Abstract
The mouse Flk1 gene is expressed in various mesodermal progenitor cells of developing embryos. Recent studies have shown that Flk1 expression marks multipotent mesodermal progenitors, giving rise to various hemato-cardiovascular cell lineages during development. Flk1 expression also marks hemato-cardiovascular cell lineages in differentiating embryonic stem (ES) cells, which may be used in transplantation decisions to treat cardiovascular diseases. Despite its developmental and clinical importance in cardiovascular tissues, the transcriptional regulatory system of Flk1 has remained unclear. Here, we report a novel enhancer of the mouse Flk1 gene directing early mesodermal expression during development as well as ES differentiation. The enhancer enriches various mesodermal progenitors, such as primitive erythropoietic progenitors, hemangioblast (BL-CFC) and cardiovascular progenitors (CV-CFC). The enhancer is activated by Bmp, Wnt and Fgf, and it contains Gata-, Cdx-, Tcf/Lef-, ER71/Etv2- and Fox-binding sites, some of which are bound specifically by each of these transcription factors. As these transcription factors are known to act under the control of the Bmp, Wnt and Fgf families, early Flk1 expression may be induced by cooperative interactions between Gata, Tcf/Lef, Cdx and ER71/Etv2 under the control of Bmp, Wnt and Fgf signaling. The enhancer is required for early Flk1 expression and for hemangioblast development during ES differentiation.
Keywords: Cardiogenesis, Flk1 (Kdr), Hemangioblasts, Hematopoiesis, VEGF, Vasculogenesis, Mouse
INTRODUCTION
Mesodermal cells give rise to blood, blood vessel, kidney, cardiac and skeletal muscle. These tissues are generated from orderly allocated mesoderm at the primitive streak stage, in accordance with the timing and site of recruitment to the mesodermal layer (Kinder et al., 1999; Kinder et al., 2001).
The mouse Flk1 gene (Kdr – Mouse Genome Informatics) encodes a receptor for vascular endothelial growth factor A (VEGF-A), which is produced in early lateral mesodermal progenitor cells at the streak stage that give rise to hematopoietic and vascular endothelial cell lineages. During later stages of embryonic development, Flk1 is expressed in endothelial cells, but not in most hematopoietic cells (Yamaguchi et al., 1993). Gene-targeting experiments have shown that Flk1-deficient mice die between 8.5 and 9.5 days postcoitum (dpc) because of the absence of blood vessels and blood islands (Shalaby et al., 1995). Therefore, Flk1 is essential to establish vascular and hematopoietic systems at the developmental stage. The existence of the hemangioblast, a bipotential precursor of hematopoietic and endothelial cells, is demonstrated by BL-CFC, a progenitor cell capable of differentiating into two lineages during ES differentiation and in mouse embryos (Choi et al., 1998; Huber et al., 2004).
Recent studies have also shown that Flk1-positive progenitor cells are multipotent, capable of differentiating into, besides hematopoietic and endothelial lineages, smooth muscle cells and cardiomyocytes; these capacities have been seen in the in vitro mouse ES cell differentiation model as well as during development (Yamashita et al., 2000; Ema et al., 2003; Motoike et al., 2003; Kattman et al., 2006; Wu et al., 2006; Moretti et al., 2006). Importantly, FLK1-positive cells derived from human ES cells are also capable of differentiating into these multiple cell types (Yang et al., 2008), indicating that they may be used to establish more useful treatment methods for human cardiovascular diseases such as ischemic diseases and infarctions. Previous studies have shown that secreted factors such as Wnt, Bmp and Fgf are important for inducing early Flk1 expression (Faloon et al., 2000; Park et al., 2004; Lindsley et al., 2006), although the molecular mechanism of the induction remains unknown. Unveiling the transcriptional regulation system of Flk1 should not only allow improved understanding of the mechanisms of mesodermal differentiation into endothelial cells, but it should also lead to novel approaches to cardiovascular regenerative medicine. Kappel and coworkers identified an enhancer activity in the first intron of the Flk1 gene, named the first intronic enhancer (Kappel et al., 1999). However, deletion of the first intronic enhancer resulted in no significant reduction of Flk1 expression in mesodermal progenitors at 7.5 dpc or in endothelial cells at 8.5 dpc (Ema et al., 2006a). These results suggest that an unknown enhancer directs Flk1 expression.
In this study, we identify a novel enhancer of the mouse Flk1 gene that directs its mesodermal expression in the early embryo. The lacZ reporter gene expression pattern observed in the 7.5 dpc embryo was very similar to that of endogenous Flk1. We also show that the reporter activity marks various hemato-cardiovascular progenitors, including EryP, BL-CFC and CV-CFC. The enhancer was activated by Bmp, Fgf and Wnt, and it contains transcription factor binding sites for GATA-binding protein (Gata), caudal type homeobox protein (Cdx), T-cell factor protein (Tcf), lymphoid enhancer binding factor protein (Lef), forkhead box protein (Fox) and ER71/Etv2. Gata4, Cdx2, Tcf4 and Lef1 bound to some of these sites specifically. Taken together, these results suggest that early mesodermal expression of Flk1 is controlled directly by Gata, Cdx, Tcf, Lef and ER71/Etv2 transcription factors, which may work downstream of Bmp, Fgf and Wnt.
MATERIALS AND METHODS
Sequence alignment for mouse and human genomes
The sequence alignment between mouse and human genomes was generated using the VISTA Browser (http://genome.lbl.gov/vista/index.shtml), with the following query conditions: ‘Base genome’=‘Mouse May 2004’, ‘Position’=‘chr5: 74,479,092-74,823,886’ (corresponds to Flk1 gene locus and its upper/lower each 150 kb sequence), and ‘VISTA tracks on UCSC Browser’. Based on the CNS (Conserved regions) data, conserved sequences corresponding to the first intronic enhancer were extracted (http://pipeline.lbl.gov/help.shtml).
Generation of transgenic and DMME-deficient mice
Enhancer candidate regions were amplified by PCR from ES genome DNA using KOD-plus DNA polymerase (TOYOBO) by PCR using the following primers: enhancer #1-forward, 5′-CGAAATGATTGGCAAAGCCAGATGT-3′; enhancer #1-reverse, 5′-ATATAAAATGCTACCACAATGCTTG-3′ (3521 bp fragment); enhancer #2-forward, 5′-AGATCATTTCACTTGCCAGTTCTAC-3′; enhancer #2-reverse, 5′-GCCTTGAGGGCTTACCTGTGTACGG-3′ (703 bp fragment); enhancer #3-forward, 5′-ACTGTCTTAGGACATTGACTCTTCT-3′; enhancer #3-reverse, 5′-TCCCAAGCCGTGTTTCCAGGGTCTG-3′ (897 bp fragment); enhancer #4-forward, 5′-GGCAAAGGAAGCCACTCTGCAGCCAC-3′; enhancer #4-reverse, 5′-TAGTTCACCTTCTGTTCCAGTCTCT-3′ (936 bp fragment).
Enhancers #1 and #2 were inserted into the 3′ side of the Flk1 1.8 kb promoter, whereas enhancers #3 and #4 were inserted into the 5′ side. Fertilized eggs collected from BDF1 females were used for pronuclear injection, and the injected eggs were transplanted into pseudo-pregnant ICR females. The embryos were collected for F0 analysis at 7.5 or 8.5 days or were allowed to develop to adulthood. Similarly, the Flk1-DMME-GFP transgene was constructed and injected into the eggs. Flk1-GFP knock-in and the Flk1-GFP BAC Tg (line #244) mice were generated as described previously (Ema et al., 2006a; Ishitobi et al., 2010). Generation of DMME-deficient mice was performed as follows: the Flk1 5′ and 3′ arms were subcloned into the PGK-neo-DT cassette vector and the resultant targeting vector was electroporated into E14 ES cells. After the germline transmission, genotyping of DMME-targeted mice was performed using the following primers: Flk1 enhancer4 check2 (wild-type allele), 5′-TCCTACTGTGGTACTGTGCCCTGCC-3′; Flk1 enhancer4 check1, 5′-CAGGTGATCAATAGGCTAGCCACCC-3′; Flk1 enhancer4 deletion1 (mutant allele), 5′-TGGGCTCTCTCCACTTGTTACC-3′; Flk1 enhancer4 deletion2, 5′-TGGTGCCCTCTAAGTTGCTCGG-3′.
The generation of ER71/Etv2 knockout (KO) animals was described elsewhere (Ferdous et al., 2009). This study was approved and conducted in accordance with the Regulations for Animal Experimentation of the University of Tsukuba.
X-gal staining
X-gal staining was performed on transgenic embryos for both F0 and lined transgenic mice analyses, as described in a previous report (Shalaby et al., 1995).
Genomic conservation analysis for multiple species and search for transcription factor binding sites
Multiple conservation analysis was performed using the UCSC Genome Browser, a program supplied by the UCSC Genome Bioinformatics site (http://genome.ucsc.edu/index.html). A search for transcription factor binding sites was targeted to extremely conserved regions using MATCH public version 1.0 (linked with the Gene Regulation site (http://www.gene-regulation.com/). After the computer search, identified transcription factor binding sites were reconfirmed based on the consensus DNA sequences reported previously (Merika and Orkin, 1993).
Nuclear extract preparation
Nuclear extracts were prepared from 293T cells transiently transfected with transcription factor expression vectors pBR-CAG-mGata4-IP, pCAG-mCdx2-IP (kindly provided by Dr H. Niwa, Riken Center for Developmental Biology, Kobe, Japan), pCAG-hTcf4 and Lef1-HA (kindly provided by Dr M. Kato, University of Tsukuba, Japan), using the Lipofectamine 2000 system (Invitrogen).
Electrophoretic mobility shift assay (EMSA)
For probe preparation, the 3′ end of each single-strand sense oligonucleotide was biotin-labeled using the Biotin 3′ End DNA Labeling Kit (Thermo Fisher Scientific) and annealed with non-labeled antisense oligonucleotide to obtain the double-stranded DNA probe. The detection of biotin-labeled DNA probes was performed using a LightShift Chemiluminescent EMSA Kit (Thermo Fisher Scientific). DNA-nuclear extract binding reactions were performed as follows: 5 μg nuclear extracts were preincubated with 10X Binding Buffer (from the LightShift Chemiluminescent EMSA Kit) and 50 ng/μl Poly (dI-dC) for 10 minutes on ice. A 200-fold molar excess of competitor oligonucleotide was then added to each reaction, and the mixtures were incubated at room temperature for 5 minutes. Biotin-labeled oligonucleotides were then added, and the reactions were incubated at room temperature for 20 minutes. The biotin-labeled DNA was detected by chemiluminescence.
The sense-strand sequences of the control Gata-, Cdx-, Tcf/Lef-, Fox-binding sites 1, 2, 3 and of the mutated (mut-) Gata-, Cdx-, Tcf/Lef-, Fox-binding sites 3, 4, and of the transcription factor binding sites in DMME used for EMSA were as follows (transcription factor binding sites are underlined). Oligonucleotide probe from DMME: Gata I/Cdx I (G1/C1), 5′-TTTCTAGGATAAAATATTTT-3′; Gata II/Cdx II (G2/C2), 5′-AAATATTTTATCTGGATTTG-3′; Gata III (G3), 5′-TTTGCAAAATCATTTCCTG-3′; Fox I/Cdx III (F1/C3), 5′-TTGACGTGTAAACATGCGCTC-3′; Fox II/Cdx IV (F2/C4), 5′-AGGCCCAATAAATATTTCAAA-3′; Cdx V (C5), 5′-TTCAAACATAAAAGGATTG-3′; Tcf/Lef I (TL1), 5′-TTGT]TCTAACAAAGGGGGTTT-3′; Gata IV (G4), 5′-CCCTGGGTTATCTGAACAGC-3′; Gata V (G5), 5′-CCGGTCCTGATAGCAAGCAA-3′; Gata VI (G6), 5′-CACAAACCTATCAGAGCACA-3′; Gata VII/Cdx VI (G7/C6), 5′-GCATCCTAGATAAGCTTTATGGAGAGT-3′; Tcf/Lef II (TL2), 5′-CTGAGTCCTTTGATGTAGCCA-3′; ER71, 5′-GCAAAATCATTTCCTGTCGTTCTG-3′; mut-ER71, 5′-GCAAAATCATAGAGTGTCGTTCTG-3′. Wild-type oligonucleotides for competition experiments: Nkx2.5 gs1 (GATA4), 5′-GGCGGGGAAGGGAGATAAGATGACATAC-3′; C2/Y1 (CDX2), 5′-AATGTGCTCCATAAATGTCACTTA-3′; Tcf/Lef-BR (Lef1), 5′-CCCTTTGATCTTACC-3′; HFH-1 #3 (FoxF1), 5′-AATTGTTTATTTA-3′. Mutated oligonucleotides for competition experiments: mut-Nkx2.5 gs1, 5′-GGCGGGGAAGGGAGCTCAGATGACATAC-3′; mut-C2/Y1, 5′-AATGTGCTCCAGAAATGTCACTTA-3′; mut-Tcf/Lef-BR, 5′-CCCTTTGGCCTTACC-3′ (referred to FOP flash); mut-HFH-1 #3, 5′-AATTCTCGATTTA-3′.
Chromatin immunoprecipitation assay
Inducible ER71/Etv2-V5 ES cells were differentiated and induced with Dox during day 1.5 to day 3.5 (Lee et al., 2008). Day 3.5 i ER71/Etv2-V5 embryoid bodies (EBs) were collected and cross-linked with 1% formaldehyde. Cells were lysed, and chromatin was fragmented by sonication. Lysates were processed using standard chromatin immunoprecipitation (ChIP) procedures using a control Rabbit IgG (Cell Signaling Technology, #2729), V5-specific antibody (ChIP grade, Abcam, ab15828). Quantitative PCR (QPCR) was performed with the SYBY Green Supermix (BioRad), and enrichment was calculated using the fold enrichment method. Primers were designed to amplify the ER71/Etv2 binding sequence in the enhancer region of the mouse Flk1 gene. Mouse IGX1A negative control primers (SABiosciences: GPM00001C(–)01A) were used to detect a specific sequence within a 900 kb open-reading-frame-free intergenic region containing no known or predicted transcription start sites on mouse chromosome 6. PCR primers designed to flank the binding site of ER71/Etv2 in the Flk1 promoter region (–174∼ –81 from TSS) were used as a positive control (Lee et al., 2008).
Primers for ChIP-PCR analysis
ER71/Etv2-binding site on Flk1 enhancer: forward, TGAACTGTCTTCAGCCCGTA; reverse, TGGTGCTTGATCTCAGAACG. Primer –174∼ –81: forward, TTGCTCTCAGATGCGACTT; reverse, GCCACTGGATACCAGGTTTGG.
Maintenance of ES cells and differentiation
Established ES cells were differentiated as described previously (Ema et al., 2003). Inhibitors for bFgf, Bmp4 and Wnt3 were added to the differentiation medium at the following concentrations: 1nM Noggin (R&D Systems, Minneapolis, Minnesota, USA), 5nM Dkk1 (R&D Systems) or 7.5 μM SU5402 (Calbiochem, Frankfurt, Germany). Dox-inducible ER71 ES cells were differentiated as described previously (Lee et al., 2008).
Quantitative and semi-quantitative RT-PCR
Total RNAs purified from the differentiated ES cells were used to generate cDNA using the Reverse Transcription Kit (Qiagen, Valencia, CA). Real-time PCR was performed using the Thermal Cycler Dice Real Time System (TaKaRa Bio, Otsu, Japan) and SYBR Premix EX Taq (TaKaRa Bio). Relative cDNA amounts were calculated using the Thermal Cycler Dice Real Time System Software and normalized against expression of Hprt (supplementary material Table S1). Oligonucleotide sequences used for real-time and semi-quantitative PCR reactions were: GFP (F), 5′-AGCTGACCCTGAAGTTCATCTG-3′; GFP (R), 5′-AAGAAGTCGTGCTGCTTCATGT-3′; Pecam1 (F), 5′-GTCATGGCCATGGTCGAGTA-3′ (30 cycle); Pecam1 (R), 5′-CTCCTCGGCATCTTGCTGAA-3′; Scl (F), 5′-ATTGCACACACGGGATTCTG-3′ (32 cycle); Scl (R), 5′-GAATTCAGGGTCTTCCTTAG-3′; Tie2 (F), 5′-TTGAAGTGACGAATGAGAT-3′ (35 cycle); Tie2 (R), 5′-ATTTAGAGCTGTCTGGCTT-3′; Gata1 (F), 5′-AGATTCCACAGGTTTCTTTTCCTCT-3′ (30 cycle); Gata1 (R), 5′-GTAGGCCTCAGCTTCTCTGTAGTAG-3′; Nkx2.5 (F), 5′-ACCTTTCTCCGATCCATCCCACTT-3′ (35 cycle); Nkx2.5 (R), 5′-GCGTTAGCGCACTCACTTTAATGG-3′; Hprt (F), 5′-TTGTTGTTGGATATGCCCTTGACTA-3′ (30 cycle); Hprt (R), 5′-AGGCAGATGGCCACAGGACTA-3′; Flk1 (F), 5′-GGGATGGTCCTTGCATCAGAA-3′ (32 cycle); Flk1 (R), 5′-ACTGGTAGCCACTGGTCTGGTTG-3′; Sma (F), 5′-TACCATGTACCCTGGCATTGCTGA-3′ (35 cycle); Sma (R), 5′-AGAAGGCCCTCTGACTTTAGAAGC-3′; Isl1 (F), 5′-TGTCAGGAGACTTGCCACTTT-3′ (27 cycle); Isl1 (R), 5′-GCCAAACGTTTATTAGTGAAATAGTCCTG-3′; Mlc2a (F), 5′-AAGGGAAGGGTCCCATCAACTTCA-3′ (30 cycle); Mlc2a (R), 5′-AACAGTTGCTCTACCTCAGCAGGA-3′; Tbx20 (F), 5′-AACAAGGTACCATACCCAGTCTGAGC-3′ (30 cycle); Tbx20 (R), 5′-GCCCTTACCCTTACTCTATGGGCATT-3′.
Flow cytometry
Embryos were incubated with 0.1% Collagenase S-1 (Nitta Gelatin Co, Osaka, Japan) at 37°C for 15 minutes. After washing, single cells were exposed to the phycoerythrin (PE) conjugated-antibody to Flk1 (clone AVAS12, BD Biosciences, San Jose, CA). Cells were analyzed by FACS Calibur or were sorted using a FACS Vantage (BD Biosciences).
Hematopoietic colony assay
Primitive hematopoietic colony assays were performed as described previously (Palis et al., 1999). The BL-CFC assay using differentiated ES cells was performed as described previously (Choi et al., 1998). The BL-CFC assay using embryo-derived cells was performed as described previously (Huber et al., 2004).
Immunohistochemistry
Embryos were dissected and staged according to the method of Downs and Davies (Downs and Davies, 1993). Immunohistochemistry was performed with anti-PECAM1 (MEC13.3; BD), anti-GATA1 (N6; Santa Cruz Biotechnology, Santa Cruz, CA), anti-SMA (clone1A4; Sigma), anti-Flk1 (clone AVAS12; BD) and anti-GFP (rabbit polyclonal; Molecular Probes, Eugene, OR) as described (Ema et al., 2006b). DiI-incorporation activity was visualized after a 4 hour incubation of the cells with 10 μM of DiI-Ac-LDL (Invitrogen).
Statistical analysis
Data are expressed as means and standard errors. Differences were considered significant at P<0.05.
RESULTS
Identification of a novel mouse Flk1 enhancer
To search for novel enhancers of mouse Flk1 that could direct multipotent mesodermal expression, we first investigated GFP reporter activity in Flk1-GFP BAC Tg mice that carried the 167 kb genomic region including the Flk1 gene (Ishitobi et al., 2010), in order to discern if the genomic region contains cis-elements sufficient for early Flk1 expression in lateral mesoderm at the primitive streak stage (supplementary material Fig. S1A,B). Consistent with GFP expression in Flk1-GFP knock-in mice, which completely reproduce early Flk1 expression in lateral mesoderm at the primitive streak stage (supplementary material Fig. S1), GFP expression in Flk1-GFP BAC Tg mice is similar to that of endogenous Flk1 protein (supplementary material Fig. S1B). This indicates that the genomic region included in the BAC is sufficient to direct early Flk1 expression in the lateral mesoderm. In order to narrow down the regulatory elements important for Flk1 expression, mouse and human genomic sequences from 150 kb upstream to 150 kb downstream of Flk1 were compared. Genomic comparison analysis among different species has become one of the most useful strategies to identify functional elements (Rojas et al., 2005; Göttgens et al., 2000; Prabhakar et al., 2006), and it identified highly conserved sequences in non-coding regions (supplementary material Fig. S2A), which are referred to as enhancer #1-#4 because cis-acting elements usually span a few hundred base pairs (da Laat et al., 2003). The lacZ reporter plasmid connected with the Flk1 1.8 kb promoter (supplementary material Fig. S2B) was used because the promoter region itself does not have a cis-element for mesodermal or endothelial expression (Kappel et al., 1999). Among these four conserved, noncoding regions within the mouse Flk1 locus, only the transgenic embryos carrying enhancer #4 showed lacZ expression in the early mesodermal region at 7.5 dpc (data not shown). As enhancer #4 recapitulates the multipotent mesodermal progenitors as shown below, it was named the Distal-Multipotent-Mesodermal-Enhancer (DMME). To further investigate the enhancer activity, transgenic animal lines carrying Flk1-DMME-lacZ Tg were generated and lacZ expression was compared to that in 6.5-8.5 dpc embryos (supplementary material Table S2) (Fig. 1) (supplementary material Fig. S3A). lacZ expression was seen in four of five transgenic lines at 7.5 dpc, and in the remaining three lines at 8.5 dpc (supplementary material Table S2). At 7.5 dpc, transgenic embryos showed strong lacZ expression in the lateral mesodermal region, but not in the primitive streak (Fig. 1C,D,F). At 8.5 dpc, lacZ expression gradually became restricted to the posterior lateral mesoderm, and it could not be observed in endothelial cells of the yolk sac or fetus (Fig. 1H).
Fig. 1.
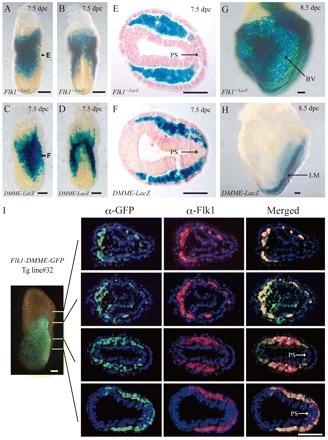
DMME recapitulated the early Flk1 expression pattern at 7.5 dpc. Whole-mount (A,B,G) and a transverse section (E) of X-gal stained Flk1-lacZ knock-in embryos. Whole-mount (C,D,H) and a transverse section (F) of X-gal stained Flk1-DMME-lacZ Tg embryos obtained from a stable transgenic line (line#711). The transverse section at 7.5 dpc showed lacZ expression in the mesoderm, but not in the primitive streak, similar to endogenous Flk1 expression (E,F: corresponds to the notation ‘E’ and ‘F’ in A,B). lacZ expression directed by Flk1-DMME gradually becomes restricted to caudal lateral mesoderm, but it was not detectable in blood vessels at 8.5 dpc (G,H). (I) DMME recapitulated early mesodermal expression of Flk1. A Flk1-DMME-GFP transgenic embryo at E7.5 (OB stage) was sectioned at different levels and subjected to immunohistochemistry with anti-Flk1 and anti-GFP antibodies. Scale bars: 100 μm in A,B,C,D,G,H; 50 μm in E,F. BV, blood vessels; LM, lateral mesoderm; PS, primitive streak.
Cells marked by Flk1-DMME contain hemato-cardiovascular progenitors
Although DMME seemed to recapitulate similar expression patterns to endogenous Flk1 in the mesoderm lineage, it is not clear whether or not their expression patterns are the same. To investigate these issues, Flk1-DMME-GFP Tg lines were created so that GFP could be used for immunohistochemical identification (Fig. 1I). Immunohistochemistry was performed on Tg embryos and GFP expression was found to overlap well with endogenous Flk1 expression (Fig. 1I) (supplementary material Fig. S3), suggesting that the Flk1-DMME element is capable of recapitulating endogenous Flk1 expression (Fig. 1I).
Although Flk1-DMME clearly recapitulated the early mesodermal Flk1 expression, it was not clear whether GFP-positive cells marked by Flk1-DMME were capable of generating hemato-cardiovascular progenitors. First, the GFP-positive and negative cells from the Flk1-DMME Tg embryos at embryonic day (E) 7.5 were sorted and gene expression was analyzed by examining hemato-cardiovascular lineage markers (Fig. 2A). The hemato-cardiovascular lineage markers such as Gata1 and Scl (Tal1 – Mouse Genome Informatics) (hematopoietic markers); VE-cadherin (Cdh5 – Mouse Genome Informatics) and Pecam1 (endothelial markers); and Isl1, Nkx2.5, Mlc2a (Myl7 – Mouse Genome Informatics) and Tbx20 (cardiac markers) were clearly enriched in GFP-positive cells but not in GFP-negative cells (Fig. 2A). Hematopoietic colony assays also clearly showed that GFP-positive cells from embryos at E7.5 contain EryP and Mac progenitors, although GFP-positive cells from embryos at E8.5 lost their progenitor properties (Fig. 2B,C). When the GFP-positive cells were cultured for 4 days on type IV collagen, they differentiated into PECAM1-positive and SMA-positive cells, whereas most of the GFP-negative cells gave rise to other cell types (Fig. 2D). As PECAM1-positive cells efficiently incorporate DiI-acetylated LDL and SMA-positive cells co-express Desmin, we concluded that they differentiated into PECAM1-positive endothelial and SMA-positive smooth muscle cells (supplementary material Fig. S4). In order to investigate if GFP-positive cells from Flk1-DMME Tg embryos at E7.5 contain multipotent progenitors such as BL-CFC and CV-CFCs, a colony assay was performed. The results indicated that two types of colonies, including BL-CFC and CV-CFC, emerged (supplementary material Fig. S2E). Consistent with a previous report (Huber et al., 2004; Kattman et al., 2006), BL-CFC expressed Gata1 (hematopoietic marker), Flk1, PECAM1, VE-cadherin (endothelial markers), and SMA (smooth muscle marker), but not Islet1, Tbx5 (cardiac markers), whereas CV-CFC expressed Islet1, Tbx5, Flk1, PECAM1, VE-cadherin and SMA, but not Gata1 (see Fig. S5 in the supplementary material). Some of the CV-CFC, but not the BL-CFC, started beating, indicating that there were functional cardiomyocytes (data not shown). Our assay clearly showed that BL-CFC and CV-CFC activities were enriched in a GFP-positive population from embryos at E7.5 (Fig. 2F). Thus, Flk1-DMME-labeled cells at E7.5 contain progenitors for hematopoietic, endothelial, cardiac and smooth muscle cells, although Flk1-DMME-labeled cells at later stages such as E8.5 have different progenitor properties.
Fig. 2.
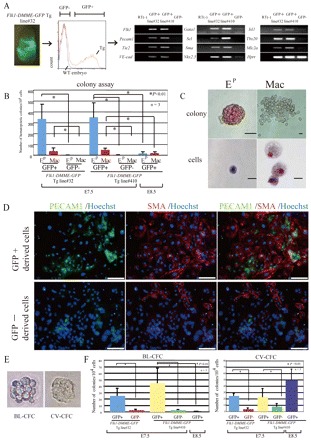
Cells labeled by Flk1-DMME-GFP contain hematopoietic, endothelial, cardiac and smooth muscle progenitors. (A) The cells marked by Flk1-DMME-GFP Tg contain hematopoietic and cardiovascular markers. GFP-positive and negative cells were sorted from Flk1-DMME-GFP Tg mouse embryos at E7.5 and semi-quantitative RT-PCR was performed to identify hematopoietic, endothelial, smooth muscle and cardiac marker genes. (B) Enrichment of hematopoietic progenitors in Flk1-DMME-GFP-positive cells. (C) EP (EryP) and Mac colonies derived from GFP-positive cells. (D) GFP-positive cells differentiate into endothelial and smooth muscle cells. GFP-positive cells from Flk1-DMME-GFP-Tg embryos differentiated into PECAM1-positive endothelial cells (shown in green) and SMA-positive smooth muscle cells (shown in red) (upper panel), whereas most of the GFP-negative derived cells were not stained with either of PECAM1 or SMA (lower panel). (E) Typical BL-CFC and cv-CFC. (F) Enrichment of BL-CFC and CV-CFC activity in the GFP-positive fraction from embryos at E7.5. Scale bars: 25 μm in C; 100 μm in D.
Previous studies showed that Flk1 expression marks progenitors for hematopoietic, endothelial, smooth muscle cells and cardiomyocytes during mouse and human ES differentiation (Yamashita et al., 2000; Kattman et al., 2006; Yang et al., 2008). Thus, the possibility that Flk1-DMME activity marks multipotent mesodermal progenitors during mouse ES differentiation was investigated. The vector described in Fig. 3A was constructed and several ES cell lines were established. When Flk1-DMME ES cell lines were differentiated into EBs along with a Flk1-GFP knock-in ES cell line carrying GFP in the Flk1 locus, the gross GFP expression pattern driven by Flk1-DMME was similar to that of the Flk1-GFP knock-in ES cell line (data not shown). EB carrying the Flk1 1.8 kb promoter does not give any signal until day 5 (data not shown), consistent with observations of Tg embryos (Kappel et al., 1999). Flow cytometry analysis showed similar expression levels for Flk1-DMME GFP Tg and Flk1-GFP knock-in ES cell lines (Fig. 3B). When GFP-positive cells were sorted and analyzed using a BL-CFC assay, BL-CFC activity was found to be enriched only in the GFP-positive cell fraction (Fig. 3C). When the cells derived from BL-CFC were cultured, they differentiated into hematopoietic, endothelial and smooth muscle cells (Fig. 3D,E), indicating that BL-CFC contains hematopoietic, endothelial and smooth muscle cells, as previously reported (Choi et al., 1998; Ema et al., 2003). Taken together, the data indicate that cells labeled by Flk1-DMME are capable of generating multiple mesodermal progenitors.
Fig. 3.
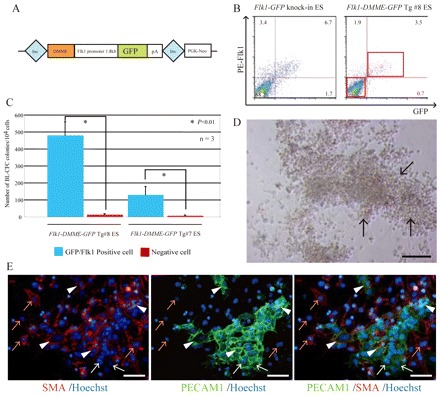
Flk1-DMME recapitulating multipotent progenitors during ES cell differentiation. (A) Schematic representation of the Flk1-DMME-GFP-pA transgene. The Flk1-DMME-GFP-pA sequence was introduced between insulator sequences. (B) Similar expression levels were seen for Flk1-GFP knock-in and Flk1-DMME-GFP-pA Tg ES cells. EBs at day 3 after LIF removal were trypsinized and incubated with PE-conjugated anti-Flk1 antibody, then analyzed by flow cytometry. (C) Enriched BL-CFC activity in Flk1-DMME-GFP-labeled cells. GFP/PE-Flk1-positive cells were sorted and subjected to a BL-CFC assay. (D,E) Differentiation of BL-CFC cells into hematopoietic, endothelial and smooth muscle cells. A BL-CFC colony was picked for subsequent culture. After 1 week, the cells contained floating hematopoietic cell clusters (D) (black arrows in upper panel). After removing the hematopoietic cells, adherent cells were subjected to immunohistochemistry for PECAM1 and SMA (E). White arrowheads indicate SMA- and PECAM1-double positive cells; orange arrows indicate SMA-single positive cells; white arrows indicate PECAM1-single positive cells. Ins, insulator sequence.
DMME activity is reduced after treatment with chemical inhibitors of Bmp, Fgf and Wnt signaling
Previous studies have shown that soluble factors such as Bmps, Fgfs and Wnts are crucial to induce hematopoietic, endothelial and cardiovascular progenitors, although it is not known how these secreted factors induce Flk1 expression. To investigate whether transcriptional activation by Flk1-DMME is mediated by Fgf, Wnt or Bmp signaling pathways, the effects of specific inhibitors against Fgf, Wnt and Bmp signaling were examined (Fig. 4A). Consistent with previous reports, chemical inhibitors against Fgf, Wnt and Bmp inhibited endogenous Flk1 expression during ES differentiation (Fig. 4B). Interestingly, GFP expression driven by Flk1-DMME Tg was similarly blocked by these inhibitors (Fig. 4B), suggesting that transcription by Flk1-DMME is regulated through Fgf, Wnt and Bmp signaling.
Fig. 4.
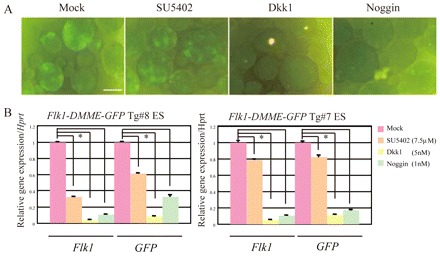
DMME is responsive to Wnt, Bmp4 and Fgf2. (A) GFP expression levels of EBs derived from Flk1-DMME-GFP-pA Tg ES cells treated with Noggin, Dkk1 or SU5402. (B) Relative Flk1 and GFP expression levels in EBs derived from Flk1-DMME-GFP-pA Tg ES cells treated with Noggin, Dkk1 or SU5402. Quantitative RT-PCR of Flk1 and GFP was performed and normalized to Hprt expression. Scale bar: 100 μm.
DMME contains highly conserved Gata-, Cdx-, Tcf/Lef- and Fox-binding sites
Chemical inhibitor analysis clearly indicated that DMME activity is controlled by Fgf, Wnt and Bmp signaling. To reveal the transcriptional basis for Fgf-, Wnt- and Bmp-induced Flk1 transcription, the sequence of DMME was investigated to see if there was conservation among multiple species and among transcription factor binding sites. The CLUSTALW multiple alignment algorithm was used to analyze DMME sequence sets from multiple species, and it identified extremely conserved sequences, including seven Gata-, six Cdx-, two Tcf/Lef- and two Fox-binding sites (Fig. 5A). The possibility that DMME is bound by these transcription factors was investigated next (Fig. 5B). Gata4 bound to all seven of the Gata-binding sites in DMME (Fig. 5B, upper panel). The binding specificity was confirmed by competition experiments using an excess amount of control oligonucleotides, as described previously (Lien et al., 1999), including one with a mutated sequence. All Gata-binding sites within the DMME bound Gata4 with high affinity. The Cdx-binding sites in DMME were also examined, and Cdx2 was shown to bind specifically to all of the expected sites (Fig. 5B). In addition to the Gata- and Cdx-binding sites, two Tcf/Lef sites were also bound by both Tcf4 and Lef1 efficiently, and the binding to Tcf/Lef II was stronger than to Tcf/Lef I (Fig. 5B). Some of the transcription factor binding sites within DMME shared parts of the consensus sequences: Gata I and Cdx I, Gata II and Cdx II, Fox I and Cdx III, Fox II and Cdx IV (Fig. 5B). Interestingly, EMSA demonstrated that only one kind of protein could bind to such binding sites, even if the sequence carries two consensus binding elements (supplementary material Table S1). Overall, these results demonstrated that the conserved seven Gata-, two Cdx- and one Tcf/Lef-binding sites are genuine binding sites for Gata4, Cdx2, Tcf4 and Lef1.
Fig. 5.
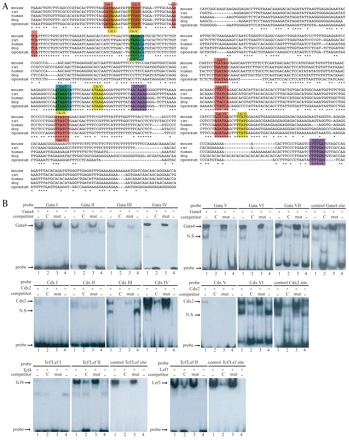
Gata, Cdx and Tcf/Lef bind to DMME. (A) DMME contains consensus binding sites for Gata, Cdx, Tcf/Lef and Fox transcription factors. Asterisks denote nucleotides that are conserved perfectly among all five species. (B) DMME is bound by Gata, Cdx, Tcf and Lef proteins. (Upper panel) Lane 1 contains Gata4 protein and a biotin-labeled probe with a Gata-binding site; lane 2 contains Gata4 protein, a probe and a 200-fold excess of the unlabeled control Gata4 site oligonucleotide (C) as a competitor; lane 3 contains Gata4 protein, a probe and a 200-fold excess of the mutated control Gata4 site (mut); lane 4 contains mock nuclear extract (represented by a minus sign) and a probe. (Middle panel) DMME contains two high-affinity Cdx-binding sites. (Lower panel) DMME contains one high-affinity Tcf/Lef-binding site.
Importance of conserved Gata-, Cdx-, Tcf/Lef- and Fox-binding sites in Flk1 expression
Conserved Gata-, Cdx-, Tcf/Lef- and Fox-binding sequences were mutated to assess the importance for Flk1 expression in multipotent mesodermal progenitors (Fig. 6A-C). Mutation of any of these sites dramatically disturbed lacZ expression at E7.5, indicating that at least one of these sites is important for efficient Flk1 expression in multipotent mesodermal progenitors.
Fig. 6.
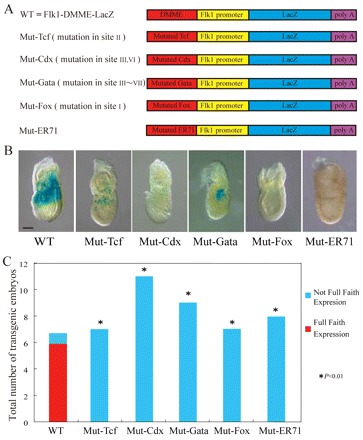
Importance of Gata-, Tcf-, Cdx-, Fox- and ER71/Etv2-binding sites in early lateral mesodermal expression of Flk1 at primitive streak stage. (A) Schematic representation of transgenes with mutations. (B) F0 analysis of DMME carrying mutations in cognate binding sites for Gata, Tcf, Cdx, ER71/Etv2 and Fox transcription factors. (C) Statistical analysis of F0 transgenic experiments. ‘Not Full Faith Expression’ indicates weak or patchy expression, whereas ‘Full Faith Expression’ indicates strong and faithful expression. Scale bar: 100 μm.
Potential involvement of ER71/Etv2 on Flk1 expression
The Ets transcription factor ER71/Etv2 regulates Flk1 gene transcription by directly activating it in response to Wnt, Bmp4 and Notch (Lee et al., 2008). ER71/Etv2 KO embryos are embryonic lethal owing to impaired hematopoiesis and vasculogenesis (Lee et al., 2008). Although ER71/Etv2 activates Flk1 transcription by binding to Flk1 promoter regions (Lee et al., 2008), it is not known whether ER71/Etv2 binds to DMME. To address this issue, we investigate whether ER71/Etv2 binds to candidate binding sites within DMME during ES cell differentiation into Flk1-positive cells using a ChIP assay (supplementary material Fig. S6A,B). The results demonstrated that ER71/Etv2 binds to DMME during ES cell differentiation. The idea was further strengthened by the fact that ER71/Etv2 was shown to bind to DMME by EMSA (supplementary material Fig. S6C). Although ER71/Etv2 was shown to bind to DMME, it is not clear whether ER71/Etv2 is important for Flk1 expression in the early lateral mesoderm at the primitive streak stage. Immunohistochemistry with anti-Flk1 antibody clearly showed that the lateral mesoderm expression of Flk1 is significantly reduced in ER71/Etv2 KO embryos at E7.5 (supplementary material Fig. S6D,E), demonstrating that its early mesodermal expression is activated by ER71/Etv2. Flk1 early mesodermal expression is reduced by only 20% in ER71/Etv2 KO embryos. Therefore it seems that the role played by ER71/Etv2 may be relatively minor. It is notable that ER71/Etv2 binds to a conserved motif directly outside the DMME region (De Val et al., 2008; Lee et al., 2008).
Importance of the DMME region for endogenous Flk1 expression and hemangioblast development
Although our data indicate that DMME is sufficient for early lateral mesoderm expression of Flk1 at the primitive streak stage, it is not clear that DMME is required for endogenous Flk1 expression. To address this issue directly, we generated Flk1-DMME KO ES cells by gene targeting (Fig. 7). Differentiated Flk1-DMME KO ES cells showed severely reduced endogenous Flk1 expression (Fig. 7C,D), clearly indicating that it is required for the embryonic expression. Furthermore, they showed a severe reduction in BL-CFC number (Fig. 7E). Together, these data indicate that DMME is required for endogenous Flk1expression and hemangioblast development during ES differentiation.
Fig. 7.
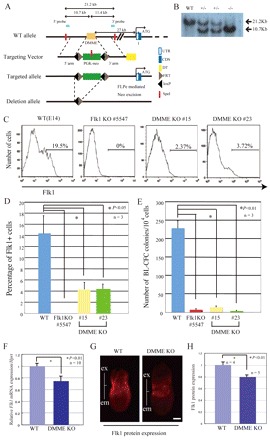
Importance of DMME on early lateral mesodermal expression of Flk1 at the primitive streak stage. (A) Schematic representation of the DMME targeting vector. (B) Verification of homologous recombination of DMME-deficient ES cells by Southern blot analysis. (C) Evaluation of endogenous Flk1 protein expression in Flk1-DMME KO ES cells. ES cells at differentiation day 3.0 were analyzed by flow cytometry using an anti-Flk1 antibody. (D) Statistical analysis of the Flk1-positive cells. (E) BL-CFC activity in Flk1-DMME KO ES cells. (F) Reduced Flk1 mRNA expression in DMME-deficient embryos at the primitive streak stage. Flk1+/ΔDMME mice were intercrossed and the embryos were dissected out at E7.5 and were used for the quantitative RT-PCR analysis. (G) Reduced Flk1 protein expression in DMME-deficient embryos at the primitive streak stage. (H) Comparison of signal intensity of Flk1 protein expression. Fluorescent image intensities were compared using NIH Image J software.
Mice deficient for DMME were born in a Mendelian ratio, but they died after few days for unknown reasons (H.I., M.E., unpublished observation). Quantitative RT-PCR analysis indicated that Flk1 mRNA expression is reduced by 25% in DMME-deficient embryos at E7.5 (Fig. 7C). Immunohistochemistry with an anti-Flk1 antibody also indicates that Flk1 protein expression is significantly lower in DMME-deficient embryos at E7.5 (Fig. 7D,E). These results demonstrated that DMME is required for animal development and in part for early lateral mesoderm expression of Flk1 at the primitive streak stage.
DISCUSSION
Flk1 requires multi-transcriptional cis-elements
The Flk1-DMME region identified in this study directed lacZ expression both in the extra- and intra-embryonic mesoderm at 7.5 dpc. As development proceeded, lacZ expression became restricted to the caudal mesoderm region at 8.5 dpc (Fig. 1). Notably, no lacZ expression was detected in endothelial or hematopoietic cells at 8.5 dpc. Thus, Flk1-DMME is sufficient for recapitulation of the early Flk1 expression pattern, but its expression is rapidly restricted after this stage. However, DMME deletion from the genome results in a significant reduction of early Flk1 expression at the primitive streak stage, demonstrating that DMME is required for early lateral mesoderm expression of Flk1 at the primitive streak stage. DMME deficiency in ES cells also severely reduced the number of Flk1-positive cells and interfered with hemangioblast development. This indicates that other cis-regulatory regions direct early Flk1 expression.
Novel aspects of the Flk1 transcriptional regulatory system
The present study demonstrates that Flk1-DMME contains Gata-, Cdx-, Tcf/Lef- and Fox-transcriptional binding sites (Fig. 5A), and that some of these are efficiently bound by Gata4, Cdx2, Tcf4 and Lef1 (Fig. 5B), suggesting that these factors may regulate Flk1 transcription directly.
Gata4 is among the earliest markers of the endoderm and the lateral mesoderm following gastrulation in the mouse embryo (Rojas et al., 2005; Burch, 2005; Heikinheimo et al., 1991; Peterkin et al., 2005), and its family protein, Gata6, is also expressed in mesodermal cell lineages (Morrisey et al., 1996). Although these proteins have been identified as direct regulators of bone morphogenetic protein 4 (Bmp4), it has also been suggested that Gata4 is regulated indirectly by Bmp4 (Rojas et al., 2005; Nemer and Nemer, 2003). Bmp family molecules such as Bmp2, Bmp4, Bmp5 and Bmp7 are strongly expressed in the lateral mesoderm, and they are known to play crucial roles in bone and cartilage development, extra-embryonic mesoderm development, blood island formation and heart development (Winnier et al., 1995; Zhao et al., 2003). Moreover, recent studies have shown that Bmp4 signaling is required for the development of Flk1- and Scl-positive cells, which give rise to endothelial and hematopoietic cells according to an in vitro ES cell differentiation model (Park et al., 2004). Bmps are expected to be involved in Flk1 regulation, and the results presented here support the hypothesized interaction of Bmp and Flk1 in the early mesoderm via Gata4 (Fig. 8).
Fig. 8.
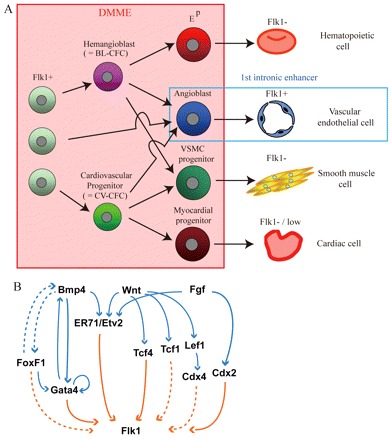
Summary of this study. (A) DMME is active in progenitors such as BL-CFCs and CV-CFCs, but not in mature cells. However, the first intronic enhancer recapitulates Flk1 expression in angioblasts and endothelial cells, as reported by Kappel and coworkers. (B) Model for Gata-, Cdx-, FoxF- and Tcf/Lef1-dependent Flk1 expression. Flk1-DMME activity is dependent on Gata-, Cdx-, FoxF- and Tcf/Lef1-binding sites identified in the present study (shown in unbroken orange arrows). The direct enhancer binding of Cdx2, Tcf1 and FoxF1 is not demonstrated and is still hypothetical (shown in broken orange arrows). This study also indicates that Bmp, Fgf and Wnt signaling is important for Flk1-DMME activity. Given that past studies show FoxF1, Gata4, Tcf4, Lef1, and Cdx2 work downstream of Bmp, Fgf and Wnt signaling (shown in blue lines), Flk1-DMME appears to be induced by Bmp, Fgf and Wnt signaling through those transcription factors.
Cdx2 binds to two of six binding sites in the DMME. It is expressed in the lateral mesoderm, neural tube and somites in the caudal portion of the embryo (Gaunt et al., 2005), and it plays essential roles in axis formation and vasculogenesis (Chawengsaksophak et al., 2004). Cdx4 is also expressed in various tissues of the caudal portion, similar to Cdx2 (Gaunt et al., 2005), and it has been identified as a direct target of the wingless (Wnt) signaling pathway (Pilon et al., 2006). Wnt signaling is required for primitive streak formation and mesoderm development (Liu et al., 1999; Huelsken et al., 2000), and inhibition of Wnt in differentiating ES cells leads to failure of Flk1-positive cell proliferation (Lindsley et al., 2006). Thus, it is possible that Wnt regulates Flk1 transcription via the Cdx family molecules during mesoderm development. The fibroblast growth factor (Fgf) signaling pathway is required for normal expression of Cdx1, Cdx2 and Cdx4 in the developing mesoderm of Xenopus (Keenan et al., 2006), and it is crucial for proliferation of Flk1- and Scl-positive cells in vitro (Faloon et al., 2000). These results suggest that Fgf is a potential regulator of Flk1 transcription, as well as Wnt. Therefore, Cdx proteins may interact with Flk1 under the direct control of both Wnt and Fgf (Fig. 8).
Both Tcf4 and Lef1 bind to one of the Tcf/Lef-binding sites. Tcf4 and Lef1 are downstream effectors of the Wnt signaling cascade (Korinek et al., 1998), and the Tcf family protein Tcf1 is also known to be a major effector of Wnt in the primitive streak and tailbud of the mouse embryo (Galceran et al., 1999; van Noort and Clevers, 2002). Null mutations in both Tcf1 and Lef1 cause a severe defect in the differentiation of paraxial mesoderm; a similar defect is seen in Wnt3a-deficient mice (Galceran et al., 1999). Thus, Wnt may regulate Flk1 expression through Cdx, Tcf and Lef (Fig. 8).
FoxF1 is a member of Forkhead expressed in lateral mesoderm at 8.5 dpc, and is necessary for mesodermal cell differentiation. The null mutation for FoxF1 results in reduced Flk1 expression (Mahlappu et al., 2001). This transcription factor is identified as the direct or indirect regulator of Gata4 and Bmp4, and it is also as the downstream target of Bmp4 (Rojas et al., 2005). These studies suggest the possibility that FoxF1 is a regulatory factor of Flk1. Overall, a number of studies have suggested that Bmp, Fgf and Wnt are involved in the transcriptional regulation of Flk1, and our results provide supporting evidence for their cooperative interactions through Gata, Cdx, Tcf, Lef and Fox proteins (Fig. 8B).
ER71/Etv2, an integral component for Wnt, Bmp4 and Notch signaling, is important for hematopoiesis and vasculogenesis (Lee et al., 2008). Our analysis clearly demonstrated that ER71/Etv2 binds to DMME during ES cell differentiation and that early mesodermal expression is activated by ER71/Etv2.
Possible role of DMME in evolution
Flk1 expression in endothelial cells is generally observed in vertebrates, such as zebrafish, Xenopus, chicken, mouse, human, and so on (Yamaguchi et al., 1993; Liao et al., 1997; Devic et al., 1999; Nimmagadda et al., 2004; Cortes et al., 1999). Consistent with this observation, the first intronic enhancer, which is sufficient for recapitulation of expression in endothelial cells, is conserved among these animals. However, genomic conservation analysis among multiple species showed that DMME has been identified in avians and mammals, but not in zebrafish or Xenopus (data not shown), suggesting an evolutionary role for DMME in homothermal animals. Our analysis clearly indicated that DMME deletion in the mouse resulted in a significant reduction of early lateral mesoderm expression of Flk1 at the primitive streak stage that was essential for survival after birth. Interestingly, in sharp contrast with Flk1 expression in mammals, Flk1 in zebrafish is expressed in endothelial cells, but not in hematopoietic or other cell types such as hemangioblasts or cardiac progenitors. On the one hand, one may imagine that acquisition of DMME in the genome allowed homothermal animals to have a broad spectrum of mesodermal expression of Flk1. On the other hand, it will be of interest in the future to investigate whether or not DMME allows zebrafish to drive a broad spectrum of Flk1 mesodermal progenitors.
It has been demonstrated that the Flk1-positive progenitor cells in differentiating ES cells have multipotential to differentiate into various hemato-cardiovascular cell lineages such as endothelial and hematopoietic cells, cardiomyocytes and vascular smooth muscle cells, according to an in vitro ES differentiation model (Yamashita et al., 2000; Ema et al., 2003; Kattman et al., 2006; Yang et al., 2008). In order to design successful therapies for injured tissues, it is essential to induce ES cells to develop into the appropriate cell types, based on an understanding of precise regulatory systems of cell differentiation. Flk1 transcription is regulated by cooperative interactions among Gata, Cdx and Tcf/Lef and Fox proteins activated by Bmp4, Fgf and Wnt (Fig. 7B), so it seems possible to establish more effective and certain ES differentiation systems to obtain target cell types. Further analyses of the Flk1 regulatory system may promote the development of novel, ES-based hemato-cardiovascular approaches to regenerative medicine.
Supplementary Material
Acknowledgments
We appreciate the technical assistance of Ms M. Ojima, M. Yanagisawa, M. Oguma and Y. Suzuki. We thank Drs P. Carlsson and H. Niwa for providing plasmids, and N. Kaneko for help with sectioning. We are also grateful to all members of the Takahashi lab.
Footnotes
Funding
This work was supported in part by Special Coordination Funds for Promoting Science and Technology (M.E.); by Grant-in-Aid for Scientific Research on Priority Areas [23122503 to M.E.]; by Princess Takamatsu Cancer Research Fund (M.E.); by PRESTO; by Japan Science and Technology Agency (JST) (M.E.); by a Grant-in-Aid for Scientific Research in Innovative Areas (‘Cellular and Molecular Basis for Neuro-vascular Wiring’) [23122503 to M.E.] of the Ministry of Education, Culture, Sports, Science, and Technology, Japan; and by an American Heart Association Postdoctoral Fellowship (F.L.).
Competing interests statements
The authors declare no competing financial interests.
Author contributions
H.I. established the transgenic animals and performed immunohistochemistry, hematopoietic colony assay and inhibitor analysis on ES differentiation. A.W. identified DMME by computer analysis, established the transgenic animals and ES cell lines, performed EMSA, prepared the figures, and co-wrote the manuscript. T.A. performed immunohistochemical analysis using a confocal microscopy. M.H. and S.T. assisted in generating transgenic animals. F.L., K.M. and H.K. provided new tools. K.C. and S.N. discussed the results. M.E. assisted with the transgene construction and ES cell establishment, guided the projects, and co-wrote the manuscript.
Supplementary material
Supplementary material available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.065565/-/DC1
References
- Burch J. B. (2005). Regulation of GATA gene expression during vertebrate development. Semin. Cell Dev. Biol. 16, 71–81 [DOI] [PubMed] [Google Scholar]
- Chawengsaksophak K., de Graaff W., Rossant J., Deschamps J., Beck F. (2004). Cdx2 is essential for axial elongation in mouse development. Proc. Natl. Acad. Sci. USA 101, 7641–7645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K., Kennedy M., Kazarov A., Papadimitriou J. C., Keller G. (1998). A common precursor for hematopoietic and endothelial cells. Development 125, 725–732 [DOI] [PubMed] [Google Scholar]
- Cortes F., Debacker C., Peault B., Labastie M. C. (1999). Differential expression of KDR/VEGFR-2 and CD34 during mesoderm development of the early human embryo. Mech. Dev. 83, 161–164 [DOI] [PubMed] [Google Scholar]
- de Laat W., Grosveld F. (2003). Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11, 447–459 [DOI] [PubMed] [Google Scholar]
- De Val S., Chi N. C., Meadows S. M., Minovitsky S., Anderson J. P., Harris I. S., Ehlers M. L., Agarwal P., Visel A., Xu S. M., et al. (2008). Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 135, 1053–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devic E., Rizzoti K., Bodin S., Knibiehler B., Audigier Y. (1999). Amino acid sequence and embryonic expression of msr/apj, the mouse homolog of Xenopus X-msr and human APJ. Mech. Dev. 84, 199–203 [DOI] [PubMed] [Google Scholar]
- Downs KM., Davies T. (1993). Staging of gastrulating mouse embryos by morphological landmarks in the dissecting microscope. Development 118, 1255–1266 [DOI] [PubMed] [Google Scholar]
- Ema M., Faloon P., Zhang W. J., Hirashima M., Reid T., Stanford W. L., Orkin S., Choi K., Rossant J. (2003). Combinatorial effects of Flk1 and Tal1 on vascular and hematopoietic development in the mouse. Genes Dev. 17, 380–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ema M., Takahashi S., Rossant J. (2006a). Deletion of the selection cassette, but not cis-acting elements, in taegeted Flk1-lacZ allele reveals Flk1 expression in multipotent mesodermal projenitors. Blood 107, 111–117 [DOI] [PubMed] [Google Scholar]
- Ema M., Yokomizo T., Wakamatsu A., Terunuma T., Yamamoto M., Takahashi S. (2006b). Primitive erythropoiesis from mesodermal precursors expressing VE-cadherin, PECAM-1, Tie2, endoglin, and CD34 in the mouse embryo. Blood 108, 4018–4024 [DOI] [PubMed] [Google Scholar]
- Faloon P., Arentson E., Kazarov A., Deng C. X., Porcher C., Orkin S., Choi K. (2000). Basic fibroblast growth factor positively regulates hematopoietic development. Development 127, 1931–1941 [DOI] [PubMed] [Google Scholar]
- Ferdous A., Caprioli A., Iacovino M., Martin C. M., Morris J., Richardson J. A., Latif S., Hammer R. E., Harvey R. P., Olson E. N., et al. 2009). Nkx2-5 transactivates the Ets-related protein 71 gene and specifies an endothelial/endocardial fate in the developing embryo. Proc. Natl. Acad. Sci. USA 106, 814–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galceran J., Farinas I., Depew M. J., Clevers H., Grosschedl R. (1999). Wnt3a–/–like phenotype and limb deficiency in Lef1(–/–)Tcf1(–/–) mice. Genes Dev. 13, 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaunt S. J., Drage D., Trubshaw R. C. (2005). cdx4/lacZ and cdx2/lacZ protein gradients formed by decay during gastrulation in the mouse. Int. J. Dev. Biol. 49, 901–908 [DOI] [PubMed] [Google Scholar]
- Göttgens B., Barton L. M., Gilbert J. G., Bench A. J., Sanchez M. J., Bahn S., Mistry S., Grafham D., McMurray A., Vaudin M., et al. (2000). Analysis of vertebrate SCL loci identifies conserved enhancers. Nat. Biotechnol. 18, 181–186 [DOI] [PubMed] [Google Scholar]
- Heikinheimo M., Scandrett J. M., Wilson D. B. (1991). Localization of transcription factor GATA-4 to regions of the mouse embryo involved in cardiac development. Dev. Biol. 1640, 361–373 [DOI] [PubMed] [Google Scholar]
- Huber T. L., Kouskoff V., Fehling H. J., Palis J., Keller G. (2004). Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature 432, 625–630 [DOI] [PubMed] [Google Scholar]
- Huelsken J., Vogel R., Brinkmann V., Erdmann B., Birchmeier C., Birchmeier W. (2000). Requirement for beta-catenin in anterior-posterior axis formation in mice. J. Cell Biol. 148, 567–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitobi H., Matsumoto K., Azami T., Itoh F., Itoh S., Takahashi S., Ema M. (2010). Flk1-GFP BAC Tg mice: an animal model for the study of blood vessel development. Exp. Anim. 59, 615–622 [DOI] [PubMed] [Google Scholar]
- Kappel A., Rönicke V., Damert A., Flamme I., Risau W., Breier G. (1999). Identification of vascular endothelial growth factor (VEGF) receptor-2 (Flk-1) promoter/enhancer sequences sufficient for angioblast and endothelial cell-specific transcription in transgenic mice. Blood 93, 4284–4292 [PubMed] [Google Scholar]
- Kattman S. J., Huber T. L., Keller G. M. (2006). Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev. Cell 11, 723–732 [DOI] [PubMed] [Google Scholar]
- Keenan I. D., Sharrard R. M., Isaacs H. V. (2006). FGF signal transduction and the regulation of Cdx gene expression. Dev. Biol. 299, 478–488 [DOI] [PubMed] [Google Scholar]
- Kinder S. J., Tsang T. E., Quinlan G. A., Hadjantonakis A. K., Nagy A., Tam P. P. (1999).The orderly allocation of mesodermal cells to the extraembryonic structures and the anteroposterior axis during gastrulation of the mouse embryo. Development 126, 4691–4701 [DOI] [PubMed] [Google Scholar]
- Kinder S. J., Loebel D. A., Tam P. P. (2001). Allocation and early differentiation of cardiovascular progenitors in the mouse embryo. Trends Cardiovasc. Med. 11, 177–184 [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker N., Willert K., Molenaar M., Roose J., Wagenaar G., Markman M., Lamers W., Destree O., Clevers H. (1998). Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol. Cell. Biol. 18, 1248–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Park C., Lee H., Lugus J. J., Kim S. H., Arentson E., Chung Y. S., Gomez G., Kyba M., Lin S., et al. (2008). ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2, 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Bisgrove B. W., Sawyer H., Hug B., Bell B., Peters K., Grunwald D. J., Stainier D. Y. (1997). The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development 124, 381–389 [DOI] [PubMed] [Google Scholar]
- Lien C. L., Wu C., Mercer B., Webb R., Richardson J. A., Olson E. N. (1999). Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development 126, 75–84 [DOI] [PubMed] [Google Scholar]
- Lindsley R. C., Gill J. G., Kyba M., Murphy T. L., Murphy K. M. (2006). Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 133, 3787–3796 [DOI] [PubMed] [Google Scholar]
- Liu P., Wakamiya M., Shea M. J., Albrecht U., Behringer R. R., Bradley A. (1999). Requirement for Wnt3 in vertebrate axis formation. Nat. Genet. 22, 361–365 [DOI] [PubMed] [Google Scholar]
- Mahlapuu M., Ormestad M., Enerback S., Carlsson P. (2001). The forkhead transcription factor Foxf1 is required for differentiation of extra-embryonic and lateral plate mesoderm. Development 128, 155–166 [DOI] [PubMed] [Google Scholar]
- Merika M., Orkin S. H. (1993). DNA-binding specificity of GATA family transcription factors. Mol. Cell. Biol. 13, 3999–4010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti A., Caron L., Nakano A., Lam J. T., Bernshausen A., Chen Y., Qyang Y., Bu L., Sasaki M., Martin-Puig S., et al. (2006). Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell 127, 1151–1165 [DOI] [PubMed] [Google Scholar]
- Morrisey E. E., Ip H. S., Lu M. M., Parmacek M. S. (1996). GATA-6: a zinc finger transcription factor that is expressed in multiple cell lineages derived from lateral mesoderm. Dev. Biol. 177, 309–322 [DOI] [PubMed] [Google Scholar]
- Motoike T., Markham D. W., Rossant J., Sato T. N. (2003). Evidence for novel fate of Flk1+ progenitor: contribution to muscle lineage. Genesis 35, 153–159 [DOI] [PubMed] [Google Scholar]
- Nemer G., Nemer M. (2003). Transcriptional activation of BMP-4 and regulation of mammalian organogenesis by GATA-4 and -6. Dev. Biol. 254, 131–148 [DOI] [PubMed] [Google Scholar]
- Nimmagadda S., Loganathan P. G., Wilting J., Christ B., Huang R. (2004). Expression pattern of VEGFR-2 (Quek1) during quail development. Anat. Embryol. 208, 219–224 [DOI] [PubMed] [Google Scholar]
- Palis J., Robertson S., Kennedy M., Wall C., Keller G. (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126, 5073–5084 [DOI] [PubMed] [Google Scholar]
- Park C., Afrikanova I., Chung Y. S., Zhang W. J., Arentson E., Fong Gh G., Rosendahl A., Choi K. (2004). A hierarchical order of factors in the generation of FLK1- and SCL-expressing hematopoietic and endothelial progenitors from embryonic stem cells. Development 131, 2749–2762 [DOI] [PubMed] [Google Scholar]
- Peterkin T., Gibson A., Loose M., Patient R. (2005). The roles of GATA-4, -5 and -6 in vertebrate heart development. Semin. Cell Dev. Biol. 16, 83–94 [DOI] [PubMed] [Google Scholar]
- Pilon N., Oh K., Sylvestre J. R., Bouchard N., Savory J., Lohnes D. (2006). Cdx4 is a direct target of the canonical Wnt pathway. Dev. Biol. 289, 55–63 [DOI] [PubMed] [Google Scholar]
- Prabhakar S., Poulin F., Shoukry M., Afzal V., Rubin E. M., Couronne O., Pennacchio L. A. (2006). Close sequence comparisons are sufficient to identify human cis-regulatory elements. Genome Res. 16, 855–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A., De Val S., Heidt A. B., Xu S. M., Bristow J., Black B. L. (2005). Gata4 expression in lateral mesoderm is downstream of BMP4 and is activated directly by Forkhead and GATA transcription factors through a distal enhancer element. Development 132, 3405–3417 [DOI] [PubMed] [Google Scholar]
- Shalaby F., Rossant J., Yamaguchi T. P., Gertsenstein M., Wu X. F., Breitman M. L., Schuh A. C. (1995). Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature 376, 62–66 [DOI] [PubMed] [Google Scholar]
- van Noort M., Clevers H. (2002). TCF transcription factors, mediators of Wnt-signaling in development and cancer. Dev. Biol. 244, 1–8 [DOI] [PubMed] [Google Scholar]
- Winnier G., Blessing M., Labosky P. A., Hogan B. L. (1995). Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 9, 2105–2116 [DOI] [PubMed] [Google Scholar]
- Wu S. M., Fujiwara Y., Cibulsky S. M., Clapham D. E., Lien C. L., Schultheiss T. M., Orkin S. H. (2006). Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 127, 1137–1150 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T. P., Dumont D. J., Conlon R. A., Breitman M. L., Rossant J. (1993). flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development 118, 489–498 [DOI] [PubMed] [Google Scholar]
- Yamashita J., Itoh H., Hirashima M., Ogawa M., Nishikawa S., Yurugi T., Naito M., Nakao K., Nishikawa S. (2000). Flk1-positive cells derived from embryonic stem cells serve as vascular progenitors. Nature 408, 92–96 [DOI] [PubMed] [Google Scholar]
- Yang L., Soonpaa M. H., Adler E. D., Roepke T. K., Kattman S. J., Kennedy M., Henckaerts E., Bonham K., Abbott G. W., Linden R. M., et al. (2008). Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature 453, 524–528 [DOI] [PubMed] [Google Scholar]
- Zhao G. Q. (2003). Consequences of knocking out BMP signaling in the mouse. Genesis 35, 43–56 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


