (Franks et al., 2002; Liu and Meyerowitz, 1995). The in situ data indicate that LUG and SEU are each required to repress miR172 expression in the sepals starting no later than stage 6.
A recent publication using a locked nucleic acid (LNA)-based probe revealed inner whorl-specific miR172 expression as early as floral stage 4 (Wollmann et al., 2010). This difference in the timing (i.e. stage) of miR172 RNA clearing in wild-type sepals might reside in the greater specificity and sensitivity of the LNA probe, which consequently is likely to detect specific miR172 species. We thus focused our analyses of miR172 expression on stage 6 and older sepals, for which both probe types detect a clearing of the miR172 signal in wild type.
LUG directly regulates miR172
The increased and ectopic miR172 expression observed above could result from ectopic carpelloid organ development in the outer floral whorls of lug-3 as carpels are known to express high levels of miR172. We therefore conducted ChIP to test whether LUG directly associates with miR172 chromatin.
We chose miR172c and miR172e, which showed the highest increase in expression in lug-3 mutants (see Fig. S1 in the supplementary material). We searched miR172c and miR172e promoters for conserved binding sites for class A genes (i.e. MADS box and AP2 domain). No MADS box binding site (CArG box) was found, but four putative AP2 binding sites (TTTGTT; T. Dinha and X.C., unpublished) were found in the promoter of miR172c and one in the promoter of miR172e. PCR and qPCR primers were designed to flank these putative AP2 binding sites (see Tables S3 and S4 in the supplementary material).
Nuclear extracts of inflorescences from 35S:GFP-LUG; lug-16 and negative control (non-transgenic wild type) were immunoprecipitated with an anti-GFP antibody. The immunoprecipitated DNA was first quantified by semi-quantitative PCR, testing a larger number of primers (see Fig. S2 and Table S3 in the supplementary material). qPCR was subsequently used to test the most relevant promoter regions of target genes (Fig. 2A). For miR172c, only binding site miR172c-2 showed over 4-fold enrichment with the GFP-LUG; lug-16 sample (Fig. 2A). The other three potential AP2 binding sites, including miR172c-1, were not enriched (Fig. 2A and see Table S3 in the supplementary material). For miR172e, the single putative AP2 binding site was enriched over 7-fold with the GFP-LUG; lug-16 sample (Fig. 2A). Neither miR172c nor miR172e was enriched with the negative control sample (Fig. 2A). A non-target control, E1F4a1, showed no enrichment (see Fig. S2 and Table S3 in the supplementary material). These data suggest that AP2, or other AP2 family members, might be involved in recruiting LUG to the promoters of miR172c and miR172e.
Fig. 2.
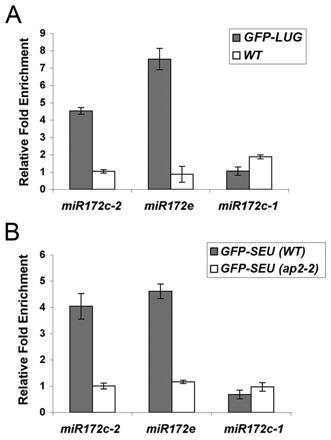
ChIP detects the in vivo association of GFP-LUG and GFP-SEU with miR172 promoter sequences. (A) ChIP assays with anti-GFP antibody and chromatin isolated from 35S:GFP-LUG; lug-16 (gray) and non-transgenic wild-type (white) A. thaliana inflorescences. Fold enrichment was quantified by qPCR using primers that flank putative AP2 binding sites. Error bars indiate s.d. of six replicates (two biological replicates with three technical repeats). (B) ChIP assays using anti-GFP antibody and chromatin from ProSEU:GFP-SEU in wild type (gray) and ap2-2 (white). Fold enrichment for the same primer sets as in A is shown. Error bars indiate s.d. of three technical replicates of one biological experiment.
Further evidence of AP2 mediating the repression of miR172 by LUG/SEU
If AP2 recruits LUG/SEU to repress miR172, ap2 mutants should exhibit expanded expression of miR172 similar to lug and seu mutants. In situ hybridization revealed ectopic miR172 expression in stage 6 and 7 ap2-2 sepals (Fig. 1G); 20 out of 20 ap2-2 sepals at floral stages 6-11 ectopically expressed miR172 (see Table S2 in the supplementary material). This was supported by northern blots (Fig. 1H) showing increased miR172 in ap2-2 flowers. By contrast, plants that express a miR172-resistant AP2 (35S:AP2m3) did not show reduced miR172 (Fig. 1H). Perhaps an overproliferation of stamens, a miR172-expressing organ, in these 35S:AP2m3 lines (Zhao et al., 2007) compensates for a reduction of miR172.
As GFP-LUG (ap2-2) plants are not currently available, GFP-SEU (ap2-2) plants were used for ChIP analysis to test the role of AP2 in mediating the association of the LUG/SEU co-repressors with miR172c and miR172e (Fig. 2B). GFP-SEU (WT) and GFP-SEU (ap2-2) inflorescences were analyzed by ChIP using the anti-GFP antibody. The same qPCR primer pairs as in Fig. 2A were used for quantifying fold enrichment. miR172c-2 and miR172e were enriched 4- and 4.8-fold, respectively, with GFP-SEU (WT) but not with GFP-SEU (ap2-2) (Fig. 2B). These data support the contention that AP2 is required for mediating the association of SEU with the miR172c-2 and miR172e promoters. Similar to GFP-LUG (Fig. 2A), GFP-SEU failed to associate with miR172c-1 (Fig. 2B). Since GFP-LUG and GFP-SEU are both associated with the same AP2 binding sites (miR172c-2 and miR172e) out of those tested (Fig. 2A,B and see Table S3 in the supplementary material), it suggests that LUG and SEU might function together in a complex, tethered by AP2, to regulate miR172. Using ChIP-seq, Yant et al. (Yant et al., 2010) reported the binding of AP2 to miR172b but not to other miR172 genes. This difference might reside in the different developmental stages at which the tissues were collected in the two studies.
SEU mediates the interaction between AP2 and LUG
The above results suggest that AP2 might directly interact with SEU and/or LUG. A yeast two-hybrid assay revealed a direct interaction of AP2-AD with SEU-BD (Fig. 3A, sector 2) but not with LUG-BD (Fig. 3A, sector 5). A truncated AP2 (AP2delta) containing the two AP2 domains (residues 124-394) failed to interact with SEU-BD (Fig. 3A, sector 3). Thus, the two AP2 domains are not sufficient for the interaction with SEU. Another AP2 domain-containing protein, AINTEGUMENTA (ANT), failed to interact with SEU (Fig. 3A, sector 4), indicating that SEU specifically interacts with AP2.
Fig. 3.
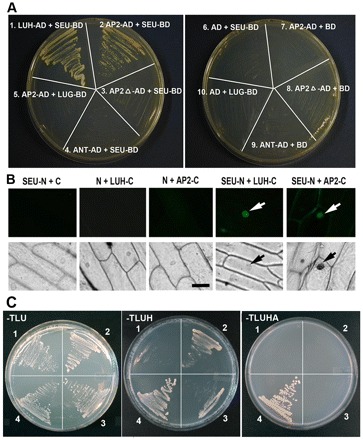
AP2 interacts with SEU but not LUG. (A) A yeast two-hybrid assay between prey (AD) and bait (BD) pairs. LEUNIG HOMOLOG (LUH)-AD against SEU-BD served as a positive control (Sitaraman et al., 2008). SEU-BD contains a truncated SEU (residues 1-563) with its C-terminal domain removed to avoid self-activation (Sridhar et al., 2006). The plate on the right shows various negative controls. The selection medium was −Trp, −Leu, −His, −Ade (−TLHA), plus 3 mM 3-amino-1,2,4-triazole. (B) A BiFC assay showing interactions in planta. Fluorescent (top row) and bright-field (bottom row) images of onion epidermal peels bombarded with BiFC plasmids. N and C represent the N- and C-terminal fragments of YFP, respectively. White arrows indicate fluorescent nuclei and black arrows indicate the same nuclei in bright field. SEU-N and LUH-C, previously shown to interact via BiFC (Hollender and Liu, 2010), serve as the positive control. Scale bar: 100μm. (C) AP2-AD and LUG-BD were tested for interaction in the presence or absence of full-length SEU expressed from the p426 vector. 1, AP2-AD + p426 vector + LUG-BD; 2, AP2-AD + p426-SEU + LUG-BD; 3, AP2delta-AD + p426-SEU + LUG-BD; 4, SEU-AD + p426 + LUG-BD (positive control). The ‘−TLU’ medium, which lacks Trp, Leu and urea, selects for all three plasmids. ‘−TLUH’ selects for His3. ‘−TLUHA’ selects for His3 and Ade2.
Bimolecular fluorescence complementation (BiFC) was used to confirm the interaction between SEU and AP2 via particle bombardment in onion cells. SEU and AP2 were each fused to the N-terminal and C-terminal fragments of YFP using the pUC-SPYNE and pUC-SPYCE vectors, respectively (Walter et al., 2004). The fusion proteins SEU-N and AP2-C were observed to interact in nuclei, whereas the negative controls did not (Fig. 3B).
Since LUG is known to interact with SEU (Sridhar et al., 2004), SEU might bridge the interaction between LUG and AP2. A yeast three-hybrid assay was employed to test whether LUG-BD interacts with AP2-AD in the presence or absence of SEU (Fig. 3C). The ‘−TLUH’ and ‘−TLUHA’ media represent two different selection stringencies. AP2-AD interacted with LUG-BD only when SEU was present (Fig. 3C, sector 2) and only in the less stringent −TLUH medium. AP2-AD failed to interact with LUG-BD when SEU was absent (sector 1). AP2delta-AD weakly interacted with LUG-BD in the presence of SEU (sector 3). The data support the proposal that SEU is capable of bridging the interaction between AP2 and LUG. The weakness of the interaction might reflect that all three proteins have to be expressed at the same time and in the same cellular compartments. This result is consistent with synergistic and semi-dominant genetic interactions among lug, seu and ap2 mutants during floral organ development (Franks et al., 2002; Liu and Meyerowitz, 1995), indicating functional interactions in vivo.
A model summarizing current understanding of A-C antagonism
miR172 regulates many important developmental processes in diverse plant species (Aukerman and Sakai, 2003; Chen, 2004; Chuck et al., 2007; Jung et al., 2007; Lauter et al., 2005; Martin et al., 2009; Wu et al., 2009). Whereas much attention has focused on finding the targets of miRNAs and on their biogenesis and metabolism, less is known about their transcriptional regulation. Our analysis provides important insights into the domain-specific transcriptional regulation of miR172.
Integral to the ABCE model is the mutual antagonism between the A and C class genes. The repression of A class activity in inner whorls is conferred by at least two mechanisms: the post-transcriptional downregulation of AP2 by miR172 (Fig. 4a) and the repression of AP1 transcription by AG (Fig. 4b) (Aukerman and Sakai, 2003; Chen, 2004; Gustafson-Brown et al., 1994; Schwab et al., 2005). However, AG and miR172 appear to act independently of each other (Wollmann et al., 2010; Zhao et al., 2007). The repression of class C genes in the outer whorls depends on the LUG/SEU co-repressors recruited by the AP1/SEP3 MADS box proteins (Sridhar et al., 2006) (Fig. 4c) as well as by AP2 (Fig. 4d). The mutual repression between AP2 and miR172 ensures positive-feedback loops that promote their own expression in the respective A and C domains (Fig. 4). It remains unresolved as to how the A-C boundary is initially established. A strict temporal order of gene activation might be one mechanism, so that earlier-acting class A genes are subsequently switched off in the inner whorls once the class C genes are switched on.
Fig. 4.
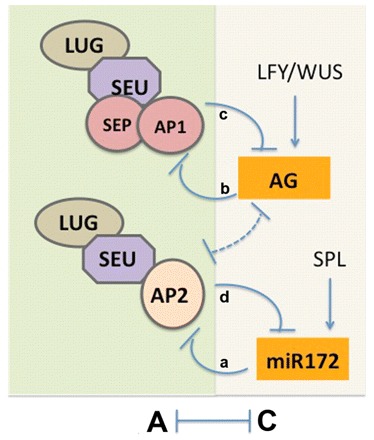
Model of A and C class gene antagonism. The negative regulation of class A gene activity in the inner two whorls is conferred by the post-transcriptional downregulation of AP2 by miR172 (a) and by the transcriptional repression of AP1 by AG (b). By contrast, the negative regulation of the class C gene AG in the outer two whorls depends on the LUG/SEU co-repressors recruited by the AP1/SEP MADS box proteins (c). The repression of miR172 also requires LUG/SEU and is mediated by AP2 (d). The dashed line represents a less well understood regulatory interaction between AG and AP2. Also indicated are the transcriptional activation of AG by LEAFY (LFY) and WUSCHEL (WUS) (Lohmann et al., 2001) and the activation of miR172 by SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) (Wu et al., 2009).
Transcriptional co-repressors are known to regulate diverse targets and developmental pathways and their target specificity is conferred by DNA-binding partners (Liu and Karmarkar, 2008). This work has reported a new regulatory target, miR172, of LUG/SEU and identified AP2 as the DNA-binding partner of LUG/SEU for miR172 regulation, providing novel insights into co-repressor function in plant development.
Supplementary Material
Acknowledgements
We thank Robert Franks for ProSEU:GFP-SEU; seu-1; Beth Krizek for ANT-pGAD424 and AP2delta-pGAD424; Detlef Weigel for pGG30; Jörg Kudla for BiFC vectors; Thanh Dinha for sharing unpublished results and the northern of 35S:AP2m3; Joann Conner for 35S:GFP-LUG; V. V. Sridhar for AP2-pGADT7 and p426-SEU; Vidyadhar Karmarkar for initiating the project; and Ranjani Iyer and Kristi Mullen for generating 35S:GFP-LUG; lug-16 lines. The work is supported by grants from the National Science Foundation (IOB0616096 and MCB0744752) to Z.L. Z.L. is partially supported by the University of Maryland Agricultural Experiment Station.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.058362/-/DC1
References
- Aukerman M. J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15, 2730-2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azhakanandam S., Nole-Wilson S., Bao F., Franks R. G. (2008). SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol. 146, 1165-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. (1991). Genetic interactions among floral homeotic genes of Arabidopsis. Development 112, 1-20. [DOI] [PubMed] [Google Scholar]
- Carr S. M., Irish V. F. (1997). Floral homeotic gene expression defines developmental arrest stages in Brassica oleracea L. vars. botrytis and italica. Planta 201, 179-188. [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G., Meeley R., Irish E., Sakai H., Hake S. (2007). The maize tasselseed4 microRNA controls sex determination and meristem cell fate by targeting Tasselseed6/indeterminate spikelet1. Nat. Genet. 39, 1517-1521. [DOI] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353, 31-37. [DOI] [PubMed] [Google Scholar]
- Conner J., Liu Z. (2000). LEUNIG, a putative transcriptional corepressor that regulates AGAMOUS expression during flower development. Proc. Natl. Acad. Sci. USA 97, 12902-12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews G. N., Bowman J. L., Meyerowitz E. M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65, 991-1002. [DOI] [PubMed] [Google Scholar]
- Franks R. G., Wang C., Levin J. Z., Liu Z. (2002). SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development 129, 253-263. [DOI] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Colombo L., Kater M. M. (2006). AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18, 1373-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregis V., Sessa A., Dorca-Fornell C., Kater M. M. (2009). The Arabidopsis floral meristem identity genes AP1, AGL24 and SVP directly repress class B and C floral homeotic genes. Plant J. 60, 626-637. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown C., Savidge B., Yanofsky M. F. (1994). Regulation of the arabidopsis floral homeotic gene APETALA1. Cell 76, 131-143. [DOI] [PubMed] [Google Scholar]
- Hollender C. A., Liu Z. (2010). Bimolecular fluorescence complementation (BiFC) assay for protein-protein interaction in onion cells using the helios gene gun. J. Vis. Exp. 40, 1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J. H., Seo Y. H., Seo P. J., Reyes J. L., Yun J., Chua N. H., Park C. M. (2007). The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 19, 2736-2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizek B. A., Fletcher J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nat. Rev. Genet. 6, 688-698. [DOI] [PubMed] [Google Scholar]
- Krizek B. A., Sulli C. (2006). Mapping sequences required for nuclear localization and the transcriptional activation function of the Arabidopsis protein AINTEGUMENTA. Planta 224, 612-621. [DOI] [PubMed] [Google Scholar]
- Lau N. C., Lim L. P., Weinstein E. G., Bartel D. P. (2001). An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294, 858-862. [DOI] [PubMed] [Google Scholar]
- Lauter N., Kampani A., Carlson S., Goebel M., Moose S. P. (2005). microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 102, 9412-9417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R. C., Ambros V. (2001). An extensive class of small RNAs in Caenorhabditis elegans. Science 294, 862-864. [DOI] [PubMed] [Google Scholar]
- Liu Z., Meyerowitz E. M. (1995). LEUNIG regulates AGAMOUS expression in Arabidopsis flowers. Development 121, 975-991. [DOI] [PubMed] [Google Scholar]
- Liu Z., Karmarkar V. (2008). Gro-Tup1 family co-repressors in plant development. Trends Plant Sci. 13, 137-144. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25, 402-408. [DOI] [PubMed] [Google Scholar]
- Lohmann J. U., Hong R. L., Hobe M., Busch M. A., Parcy F., Simon R., Weigel D. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105, 793-803. [DOI] [PubMed] [Google Scholar]
- Mandel M. A., Gustafson-Brown C., Savidge B., Yanofsky M. F. (1992). Molecular characterization of the Arabidopsis floral homeotic gene APETALA1. Nature 360, 273-277. [DOI] [PubMed] [Google Scholar]
- Martin A., Adam H., Diaz-Mendoza M., Zurczak M., Gonzalez-Schain N. D., Suarez-Lopez P. (2009). Graft-transmissible induction of potato tuberization by the microRNA miR172. Development 136, 2873-2881. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A., Deplancke B., Walhout A. J., Tissenbaum H. A. (2008). Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat. Protoc. 3, 698-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pall G. S., Hamilton A. J. (2008). Improved northern blot method for enhanced detection of small RNA. Nat. Protoc. 3, 1077-1084. [DOI] [PubMed] [Google Scholar]
- Pang M., Woodward A. W., Agarwal V., Guan X., Ha M., Ramachandran V., Chen X., Triplett B. A., Stelly D. M., Chen Z. J. (2009). Genome-wide analysis reveals rapid and dynamic changes in miRNA and siRNA sequence and expression during ovule and fiber development in allotetraploid cotton (Gossypium hirsutum L.). Genome Biol. 10, R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R., Palatnik J. F., Riester M., Schommer C., Schmid M., Weigel D. (2005). Specific effects of microRNAs on the plant transcriptome. Dev. Cell 8, 517-527. [DOI] [PubMed] [Google Scholar]
- Sitaraman J., Bui M., Liu Z. (2008). LEUNIG_HOMOLOG and LEUNIG perform partially redundant functions during Arabidopsis embryo and floral development. Plant Physiol. 147, 672-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V. V., Surendrarao A., Gonzalez D., Conlan R. S., Liu Z. (2004). Transcriptional repression of target genes by LEUNIG and SEUSS, two interacting regulatory proteins for Arabidopsis flower development. Proc. Natl. Acad. Sci. USA 101, 11494-11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V. V., Surendrarao A., Liu Z. (2006). APETALA1 and SEPALLATA3 interact with SEUSS to mediate transcription repression during flower development. Development 133, 3159-3166. [DOI] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Plant biology. Floral quartets. Nature 409, 469-471. [DOI] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schutze K., Batistic O., Weckermann K., Nake C., Blazevic D., Grefen C., Schumacher K., Oecking C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428-438. [DOI] [PubMed] [Google Scholar]
- Wollmann H., Mica E., Todesco M., Long J. A., Weigel D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137, 3633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Park M. Y., Conway S. R., Wang J. W., Weigel D., Poethig R. S. (2009). The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138, 750-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T. T., Ott F., Lanz C., Wollmann H., Chen X., Schmid M. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22, 2156-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Kim Y., Dinh T. T., Chen X. (2007). miR172 regulates stem cell fate and defines the inner boundary of APETALA3 and PISTILLATA expression domain in Arabidopsis floral meristems. Plant J. 51, 840-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


