Abstract
Comparative studies of the tetrapod raldh2 (aldh1a2) gene, which encodes a retinoic acid (RA) synthesis enzyme, have led to the identification of a dorsal spinal cord enhancer. Enhancer activity is directed dorsally to the roof plate and dorsal-most (dI1) interneurons through predicted Tcf- and Cdx-homeodomain binding sites and is repressed ventrally via predicted Tgif homeobox and ventral Lim-homeodomain binding sites. Raldh2 and Math1/Cath1 expression in mouse and chicken highlights a novel, transient, endogenous Raldh2 expression domain in dI1 interneurons, which give rise to ascending circuits and intraspinal commissural interneurons, suggesting roles for RA in the ontogeny of spinocerebellar and intraspinal proprioceptive circuits. Consistent with expression of raldh2 in the dorsal interneurons of tetrapods, we also found that raldh2 is expressed in dorsal interneurons throughout the agnathan spinal cord, suggesting ancestral roles for RA signaling in the ontogenesis of intraspinal proprioception.
Keywords: Retinoic acid, Spinal cord, Roof plate, Commissural interneurons, Proprioception, Paired fin loss, Mouse, Chicken, Xenopus, Zebrafish, Lamprey, Medaka
INTRODUCTION
The vertebrate central nervous system (CNS) forms along the embryonic anteroposterior (AP) axis from the neural plate, a sheet of ectodermal cells that invaginates and transforms into a cylindrical neural tube. Along the dorsoventral (DV) axis, the closed neural tube divides into sensory and motor domains in dorsal and ventral regions, respectively, whereas interneurons distribute between sensory and motor regions.
Spinal cord (SC) DV patterning occurs through a balance of Sonic hedgehog signals emanating ventrally from the notochord/floor plate, together with dorsally derived bone morphogenetic protein (BMP) and Wnt from the roof plate (RP). These opposing influences activate homeodomain and basic helix-loop-helix transcription factors that set in motion competing programs of ventral and dorsal neuron specification. In this model, neural progenitors obtain positional information according to the strength of the ventralizing and dorsalizing signals they receive (Wilson and Maden, 2005).
Signaling via vitamin A-derived retinoic acid (RA) is one of the mechanisms that shape the vertebrate neural tube. RA signaling participates in four major developmental programs of the CNS: neural differentiation, AP patterning, specification of motoneurons and DV organization (Diez del Corral et al., 2003; Muhr et al., 1999; Nieuwkoop, 1952; Novitch et al., 2003; Sockanathan and Jessell, 1998; Wilson et al., 2004). RA produced by paraxial mesoderm controls early programs of neural differentiation and AP patterning, whereas mesoderm, together with ventral SC-derived RA, influence motoneuron specification (Vermot et al., 2005). This contrasts with the much less well understood roles in DV organization that are performed by the RA that is produced inside the dorsal neural tube (Wilson et al., 2004). In the dorsal neural tube, RA synthesis is associated with the RP (Berggren et al., 1999), a dorsal-most group of glial cells that plays a pivotal role in the specification of adjacent dorsal progenitors of sensory neurons and dorsal interneurons (dIs) (Chizhikov and Millen, 2004).
In vertebrates, retinoid signaling is triggered by the synthesis of RA by two enzyme families. raldh1-3 (aldh1a1-3) are paralogs that belong to the Aldh1a family of all-trans and 9-cis retinaldehyde dehydrogenases, whereas raldh4 (aldh8a1) is a member of the Aldh8 family, which represents enzymes with preference for 9-cis retinaldehyde (Simões-Costa et al., 2008). Among Raldh genes, raldh2 plays the most important developmental role. Raldh2 displays the highest affinity for retinaldehyde, is the first Raldh to appear in mouse and chick embryos in early development, and is regulated in patterns that overlap with the activation of RA signaling (Blentic et al., 2003; Lin et al., 2003; Moss et al., 1998; Niederreither et al., 1997; Ulven et al., 2000; Wang et al., 1996; Xavier-Neto et al., 2000; Zhao et al., 1996).
Here we identify a conserved intronic enhancer that drives raldh2 expression in the RP and dIs of the frog, mouse and chicken dorsal SC, suggesting that DV programs of neural tube development regulated by RA are encoded by evolutionarily conserved cis-regulatory modules. The combined activation of raldh2 in the RP and dIs by a single enhancer suggests that these two RA signaling domains play related roles and this prompted us to utilize a comparative approach to gain insight into these functions. By investigating the patterns of raldh2 expression in the SC of agnathan, teleost and tetrapod embryos, we discovered a novel, transient, field of RA synthesis in dI precursors. By comparing the patterns of raldh2 expression in agnathans and teleosts with those of amniotes, we provide evidence that RA signaling might be involved in the ontogeny of two specific SC sensory functions: proprioception from vertebrate paired appendages and modulation of the intrinsic SC locomotor circuitry.
MATERIALS AND METHODS
Bioinformatics
Aldh gene sequences were obtained from NCBI, the Ensembl genome browser, JGI Eukaryotic Genomes and through the method of Sobreira and Gruber (Sobreira and Gruber, 2008). Conserved non-coding elements (CNEs) were identified using the ECR Browser and BLAST 2 sequences. The CNE search was limited to between 30 kb upstream of the transcription start site and 30 kb downstream of the stop codon. Transcription factor binding site (TFBS) prediction was performed using a locally available software that implements the MatInspector algorithm (Quandt et al., 1995) with TFBS matrices from TRANSFAC 6.0 (Wingender et al., 1996). TFBSs were accepted if at least 90% similar to a matrix core/whole matrix score.
Phylogenetic analysis
Thirty Aldh protein sequences from six vertebrate species ranging from agnathans to primates were aligned using MUSCLE (Edgar, 2004). The alignment (available on request) consisted of 464 amino acid positions, manually refined to eliminate gaps. Phylogenetic trees were generated using neighbor-joining (NJ) (Saitou and Nei, 1987), maximum parsimony (MP) (Swofford, 2000), maximum likelihood (ML) (Schmidt et al., 2002) and Bayesian inference (BI) (Ronquist and Huelsenbeck, 2003) methods. The WAG model was selected by ProtTest (Abascal et al., 2005). Node support was assessed in NJ and MP trees by 2000 bootstrap replicates. ML was performed with 100,000 puzzling steps. For BI we used two runs of 5,000,000 generations. Convergence was verified and an appropriate burn-in period of 2000 was determined. Consensus trees and posterior probabilities were calculated using the 50% majority rule.
Plasmids and constructs
raldh2 intron 1G CNEs from mouse, chicken and X. laevis were PCR amplified from genomic DNA and cloned into HSP68-lacZ and pTkeGFP vectors (Kothary et al., 1989; Rossant et al., 1991).
Mutagenesis
Nested deletion mutants of the chicken Raldh2 intron 1G enhancer with 734 bp, 450 bp, 355 bp and 149 bp fragments, as well as a 279 bp fragment containing RP and interneuron activator elements (RPIE), plus overlapping fragments A (62 bp), B (91 bp), C (92 bp) and D (114 bp), were PCR amplified and cloned into pTkeGFP. For site-directed mutagenesis we utilized QuickChange II (Stratagene, 200524). Nucleotides belonging to a predicted binding site for the Lim-homeodomain protein Lhx3 (116-126 bp) in the enhancer were substituted by an AscI restriction site. The Tcf/Cdx site (207-221 bp) was substituted by an AscI restriction site. A second Tcf/Cdx motif (353-367 bp) was substituted by an FseI restriction site. A Tcf/Tgif site (427-441 bp) was substituted by an AscI restriction site. All constructs were confirmed by DNA sequencing.
Transgenic mice
The mouse Raldh2 intron 1G CNE-HSP68-lacZ reporter was excised by SalI digestion. Pronuclear injection was as described (Xavier-Neto et al., 1999).
Electroporation
Enhancer-eGFP plasmid (2 μg/ml) plus pCAβ-RFP DNA (0.5-1.0 μg/ml) in 0.5% Fast Green in water were injected into the lumen of the chicken neural tube (HH17-18). In ovo electroporation was performed by applying six 60- to 100-millisecond pulses at 15-18 V with a 0.5 mm platinum electrode.
Xenopus laevis embryo injections
The Xenopus intronic enhancer, or the negative control pTkeGFP plasmid (30 pg), were co-injected with β-galactosidase mRNA (250 pg total) at the 4-cell stage into both dorsal blastomeres. Injections were performed at the animal pole ~45° from the top.
Staining and in situ hybridization
Stains were performed as described previously (Xavier-Neto et al., 1999) using an anti-GFP rabbit polyclonal antibody (1:500; Abcam, ab6556) and donkey anti-rabbit IgG conjugated with Alexa Fluor 488 (1:1000; Molecular Probes, A21206). In situ hybridization (ISH) (Wilkinson, 1992) and double ISH (Stern, 1998) were preformed as described previously, using mouse (Zhao et al., 1996), chicken, Xenopus (Chen et al., 2001), zebrafish (via Deborah Yelon, New York University School of Medicine, New York, USA) and lamprey raldh2 probes. Other probes included lamprey zic-1 (Sauka-Spengler et al., 2007), mouse Math1 (Helms and Johnson, 1998) and chicken Cath1 (Wilson and Wingate, 2006). Lamprey raldh2 was cloned by low-stringency hybridization with a 32P-labeled zebrafish raldh2 probe (Sauka-Spengler et al., 2007). The lamprey raldh2 sequence was deposited in GenBank as FJ536260. An in situ probe for medaka raldh2 was PCR amplified using medaka cDNA.
RESULTS
raldh2 intron 1G is a conserved non-coding element enriched with highly conserved transcription factor binding sites
To identify conserved non-coding elements (CNEs) potentially carrying raldh2 regulatory sequences, we aligned raldh2 orthologs from vertebrates including fugu, zebrafish, frog, chick and mouse, using human as baseline. This analysis revealed 72 CNEs displaying more than 75% identity over 183±16 bp (range 32-905 bp) in 5′, intronic and 3′ regions (Fig. 1). To identify sequence modules that regulate raldh2 expression, we screened vertebrate raldh2 CNEs for enhancer function. We selected three CNEs: a 5′ and a 3′ CNE conserved in all species (Fig. 1, gray arrowheads), plus raldh2 intron 1G, the largest raldh2 CNE (Fig. 1, red arrowhead). Of those, only mouse Raldh2 intron 1G displayed enhancer activity in 10.5 days post-coitum (dpc) transient transgenic mice. raldh2 intron 1G is conserved in amphibians, avians, rodents and primates and spans an average of 843±47 bp (702-905 bp) (Fig. 1, red box; see Fig. S1 in the supplementary material), but could not be detected in teleosts (see Fig. S2 in the supplementary material).
Fig. 1.
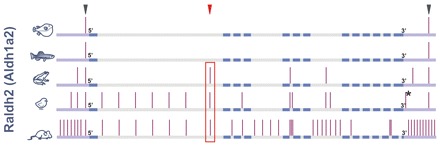
Evolutionary conservation of raldh2. Five raldh2 orthologs (fugu, zebrafish, frog, chick and mouse) were aligned using Homo sapiens as baseline. Vertical bars, conserved non-coding elements (CNEs). The red box highlights raldh2 intron 1G conservation. Arrowheads indicate the three CNEs chosen for transient transgenesis. Exons, blue bars. Introns, gray bars. 5′ and 3′ regions, violet bars. The asterisk indicates the CNE conserved between chicken and human, but lost from mouse.
For a list of transcription factor binding sites (TFBSs) that display matrix identities to the matrix core that are higher than 90% and that are conserved across amphibian, avian, marsupial, rodent and primate raldh2 intron 1G enhancers, see Fig. S3 in the supplementary material. There is deep conservation for TFBSs associated with Wnt signaling (i.e. Tcf binding sites), for homeodomain and Lim-homeodomain factors, and for factors such as Pax, Pou and basic helix-loop-helix. Some predicted sites for Forkhead factors, Vsx2 (Chx10), Lhx3, Pou3f1 and Klf4 are so rare that their presence in the enhancer reflects a statistically significant event when compared with their occurrence in a billion-bp random set.
The mouse Raldh2 intron 1G CNE directs a dorsal subset of the Raldh2 expression domain in transgenic mice
Transient transgenic mice revealed transcription driven by the mouse Raldh2 intron 1G CNE in embryonic tissues within the endogenous domains of Raldh2 expression (compare Fig. 2A-D with 2E). β-galactosidase activity was observed in the dorsal neural tube, running from the brachial level down to the tail bud of 10.5 dpc founder (F0) embryos (Fig. 2A-D). Age-matched embryos processed for Raldh2 in situ hybridization (ISH) indicated that the intron 1G CNE directs a dorsal-most subset of the endogenous Raldh2 expression pattern in the posterior embryonic quarter (Fig. 2E). Two stable transgenic lines, #191 and #583, confirmed that the neural tube is a prime target for this mouse Raldh2 intron 1G enhancer.
Fig. 2.
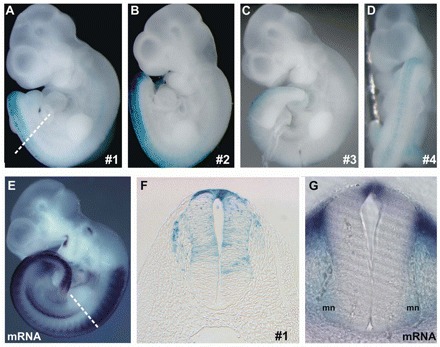
Mouse Raldh2 intron 1G CNE activity in transient transgenic mice. (A-D) The CNE drives lacZ expression (blue) at the posterior dorsum in all (4/4) transient transgenic mice harvested at 10.5 dpc. (E) This β-galactosidase field is a subset of the endogenous Raldh2 expression domain, as indicated by Raldh2 in situ hybridization (ISH). (F) Thoracic transverse section depicts dorsal spinal cord (SC) and dorsal root ganglia enhancer activation. (G) Brachial transverse section shows Raldh2 expression in the roof plate (RP) and motoneurons (mn). Dashed lines in A and E indicate the plane of the sections in F and G, respectively.
The intron 1G enhancer mirrors Raldh2 expression in the mouse dorsal spinal cord, but some features of its activity differ from the endogenous gene
The Raldh2 intron 1G enhancer regulates subsets of the endogenous Raldh2 expression domain, and some aspects of its activity differ from the endogenous gene. For example, stable intron 1G transgenic mouse lines display an early lacZ domain at the midbrain/hindbrain boundary (see Fig. S4 in the supplementary material). The midbrain/hindbrain region contains cerebellum and tectum progenitors that normally activate Raldh2, but only from 15.5 dpc onwards (Zhang et al., 2003). Thus, it is possible that the two stable transgenic mouse lines (#191 and #583) display heterochronic acceleration of an endogenous Raldh2 domain (see Fig. S4G-I in the supplementary material), suggesting that some important Raldh2 repressors are missing from the intron 1G CNE. Therefore, the intronic enhancer is not the sole regulator of Raldh2 expression in all tissues, but drives Raldh2 in the developing dorsal SC, which we validated as a natural domain of mouse Raldh2 expression (see below).
The mouse Raldh2 intron 1G enhancer activates expression in the roof plate and dorsal interneurons of the spinal cord
The mouse Raldh2 intron 1G enhancer activates lacZ in the neural tube of transgenic mouse embryos (Fig. 2A-D). The major territory of enhancer activation is a broad domain of the dorsal SC (Fig. 2F). Brachial sections of 10.5-11.5 dpc mouse embryos revealed intense β-galactosidase activity in the RP (Fig. 3B,F, arrowhead). From the RP, a second field of β-galactosidase was found in dI1 interneuron progenitors (Fig. 3B,F, arrow). These progenitors migrate ventrally, giving rise to neurons that receive proprioceptive afference and project to the cerebellum and to commissural interneurons (CIs), which cross the floor plate and project to the contralateral SC (Bermingham et al., 2001; Helms and Johnson, 1998; Lewis, 2006). A trail of diffuse β-galactosidase activity in the mantle region highlighted the ventral migration of dIs (Fig. 3B-F, asterisk), similar to the expression of a Math1 (Atoh1) transgene and of Dcc and Cntn2 (Tag-1) (Bermingham et al., 2001; Dodd et al., 1988; Furley et al., 1990; Helms and Johnson, 1998; Keino-Masu et al., 1996). Ventrally, β-galactosidase activity was found in CI axonal tracts (Fig. 3B-F, white arrows), reminiscent of Math1-expressing CIs (Altman and Bayer, 1997; Helms and Johnson, 1998).
Fig. 3.
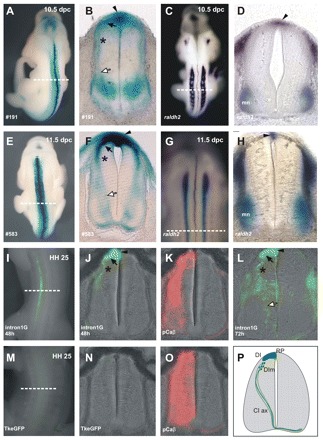
The Raldh2 intron 1G enhancer is a roof plate and dorsal interneuron enhancer in amniotes. (A,E) Dorsal views of stable transgenic mice. (B,F) Transverse sections at the brachial SC of the stable transgenic mice shown in A and E, respectively. The enhancer is active in the RP (arrowhead), in dorsal interneurons (DI) (black arrow) and in ventrally migrating dorsal interneurons (DIm) (asterisk). β-galactosidase expression is also observed in axons of commissural interneurons (CI ax) (white arrow). (C,D,G,H) ISH shows Raldh2 expression in the 10.5-11.5 dpc mouse brachial RP. (C,G) Dorsal views. (D,H) Transverse sections. (I-O) Enhancer activity is conserved in chicken. (I) Chicken thoracic SC 48 hours after Raldh2 intron 1G electroporation. (J) Section of the embryo in I showing RP (arrowhead), dorsal interneuron (black arrow) and migrating dorsal interneuron expression (asterisk). (L) Chicken thoracic SC 72 hours after Raldh2 intron 1G electroporation showing enhancer activation in the RP (arrowhead), dorsal interneuron (black arrow) and migrating dorsal interneurons (asterisk), as well as reporter expression in CI ax (white arrow). (K,O) RFP expression (red) driven by the chicken beta-actin promoter (positive control). (M,N) eGFP expression driven by the minimal Tk promoter. Dashed lines indicate plane of section in adjacent panels. (P) Scheme of enhancer-driven expression in mouse and chicken embryonic SC. mn, motoneurons.
Roof plate and interneuron functions are conserved in tetrapod enhancers
To determine whether RP and dI enhancer functions are conserved in amniotes and amphibians, we cloned chicken and X. laevis raldh2 intron 1G CNEs. The chicken CNE drove eGFP expression in the RP and dI1 (Fig. 3J,L). Expression of eGFP was also observed in dIs migrating towards the ventral thoracic SC (Fig. 3J,L), as well as in ventral axonal projections of CIs (Fig. 3L), reminiscent of lacZ patterns in transgenic mice (compare Fig. 3F with 3L). Injection of the X. laevis enhancer reporter construct into X. laevis dorsal blastomeres produced restricted eGFP expression in the RP and dIs (Fig. 4A,B), indicating that the dorsal SC function associated with the enhancer has been conserved for at least 370 million years since the divergence of amphibians and amniotes (Hedges and Kumar, 2003).
Fig. 4.
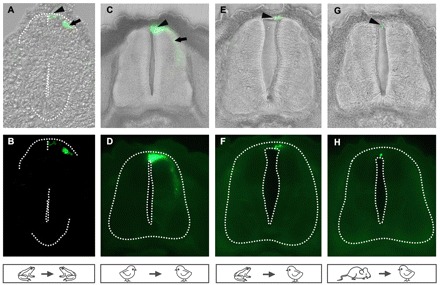
The raldh2 intron 1G enhancer is a tetrapod dorsal spinal cord enhancer. (A,B) Injection of the Xenopus raldh2 intron 1G CNE into Xenopus laevis blastomeres activates eGFP expression in the RP (arrowhead) and in dIs (arrow) at NF25. (C-H) Electroporation of chicken embryos. Frog (E,F) and mouse (G,H) enhancers retain RP, but not interneuron, activity in the electroporated chicken thoracic SC at HH25, contrasting with robust activity of the chicken enhancer in the chicken RP (arrowhead) and dIs (arrow) (C,D). The neural tube boundary is outlined.
In heterologous assays in chicken embryos, the mouse and X. laevis enhancers activated reporter transcription in the RP of the SC (Fig. 4E-H), albeit with differences in domain extension when compared with the chicken enhancer (Fig. 4C,D). Neither the mouse nor the X. laevis enhancer was activated in chicken dIs (Fig. 4E-H), suggesting that the RP function, rather than the dI role, has been conserved across tetrapod enhancers.
The enhancer is activated by three Tcf-homeodomain sites and is repressed by Lim-homeodomain and Tgif sites
We generated deletion mutants of the chicken Raldh2 intron 1G enhancer and electroporated constructs into the chicken SC (Fig. 5A). Removal of the 5′ 144 bp did not interfere with RP or dI expression, but led to eGFP expression throughout the SC, indicating that repression of ventral interneuron expression was lost in the 734 bp construct (Fig. 5B). Because this 5′ 144 bp region contains a predicted Lim-homeodomain binding site that is conserved from amphibians to primates, we used site-specific mutagenesis to modify this sequence in the context of the full intron 1G enhancer. Electroporation of this Lim mutant resulted in eGFP expression in SC domains ventral to the RP and dI1, indicating that this sequence is one of the repressors in the 5′ 144 bp fragment (Fig. 5G).
Fig. 5.
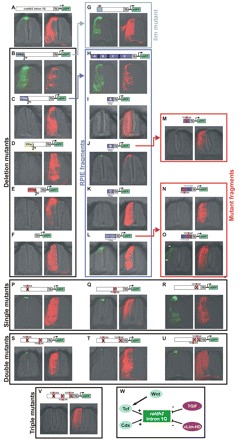
Dissection of the chicken Raldh2 intron 1G enhancer. (A) The wild-type chicken enhancer drives RP and dorsal-most interneuron expression. (B) Removal of the 5′ 144 bp expands expression throughout the SC, highlighting the presence of strong inhibitors of ventral interneuron expression. (C) Further removal of 279 bp abrogates RP and interneuron expression, indicating that this fragment contains a RP and interneuron activator element (RPIE). (D,E) Further removal of 95 bp (D) and 207 bp (E) does not reveal relevant cis regulators. (F) A minimal Tk promoter does not drive significant eGFP expression. (G) Mutation of a Lim-homeodomain site within the 5′ 144 bp repressor releases interneuron expression. (H) The isolated RPIE drives RP and interneuron expression. (I-L) RPIE dissection into four overlapping fragments (A-D) indicates that RP and interneuron activities reside in fragments B and D (J,L). (M) Mutation of a double Tcf/Cdx site in fragment B abolishes RP and interneuron expression. (N) Mutation of a double Tcf/Cdx site in D1 abrogates RP and interneuron expression. (O) Mutation of a double Tcf/Tgif site (D2) does not change RP expression, but expands expression into more-ventral interneurons (arrows). (P-R) Single mutations in double Tcf/homeodomain sites in the context of the full chicken Raldh2 intron 1G enhancer. (P) 5′ Tcf/Cdx mutation limits RP expression and restricts interneuron expression. (Q) 3′ Tcf/Cdx mutation limits RP expression and eliminates interneuron expression. (R) Tcf/Tgif mutation derepresses ventral interneuron expression. (S-U) Double mutants B+D1, B+D2 and D1+D2 do not eliminate RP expression. (V) Dorsal SC expression is eliminated in the triple mutant. (W) Model of Raldh2 intron 1G regulation in the SC. HD, homeodomain.
Further removal of 279 bp from the 734 bp sequence abrogated RP and interneuron expression in the 450 bp construct (Fig. 5C), indicating that the cis-regulatory elements directing RP and interneuron expression are contained in a 279 bp fragment. This fragment, termed the RP/interneuron activator element (RPIE), was isolated and shown to drive RP and interneuron expression throughout the SC (Fig. 5H). The RPIE was divided into four consecutive overlapping fragments, A-D, and we determined that RP and interneuron activities reside in fragments B and D (Fig. 5J,L). To identify the cis-elements controlling RP and interneuron expression, we aligned chicken and mouse B and D regions to detect sequences displaying full conservation. These were then altered by site-directed mutagenesis, creating one mutant for fragment B and two mutants for fragment D (D1 and D2). Sites B and D1 contain overlapping nested binding sites for the Wnt pathway transcription factor Tcf and for the Cdx-homeodomain protein. These sites display matrix identities to the matrix core that range between 86 and 90% and are conserved across amphibian, avian, marsupial, rodent and primate enhancers. Site D2 contains another, overlapping Tcf-homeodomain binding site, this time for the repressor homeodomain transcription factor 5′-TG-3′ interacting factor (Tgif; also known as TGFB-induced factor homeobox 1) (Knepper et al., 2006). These binding sites display matrix identities of 90% and are conserved in amphibian, avian, marsupial, rodent and primate enhancers. We showed that RP and interneuron expression was eliminated when fragments B and D were mutated at sites B and D1 (Fig. 5M,N), indicating that they contain RP and dI activators. By contrast, mutation of fragment D at site D2 (Fig. 5O) expanded expression into more-ventral interneurons (Fig. 5O, arrows), suggesting that it contains a repressor element.
Next, we mutated sites B, D1 and D2 in the context of the full intron 1G enhancer. RP expression was not eliminated by individual mutations (Fig. 5P-R), although dI expression was sharply restricted in B and D1 mutants (Fig. 5P,Q). Interestingly, D2 mutants displayed increased ventral interneuron expression, confirming that the mutated Tcf/Tgif sequence contains an interneuron-specific repressor (Fig. 5R). Complete inhibition of RP expression was not observed in double mutants (Fig. 5S-U), but was achieved in the triple mutant (B, D1 and D2), highlighting redundant mechanisms of RP expression (Fig. 5V). Thus, the dorsal SC activity of the chicken Raldh2 intron 1G enhancer is achieved by three elements that induce activation in the RP and dorsal-most interneurons, as well as by the combined inhibition of ventral expression by at least two repressors: a Lim-homeodomain site in the 5′ 144 bp fragment and a Tcf/Tgif motif in D2 (Fig. 5W).
Enhancer activation in the spinal cord reveals a novel transient domain of endogenous Raldh2 expression in amniotes
Prompted by the expression patterns driven by the Raldh2 intron 1G enhancer, we re-examined the profiles of Raldh2 expression in the amniote SC. Besides being strongly expressed in the mouse RP, the endogenous Raldh2 domain was seen to extend ventrally in the 9.0-10.5 dpc lumbar SC, reaching DV levels occupied by dI1 precursors (Fig. 6E,G,H; Fig. 7R; see Fig. S5C in the supplementary material). This novel Raldh2 domain might have previously escaped detection because of its transient nature. Indeed, at 11.5 dpc, Raldh2 expression rapidly receded from interneuron precursors, remaining highest in the RP (see Fig. S5E in the supplementary material, arrowhead), although residual expression was observed as a faint labeling in dI precursors (see Fig. S5E in the supplementary material, bracket). In chicken, Raldh2 expression initiated bilaterally in dIs of the brachial SC (Fig. 6N). Later, expression receded in dIs, becoming restricted to the RP (Fig. 6J, arrowhead; Fig. 7N,O, arrowhead).
Fig. 6.
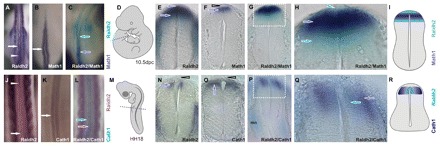
A novel spinal cord Raldh2 domain in dorsal interneurons. (A,J) Bilateral Raldh2 expression in dIs of 10.5 dpc mouse (A) and chicken (J) embryos. Arrows and arrowheads point to interneuron and RP domains, respectively. Raldh2 expression in the chicken dorsal SC shifts from bilateral interneuron fields at brachial levels to a single midline RP domain at cervical levels. (B,K) Mouse Math1/chicken Cath1 expression marks dorsal-most (dI1) interneurons. (C,L) Double ISH for Raldh2 and Math1/Cath1 indicates overlapping Raldh2 and Math1/Cath1 domains in dI1. (D,M) Diagrams depicting embryo position in A-C and J-L and section levels (dashed lines) in E-H and N-Q. (E) Raldh2 expression in the 10.5 dpc mouse lumbar SC from the RP (arrowhead) to adjacent dIs (arrow). (F) Math1 expression labels mouse dI1 (arrow). Note the lack of staining in the RP (open arrowhead). (G) Double ISH for Raldh2 (light blue) and Math1 (dark blue). (H) Enlargement of the boxed region in G. Raldh2 expression spreads from the RP (arrowhead) through the Math1 domain (dark-blue arrow), emerging ventral to dI1 (light-blue arrow). (I) Scheme of Raldh2 and Math1 expression in the mouse dorsal lumbar SC. (N) Raldh2 expression in the HH18 chicken brachial SC. Raldh2 is expressed in the ventricular zone of dIs, but not in the RP (open arrowhead). (O) Cath1 expression labels dI1 (arrow). Note the lack of staining in the RP (open arrowhead). (P) Double ISH for Raldh2 (light blue) and Cath1 (dark blue). Note the dorsal expression of Raldh2 and Cath1 and ventral expression of Raldh2 in motoneurons. (Q) Enlargement of the boxed region in P. Raldh2 is expressed in a ventricular (medial) subset of the Cath1 interneuron (dI1) domain. (R) Scheme of Raldh2 and Cath1 expression.
Fig. 7.
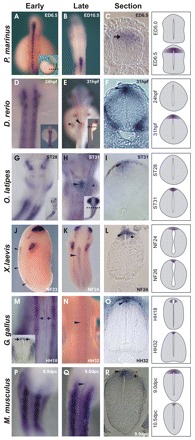
Ontogeny of raldh2 expression in the vertebrate dorsal spinal cord. (A-C) In the lamprey (Petromyzon marinus) embryo, raldh2 is expressed in dIs (C, arrow), but not in the RP. (D-F) raldh2 expression in zebrafish embryos. (D) No raldh2 expression is detected in the zebrafish neural tube 24 hours post-fertilization (hpf), but at 31 hpf raldh2 expression is detected in a small domain in the hindbrain/SC transition at the pectoral fin bud level (E, asterisk; E,F, arrowheads). (G-I) raldh2 ISH in stage-matched medaka (Oryzias latipes) embryos indicates onset of expression in the same hindbrain/SC domain as in zebrafish (H,I). (J-R) Frog (J-L), chicken (M-O) and mouse (P-R) embryos express raldh2 in the RP (arrowheads) and in dIs (arrows). (M-O) SC Raldh2 expression starts in chicken interneurons at HH18, at the forelimb bud level (M, arrows). Raldh2 expression is restricted to the RP in HH32 chicken SC (M-O). In frogs (J-L) and mice (P-R), raldh2 is initially expressed in RP and in dIs, but is subsequently restricted to the RP. Insets show whole embryos, with plane of section indicated (dashed line). Inset in M shows Raldh2 staining (arrows) in chicken brachial sections. Expression patterns are represented schematically on the right.
To directly demonstrate that Raldh2 is expressed in SC dIs we performed single and double ISH for Raldh2 and Math1/Cath1 (Cath1 is the chicken homolog of mouse Math1), a dI1 marker. Fig. 6A,B display Raldh2 and Math1 interneuron domains along the mouse SC and Fig. 6C shows that these two domains overlap at the hindlimb level. Sections of the lumbar SC showed that Raldh2 expression extends from the RP, overlaps with Math1 expression and extends beyond it, reaching interneurons ventral to the Math1 domain (Fig. 6G,H and see Fig. S5D in the supplementary material). In chicken, Cath1 was expressed in dIs at the same AP level as those that express Raldh2 (Fig. 6J,K,N,O) and double-marker analysis indicated that Raldh2 and Cath1 overlap in dI1 and that the Raldh2 domain constitutes a medial, ventricular subset of the Cath1 territory (Fig. 6L,P,Q).
RA signaling is an ancestral mechanism associated with dorsal interneuron ontogenesis in the vertebrate spinal cord
To establish whether expression of raldh2 in the RP and in dIs is a tetrapod feature or an ancestral vertebrate trait, we cloned raldh2 from the agnathan lamprey Petromyzon marinus (Fig. 7, Fig. 8). The raldh2 identity of the lamprey clone was confirmed by phylogenetic and expression analyses (Fig. 8) (M.S.S.-C., unpublished). Lamprey embryos displayed strong raldh2 activation throughout the early dorsal SC (Fig. 7A, Fig. 8C), similar to tetrapods, which exhibit expression along most of the dorsal SC (Fig. 7J,K,N,Q). Although regional AP activation of raldh2 in the dorsal neural tube was similar in lampreys and tetrapods (Fig. 7A,J,N,Q), there were DV differences. In lampreys, activation of raldh2 was observed in dIs, but not in the RP (Fig. 7C; Fig. 8B,D,F). This selective activation of lamprey raldh2 in dIs contrasts with expression of the dorsal marker zic-1 in the lamprey RP and dIs, indicating that a RP is present in this agnathan (Fig. 8G-L). Thus, the absence of raldh2 expression in the RP of limbless lampreys contrasts with raldh2 expression in the RP and, transiently, in the dIs of tetrapods (Fig. 7J-R). In connection with this, we identified a short sequence homologous to the tetrapod raldh2 intron 1G enhancer among lamprey genome traces (see Fig. S1 in the supplementary material). This sequence (gnl|ti|1386265511) aligns to a stretch of 230 bp in the 3′ region of the raldh2 intron 1G enhancer with 69.1% identity, and a reciprocal BLAST search against the non-redundant database identified the H. sapiens RALDH2 intron 1G enhancer as a high-score match for the lamprey sequence (score 60.8, E-value 3.0×10−6), supporting the idea that activation of RA signaling in dIs is an ancestral vertebrate feature.
Fig. 8.
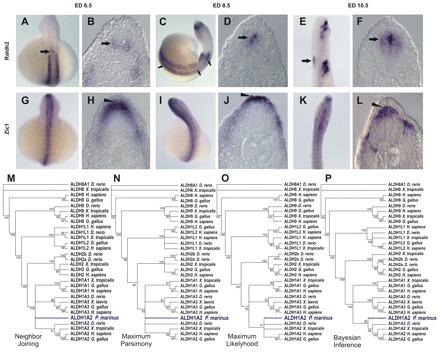
raldh2 expression in the lamprey spinal cord. (A-F) Time course of raldh2 SC expression in the lamprey embryo. SC raldh2 expression starts at embryonic day (ED) 6.5 in dIs (arrow in B), in a small anterior domain (arrow in A). At ED 8.5, raldh2 expression extends to encompass the whole AP axis of the embryonic SC (C, arrows), where it is restricted to dIs (D, arrow). Later (ED 10.5), raldh2 expression is restricted to dIs of the anterior SC domain (E,F, arrows). (G-L) Expression of the dorsal marker zic-1 labels RP and dIs throughout the lamprey neural tube, indicating that a RP is present in lamprey (arrowhead) and highlighting the restriction of lamprey raldh2 expression to dIs. (M-P) Phylogenetic trees constructed from the alignment of vertebrate Aldh protein sequences. Nodes with bootstraps inferior to 80%, 80%, 80% and 90% were collapsed in neighbor-joining (NJ), maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) trees, respectively.
Comparative ontogeny of raldh2 expression suggests that RA functions in the innervation of vertebrate appendages
In contrast to the conserved AP patterns of raldh2 expression in the agnathan and tetrapod SC, the teleost D. rerio exhibits RP expression of raldh2 only in a small region between the caudal hindbrain and SC, adjacent to somites 2 to 3 (Skromne et al., 2007). This restricted cranial domain of zebrafish raldh2 RP expression corresponds to the AP levels at which the pectoral fin buds form in the lateral mesoderm (Fig. 7E,F). Thus, raldh2 expression in the zebrafish RP is restricted to this cranial area, indicating posterior truncation of most of the SC domain (Fig. 7E,F). To establish whether this raldh2 pattern is specific for D. rerio, or is a general teleost characteristic, we cloned medaka (Oryzias latipes) raldh2 and characterized its expression pattern in carefully stage-matched embryos (Signore et al., 2009). As shown in Fig. 7G-I, medaka embryos also display the restricted cranial domain of raldh2 expression at the level of the pectoral fin buds. Therefore, the divergent patterns of raldh2 expression in the dorsal neural tube displayed by D. rerio and O. latipes are consistent with our inability to find an ortholog of the tetrapod raldh2 intron 1G enhancer in any of the sequenced teleost genomes. Perhaps, another regulatory sequence drives the divergent teleost raldh2 domain. However, there is an important parallel between the onset of raldh2 expression in the dorsal SC of chicken, zebrafish and medaka. In chicken and teleost embryos, dorsal SC expression of raldh2 begins in restricted AP domains at the level of the forelimb/pectoral fin buds (Fig. 7E,H,M), suggesting that raldh2 expression in the dorsal SC is functionally related to the innervation of vertebrate forelimbs/fins.
DISCUSSION
We describe an intronic enhancer of the raldh2 gene that is activated in the RP of the SC and adjacent dIs, consistent with activation of endogenous raldh2 in the RP and, transiently, in the amniote dI1 domain. Enhancer analysis suggests a model for raldh2 regulation in the developing neural tube whereby raldh2 expression in the caudal SC results from the interaction of positive influences, represented by an interaction with Cdx and Wnt signaling, and inhibitory influences, represented by Lim-homeodomain and Tgif factors. In this model, the intersection between Cdx and Wnt family activation focuses raldh2 expression to the dorsal SC and this dorsal expression domain is refined through inhibition by Lim-homeodomain and Tgif factors that are expressed ventral to the dorsal-most interneuron domains (i.e. dI1), restricting raldh2 expression to the RP and dI1 (Fig. 9) (Wine-Lee et al., 2004; Zhou et al., 2003; Iulianella et al., 2003; Knepper et al., 2006; Sheng et al., 1997).
Fig. 9.
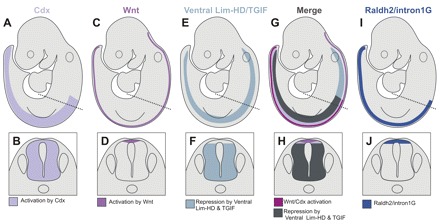
Model for dorsal spinal cord raldh2 regulation. (A,B) Cdx genes are expressed throughout the posterior SC. (C,D) Wnt genes are expressed in the RP throughout the anterior neural tube and the SC. (E,F) Combined expression of Lim-homeodomain genes, such as Lhx1-3,9 and Tgif, define a SC domain ventral to the RP and dorsal-most interneurons. (G,H) Intersection of positive (Cdx and Wnt) and negative Lim-homeodomain and Tgif regulators of the raldh2 intron 1G enhancer in the embryo (G) and SC (H). (I,J) raldh2 intron 1G enhancer and endogenous raldh2 domains in the SC are defined by a positive layer of regulation due to Cdx and by autocrine and paracrine activation by the Wnt pathway in the RP and adjacent dIs. Enhancer and gene expression domains are refined through inhibition via Tgif homeoboxes and ventrally expressed repressors such as Lim-homeodomain (HD) transcription factors. The dashed line in the top row indicates the plane of section in the bottom row.
The activity of the raldh2 intron 1G enhancer suggests a role for RA signaling in the organization of dorsal spinal cord pathways
The raldh2 intron 1G enhancer activates transcription in the dorsal SC when dI identities are being determined and when the ventralizing effects of vitamin A deprivation are pronounced (Berggren et al., 1999; Wilson et al., 2004; Wilson and Maden, 2005). This suggests that the roles of retinoid signaling in DV organization of the SC are played by transient, autocrine RA signaling in dI progenitors and by ventral diffusion of the RP-derived RA. This is consistent with the fact that dIs are within range of activation by the RA diffusing from the RP or dI1, as established by expansion of ventral SC markers in vitamin A-deprived avian embryos (Wilson et al., 2004).
raldh2 expression suggests that paracrine RA from the roof plate plays roles in spinocerebellar proprioception
The onset of raldh2 expression in the dorsal SC in register with forelimb buds in chicken, and the restricted RP and dI expression of raldh2 at the teleost pectoral fin level, suggest that raldh2 expression in the dorsal SC is associated with the formation of circuits that convey sensory information from vertebrate fins/limbs. Because the RP is glial and does not take part in neural circuits, it is likely that RP-derived RA signaling plays a role in the development of the adjacent dI1 progenitors, which give rise to SC circuits and ascending pathways (Bermingham et al., 2001; Helms and Johnson, 1998). Activation of the mouse Raldh2 intron 1G enhancer and of Raldh2 expression in the RP, coupled with transient Raldh2 expression in dIs, suggests that RA signaling is involved in dI1 development. Math1-expressing dI1 interneurons migrate to ventral locations in the gray matter, coming to rest between laminae VI and VIII to form integrative centers that receive proprioceptive input from fore and hindlimbs, as well as from trunk, neck and thorax, relaying it to the cerebellum via dorsal and ventral spinocerebellar tracts, which are depleted in Math1-null mice (Bermingham et al., 2001; Bloedel and Courville, 1981; Brodal, 1981; Helms and Johnson, 1998; Lewis, 2006). The integrative SC center that serves the dorsal spinocerebellar system is Clarke's nucleus. These interneuron cells receive afferents from lower trunk and hindlimbs and are concentrated in lamina VII, which also receives information from posterior trunk, hindlimbs and forelimbs and projects to the cerebellum via ventral spinocerebellar and rostral spinocerebellar tracts (Bloedel and Courville, 1981; Brodal, 1981). Therefore, RP-derived RA is poised to influence dI1 interneurons, which give rise to proprioceptive spinocerebellar circuits.
raldh2 expression suggests that autocrine RA in dorsal interneurons plays roles in crossed spinal cord proprioception
The exclusive expression of raldh2 in interneurons of the developing lamprey SC provides clues to raldh2 function in tetrapod SC interneurons. The lamprey SC contains neurons that are responsible for locomotion (Grillner, 2003). The basic SC locomotor network is modulated by sensory signals from spinal stretch receptors. During lamprey locomotion, segmental muscular contractions on one side activate intraspinal stretch receptors on the contralateral, distended side. These glycinergic stretch receptors send axons to synapse with, and inhibit, pattern generator neurons on the active side (Grillner et al., 1984). The regular activity of these crossed inhibitory interneurons is crucial for motor coordination during locomotion and it is likely that crossed circuits of this sort are present in most vertebrates (Grillner, 2003). The expression patterns directed by the raldh2 intron 1G enhancer in the tetrapod dorsal SC suggest that RA signaling serves a regulatory network that supports the development of CIs belonging to sensory pathways that are akin to the crossed inhibitory pathway of lampreys. A subset of these CIs descends from Math1-expressing dI1 progenitors that are underrepresented in the SC of Math1-null mice (Bermingham et al., 2001). As demonstrated in Figs 3 and 6 and Fig. S5 in the supplementary material, activation of the Raldh2 intron 1G enhancer and of Raldh2 at the early stages of dI development supports a role for RA signaling in dI1 ontogeny. Moreover, β-galactosidase activity driven by the mouse Raldh2 intron 1G enhancer specifically labels CIs, indicating that the enhancer is also activated in commissural progeny of dI1 (Fig. 3F), consistent with demonstrations of cellular RA-binding protein (Crabp) expression in these cells (Colbert et al., 1995; Maden et al., 1989).
Proprioceptive pathways from vertebrate fins/limbs: RA signaling and the simplification of teleost fins
When contrasted with agnathan and tetrapod dorsal SC domains, the diminutive raldh2 cranial domain of zebrafish and medaka suggests that the absence of the raldh2 intron 1G enhancer in teleosts is secondary to regulatory simplifications associated with anatomic reductions/losses of fins in these actinopterygians (Davis et al., 2007; Freitas et al., 2007; Santini and Tyler, 2003; Shapiro et al., 2004; Tanaka et al., 2005). Zebrafish fins are filled with dermal rays and have only small proximal radials, which provide support for actuation by only two muscles that adduct or abduct the fin (Thorsen and Hale, 2007). Moreover, teleost pelvic fins are lost in freshwater populations of stickleback (Gasterosteus aculeatus), or lost outright in Tetraodon and fugu, contrasting with the fins/limbs of basal actinopterygeans, sarcopterygeans and tetrapods that display complex endochondral bone elements, tendons and articular surfaces innervated with afferents conveying information about position, muscular contraction and tendon tension to the SC (Kandel et al., 1991). The complex traffic of motor and sensory information to and from the muscular limbs of cartilaginous fish, basal actinopterygeans and sarcopterygeans is associated with a higher number of nerves servicing these appendages than in the simpler fins of D. rerio (Thorsen and Hale, 2007). Therefore, it is possible that the volume of information from sarcopterygean limbs requires a denser network of SC interneuron circuits and ascending fibers than is required for the comparatively simpler teleost fins.
Afferent information from amniote limbs is routed to SC segments level with the emergence of fore/hindlimbs, where they are grouped as Clarke's nucleus. However, a great deal of limb afferent information is also distributed to SC segments above or below the limb buds (Brodal, 1981). As such, amniotes display an extensive SC interneuron column, termed Clarke's column, which is distributed along neck, trunk and lumbar segments. The restriction of RP raldh2 expression to a cranial SC domain that is level with teleost pectoral fin buds brings into question the existence of a teleost homolog of the amniote Clarke's column. It is possible that the relatively small amount of afferent information from teleost fins is handled exclusively by SC segments in register with, or adjacent to, the fins, and for this reason no structure homologous to the Clarke's column has been reported in teleosts. The distribution of SC nuclei is indeed plastic in vertebrates and new spinal nuclei/columns are formed when sensory input from peripheral receptors increases after the emergence of complex peripheral structures, or novel sensory capabilities, as indicated by the development of specific SC nuclei/columns associated with the evolution of chemosensation in the pectoral fins of the teleost northern sea robin (Prionotus carolinus) (Finger, 2000). In summary, the absence of an raldh2 intron 1G enhancer and of raldh2 expression throughout most of the SC in zebrafish and medaka are consistent with the loss of cis and trans factor components of regulatory networks associated with the simplification of teleost fins (Hildebrand and Goslow, 1998; Shapiro et al., 2004; Tanaka et al., 2005). This is further supported by the absence of expression of raldh paralogs in the embryonic teleost SC (Liang et al., 2008; Pittlik et al., 2008).
The ontogeny and phylogeny of RA signaling in the dorsal spinal cord
The raldh2 intron 1G enhancer is a module that controls RA signaling in dIs and RP. This module is ontogenetically and phylogenetically associated with the development of two proprioception modes: one launched by the early activation of autocrine RA signaling in dI progenitors and linked to the emergence of intraspinal proprioceptive circuits responsible for motor coordination across both sides of the SC during locomotion (Fetcho, 1992); and another represented by a later paracrine signaling from the RP to a subset of interneuron progenitors that is linked to the development of spinocerebellar neural circuits conveying fin/limb proprioception. The first mode is probably older and might trace back to the ancestral chordate, presumably a finless cephalochordate-like animal capable of bending its body to swim, whereas the second mode probably evolved with the appearance of vertebrate paired appendages. Thus, RA signaling is an ancestral mechanism of DV organization of the SC that seems to be plastic, allowing for changes in genetic regulation associated with the evolution of the diverse locomotor patterns and morphologies of vertebrate paired appendages (Goulding, 2009). In this sense, it is likely that the lack of raldh2 expression in the lamprey RP is a derived feature. Although lampreys display ancestral vertebrate characteristics, Haikouichthys, Myllokunmingia and other agnathan fossils indicate that primitive vertebrates already had prototypes of bilateral fins represented by a pair of continuous mediolateral fin-folds spanning the AP axis, implying that fin absence in extant agnathans is a derived feature of lampreys (Forey, 1995). Thus, it is possible that raldh2 expression was lost from the RP owing to a secondary loss of paired fins in lampreys (Forey, 1995).
A comparative approach to understanding signaling in spinal cord development and evolution
Combining comparative genomic and developmental methods is a fruitful approach to investigating the ontogeny/phylogeny of developmental mechanisms that are controlled by signaling systems in the vertebrate CNS. A considerable amount of non-coding sequence conservation between distantly related vertebrates is represented around genes that play essential roles in CNS and heart development (Woolfe et al., 2005), organs that harbor key vertebrate-specific innovations (Gans and Northcutt, 1983; Simoes-Costa et al., 2005; Xavier-Neto et al., 2007). Thus, it is feasible to search for core, conserved vertebrate developmental gene regulatory networks that can be used, in selected cases, to infer how specific vertebrate adaptions have emerged and to understand how class-specific vertebrate body plans differ from each other.
Supplementary Material
Acknowledgements
We are indebted to Ursula Dräger, Peter McCaffery and Marcus Vinicius Baldo for comments and suggestions; to Richard Behringer and Wellington Cardoso for comments on the manuscript; to Masanori Uchikawa and Jane Johnson for reagents; and to the Faculty of Medicine of the University of São Paulo for access to its high-performance computing cluster. This work was supported by grants from FAPESP (02/11340-2; 04/11569-5; 04/15704-4; 05/60637-6; 06/50843-0; 06/61317-8), CNPq 305260/2007-3 and by a Development Travelling Fellowship from The Company of Biologists.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.043257/-/DC1
References
- Abascal F., Zardoya R., Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21, 21-2104 [DOI] [PubMed] [Google Scholar]
- Altman J., Bayer S. A. (1997). Development of the Cerebellar System In Relation to Its Evolution, Structure, and Functions. New York: CRC Press; [Google Scholar]
- Berggren K., McCaffery P., Drager U., Forehand C. J. (1999). Differential distribution of retinoic acid synthesis in the chicken embryo as determined by immunolocalization of the retinoic acid synthetic enzyme, RALDH-2. Dev. Biol. 210, 210-288 [DOI] [PubMed] [Google Scholar]
- Bermingham N. A., Hassan B. A., Wang V. Y., Fernandez M., Banfi S., Bellen H. J., Fritzsch B., Zoghbi H. Y. (2001). Proprioceptor pathway development is dependent on Math1. Neuron 30, 30-411 [DOI] [PubMed] [Google Scholar]
- Blentic A., Gale E., Maden M. (2003). Retinoic acid signalling centres in the avian embryo identified by sites of expression of synthesising and catabolising enzymes. Dev. Dyn. 227, 227-114 [DOI] [PubMed] [Google Scholar]
- Bloedel J. R., Courville J. (1981). Cerebellar afferent systems. In Handbook of Physiology, vol. 2 Baltimore: Waverly Press; [Google Scholar]
- Brodal A. (1981). Neurological Anatomy in Relation to Clinical Medicine. New York: Oxford University Press; [Google Scholar]
- Chen Y., Pollet N., Niehrs C., Pieler T. (2001). Increased XRALDH2 activity has a posteriorizing effect on the central nervous system of Xenopus embryos. Mech. Dev. 101, 101-91 [DOI] [PubMed] [Google Scholar]
- Chizhikov V. V., Millen K. J. (2004). Mechanisms of roof plate formation in the vertebrate CNS. Nat. Rev. Neurosci. 5, 5-808 [DOI] [PubMed] [Google Scholar]
- Colbert M. C., Rubin W. W., Linney E., LaMantia A. S. (1995). Retinoid signaling and the generation of regional and cellular diversity in the embryonic mouse spinal cord. Dev. Dyn. 204, 204-1 [DOI] [PubMed] [Google Scholar]
- Davis M. C., Dahn R. D., Shubin N. H. (2007). An autopodial-like pattern of Hox expression in the fins of a basal actinopterygian fish. Nature 447, 473-476 [DOI] [PubMed] [Google Scholar]
- Diez del Corral R., Olivera-Martinez I., Goriely A., Gale E., Maden M., Storey K. (2003). Opposing FGF and retinoid pathways control ventral neural pattern, neuronal differentiation, and segmentation during body axis extension. Neuron 40, 40-65 [DOI] [PubMed] [Google Scholar]
- Dodd J., Morton S. B., Karagogeos D., Yamamoto M., Jessell T. M. (1988). Spatial regulation of axonal glycoprotein expression on subsets of embryonic spinal neurons. Neuron 1, 1-105 [DOI] [PubMed] [Google Scholar]
- Edgar R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetcho J. R. (1992). The spinal motor system in early vertebrates and some of its evolutionary changes. Brain Behav. Evol. 40, 40-82 [DOI] [PubMed] [Google Scholar]
- Finger T. E. (2000). Ascending spinal systems in the fish, Prionotus carolinus. J. Comp. Neurol. 422, 422-106 [DOI] [PubMed] [Google Scholar]
- Forey P. L. (1995). Agnathans recent and fossil, and the origin of jawed vertebrates. Rev. Fish Biol. Fisheries 5, 5-267 [Google Scholar]
- Freitas R., Zhang G., Cohn M. J. (2007). Biphasic Hoxd gene expression in shark paired fins reveals an ancient origin of the distal limb domain. PLoS ONE 2, e754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furley A. J., Morton S. B., Manalo D., Karagogeos D., Dodd J., Jessell T. M. (1990). The axonal glycoprotein TAG-1 is an immunoglobulin superfamily member with neurite outgrowth-promoting activity. Cell 61, 157-170 [DOI] [PubMed] [Google Scholar]
- Gans C., Northcutt R. G. (1983). Neural crest and the origin of vertebrates: a new head. Science 220, 220-268 [DOI] [PubMed] [Google Scholar]
- Goulding M. (2009). Circuits controlling vertebrate locomotion: moving in a new direction. Nat. Rev. Neurosci. 10, 10-507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillner S. (2003). The motor infrastructure: from ion channels to neuronal networks. Nat. Rev. Neurosci. 4, 4-573 [DOI] [PubMed] [Google Scholar]
- Grillner S., Williams T., Lagerback P. A. (1984). The edge cell, a possible intraspinal mechanoreceptor. Science 223, 223-500 [DOI] [PubMed] [Google Scholar]
- Hedges S. B., Kumar S. (2003). Genomic clocks and evolutionary timescales. Trends Genet. 19, 19-200 [DOI] [PubMed] [Google Scholar]
- Helms A. W., Johnson J. E. (1998). Progenitors of dorsal commissural interneurons are defined by MATH1 expression. Development 125, 125-919 [DOI] [PubMed] [Google Scholar]
- Hildebrand M., Goslow G. (1998). Analysis of Vertebrate Structure. New York: Wiley; [Google Scholar]
- Iulianella A., Vanden Heuvel G., Trainor P. (2003). Dynamic expression of murine Cux2 in craniofacial, limb, urogenital and neuronal primordia. Gene Expr. Patterns 3, 3-571 [DOI] [PubMed] [Google Scholar]
- Kandel E. R., Schwartz J. H., Jessell T. M. (1991). Principles of Neural Science. New York: Elsevier; [Google Scholar]
- Keino-Masu K., Masu M., Hinck L., Leonardo E. D., Chan S. S., Culotti J. G., Tessier-Lavigne M. (1996). Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell 87, 87-175 [DOI] [PubMed] [Google Scholar]
- Knepper J. L., James A. C., Ming J. E. (2006). TGIF, a gene associated with human brain defects, regulates neuronal development. Dev. Dyn. 235, 235-1482 [DOI] [PubMed] [Google Scholar]
- Kothary R., Clapoff S., Darling S., Perry M. D., Moran L. A., Rossant J. (1989). Inducible expression of an hsp68-lacZ hybrid gene in transgenic mice. Development 105, 105-707 [DOI] [PubMed] [Google Scholar]
- Lewis K. E. (2006). How do genes regulate simple behaviours? Understanding how different neurons in the vertebrate spinal cord are genetically specified. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 361-45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D., Zhang M., Bao J., Zhang L., Xu X., Gao X., Zhao Q. (2008). Expressions of Raldh3 and Raldh4 during zebrafish early development. Gene Expr. Patterns 8, 8-248 [DOI] [PubMed] [Google Scholar]
- Lin J. T., Wu M. S., Wang W. S., Yen C. C., Chiou T. J., Liu J. H., Yang M. H., Chao T. C., Chou S. C., Chen P. M. (2003). All-trans retinoid acid increases Notch1 transcript expression in acute promyelocytic leukemia. Adv. Ther. 20, 20-337 [DOI] [PubMed] [Google Scholar]
- Maden M., Ong D. E., Summerbell D., Chytil F., Hirst E. A. (1989). Cellular retinoic acid-binding protein and the role of retinoic acid in the development of the chick embryo. Dev. Biol. 135, 135-124 [DOI] [PubMed] [Google Scholar]
- Moss J. B., Xavier-Neto J., Shapiro M. D., Nayeem S. M., McCaffery P., Drager U. C., Rosenthal N. (1998). Dynamic patterns of retinoic acid synthesis and response in the developing mammalian heart. Dev. Biol. 199, 199-55 [DOI] [PubMed] [Google Scholar]
- Muhr J., Graziano E., Wilson S., Jessell T. M., Edlund T. (1999). Convergent inductive signals specify midbrain, hindbrain, and spinal cord identity in gastrula stage chick embryos. Neuron 23, 23-689 [DOI] [PubMed] [Google Scholar]
- Niederreither K., McCaffery P., Drager U. C., Chambon P., Dolle P. (1997). Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech. Dev. 62, 62-67 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P. D. (1952). Activation and organization of the central nervous system in amphibians. Part III. Synthesis of a new working hypothesis. J. Exp. Zool. 120, 120-83 [Google Scholar]
- Novitch B. G., Wichterle H., Jessell T. M., Sockanathan S. (2003). A requirement for retinoic acid-mediated transcriptional activation in ventral neural patterning and motor neuron specification. Neuron 40, 40-81 [DOI] [PubMed] [Google Scholar]
- Pittlik S., Domingues S., Meyer A., Begemann G. (2008). Expression of zebrafish aldh1a3 (raldh3) and absence of aldh1a1 in teleosts. Gene Expr. Patterns 8, 8-141 [DOI] [PubMed] [Google Scholar]
- Quandt K., Frech K., Karas H., Wingender E., Werner T. (1995). MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23, 4878-4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F., Huelsenbeck J. P. (2003). MrBayes 3, Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 19-1572 [DOI] [PubMed] [Google Scholar]
- Rossant J., Zirngibl R., Cado D., Shago M., Giguere V. (1991). Expression of a retinoic acid response element-hsplacZ transgene defines specific domains of transcriptional activity during mouse embryogenesis. Genes Dev. 5, 5-1333 [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 4-406 [DOI] [PubMed] [Google Scholar]
- Santini F., Tyler J. C. (2003). A phylogeny of the families of fossil and extant tetraodontiform fishes (Acanthomorpha, Tetraodontiformes), upper cretaceous to recent. Zool. J. Linn. Soc. 139, 139-565 [Google Scholar]
- Sauka-Spengler T., Meulemans D., Jones M., Bronner-Fraser M. (2007). Ancient evolutionary origin of the neural crest gene regulatory network. Dev. Cell 13, 13-405 [DOI] [PubMed] [Google Scholar]
- Schmidt H. A., Strimmer K., Vingron M., von Haeseler A. (2002). TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18, 18-502 [DOI] [PubMed] [Google Scholar]
- Shapiro M. D., Marks M. E., Peichel C. L., Blackman B. K., Nereng K. S., Jonsson B., Schluter D., Kingsley D. M. (2004). Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature 428, 428-717 [DOI] [PubMed] [Google Scholar]
- Sheng H. Z., Bertuzzi S., Chiang C., Shawlot W., Taira M., Dawid I., Westphal H. (1997). Expression of murine Lhx5 suggests a role in specifying the forebrain. Dev. Dyn. 208, 208-266 [DOI] [PubMed] [Google Scholar]
- Signore I. A., Guerrero N., Loosli F., Colombo A., Villalon A., Wittbrodt J., Concha M. L. (2009). Zebrafish and medaka: model organisms for a comparative developmental approach of brain asymmetry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 364-991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoes-Costa M. S., Vasconcelos M., Sampaio A. C., Cravo R. M., Linhares V. L., Hochgreb T., Yan C. Y., Davidson B., Xavier-Neto J. (2005). The evolutionary origin of cardiac chambers. Dev. Biol. 277, 277-1 [DOI] [PubMed] [Google Scholar]
- Simoes-Costa M. S., Azambuja A. P., Xavier-Neto J. (2008). The search for non-chordate retinoic acid signaling: lessons from chordates. J. Exp. Zoolog. B Mol. Dev. Evol. 310, 310-54 [DOI] [PubMed] [Google Scholar]
- Skromne I., Thorsen D., Hale M., Prince V. E., Ho R. K. (2007). Repression of the hindbrain developmental program by Cdx factors is required for the specification of the vertebrate spinal cord. Development 134, 2147-2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobreira T. J., Gruber A. (2008). Sequence-specific reconstruction from fragmentary databases using seed sequences: implementation and validation on SAGE, proteome and generic sequencing data. Bioinformatics 24, 1676-1680 [DOI] [PubMed] [Google Scholar]
- Sockanathan S., Jessell T. M. (1998). Motor neuron-derived retinoid signaling specifies the subtype identity of spinal motor neurons. Cell 94, 503-514 [DOI] [PubMed] [Google Scholar]
- Stern C. D. (1998). Detection of multiple gene products simultaneously by in situ hybridization and immunohistochemistry in whole mounts of avian embryos. Curr. Top. Dev. Biol. 36, 36-223 [DOI] [PubMed] [Google Scholar]
- Swofford D. L. (2000). PAUP: Phylogenetic Analyses Using Parsimony and Other Methods. Sunderland, MA: Sinauer; [Google Scholar]
- Tanaka M., Hale L. A., Amores A., Yan Y. L., Cresko W. A., Suzuki T., Postlethwait J. H. (2005). Developmental genetic basis for the evolution of pelvic fin loss in the pufferfish Takifugu rubripes. Dev. Biol. 281, 281-227 [DOI] [PubMed] [Google Scholar]
- Thorsen D. H., Hale M. E. (2007). Neural development of the zebrafish (Danio rerio) pectoral fin. J. Comp. Neurol. 504, 504-168 [DOI] [PubMed] [Google Scholar]
- Ulven S. M., Gundersen T. E., Weedon M. S., Landaas V. O., Sakhi A. K., Fromm S. H., Geronimo B. A., Moskaug J. O., Blomhoff R. (2000). Identification of endogenous retinoids, enzymes, binding proteins, and receptors during early postimplantation development in mouse: important role of retinal dehydrogenase type 2 in synthesis of all-trans-retinoic acid. Dev. Biol. 220, 220-379 [DOI] [PubMed] [Google Scholar]
- Vermot J., Schuhbaur B., Le Mouellic H., McCaffery P., Garnier J. M., Hentsch D., Brulet P., Niederreither K., Chambon P., Dolle P., et al. (2005). Retinaldehyde dehydrogenase 2 and Hoxc8 are required in the murine brachial spinal cord for the specification of Lim1+ motoneurons and the correct distribution of Islet1+ motoneurons. Development 132, 132-1611 [DOI] [PubMed] [Google Scholar]
- Wang X., Penzes P., Napoli J. L. (1996). Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. Recognition of retinal as substrate. J. Biol. Chem. 271, 271-16288 [DOI] [PubMed] [Google Scholar]
- Wilkinson D. (1992). Whole Mount In Situ Hybridization: A Practical Approach. Oxford: IRL Press; [Google Scholar]
- Wilson L., Maden M. (2005). The mechanisms of dorsoventral patterning in the vertebrate neural tube. Dev. Biol. 282, 282-1 [DOI] [PubMed] [Google Scholar]
- Wilson L. J., Wingate R. J. (2006). Temporal identity transition in the avian cerebellar rhombic lip. Dev. Biol. 297, 297-508 [DOI] [PubMed] [Google Scholar]
- Wilson L., Gale E., Chambers D., Maden M. (2004). Retinoic acid and the control of dorsoventral patterning in the avian spinal cord. Dev. Biol. 269, 269-433 [DOI] [PubMed] [Google Scholar]
- Wine-Lee L., Ahn K. J., Richardson R. D., Mishina Y., Lyons K. M., Crenshaw E. B., 3rd (2004). Signaling through BMP type 1 receptors is required for development of interneuron cell types in the dorsal spinal cord. Development 131, 131-5393 [DOI] [PubMed] [Google Scholar]
- Wingender E., Dietze P., Karas H., Knuppel R. (1996). TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 24, 24-238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D. K., Snell P., McEwen G. K., Vavouri T., Smith S. F., North P., Callaway H., Kelly K., et al. (2005). Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier-Neto J., Neville C. M., Shapiro M. D., Houghton L., Wang G. F., Nikovits W., Jr, Stockdale F. E., Rosenthal N. (1999). A retinoic acid-inducible transgenic marker of sino-atrial development in the mouse heart. Development 126, 126-2677 [DOI] [PubMed] [Google Scholar]
- Xavier-Neto J., Shapiro M. D., Houghton L., Rosenthal N. (2000). Sequential programs of retinoic acid synthesis in the myocardial and epicardial layers of the developing avian heart. Dev. Biol. 219, 219-129 [DOI] [PubMed] [Google Scholar]
- Xavier-Neto J., Castro R. A., Sampaio A. C., Azambuja A. P., Castillo H. A., Cravo R. M., Simoes-Costa M. S. (2007). Parallel avenues in the evolution of hearts and pumping organs. Cell Mol. Life Sci. 64, 64-719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Smith D., Yamamoto M., Ma L., McCaffery P. (2003). The meninges is a source of retinoic acid for the late-developing hindbrain. J. Neurosci. 23, 23-7610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., McCaffery P., Ivins K. J., Neve R. L., Hogan P., Chin W. W., Drager U. C. (1996). Molecular identification of a major retinoic-acid-synthesizing enzyme, a retinaldehyde-specific dehydrogenase. Eur. J. Biochem. 240, 240-15 [DOI] [PubMed] [Google Scholar]
- Zhou X. H., Brandau O., Feng K., Oohashi T., Ninomiya Y., Rauch U., Fassler R. (2003). The murine Ten-m/Odz genes show distinct but overlapping expression patterns during development and in adult brain. Gene Expr. Patterns 3, 3-397 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


