Abstract
Telomerase is a ribonucleoprotein complex that is required for maintenance of linear chromosome ends (telomeres). In yeast, the Est2 protein reverse transcribes a short template region of the TLC1 RNA using the chromosome terminus to prime replication. Yeast telomeres contain heterogeneous G1–3T sequences that arise from incomplete reverse transcription of the TLC1 template and alignment of the DNA primer at multiple sites within the template region. We have previously described mutations in the essential N-terminal TEN domain of Est2p that alter telomere sequences. Here, we demonstrate that one of these mutants, glutamic acid 76 to lysine (est2-LTE76K), restricts possible alignments between the DNA primer and the TLC1 template. In addition, this mutant exhibits increased processivity in vivo. Within the context of the telomerase enzyme, the Est2p TEN domain is thought to contribute to enzyme processivity by mediating an anchor-site interaction with the DNA primer. We show that binding of the purified TEN domain (residues 1–161) to telomeric DNA is enhanced by the E76K mutation. These results support the idea that the anchor-site interaction contributes to telomerase processivity and suggest a role for the anchor site of yeast telomerase in mediating primer–template alignment within the active site.
Key words: Anchor site, Est2p, Telomerase, TLC1
Introduction
Chemical constraints of DNA replication prevent complete duplication of linear eukaryotic chromosomes. In most eukaryotic organisms, chromosome shortening is counteracted by the enzyme telomerase, a ribonucleoprotein complex that uses a portion of its intrinsic RNA molecule as a template for nucleotide addition to the end of the chromosome (Greider and Blackburn, 1985). Telomeres consist of a variable number of double-stranded G-rich repeats, terminating in a short 3′ overhang (Shampay et al., 1984). The TG-rich telomere forms the basis of the telosome by providing binding sites for proteins that protect the chromosome terminus from nucleolytic digestion and inappropriate DNA repair.
The telomerase enzyme is minimally composed of a catalytic subunit (telomerase reverse transcriptase; TERT) and an RNA containing a short template region complementary to the telomeric repeat. Base pairing between the telomerase RNA and the single-stranded 3′ telomeric overhang allows nucleotide addition to the chromosome terminus by reverse transcription of template nucleotides (de Lange et al., 2006). The yeast Saccharomyces cerevisiae TERT is encoded by the EST2 gene (Lendvay et al., 1996; Lingner et al., 1997b) and the RNA moiety is encoded by TLC1 (Lendvay et al., 1996; Singer and Gottschling, 1994). These components constitute the catalytic core of the enzyme and are required for in vitro and in vivo activity (Lingner et al., 1997a). Similarly to other TERT homologs, Est2p contains a catalytic domain that is conserved with reverse transcriptases of retroviruses and non-LTR retrotransposons (Lingner et al., 1997b; Nakamura et al., 1997).
Telomerase displays two types of processivity during telomere synthesis. Type I (nucleotide addition) processivity refers to the propensity of the enzyme to synthesize a full repeat extending to the end of the RNA template region. By contrast, Type II (repeat addition) processivity describes the ability of telomerase to reposition the 3′ end of a newly synthesized repeat within the active site for a second round of reverse transcription (Greider, 1991; Lue, 2004). Repeat addition processivity of both human and Tetrahymena telomerase is affected by sequences at the 5′ end of the primer (Finger and Bryan, 2008; Jacobs et al., 2006; Lee and Blackburn, 1993). Multiple experiments have suggested that a region of telomerase outside of the active site mediates an ‘anchor-site’ interaction with these upstream primer nucleotides that is important for the ability of telomerase to undergo reiterative copying of the RNA template (Autexier and Lue, 2006).
Assays of telomerase activity implicate an N-terminal domain of TERT [called telomerase essential N-terminus (TEN), GQ or Region I (Friedman and Cech, 1999; Jacobs et al., 2006; O'Connor et al., 2005; Xia et al., 2000)] in anchor-site function (Lue, 2004; Lue, 2005; Moriarty et al., 2005). Mutations within the Tetrahymena TEN domain decrease interaction with the DNA primer (Jacobs et al., 2006) and the primer can be photo-crosslinked to a fragment containing the Tetrahymena and S. cerevisiae TEN domains (Lue, 2005; Romi et al., 2007). Direct binding assays have demonstrated that the isolated TEN domain from human and Tetrahymena binds telomeric DNA (Finger and Bryan, 2008; Sealey et al., 2010; Wyatt et al., 2007; Wyatt et al., 2009). In S. cerevisiae, a fragment of Est2p containing the TEN domain interacts with full-length TLC1 RNA and increasing amounts of DNA compete with the RNA for protein binding in vitro (Xia et al., 2000). Mutations thought to disrupt the S. cerevisiae anchor-site interaction impair catalytic function and processivity on specific primers in vitro (Lue and Li, 2007). The TEN domain of human TERT might also mediate positioning of the 3′ end of the primer in the active site in a manner that is independent of the anchor-site interaction defined above (Jurczyluk et al., 2011).
In contrast to human telomeres, which contain perfect 5′-TTAGGG-3′ repeats, S. cerevisiae telomeres contain 300 base pairs (±50) of heterogeneous C1–3A/TG1–3 tracts (Shampay et al., 1984). Heterogeneity arises in part because of low nucleotide addition processivity. Synthesis often terminates before the 5′ end of the template, resulting in the generation of variable primer sequences for the next round of synthesis (Förstemann and Lingner, 2001). In addition, yeast telomerase tolerates multiple primer–template alignment registers (Förstemann and Lingner, 2001; Teixeira et al., 2004). Although the general phenomena that generate telomere heterogeneity have been described, it is less well understood how the catalytic core modulates telomere sequence.
We have previously described mutants in the TEN domain of Est2p (the est2-LT alleles) that alter the sequence of the telomeric repeat, but were unable to determine the mechanism that gives rise to this change. Interestingly, although these mutations increase telomere length by about 30% (~100 base pairs), this telomere over-elongation does not appear to be directly related to the change in telomere sequence (Ji et al., 2008). The est2-LT mutations do not affect nucleotide addition processivity in vitro (Ji et al., 2005), but the est2-LTE76K allele increases repeat addition processivity under specific primer extension conditions (Lue and Li, 2007).
Here, we investigate the telomere sequence alteration and processivity phenotypes of the est2-LTE76K allele. Analysis of both de novo telomere addition and endogenous telomere sequences supports the hypothesis that this mutation alters telomere sequences by affecting the alignment of the DNA primer with the telomerase RNA template. We also use expression of two distinguishable TLC1 template alleles to show that the est2-LTE76K mutation slightly increases processivity of telomerase in vivo, similar to the effect previously reported in vitro. Recombinant Est2p TEN domain (residues 1–161) associates with a single-stranded oligonucleotide containing the yeast telomere sequence. Mutations previously suggested to disrupt anchor-site interaction eliminate DNA binding in this assay, and introduction of the E76K mutation into the purified TEN domain increases its ability to bind telomeric DNA. These results support the idea that the anchor-site interaction in yeast contributes to repeat addition processivity and suggest a role in guiding alignment of the DNA primer with the RNA template.
Results
Telomere over-elongation does not correlate with a specific change in telomere sequence
Our previous work showed that mutations in EST2 that cause telomere lengthening are associated with changes in the telomere sequence (Ji et al., 2008). Both est2-LTE76K and est2-up34 [D460N; located within the reverse transcriptase domain (Eugster et al., 2006)] cause the same telomere sequence changes despite requiring different genetic pathways for over-elongation (Ji et al., 2008). Based on these data, it remained possible that telomere sequence changes observed in these est2 mutant strains are an indirect consequence of disrupted telomere length homeostasis.
Although the yeast telomere sequence is heterogeneous, a heptamer motif (5′-TGGGTGT-3′) complementary to the central portion of the TLC1 template (underlined region in 3′-ACACACACCCACACCAC-5′) can be used to define individual repeats (Förstemann and Lingner, 2001). In some cases, ‘core motifs’ occur in groups of two or more, where the last T of the first motif corresponds to the first T of the next motif (TGGGTGTGGGTGT; called an ‘overlap’). In other cases, core motifs are separated by (TG)n or GG(TG)n sequences of variable length. Rare variations of the spacer sequence (<10% of repeats) also occur. Because telomeres in both wild-type and mutant strains contain core motifs, we have analyzed two properties of ‘inter-core regions’ that lie between core motifs. First, the frequency of core motif overlap is calculated as the number of inter-core regions that consist of overlaps divided by the total number of inter-core regions (Ji et al., 2008). Second, the frequency with which the GG dinucleotide occurs within the telomeric sequence is calculated as the number of inter-core regions containing the GG dinucleotide divided by the total number of core motifs (Ji et al., 2008). By analyzing the sequence of an ADE2-marked telomere lacking subtelomeric repeats in the GA426 EST2 strain, we previously showed that 53% of core motifs are followed by a GG dinucleotide, whereas 29% of the inter-core regions are overlaps. By contrast, GA426 est2-LTE76K has telomeres in which 67% of core motifs are followed by a GG dinucleotide and only 18% of the inter-core regions are overlaps (Ji et al., 2008). As described in the Materials and Methods, sequence calculations are confined to those telomeric repeats synthesized during clonal growth from a single cell, thus representing the sequence generated after introduction of the mutant allele.
We first verified that sequence phenotypes are reproducible by analyzing different chromosome ends and wild-type strain backgrounds. To address chromosome arm specificity, we analyzed telomere sequences from chromosome XV-L. As shown in Fig. 1A, the overlap and GG-dinucleotide frequencies were extremely similar to those at the ADE2-marked telomere. To address strain specificity, telomeres from chromosome XV-L were cloned and sequenced from three additional EST2 strains. Telomere sequence patterns were very similar between these strains with GG dinucleotide incorporation of 47–55% and overlap generation of 26–29% (Fig. 1A). Telomeres from a UCC5706 est2-LTE76K strain show an indistinguishable sequence pattern from those obtained in the GA426 est2-LTE76K background (Fig. 1A). Together, these results suggest that the observed telomere sequence differences between EST2 and est2-LTE76K strains are not dependent on chromosome arms or the strain. Given that cloning and sequencing of individual telomeres is both time consuming and cost intensive, these data also demonstrate that analysis of approximately 80 core motifs per strain gives a reliable and reproducible measure of the telomeric sequence pattern.
Fig. 1.
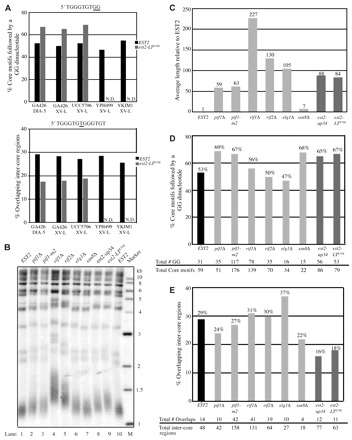
Disruption of negative telomere length regulation does not result in a specific telomere sequence phenotype. The indicated strains were grown for a minimum of 75 generations before cloning of telomeres by ligation-mediated PCR from the ADE2 marked chromosome V-R (DIA-5) or from chromosome XV-L. Chromosome V-R data for the EST2, est2-up34 and est2-LTE76K GA426 strains were previously published (Ji et al., 2008) and are included for comparison. (A) Telomere sequence characteristics of the indicated chromosome ends expressing either EST2 (black bar) or est2-LTE76K (gray bar). N.D., not determined. (B) Telomere length of GA426 strains determined by Southern blotting. Genomic DNA from the indicated strains was cleaved with XhoI, Southern blotted, and probed with a radiolabeled telomeric fragment. Marker sizes are in kilobases (kb). (C) Average length of 9–21 cloned telomeres used to generate data in D and E. Telomere length is normalized to the GA426 EST2 strain. Black bar, EST2; dark gray bars, est2-up34 and est2-LTE76K; light gray bars, six additional mutants. (D) Percentage of core motifs followed by a GG dinucleotide in telomeres from the indicated GA426 strains. Primary data for each strain are indicated (bottom). Gray-scale categories are the same as in C. (E) Percentage of inter-core region overlaps in the same telomeres analyzed in D. Primary data for each strain are indicated (bottom). Gray-scale categories are the same as in C and D.
Having established the robustness of this assay, we tested whether other strains with over-elongated telomeres cause similar telomere sequence phenotypes using the ADE2-marked chromosome V-R from strain GA426. Six different mutant strains were analyzed: rif1Δ, rif2Δ, pif1Δ, pif1-m2, elg1Δ and ssn8Δ. The Rif1, Rif2 and Pif1 proteins have well characterized roles in the negative regulation of telomere length. Rif1p and Rif2p negatively regulate telomere length through interactions with Rap1p (Hardy et al., 1992; Wotton and Shore, 1997). Telomere lengthening by the est2-up34 mutation requires the function of Pif1p (Eugster et al., 2006). This DNA helicase negatively regulates telomere length by dissociating telomerase from the chromosome terminus (Boulé et al., 2005; Zhou et al., 2000) and has a second function in the maintenance of mitochondrial DNA (Lahaye et al., 1991). To avoid complications of this pleiotrophic phenotype, we analyzed telomere sequences in GA426 strains that contained either pif1Δ or pif1-m2, an allele that retains mitochondrial function but lacks most telomere function (Schulz and Zakian, 1994). ELG1 and SSN8 were described as negative regulators of telomere length in a genome-wide telomere maintenance screen (Askree et al., 2004). ELG1 plays a role in genome stability (Ben-Aroya et al., 2003) and transcriptional silencing (Smolikov et al., 2004). SSN8 encodes a cyclin homolog with 35% identity to human cyclin C, but its role in telomere length regulation is unclear (Kuchin et al., 1995).
All strains except ssn8Δ caused telomere over-elongation to an extent similar or greater than that observed in the est2-LTE76K and est2-up34 strains [measured by Southern blot (Fig. 1C) and average length of cloned telomeres (Fig. 1D)]. In this light, it is interesting that only ssn8Δ has a telomere sequence phenotype similar to that of est2-LTE76K (Fig. 1D,E; 22% overlaps and 68% GG dinucleotides) whereas the other mutant strains either resemble EST2 (rif1Δ, rif2Δ), have an intermediate phenotype (pif1Δ, pif1-m2), or display a telomeric sequence pattern not previously observed (elg1Δ). Taken together, these data suggest the est2-LTE76K sequence phenotypes are not a general consequence of overly long telomeres.
The Est2p E76K mutation alters the alignment of telomerase with the telomeric primer
The heterogeneity of yeast telomeres has been proposed to arise through a combination of poor processivity and multiple sites of primer–template alignment (Förstemann and Lingner, 2001). Using an allele of TLC1 that contains a mutation near the 5′ end of the template region (A469U), we found that the rate at which the complementary A is incorporated into the telomere by est2-LTE76K telomerase is indistinguishable from that of EST2 telomerase (Ji et al., 2008). These results suggest that altered nucleotide-addition processivity alone is unlikely to explain the differences in telomere sequence; however, subtle changes in telomerase processivity at nucleotides proximal to the core motif cannot be ruled out.
We hypothesized that the est2-LTE76K mutation instead alters the position at which the telomerase template aligns with the DNA substrate. Because terminal primers generated during normal telomere replication can have different 3′ termini, determining the site at which the 3′ end of the primer aligns with the TLC1 template during the next round of addition is difficult. Instead, we took advantage of a strain that allows extension of a known telomeric seed by telomerase in vivo (Diede and Gottschling, 1999). In this de novo telomere assay, the non-essential terminus of chromosome VII-L is modified to contain the ADE2 gene, an 81 base pair telomeric ‘seed’ sequence, and a telomere-proximal HO endonuclease cleavage site (Fig. 2A). A LYS2 marker integrated distal to the HO cleavage site allows selection for cells that retain the chromosome terminus before induction of the HO endonuclease.
Fig. 2.
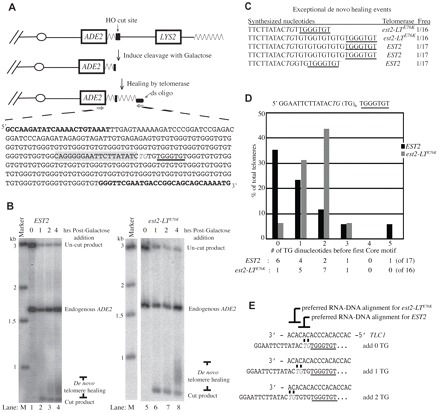
The est2-LTE76K mutation alters the alignment of telomerase with a short telomeric primer. (A) Illustration of the de novo telomere addition system developed by Diede and Gottschling (Diede and Gottschling, 1999). The left arm of chromosome VII is modified by addition (from centromere to telomere) of an ADE2 gene, 81 base pairs of telomere sequence, an HO endonuclease cleavage site and the LYS2 gene. Upon addition of galactose, expression of the HO endonuclease is induced and cleavage exposes a single-stranded overhang. De novo telomeres are isolated by ligation-mediated PCR using primers specific to the ligated oligonucleotide and to the ADE2 gene (bold). An example telomere synthesized by est2-LTE76K telomerase is shown. The 5′-TG-3′ sequence assumed to facilitate primer–template alignment is italicized and the remainder of the HO endonuclease recognition site is highlighted in gray. The first core motif of the de novo telomere is underlined. (B) Time course of de novo telomere addition by EST2 (lanes 1–4) and est2-LTE76K (lanes 5–8) telomerase. Cells were harvested immediately before HO induction (0 hours) or 1, 2 and 4 hours after galactose addition, as indicated. Marker sizes are in kilobases (lane M). Positions of the uncut product (no HO endonuclease cleavage) and endogenous ADE2 fragment are indicated. Cleavage by HO endonuclease releases a short fragment (cut product) that is elongated by telomerase (de novo telomere healing). (C) Exceptional de novo telomere addition events. Sequences added to the 5′-TG-3′ primer within the HO endonuclease cleavage site are shown in a 5′ to 3′ orientation. The first core motif is underlined and the TG sequence generated by HO endonuclease cleavage is italicized. (D) Number of TG dinucleotide sequences synthesized between the 5′-TG-3′ HO cleavage site and the first core motif by EST2 or est2-LTE76K telomerase. Total de novo telomeres analyzed were 17 for EST2 and 16 for est2-LTE76K. (E) Model of the preferred alignment for EST2 and est2-LTE76K telomerase with the primer remaining after HO endonuclease cleavage (indicated in gray italics). Newly synthesized sequence extends to the right. The first core motif is underlined.
EST2 and est2-LTE76K strains were grown as described in the Materials and Methods. The de novo telomeres present at the 4 hour time point were amplified from the ADE2-marked telomere by ligation-mediated PCR, cloned and sequenced. Consistent with the telomere length measured by Southern blot (Fig. 2B), the average de novo telomere synthesized by EST2 was 154 base pairs compared with the average de novo telomere of 263 base pairs synthesized by est2-LTE76K telomerase. The HO endonuclease generates a four-base 3′ overhang ending in 5′-TGTT-3′ (Kostriken et al., 1983). With one exception, telomere addition was initiated on this site and not within the telomeric seed sequence. Because only a single T remains at the junction of de novo telomere addition, the terminal T must be removed before telomerase action. For the remaining 5′-TGT-3′ sequence, it is unclear which nucleotides were present in the overhang and which were added by telomerase during de novo telomere synthesis. For simplicity, we assumed that synthesis initiated on a primer ending in 5′-TG-3′ (Fig. 2A, italics).
Analysis was conducted on 17 EST2 telomeres and 16 est2-LTE76K telomeres. With few exceptions (three in the EST2 strain and two in the est2-LTE76K strain; Fig. 2C), telomerase synthesized 0–5 TG dinucleotides before the first core motif (5′-TG0–5TGGGTGT; Fig. 2D). Addition of 0–2 TG dinucleotides is most simply explained as a single annealing event, followed by synthesis through the central region of the template (Fig. 2E). Addition of three or more TG dinucleotides requires more than one round of annealing and synthesis, making the specific site of primer–template annealing unclear. Several anomalous cases were observed in which a GG dinucleotide interrupted the TG repeats, or in which the TT dinucleotide present in the HO cleavage site was retained (Fig. 2C). These events did not appear to differ in type or frequency between the EST2 and est2-LTE76K strains and were not analyzed further.
Among those telomeres in which 0–2 TG dinucleotides were added before the first core motif, there was a striking difference between the EST2 and est2-LTE76K strains. As shown in Fig. 2D, 35% of the telomeres synthesized in the EST2 strain contained a core motif immediately adjacent to the 5′-TG-3′ primer (0 TG), whereas the est2-LTE76K strain exhibited this pattern in only 6% of de novo telomeres. Incorporation of one TG dinucleotide was approximately equal in the two strains. By contrast, 44% of est2-LTE76K telomeres contained two TG dinucleotides before the first core motif, whereas the same pattern was observed in only 12% of telomeres synthesized in the EST2 strain (Fig. 2D). These differences are statistically significant (Chi-square, P=0.04) and suggest that the est2-LTE76K mutation alters the preferred sites at which telomerase aligns with a short TG primer. EST2 telomerase prefers to align with template nucleotides 481CAC479 (immediately 3′ of the core motif template) whereas est2-LTE76K prefers to align at a more 3′ position of the template with nucleotides 484ACA482 (Fig. 2E).
Improved primer–template complementarity eliminates the difference in telomere sequences from EST2 and est2-LTE76K strains
Because the activity of telomerase in the de novo telomere-healing assay might not fully recapitulate the behavior of the enzyme during normal telomere replication, we sought to verify the results shown in Fig. 2 by analyzing the pattern of telomere addition at normal chromosome ends. As described above, analysis within endogenous telomeres is difficult because the nature of the 3′ telomeric primer is ambiguous. Potential exceptions to this generalization are repeats in which a GG dinucleotide is incorporated following the core motif. We and others have shown that the GG dinucleotides arise exclusively from reverse transcription of template nucleotides 471CC470 by EST2 (Förstemann and Lingner, 2001) and est2-LTE76K (Ji et al.,2008). Also, est2-LTE76K telomerase incorporates the nucleotide complementary to TLC1 position A469 at the same rate as the EST2 enzyme (Ji et al., 2008). Following a GG dinucleotide, a maximum of one TG dinucleotide can be added by reverse transcription at the 5′ end of the RNA template. Additional TG dinucleotides must arise through alignment of the primer terminus with the 3′ end of the RNA template (Fig. 3A, top). We therefore reasoned that differences in primer alignment contributed by the est2-LTE76K mutation might be detected in endogenous telomeres as a difference in the number of TG dinucleotides incorporated between each GG dinucleotide and the subsequent core motif.
Fig. 3.
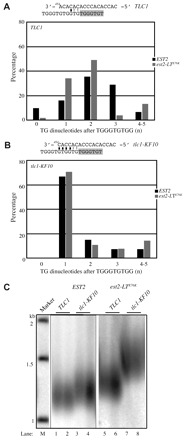
A mutation in the TLC1 template eliminates sequence pattern differences. (A) Top panel shows alignment of a telomeric primer ending in 5′-TGGGTGTGG-3′ with the TLC1 template to generate a single TG dinucleotide prior to the following core motif. The black horizontal line indicates the position of complementarity. Complementarity might continue through the following two template positions (gray bars) if synthesis in the previous round extends to the end of the TLC1 template (see text for detail). Newly synthesized nucleotides are highlighted in gray. Bottom panel shows percentage of core motifs associated with a GG dinucleotide (5′-TGGGTGTGG-3′) that is followed by the indicated number of TG dinucleotides before the subsequent core motif. Black bars, EST2 telomeres; gray bars, est2-LTE76K telomeres. (B) Same as (A) except the template used is from the tlc1-KF10 allele. (C) Southern blot of telomere length in GA426 tlc1::KAN strains expressing EST2 (lanes 1–4) or est2-LTE76K (lanes 5–8) from the endogenous locus and complemented with plasmids expressing TLC1 (lanes 1,2,5,6) or tlc1-KF10 (3,4,7,8). Marker sizes are in kilobases (kb).
In the divergent 39 end-portions of telomeres synthesized by EST2 with TLC1, the GG dinucleotide and adjacent core motif are separated by 1–3 TG dinucleotides, although rare instances of zero or four TG repeats are observed (Fig. 3A). The exact position at which the 3′ end of the primer aligns with the template is unknown because the first TG sequence might be derived from copying of the 5′ template nucleotides (469AC468) or from realignment of a GG-terminated primer at the 3′ end of the template (Fig. 3A, top). Despite this ambiguity, it appears that EST2 telomerase uses several different alignment registers, with one example being that depicted in Fig. 3A. By contrast, the divergent 3′ end-portions of telomeres synthesized in an est2-LTE76K TLC1 strain are more homogeneous, with 83% of GG dinucleotides followed by 1–2 TG repeats (Fig. 3A). The differences in TG repeat distribution between the two strains is statistically significant (Chi-square, P=0.0029). These results at endogenous telomeres are consistent with the conclusion from the de novo telomere addition assay that alignment between primer and template is changed by the est2-LTE76K mutation.
If differences in primer–template alignment contribute to the telomere sequence changes observed at endogenous telomeres in the est2-LTE76K strain, then a mutation in the TLC1 template that increases complementarity between the DNA primer and the 3′ end of the RNA template might overcome, or be epistatic to, any difference in alignment caused by the est2-LTE76K mutation. We took advantage of a TLC1 allele previously created and characterized by the Lingner lab (tlc1-KF10) (Förstemann and Lingner, 2001). This allele alters template nucleotides 484ACA482 to CAC. As a result, a telomeric primer that terminates in 5′-TGGGTGTGG(TG)-3′ can base pair perfectly with four (or six) of the 3′ nucleotides of the TLC1 template (Fig. 3B, top).
By Southern blot, expression of tlc1-KF10 increases telomere length in both the EST2 and est2-LTE76K strains (Fig. 3C). The increase in telomere length for the EST2 tlc1-KF10 strain has also been reported by the Lingner group (Förstemann and Lingner, 2001) and probably results from the reduced number of predicted Rap1p binding sites within this altered telomere sequence. When using this template, EST2 telomerase almost always synthesizes a GGTG inter-core region (TGGGTGTGGTGTGGGTGT), which is consistent with the alignment predicted for this template mutant (Fig. 3B). Telomeres in the est2-LTE76K tlc1-KF10 strain show a nearly identical pattern (Chi-square, P=0.6372). The ability of the improved base pairing interaction between the DNA primer and the tlc1-KF10 RNA template to eliminate the observed difference between EST2 and est2-LTE76K telomeres is consistent with the proposal that altered primer–template alignment underlies sequence changes within the est2-LTE76K telomeres. However, because the tlc1-KF10 allele also affects the sequence of telomeres synthesized by EST2 telomerase, we cannot exclude the possibility that an unrelated defect of the est2-LTE76K allele is masked under these conditions.
The E76K mutation in Est2p increases the repeat-addition processivity of telomerase in vivo
Yeast telomerase generally has low repeat-addition processivity, except at telomeres shorter than 125 base pairs (Chang et al., 2007). The est2-LTE76K mutation has been reported to moderately increase repeat-addition processivity of telomerase in vitro (Lue and Li, 2007). However, the relevance of this observation for the behavior of telomerase in vivo has been unclear. To test whether the mutant enzyme also demonstrates increased processivity in cells, we co-expressed a wild-type and a mutant TLC1 allele in GA426 EST2 and GA426 est2-LTE76K strains, cloned and sequenced the newly synthesized telomeric DNA, and analyzed the pattern of interspersion of the two repeat types. The tlc1-tm allele contains a highly altered template sequence (3′-ACACCUAAACCACACACAC-5′; Fig. 4A), but has a minor effect on telomere length when expressed alone (Chang et al., 2007). Importantly, when TLC1 and tlc1-tm are co-expressed and telomere addition is monitored during a single S phase, a fraction of the shortest telomeres that undergo processive elongation contain strings of mutant repeats, indicating that tlc1-tm can be used processively by the EST2 enzyme (Chang et al., 2007).
Fig. 4.
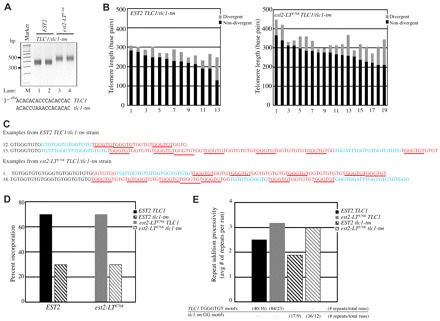
est2-LTE76K telomerase demonstrates increased processivity in vivo. (A) Ligation-mediated telomere PCR of the ADE2-marked chromosome V-R was used to amplify telomeres from asynchronous cultures of EST2 and est2-LTE76K GA426 strains expressing both TLC1 and tlc1-tm RNAs. PCR products were separated by agarose gel electrophoresis. Marker sizes are in base pairs (bp). The template sequences of TLC1 and tlc1-tm are shown (bottom). (B) Length of 13 EST2 TLC1/tlc1-tm and 19 est2-LTE76K TLC1/tlc1-tm cloned telomeres. Black bars, non-divergent TG sequences; gray bars, divergent TG sequences. (C) Two representative telomeric sequences from EST2 (telomeres 12 and 13) and est2-LTE76K (telomeres 1 and 18) strains. Numbers correspond to the telomeres shown in B. Black text, a small portion of the non-divergent sequence; red text, sequence inferred to derive from the TLC1 template with core motifs underlined; blue text, sequence inferred to derive from the tlc1-tm template. (D) The percentage of total telomeric repeats synthesized from the TLC1 (solid bar) or tlc1-tm (hatched bar) template is indicated. Strains expressed EST2 (black) or est2-LTE76K (gray). (E) The mean number of consecutive repeats derived from a single template (TLC1 or tlc1-tm) for strains expressing EST2 or est2-LTE76K. Consecutive repeats of the same type are defined as a ‘run’. The average run length was obtained from the total number of either TLC1 or tlc1-tm repeats divided by the total number of runs for each template (data are indicated below each sample in the graph). Samples are labeled as in D.
PCR amplification of the ADE2-marked chromosome V-R telomere confirmed that co-expression of TLC1 and tlc1-tm does not alter the long-telomere phenotype of est2-LTE76K (Fig. 4A). The lengths of identical and divergent portions of each cloned telomere are shown in Fig. 4B. The average total length of telomeres from the est2-LTE76K strain was 63 base pairs longer than EST2 telomeres, with an average of 15 base pairs more divergent telomeric sequence per telomere than in the EST2 strain (Fig. 4B).
An EST2 tlc1-tm strain generates a telomeric consensus repeat of (TG0–4TGG)n (Chang et al., 2007). The tlc1-tm sequences are distinguishable from those synthesized using TLC1 as a template because of the frequent incorporation of the GG dinucleotide without an intervening GGG sequence and by the occasional synthesis of an ATTTGG repeat (Chang et al., 2007). The number of consecutive tlc1-tm repeats was defined as the number of consecutive GG motifs in the absence of an intervening core motif. TLC1 repeats were defined by the number of consecutive core motifs (5′ TGGGTGT). Because the TLC1 template can also be used to synthesize GG dinucleotides, we made the assumption that any GG dinucleotide immediately following a wild-type core motif was synthesized by TLC1 (see the example sequences in Fig. 4C).
Analysis was conducted on the divergent portions of 13 telomeres from an EST2 TLC1/tlc1-tm strain and 19 telomeres from an est2-LTE76K TLC1/tlc1-tm strain (Fig. 4B). EST2 telomerase incorporated both TLC1 and tlc1-tm repeats, although the majority of the repeats are derived from TLC1 (70.2%; Fig. 4D). This value is identical to that previously reported for telomere addition during a single cell cycle in the presence of both RNAs (Chang et al., 2007). In our assay, EST2 telomeres contain an average of 2.5 TLC1 repeats and 1.9 tlc1-tm repeats per run (where a run is defined as a series of repeats of a single type). The est2-LTE76K telomerase does not alter the relative use of the wild-type TLC1 template (70.0%; Fig. 4D). However, est2-LTE76K telomerase generates longer run lengths than EST2 telomerase, with an average of 3.2 consecutive TLC1 core motifs and 3.0 consecutive tlc1-tm GG dinucleotide repeats (Fig. 4E). These data suggest that est2-LTE76K telomerase has slightly increased repeat-addition processivity compared with the EST2 enzyme in vivo.
Glu76 enhances DNA binding by the Est2p TEN domain
Given that repeat addition processivity is moderately increased by the est2-LTE76K mutation (Fig. 4), we speculated that Glu76 might influence the strength of DNA interaction by the TEN domain anchor site. Crosslinking studies in vivo have implicated this N-terminal domain of Est2p in interactions with the telomeric primer (Lue, 2005) and the recombinant TEN domain has been previously shown to have a non-sequence-specific nucleic acid binding activity (Xia et al., 2000). To test whether the E76K mutation affects the ability of the TEN domain to interact with TG-rich single-stranded DNA, a biotin pull-down assay, similar to that used to characterize the human TEN-domain–DNA interaction (Wyatt et al., 2007), was developed.
The first 161 amino acids of Est2p were N-terminally tagged with MBP (MBP–Est2pTEN) and both wild-type and mutant variants were sequentially purified from BL21 E. coli cells by amylose affinity and size-exclusion chromatography (Fig. 5A). A fourfold molar excess of MBP–Est2pTEN was incubated with 5′ biotinylated TG-rich, AC-rich or random sequence single-stranded DNA oligonucleotides (Fig. 5). TG-rich oligonucleotides of two different lengths were used to explore the length requirements of the MBP–Est2pTEN binding interaction. DNA was bound to streptavidin beads and western analysis was used to detect the presence of the MBP-tagged proteins (Fig. 5B). Results from a minimum of four replicates were quantified and normalized to values obtained for the 16-mer AC-rich (AC16) control oligonucleotide. MBP alone shows no detectable binding to the biotinylated oligonucleotides (Fig. 5B, lanes 1 and 2). However, as shown in Fig. 5B, lanes 5–10, MBP–Est2pTEN shows an increased affinity for the 16- and 28-mer TG-rich oligonucleotides (TG16 and TG28 respectively) compared with the AC16 control (Student's t-test, P=0.0004). Interaction of MBP–Est2pTEN with an oligonucleotide of random sequence (Random) was not enriched over the AC16 background (Fig. 5B). There was no statistical difference in binding by MBP–Est2pTEN to the two TG oligonucleotides of different length (Student's t-test, P=0.73). RNase treatment before DNA incubation did not alter protein–DNA binding (Fig. 5B, lanes 11 and 12). Together, these data show that the TEN domain of yeast TERT interacts with a single-stranded TG-rich oligonucleotide of at least 16 base pairs in length in the absence of the TLC1 RNA.
Fig. 5.
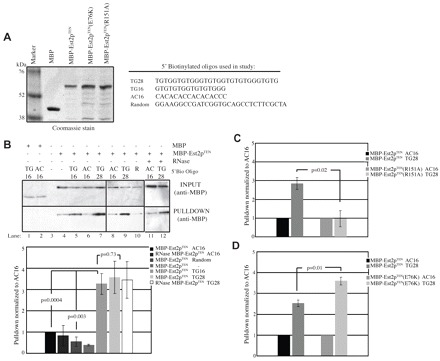
Binding of recombinant MBP–Est2pTEN to TG-rich DNA is enhanced by the E76K mutation. (A) Left panel shows Coomassie-stained gel of maltose-binding protein (MBP), MBP–Est2pTEN, MBP–Est2pTEN(E76K) and MBP–Est2pTEN(R151A). MBP–Est2p variants were expressed in E. coli and purified by amylose-affinity and size-exclusion chromatography. Right panel shows single-stranded 5′-biotinylated DNA oligonucleotides used for the binding assay. (B) Purification of MBP–Est2pTEN with the TG16 and TG28 oligonucleotides is enhanced over the AC16 and random-sequence (R) oligonucleotides. Input samples (INPUT) and samples following binding of the biotin-labeled primers to streptavidin beads (PULLDOWN) were separated by polyacrylamide gel electrophoresis, western blotted and probed with anti-MBP antibody (top panel). Binding reactions contained MBP alone or MBP–Est2pTEN as indicated. Lane 4 is a binding reaction in the absence of biotinylated oligonucleotide. Lanes 11 and 12 are binding reactions of RNase-treated MBP–Est2pTEN. The average binding efficiency of MBP–Est2pTEN to the indicated oligonucleotide is shown from at least four replicates (bottom panel). Values are normalized to that of the AC16 oligonucleotide. Error bars represent s.e.m. (C) Association of the MBP–Est2pTEN(R151A) protein with the TG28 oligonucleotide is not above background. Binding was determined and quantified as in B from at least three replicates. (D) MBP–Est2pTEN(E76K) significantly enhances binding to the TG28 oligonucleotide as compared with MBP–Est2pTEN. The average binding efficiency is calculated as in B from at least four replicates.
Est2p containing a mutation of R151A maintains normal levels of association with TLC1 RNA, but is defective in maintenance of telomere length and in vitro repeat-addition processivity (Lue and Li, 2007), which is consistent with an anchor-site defect. The R151A mutation was introduced into the pET MBP–Est2pTEN vector by site-directed mutagenesis and purified as described above (Fig. 5A). The interaction of MBP–Est2pTEN(R151A) with the TG16 oligonucleotide is not enriched above the AC16 background, and shows significantly less binding than MBP–Est2pTEN (Fig. 5C; Student's t-test, P=0.02), supporting the specificity of the assay. The E76K mutation was introduced into the pET MBP–Est2pTEN vector by site-directed mutagenesis and recombinant protein was produced (Fig. 5A). Importantly, the interaction of MBP–Est2pTEN(E76K) with the TG28 oligonucleotide is significantly greater than that of MBP–Est2TEN (Student's t-test, P=0.013). Collectively, these data confirm that the TEN domain of yeast Est2p functions as an anchor site for yeast telomerase and show that mutation of E76K enhances this anchor-site interaction.
Discussion
Here, we demonstrate that the isolated yeast Est2p TEN domain is sufficient to bind telomeric DNA in a sequence-specific manner. A mutation in this domain (E76K) increases the strength of the interaction (Fig. 5). Because this mutant supports robust telomere elongation, it affords a unique opportunity to characterize the functional importance of the anchor-site interaction. We show that the est2-LTE76K mutation alters the spectrum of sites at which the telomerase template aligns with the chromosome terminus (Figs 2, 3), suggesting that residues within the TEN domain might influence positioning of the primer within the enzyme active site. This mutation also slightly increases the processivity of yeast telomerase in vivo (Fig. 4), a phenotype previously only reported in vitro.
Three observations argue that est2-LTE76K alters primer–template alignment in vivo. First, EST2 and est2-LTE76K telomerase differed markedly in the distribution of sequences added to a short telomeric seed generated by HO endonuclease cleavage (Fig. 2). Second, analysis of a subset of endogenous telomeric repeats reveals sequence alterations consistent with a restriction of the number of possible alignment registers by est2-LTE76K telomerase (Fig. 3). Third, expression of a TLC1 RNA variant that improves primer–template complementarity forces both EST2 and est2-LTE76K telomerase into the same alignment register (compare Fig. 3A,B). These results suggest that interactions between the DNA primer and the TEN domain have the most influence on primer–template alignment when base-pairing interactions are weak. These data are also consistent with the observation that both EST2 and est2-LTE76K telomerase synthesize perfect repeats when using a ‘humanized’ TLC1 template with a single, highly preferred alignment register (Ji et al., 2008).
The observed changes in primer–template alignment can account for the decreased occurrence of core motif overlaps and the increased frequency of GG dinucleotides in est2-LTE76K telomeres (Fig. 1). Overlapping core motifs can be generated by alignment of a 3′-GG-terminated primer at template residue 476C such that synthesis through the template converts the GG dinucleotide into a GGG trinucleotide. Although we cannot directly monitor such events within endogenous telomeres, the trend shown in Fig. 3 suggests that this alignment occurs infrequently in the est2-LTE76K strain.
It is tempting to speculate that the tighter anchor-site interaction of the Est2-LTE76K protein (Fig. 5) directly affects primer–template alignment. Mutation of residues 170–175 in the TEN domain of human TERT has recently been reported to alter positioning of the 3′ end of the primer in the enzyme active site, perhaps by affecting required conformational changes (Jurczyluk et al., 2011). However, in that case, the catalytic defect is independent of alterations in primer binding. Therefore, we cannot rule out the possibility that changes in DNA association and primer alignments by est2-LTE76K telomerase are independent manifestations of the same amino acid change.
Using an Oxytricha telomeric primer, the est2-LTE76K telomerase was previously shown to slightly increase repeat-addition processivity in vitro (Lue and Li, 2007). By co-expressing wild-type and mutant TLC1 (tlc1-tm) alleles, we found that the number of consecutive telomeric repeats generated from the same template increased in an est2-LTE76K strain (Fig. 4), consistent with increased processivity. This observation raises the intriguing possibility that the TEN domain, and perhaps residue E76 specifically, might be a target of regulation at critically short telomeres where enzyme processivity is markedly increased (Chang et al., 2007). The increase in processivity seen in the est2-LTE76K strain is less pronounced than is observed at critically short telomeres (where the average number of consecutive TLC1 repeats increases to ~7). A moderate effect on enzyme activity is not surprising given the relatively small increase in processivity in vitro (Lue and Li, 2007) and suggests that additional determinants of processivity might exist. Alternatively, the multiple rounds of extension and shortening that telomeres experience during culture outgrowth might mask greater increases in processivity.
The anchor-site of human TERT has been shown to bind directly to a telomeric oligonucleotide through its N-terminal TEN domain in the absence of the RNA (Sealey et al., 2010; Wyatt et al., 2007; Wyatt et al., 2009). We developed a similar assay for the yeast TEN domain and found that MBP–Est2pTEN associates in a sequence-specific manner with telomeric DNA. Importantly, R151A, a mutation previously suggested to decrease anchor-site function in yeast telomerase (Lue and Li, 2007) abolished this interaction (Fig. 5C). By contrast, the E76K mutation increases interaction of the isolated TEN domain with telomeric DNA (Fig. 5D). One possibility is that loss of the negative charge and/or introduction of a positive charge at residue 76 alters electrostatic interactions with the DNA to increase DNA binding. However, a structural model of the yeast TEN domain proposed by the Lue lab places residue E76 distal to the proposed site of DNA interaction (which includes R151) (Lue and Li, 2007) suggesting that the effect of E76 might be indirect.
A nearly identical N-terminal peptide of Est2p was previously shown by filter binding to interact in a non-sequence-specific manner with both RNA and DNA. Interestingly, the first 50 amino acids of the protein is sufficient to mediate this interaction, with no apparent contribution by the remainder of the TEN domain (Xia et al., 2000). The requirement of residue R151 for interaction in our assay suggests that we are monitoring a different binding activity. We were unable to obtain a DNA binding affinity for the TEN domain using fluorescence anisotropy (data not shown), which is similar to results reported for both the Tetrahymena and human TEN domains using filter binding and electrophoretic mobility shift assays (Jacobs et al., 2006; Moriarty et al., 2005; Sealey et al., 2010). This result is not unexpected because a weak or rapid exchange between the DNA and the anchor-site would aid in translocation of the DNA substrate during telomeric synthesis.
We propose that strengthening of the anchor-site interaction in est2-LTE76K telomerase leads to increased enzyme processivity, but simultaneously restricts the manner in which the DNA primer can be positioned relative to the active site, causing the observed changes in alignment during telomere synthesis. The anchor-site interaction might play a disproportionately large role in dictating primer alignment in yeast because primer and template sequences can have low levels of complementarity. We speculate that yeast might have evolved a non-optimal anchor-site interaction to facilitate the generation of heterogeneous telomeric repeats, a phenomenon that may be important for reducing homology between telomeres in this highly recombinogenic organism.
Materials and Methods
Strains and plasmids
See supplementary material Table S1 for the strains and plasmids and supplementary material Table S2 for the primers used in this work. YKJM1 (Banerjee et al., 2008) and YPH499 (Schulz and Zakian, 1994) were gifts from Kyungjae Myung (Genetics and Molecular Biology Branch, National Institutes of Health, Bethesda, MD) and Virginia Zakian (Princeton University, Princeton, NJ), respectively. YKF500 (GA426 est2-LTE76K) has been previously described (Ji et al., 2008). Two-step gene replacement was used to generate strain YKF511 (GA426 est2-up34). Gene disruptions of RIF1, RIF2, PIF1 (YKF600–YKF602, respectively), ELG1 (YKF604) and SSN8 (YKF605) were obtained by amplification of the KanR gene and flanking DNA from the appropriate gene knockout strain (Open Biosystems). PCR products were transformed into GA426 and creation of the correct strains was verified by PCR. To make GA426 pif1-m2 (YKF603), the pif1-m2 allele was integrated by two-step gene replacement using plasmid pVS31. The est2-LTE76K mutant was integrated as described (Ji et al., 2008) into UCC5706 previously transformed with a RAD52 TRP1 CEN plasmid to create YKF512. After integration of the mutant allele, colonies were screened for loss of the RAD52 TRP1 plasmid. To create strains expressing EST2 or est2-LTE76K with either TLC1 or tlc1-KF10, the endogenous TLC1 gene was disrupted by KanR in strains GA426 and YKF500. Plasmids pKF5 (TLC1) or pKF10 (tlc1-KF10) (gift from Joachim Lingner; Ecole Polytechnique Federale de Lausanne, Lausanne, Switzerland) were transformed and RAD52 was subsequently disrupted by transformation with a rad52::LEU2 fragment (Friedman and Cech, 1999). To make EST2 or est2-LTE76K strains expressing tlc1-tm, the pLIB17 plasmid (Förstemann et al., 2003) was digested with HindIII and HpaI, resulting in 2.3 kb fragment that was ligated into pRS306 (URA3) (Sikorski and Hieter, 1989) to make pRS306 tlc1-tm. pRS306 tlc1-tm was digested with NruI and transformed into GA426 and YKF500 to create YKF610 and YKF611, respectively. The TEN domain mutants (E76K and R151A) were made by site-directed PCR mutagenesis as described (Landt et al., 1990) from pET Duet MBP–EST2TEN (pKF1201) (Talley et al., 2011) to make pKF1202 and pKF1203, respectively. The point mutants were confirmed by PCR and sequencing.
Telomere DNA cloning and sequence analysis
Telomeric DNA was cloned as previously described with minor modifications (Kramer and Haber, 1993). All primer sequences are listed in supplementary material Table S2. Genomic DNA was extracted by glass bead lysis. RNase-treated DNA samples (~20–40 μg) were blunted by treatment with 4.5 U of T4 DNA polymerase (NEB) in the presence of 1 mM dNTPs at 12°C. The samples were ethanol precipitated and 40 ng of a double-stranded oligonucleotide (created by annealing primers ds oligo 1 and ds oligo 2; supplementary material Table S2) was incubated with the genomic DNA and 20 U of T4 DNA ligase (NEB) overnight at 17°C. Ligation products were ethanol precipitated and telomeres were amplified using one primer specific to the double-stranded oligonucleotide (ds oligo 1 or ds oligo 2) and another primer designed to anneal internal to the telomere on the chromosome of interest. The internal primer for the GA426 strains (primer DIA5-1) anneals to the ADE2 gene integrated at chromosome V-R (Förstemann et al., 2000). The internal primer used in strains UCC5706, YKJM1 and YPH499 is specific to chromosome XV-L (primer XV-L). PCR products were separated in 1.8% agarose gels and the telomeric smear was purified from the gel (Qiagen). Products were ligated into pGEM T Easy vector (Promega) and transformed in DH5α E. coli. Inserts were sequenced by GENEWIZ (South Plains, NJ) using the primer M13R. Analysis of telomeric sequence data was performed as described (Ji et al., 2008) using JMP software unless otherwise noted in the text.
De novo telomere formation
Telomere formation events occurring at the site of HO cleavage were generated as previously described (Diede and Gottschling, 1999) with minor modifications. Fresh colonies from UCC5706 strains were grown in medium lacking lysine and used to inoculate a 100 ml culture of YEP with 2.5% Raffinose and grown to an OD600 of approximately 0.5. Cultures were blocked with nocodazole (10 μg/ml) for 4 hours or until >90% of the cells had a single large bud. A 15 ml sample was isolated as the ‘pre-healing’ control. The pellet from an additional 15 ml sample was washed twice with cold ddH2O, resuspended in 100 ml of pre-warmed YEP with 3% galactose to allow release from the nocodozole block, and harvested after incubation for 6 hours. The pellet derived from the remaining culture was washed twice as described above and resuspended in 100 ml of pre-warmed YEP with 3% galactose and nocodazole (10 μg/ml). 15 ml samples were removed every 2 hours over a 6-hour time period. The de novo telomeres were cloned and sequenced as described above with internal primer Inv 5′ADE2 (Diede and Gottschling, 1999). FACS analysis confirmed the nocodazole block efficiency (data not shown).
Protein purification and biotin pull-down assay
Wild-type or mutant variants of MBP-tagged Est2 TEN domain (residues 1-161; pKF1201–pKF1203) were expressed in BL21 E. coli. The protein was induced with 500 mM IPTG followed by growth for 16–20 hours in LB medium supplemented with ampicillin (50 mg/ml) at 17°C. Cells were lysed by emulsification in gel filtration buffer (20 mM Tris-HCl, pH 7.5, 200 mM sodium chloride, 1 mM EDTA, pH 8, 10% glycerol) plus protease inhibitors (one Roche complete protease inhibitor tablet per 10 ml). The MBP-tagged protein was isolated from the resulting supernatant with amylose resin (NEB). The resin was washed with wash buffer A (20 mM Tris-HCl, pH 7.4, 500 mM sodium chloride, 1 mM EDTA, pH 8, one Roche complete protease inhibitor tablet per 10 ml) and eluted with elution buffer B (20 mM Tris-HCl, pH 7.4, 500 mM sodium chloride, 1 mM EDTA, pH 8, 5 mM maltose, one Roche complete protease inhibitor tablet per 10 ml). The eluate was concentrated and purified by size-exclusion chromatography. Protein concentrations were verified by a Nanodrop spectrometer. For the pull-down assay, 19.2 fmoles of MBP-tagged proteins (with or without 5 μg RNase) were incubated with 5 fmoles 5′ biotinylated oligo (supplementary material Table S2) on ice for 30 minutes in 75 μl of gel filtration buffer. Steptavidin UltraLink Resin (Pierce) was equilibrated with four washes of 1 ml each with equilibration solution (gel filtration buffer plus one Roche complete protease inhibitor tablet per 10 ml, 0.1% Tween, 0.01% 1M DTT) and 5 μl equilibrated resin (resuspended in half the original volume) was incubated with each sample for 45 minutes at 4°C with gentle agitation. Bound resin was washed twice for 10 minutes with 500 μl cold wash solution (wash buffer A plus one Roche complete protease inhibitor tablet per 10 ml, 0.1% Tween, 0.01% 1M DTT) at 4°C, heated in SDS reducing buffer (Bio-Rad) at 100°C for 4 minutes and the supernatant was separated in a 10% Bio-Rad SDS-PAGE gel for 1 hour at 200 V. The proteins were wet-transferred to PVDF membrane (Amersham) at 30 V for 2.5 hours and blocked overnight at 4°C in PBST with milk (1× PBS, 0.05% Tween, 5% dried milk). MBP protein was detected using a horseradish peroxidase (HRP)-conjugated anti-MBP monoclonal antibody from NEB at a 1:50,000 dilution. After washing the membrane with a PBST solution (16 PBS, 0.05% Tween), the ECL Plus western blotting detection system and Hyperfilm ECL (GE Healthcare) was used. The sequences of 5′ biotinylated oligonucleotides TG28, TG16, AC16 and Random are listed in supplementary material Table S2.
Southern analysis of telomere length
Southern blot analysis of endogenous telomeres was done as previously described (Friedman et al., 2003). For Southern blot analysis of de novo telomere formation in strains UCC5706 and YKF512, genomic DNA was digested with SpeI and the blot was probed with a fragment of ADE2 as previously described (Diede and Gottschling, 1999).
Supplementary Material
Acknowledgments
We thank T. Graham, D. Gottschling, J. Lingner, K. Myung, J. Patton, and V. Zakian, for strains and reagents, J. Talley for technical assistance during the protein purification, B. Eichman and T. Graham for specialized equipment access, as well as J. Miles for aid in the tlc1-tm strain and plasmid creations. We appreciate critical comments on the manuscript from L. Berchard, C. Hawkins, M. Platts and J. Talley.
Footnotes
Funding
This work was funded by the National Science Foundation [grant number MCB-0721595] to K.L.F.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.090761/-/DC1
References
- Askree S. H., Yehuda T., Smolikov S., Gurevich R., Hawk J., Coker C., Krauskopf A., Kupiec M., McEachern M. J. (2004). A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 101, 8658-8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Autexier C., Lue N. F. (2006). The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 75, 493-517 [DOI] [PubMed] [Google Scholar]
- Banerjee S., Smith S., Oum J. H., Liaw H. J., Hwang J. Y., Sikdar N., Motegi A., Lee S. E., Myung K. (2008). Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J. Cell Biol. 181, 1083-1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Aroya S., Koren A., Liefshitz B., Steinlauf R., Kupiec M. (2003). ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc. Natl. Acad. Sci. USA 100, 9906-9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulé J. B., Vega L. R., Zakian V. A. (2005). The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature 438, 57-61 [DOI] [PubMed] [Google Scholar]
- Chang M., Arneric M., Lingner J. (2007). Telomerase repeat addition processivity is increased at critically short telomeres in a Tel1-dependent manner in Saccharomyces cerevisiae. Genes Dev. 21, 2485-2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T., Blackburn E. H., Lundblad V. (2006). Telomeres, pp. 1-576 Cold Spring HarborNY: Cold Spring Harbor Laboratory Press; [Google Scholar]
- Diede S. J., Gottschling D. E. (1999). Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases alpha and delta. Cell 99, 723-733 [DOI] [PubMed] [Google Scholar]
- Eugster A., Lanzuolo C., Bonneton M., Luciano P., Pollice A., Pulitzer J. F., Stegberg E., Berthiau A. S., Forstemann K., Corda Y., et al. (2006). The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat. Struct. Mol. Biol. 13, 734-739 [DOI] [PubMed] [Google Scholar]
- Finger S. N., Bryan T. M. (2008). Multiple DNA-binding sites in Tetrahymena telomerase. Nucleic Acids Res. 36, 1260-1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K., Lingner J. (2001). Molecular basis for telomere repeat divergence in budding yeast. Mol. Cell. Biol. 21, 7277-7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K., Hoss M., Lingner J. (2000). Telomerase-dependent repeat divergence at the 3’ ends of yeast telomeres. Nucleic Acids Res. 28, 2690-2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstemann K., Zaug A. J., Cech T. R., Lingner J. (2003). Yeast telomerase is specialized for C/A-rich RNA templates. Nucleic Acids Res. 31, 1646-1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K. L., Cech T. R. (1999). Essential functions of N-terminal domains in the yeast telomerase catalytic subunit revealed by selection for viable mutants. Genes Dev. 13, 2863-2874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K. L., Heit J. J., Long D., Cech T. R. (2003). N-terminal domain of yeast telomerase reverse transcriptase: recruitment of Est3p to the telomerase complex. Mol. Biol. Cell 14, 1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C. W. (1991). Telomerase is processive. Mol. Cell. Biol. 11, 4572-4580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider C. W., Blackburn E. H. (1985). Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell 43, 405-413 [DOI] [PubMed] [Google Scholar]
- Hardy C. F. J., Sussel L., Shore D. (1992). A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 6, 801-814 [DOI] [PubMed] [Google Scholar]
- Jacobs S. A., Podell E. R., Cech T. R. (2006). Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 13, 218-225 [DOI] [PubMed] [Google Scholar]
- Ji H., Platts M. H., Dharamsi L. M., Friedman K. L. (2005). Regulation of telomere length by an N-terminal region of the yeast telomerase reverse transcriptase. Mol. Cell. Biol. 25, 9103-9114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H., Adkins C. J., Carthwright B. R., Friedman K. L. (2008). Yeast Est2p affects telomere length by influencing association of Rap1p with telomeric chromatin. Mol. Cell Biol. 28, 2380-2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurczyluk J., Nouwens A. S., Holien J. K., Adams T. E., Lovrecz G. O., Parker M. W., Cohen S. B., Bryan T. M. (2011). Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 39, 1774-1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostriken R., Strathern J. N., Klar A. J., Hicks J. B., Heffron F. (1983). A site-specific endonuclease essential for mating-type switching in Saccharomyces cerevisiae. Cell 35, 167-174 [DOI] [PubMed] [Google Scholar]
- Kramer K. M., Haber J. E. (1993). New telomeres in yeast are initiated with a highly selected subset of TG1-3 repeats. Genes Dev. 7, 2345-2356 [DOI] [PubMed] [Google Scholar]
- Kuchin S., Yeghiayan P., Carlson M. (1995). Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 92, 4006-4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye A., Stahl H., Thines-Sempoux D., Foury F. (1991). PIF1: a DNA helicase in yeast mitrochondria. EMBO J. 10, 997-1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landt O., Grunert H. P., Hahn U. (1990). A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene 96, 125-128 [DOI] [PubMed] [Google Scholar]
- Lee M. S., Blackburn E. H. (1993). Sequence-specific DNA primer effects on telomerase polymerization activity. Mol. Cell. Biol. 13, 6586-6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lendvay T. S., Morris D. K., Sah J., Balasubramanian B., Lundblad V. (1996). Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics 144, 1399-1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Cech T. R., Hughes T. R., Lundblad V. (1997a). Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. USA 94, 11190-11195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J., Hughes T. R., Shevchenko A., Mann M., Lundblad V., Cech T. R. (1997b). Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 276, 561-567 [DOI] [PubMed] [Google Scholar]
- Lue N. F. (2004). Adding to the ends: what makes telomerase processive and how important is it? Bioessays 26, 955-962 [DOI] [PubMed] [Google Scholar]
- Lue N. F. (2005). A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 280, 26586-26591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lue N. F., Li Z. (2007). Modeling and structure function analysis of the putative anchor site of yeast telomerase. Nucleic Acids. Res. 35, 5213-5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriarty T. J., Ward R. J., Taboski M. A., Autexier C. (2005). An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell 16, 3152-3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T. M., Morin G. B., Chapman K. B., Weinrich S. L., Andrews W. H., Lingner J., Harley C. B., Cech T. R. (1997). Telomerase catalytic subunit homologs from fission yeast and human. Science 277, 955-959 [DOI] [PubMed] [Google Scholar]
- O'Connor C. M., Lai C. K., Collins K. (2005). Telomerase RNA mutations in Saccharomyces cerevisae alter telomerase action and reveal nonprocessivity in vivo and in vitro. Genes Dev. 11, 528-540 [DOI] [PubMed] [Google Scholar]
- Romi E., Baran N., Gantman M., Shmoish M., Min B., Collins K., Manor H. (2007). High-resolution physical and functional mapping of the template adjacent DNA binding site in catalytically active telomerase. Proc. Natl. Acad. Sci. USA 104, 8791-8799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz V. P., Zakian V. A. (1994). The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell 76, 145-155 [DOI] [PubMed] [Google Scholar]
- Sealey D. C. F., Zheng L., Taboski M. A. S., Cruickshank J., Ikura M., Harringtion L. A. (2010). The N-terminus of hTERT contains a DNA-binding domain and is required for telomerase activity and cellular immortalization. Nucleic Acids. Res. 38, 2019-2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J., Szostak J. W., Blackburn E. H. (1984). DNA sequences of telomeres maintained in yeast. Nature 310, 154-157 [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. S., Gottschling D. E. (1994). TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 266, 404-409 [DOI] [PubMed] [Google Scholar]
- Smolikov S., Mazor Y., Krauskopf A. (2004). ELG1, a regulator of genome stability, has a role in telomere length regulation and in silencing. Proc. Natl. Acad. Sci. USA 101, 1656-1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley J. M., DeZwann D. C., Maness L. D., Freeman B. C., Friedman K. L. (2011). Stimulation of yeast telomerase activity by the Ever Shorter Telomeres 3 (Est3) subunit is dependent on direct interaction with the catalytic protein Est2. J. Biol. Chem. 286, 26431-26439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira M. T., Arneric M., Sperisen P., Lingner J. (2004). Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell 117, 323-335 [DOI] [PubMed] [Google Scholar]
- Wotton D., Shore D. (1997). A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 11, 748-760 [DOI] [PubMed] [Google Scholar]
- Wyatt H. D. M., Lobb D. A., Beattie T. L. (2007). Characterization of physical and functional anchor site interactions in human telomerase. Mol. Cell Biol. 27, 3226-3240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt H. D. M., Tsang A. R., Lobb D. A., Beattie T. L. (2009). Human telomerase reverse transcriptase (hTERT) Q169 is essential for telomerase function in vitro and in vivo. PLoS One 4, 1-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Peng Y., Mian I. S., Lue N. F. (2000). Identification of functionally important domains in the N-terminal region of telomerase reverse transcriptase. Mol. Cell. Biol. 20, 5196-5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Monson E. K., Teng S., Schulz V. P., Zakian V. A. (2000). Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science 289, 771-774 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


