Summary
Modulating chromatin through histone methylation orchestrates numerous cellular processes. SETD2-dependent trimethylation of histone H3K36 is associated with active transcription. Here, we define a role for H3K36 trimethylation in homologous recombination (HR) repair in human cells. We find that depleting SETD2 generates a mutation signature resembling RAD51 depletion at I-SceI-induced DNA double-strand break (DSB) sites, with significantly increased deletions arising through microhomology-mediated end-joining. We establish a presynaptic role for SETD2 methyltransferase in HR, where it facilitates the recruitment of C-terminal binding protein interacting protein (CtIP) and promotes DSB resection, allowing Replication Protein A (RPA) and RAD51 binding to DNA damage sites. Furthermore, reducing H3K36me3 levels by overexpressing KDM4A/JMJD2A, an oncogene and H3K36me3/2 demethylase, or an H3.3K36M transgene also reduces HR repair events. We propose that error-free HR repair within H3K36me3-decorated transcriptionally active genomic regions promotes cell homeostasis. Moreover, these findings provide insights as to why oncogenic mutations cluster within the H3K36me3 axis.
Graphical Abstract
Highlights
-
•
A role for SETD2 in DSB resection and homologous recombination repair
-
•
Histone H3K36me3 is required for homologous recombination
-
•
SETD2 and RAD51 suppress mutations arising from microhomology-mediated end-joining
-
•
Mutations affecting H3K36me3 levels may promote tumorigenesis
The SETD2 gene encodes the histone H3K36 trimethyltransferase. Pfister et al. now show that human SETD2-dependent H3K36me3 maintains genome stability by promoting error-free DNA repair through homologous recombination (HR). Upon DNA damage, SETD2-depleted cells exhibit reduced DNA resection, impaired recruitment of early HR factors, and increased utilization of the error-prone microhomology-mediated end-joining repair pathway. Eliminating H3K36me3 by overexpressing the oncogene KDM4A also impairs HR. Thus, H3K36me3 suppresses tumorigenesis by promoting accurate DNA repair.
Introduction
DNA double-stranded breaks (DSBs) are potentially lethal lesions if unrepaired, and their misrepair can give rise to genome instability, a hallmark of cancer (Jeggo and Lavin, 2009). To maintain genome stability in response to such lesions, cells employ either homologous recombination (HR) or nonhomologous end-joining (NHEJ) pathways to repair DSBs. HR is initiated by 5′ end resection to generate a 3′ single-stranded DNA (ssDNA) overhang. Resection is a two-step process initiated by removing a short oligonucleotide through the activities of the Mre11-Rad50-Nbs1 (MRN) complex and CtIP. Extensive resection is performed by Exo1 or DNA2-BLM in conjunction with Replication Protein A (RPA), which binds ssDNA and removes secondary structures (Sugiyama et al., 1998, Symington and Gautier, 2011). This, in turn, leads to RAD51 nucleofilament formation, which promotes strand invasion of a homologous chromatid, leading to accurate repair (Heyer et al., 2010). During classical NHEJ (C-NHEJ), the broken ends are rapidly bound and protected by the Ku70/Ku80 heterodimer, which acts as a platform to recruit the DNA-PK catalytic subunit (DNA-PKcs). Damaged ends are then processed and subsequently joined by the Ligase 4 (Lig4), XRCC4, XLF complex in a template-independent manner, which can lead to inaccurate repair (Lieber, 2010). DSBs may also be repaired through alternative end-joining pathways, such as microhomology-mediated end-joining (MMEJ), which do not require Ku or Lig4. Like HR, MMEJ is initiated by resection, and end-joining is mediated through annealing of short direct repeats of microhomology. MMEJ leads to deletions and is frequently associated with chromosomal rearrangements (McVey and Lee, 2008).
DSB repair is further facilitated through chromatin modification by chromatin remodeling complexes, by incorporation of histone variants, and by histone modification (Smeenk and van Attikum, 2013). Histone H3K36 dimethylation has recently been proposed to facilitate NHEJ, where Metnase (SETMAR) directly mediates H3K36 dimethylation near the break site, leading to recruitment and stabilization of NBS1 and Ku70 (Fnu et al., 2011). Histone H3K36 is also trimethylated, which in mammalian cells is performed uniquely by the SETD2/HYPB methyltransferase (Edmunds et al., 2008). H3K36me3 is associated with transcriptional elongation and is found in gene coding regions, peaking at 3′ ends (Edmunds et al., 2008). SETD2-dependent H3K36 trimethylation facilitates a number of processes within the cell, including splicing, repression of intragenic transcripts, and chromatin accessibility (Li et al., 2013, Wagner and Carpenter, 2012). SETD2 is also mutated in a number of cancer types, including breast, lung, acute lymphoblastic leukemia, clear cell renal cell carcinoma, and glioma, supporting its role as a tumor suppressor (Al Sarakbi et al., 2009, Dalgliesh et al., 2010, Fontebasso et al., 2013, Newbold and Mokbel, 2010, Zhang et al., 2012). SETD2-dependent H3K36 trimethylation has recently been shown to regulate DNA mismatch repair (Li et al., 2013). However, SETD2-deficient cancers exhibit a wide range of mutations, including insertions, deletions (indels), and chromosomal aberrations (Sato et al., 2013, Zhu et al., 2014), suggesting an additional role for SETD2 in genome stability.
Additionally, H3K36 methylation is regulated by the KDM4/JMJD2 family of histone demethylases. These contain JmjN-JmjC and tandem-Tudor domains that specifically remove the tri- and dimethyl forms of both H3K9 and H3K36, with preference for the trimethyl form being observed in the case of KDM4A/JMJD2A (Couture et al., 2007, Klose et al., 2006, Whetstine et al., 2006). KDM4 family proteins are frequently overexpressed in cancers and are associated with poor patient survival (Berry and Janknecht, 2013, Black et al., 2013).
Here, we investigate the role of SETD2-dependent H3K36me3 in maintaining genome stability. We establish a role for H3K36me3 in HR repair by facilitating resection. In addition, we define a role for SETD2 and RAD51 in maintaining genome stability through suppressing MMEJ at break sites.
Results
SETD2 Suppresses Break-Induced Mutations
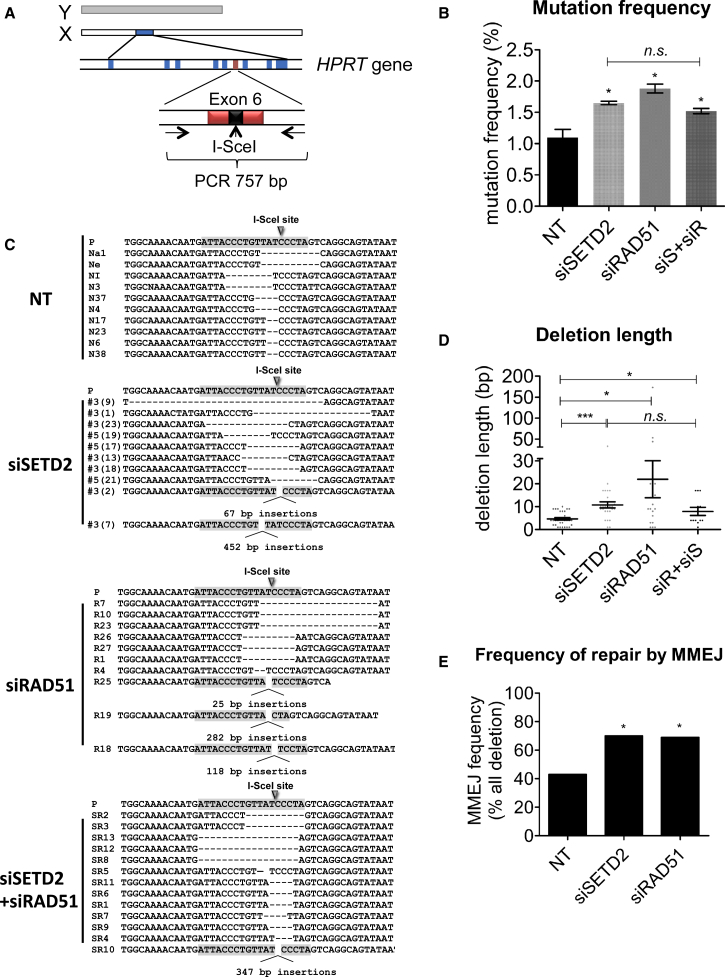
Tumor suppressors can suppress carcinogenesis by preventing genome instability, a hallmark of cancer. To investigate the impact of loss of SETD2, a tumor suppressor, on genome stability following DSB induction, we developed an I-SceI-induced loss of function assay (HPRT+:I-SceI). Intron-encoded endonuclease 1 from Saccharomyces cerevisiae (I-SceI) is a rare-cutting endonuclease that has no predicted recognition site in the mammalian genome (Jasin, 1996). An I-SceI recognition sequence was inserted into the endogenous HPRT exon 6 in HT1080 (fibrosarcoma) cells. The I-SceI site maintains the HPRT reading frame and makes only a single amino acid change that does not impair the function of HPRT (Figure 1A). Following I-SceI induction, inaccurate DSB repair generates HPRT-negative mutants, which can be selected for using 6-thioguanine (6-TG), and indels can be detected by PCR amplification using primers flanking the break site (Figure 1A). Following small interfering RNA (siRNA) knockdown and I-SceI induction, the frequency of HPRT loss (mutation frequency) in SETD2-depleted cells was 1.65%, significantly higher (p = 0.0014) than that of nontargeting controls (1.1%). These findings resembled those in RAD51-depleted cells in which the frequency of HPRT loss was also significantly increased to 1.88% (p = 0.0022) compared to nontargeting controls (Figure 1B). SETD2 and RAD51 codepletion (siS+SiR) also significantly increased the mutation frequency to 1.52% (p = 0.037), which was similar to the frequency observed following either SETD2 or RAD51 depletion (Figure 1B). We confirmed that I-SceI protein expression and cleavage efficiency were the same in cells transfected with either control siRNA or SETD2 siRNA (Figures S1A–S1C). In addition, HPRT gene transcription was not affected by SETD2 knockdown, as shown by quantitative RT-PCR (qRT-PCR) of spliced and unspliced HPRT mRNA (Figures S1D and S1E). Therefore, the mutation frequency in this system is likely to be directly associated with misrepair of the DSB at the I-SceI site.
Figure 1.
SETD2- and RAD51-Depleted Cells Exhibit a Common Break-Induced Mutation Signature
(A) Schematic map of the HPRT+:I-SceI assay, where arrows indicate PCR primers used to amplify genomic DNA to allow sequencing across the break site (see Supplemental Experimental Procedures).
(B) Mutation frequency of nontargeting control (NT), SETD2-depleted, RAD51-depleted, and SETD2/RAD51-codepleted cells (siS+siR) after I-SceI induced DSB. Error bars represent SEM and ∗p < 0.05; n.s., not significant.
(C) Representative sequence alignments of the above-mentioned PCR products in nontargeting control (NT), SETD2-depleted, RAD51-depleted, and SETD2/RAD51-codepleted backgrounds, respectively.
(D) Average length of deletions (bp) in different backgrounds, where each dot represents an independent clone. The lines represent mean and SEM, ∗p < 0.05, ∗∗∗p < 0.001.
(E) Frequency of repair by MMEJ in deletion mutants isolated from different backgrounds. p values calculated by statistical analysis “difference between proportions,” ∗p < 0.05. See also Figure S1.
Further, we studied the mutation patterns across the break site of 30 individually isolated HPRT-negative clones from each background by PCR amplification and sequencing. HPRT-negative clones from cells treated with nontargeting control siRNA indicated the presence of microdeletions of 2–5 bp, consistent with DSB repair by C-NHEJ. Larger microdeletions were also observed that were associated with regions of microhomology located either side of the break site, consistent with DSB repair through MMEJ (Figures 1C and S1F).
Sequence analysis indicated that SETD2-depleted cells exhibited a significant increase in average deletion length (11 bp, p < 0.0001) compared to control cells (5 bp) (Figures 1C and 1D). RAD51-depleted cells exhibited a further significant increase in the average deletion length (22 bp, p = 0.014) (Figures 1C and 1D). RAD51 and SETD2 codepletion generated deletion lengths (an average of 8 bp) comparable to those observed following SETD2 knockdown, which in turn was significantly higher than the nontargeting control (p = 0.026) (Figures 1C and 1D). Insertions at the HPRT break site were also detected. However, the frequency and size of the insertions was not significantly altered between SETD2-depleted cells, SETD2/RAD51 codepleted and nontargeting controls.
We next examined the mechanism for misrepair in cells exhibiting deletions. We found that the frequency of microhomology-mediated end-joining (MMEJ) was significantly increased in SETD2 and RAD51-depleted cells (p < 0.05) (Figures 1E and S1G). This suggested that HR-deficient cells use MMEJ as an alternative repair mechanism, which gives rise to a mutation signature of sequence loss between regions of microhomology. These results support a role for SETD2 in tumor suppression by maintaining genome stability through preventing deleterious mutations arising in response to DSBs. These findings further identify a similar break-induced mutation signature between SETD2- and RAD51-depleted cells, suggesting an early role for SETD2 in HR repair.
SETD2 Is Required for Homologous Recombination Repair
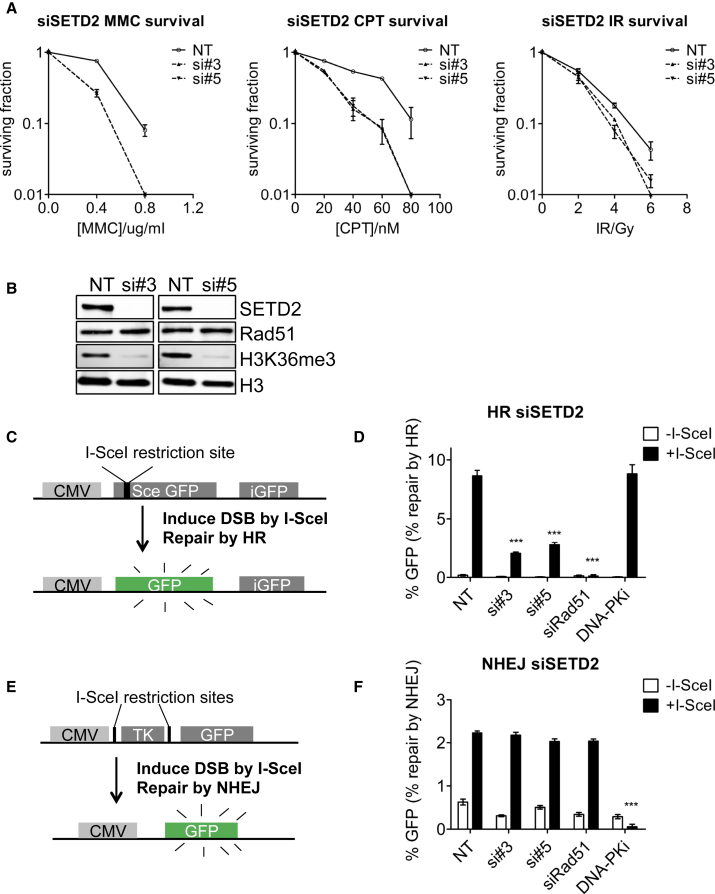
To test the role for SETD2 in DSB repair, we depleted SETD2 in U2OS and HeLa cells using two independent siRNAs and measured the clonogenic survival following exposure to DNA-damaging agents. SETD2 knockdown significantly reduced survival after treatment with mitomycin C (MMC), camptothecin (CPT), or ionizing radiation (IR) in U2OS cells and HeLa cells compared to nontargeting controls (Figures 2A and S2A). These results identify a role for SETD2 in the cellular response to DNA damage and are consistent with a role for SETD2 in DSB repair.
Figure 2.
SETD2 Is Required for Homologous Recombination
(A) Clonogenic survival of SETD2 knockdown (si#3 and si#5) or nontargeting control (NT) U2OS cells treated with indicated concentrations of MMC, CPT, and IR. Error bars show SEM from three independent experiments.
(B) Western blots showing levels of SETD2, RAD51, and H3K36me3 72 hr after siRNA transfection.
(C) Schematic map of the DR-GFP cassette for assessing HR efficacy.
(D) HR repair efficacy of reporter cells treated with nontargeting control siRNA (NT), SETD2 siRNAs (si#3 and si#5), RAD51 siRNA (siRad51), or DNA-PK inhibitor NU7441 (Axon), indicated by the percentage of GFP-positive cells. Error bars show SEM from three independent experiments. ∗∗∗p < 0.001.
(E) Schematic map of the NHEJ cassette.
(F) NHEJ repair efficacy of reporter cells treated with nontargeting control siRNA (NT), SETD2 siRNAs (si#3 and si#5), RAD51 siRNA (siRAD51), or DNA-PK inhibitor NU7441, indicated by the percentage of GFP-positive cells. Error bars show SEM from three independent experiments. ∗∗∗p < 0.001. See also Figure S2.
SETD2 knockdown was validated by western blotting (Figure 2B). Western blotting also showed that SETD2 knockdown significantly reduced global levels of H3K36me3, confirming SETD2 as the major H3K36me3 methyltransferase in human cells (Figure 2B). Because RAD51 is more likely to be affected among DNA repair genes by siRNA off-target effects (Adamson et al., 2012), we also confirmed that SETD2 knockdown by siRNA did not affect the RAD51 protein levels (Figure 2B). We confirmed that SETD2 knockdown had no effect on the expression of major HR proteins, including BRCA2, RAD50, CtIP, MRE11, and RPA and the NHEJ protein KU80 (Figure S2B). We also observed no changes to the cell cycle after SETD2 knockdown (Figure S2C).
Both MMC and CPT induce S phase-specific DSBs, which are predominantly repaired by HR (Arnaudeau et al., 2001, Moynahan et al., 2001). To test a possible role for SETD2 in HR repair, we used a well-characterized GFP-based reporter for HR (DR-GFP). The HR reporter contains an I-SceI recognition sequence, which upon I-SceI expression is cleaved to generate a DSB. DSB repair by HR using a direct repeat within the reporter cassette as a template results in an intact GFP gene (Figure 2C)(Pierce et al., 1999). SETD2 knockdown by two independent siRNAs significantly reduced HR repair of an I-SceI induced DSB by 67%–77% compared to nontargeting control cells (p < 0.0003) (Figure 2D). As a control for the HR reporter, knockdown of RAD51 reduced HR activity by 97% (p < 0.0001), whereas DNA-PKcs inhibitor NU7441 had no effect on HR (Figure 2D). In contrast, SETD2 knockdown had no effect on repair of an I-SceI induced DSB in an NHEJ reporter (IRES-TK-EGFP) in which joining of two I-SceI sites separated by a thymidine kinase gene (TK) results in GFP expression (Figures 2E and 2F)(Ogiwara et al., 2011). As a control, RAD51 had no effect on the same NHEJ reporter, whereas the DNA-PK inhibitor NU7441 reduced NHEJ activity by 98% (p < 0.0001) (Figure 2F). These results identify a role for SETD2 in HR repair.
An Early Role for SETD2 in Homologous Recombination
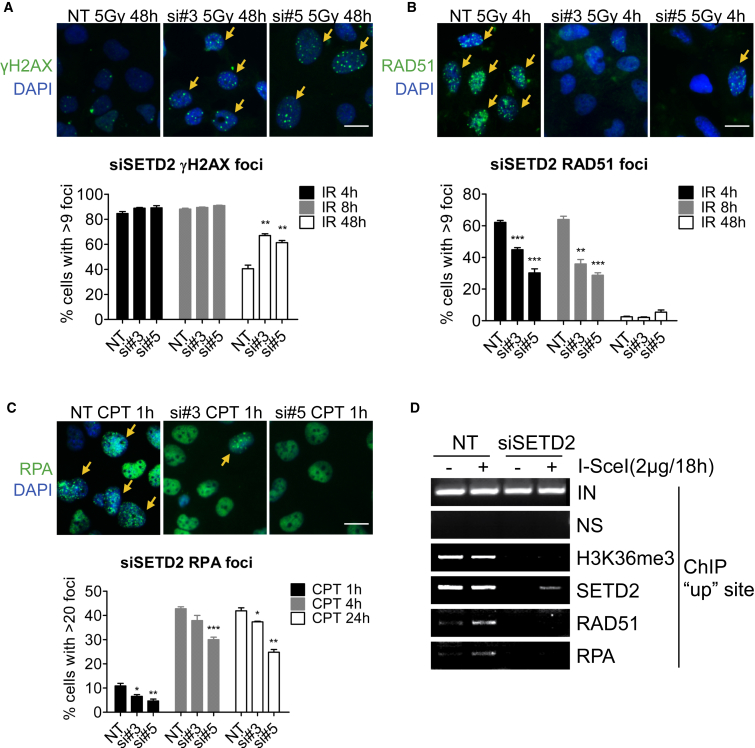
To further investigate the role of SETD2 in DSB repair, we examined the rate of repair following IR. We used γH2AX foci as a marker of DNA damage and found that following exposure to 5 Gy IR, SETD2 knockdown significantly delayed removal of γH2AX foci at 48 hr (p < 0.003) compared to nontargeting controls, supporting a role for SETD2 in efficient DSB repair (Figure 3A). We next investigated the effect of SETD2 depletion on the recruitment of HR repair proteins following DNA damage. SETD2 knockdown resulted in significantly reduced RAD51 foci formation 4 and 8 hr after exposure to IR compared to nontargeting controls (p < 0.0013) (Figure 3B). SETD2 knockdown also significantly reduced RAD51 foci formation after exposure to CPT (p < 0.05) (Figure S2D).
Figure 3.
SETD2 Is Required for the Recruitment of RPA and RAD51
(A and B) γH2AX (A) or RAD51 (B) foci formation at indicated times after IR (5 Gy) in U2OS cells treated with nontargeting control siRNA (NT) or SETD2 siRNAs (si#3 and si#5). Error bars represent SEM from three independent experiments.
(C) RPA32 foci formation at indicated times after treatment with CPT (10 μM) in U2OS cells transfected with control siRNA (NT) or SETD2 siRNAs (si#3 and si#5). Representative fluorescent images are shown; scale bar, 20 μm. For each condition, 45 fields and at least 500 cells were examined by Incell (GE Healthcare). Error bars show SEM from three independent experiments. ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05.
(D) ChIP analysis on DR-GFP U2OS cells transfected with nontargeting control (NT) or SETD2 siRNA for 72 hr, followed by transfection of either vector or I-SceI plasmid for a further 18 hr, as indicated. ChIP on the “up” DNA site of the DR-GFP cassette was performed on the lysate using antibodies against nonspecific Ig (NS), H3K36me3 (Abcam), SETD2 (Abcam), RAD51 (Santa Cruz Biotechnology), or RPA (Millipore); inputs (IN) are also indicated.
See also Figures S2 and S3.
We also examined the effect of SETD2 depletion on RPA recruitment, where we induced DSBs in S phase cells using CPT or MMC. A significant reduction in RPA foci formation was observed after CPT or MMC treatment following SETD2 knockdown compared to nontargeting controls (p < 0.023) (Figures 3C and S2E). SETD2 knockdown leads to reduced H3K36me3 in the nuclei as confirmed by costaining the cells with antibodies against H3K36me3 and RPA (Figure S2F). Together, these findings support a role for SETD2 in promoting HR through facilitating recruitment of RPA and RAD51 to DNA damage sites.
To further confirm the impact of SETD2 loss on the recruitment of HR factors to the site of DSBs, we performed chromatin immunoprecipitation (ChIP) near the I-SceI site in the DR-GFP cassette (Figure S3A). The I-SceI site is located within the DR-GFP cassette under the control of the highly active CMV promoter, and thus H3K36me3, which is associated with active transcription, is predicted to be present throughout the cassette. After induction of DSBs by I-SceI, no change in H3K36me3 or SETD2 levels was detected proximal or distal to the break site after 8, 24, or 48 hr following I-SceI transfection (Figure S3B). The results were also quantified by ChIP-qPCR (Figure S3C). Although other groups also observed no changes to H3K36me3 level after DSB, globally (Fnu et al., 2011) or locally at the break site (Pei et al., 2011, Aymard et al., 2014), we were surprised by the lack of reduction in H3K36me3 or SETD2 knowing that regional transcriptional silencing has been observed at sites of DSBs (Shanbhag et al., 2010). We speculate that the unexpected maintenance of H3K36me3 at the DSB site by SETD2 may serve as an anchor to mediate HR repair.
We monitored recruitment of major HR factors to the DSB site in the ChIP system. RPA and RAD51 were recruited to the DSB site, resulting in increased binding to the “up” site 18 hr after I-SceI transfection, compared to uncut controls (Figure 3D). SETD2 depletion substantially reduced the recruitment of RPA and RAD51 after the I-SceI cut (Figure 3D). SETD2 protein depletion was validated by western blotting (Figure S3D), and was further confirmed by the absence of SETD2 or H3K36me3 at the break site (Figure 3D). These findings are consistent with the RPA and RAD51 foci data, indicating that SETD2 is required to recruit RPA and RAD51 to DSB sites. Together, these findings confirm a presynaptic role for SETD2 in facilitating HR repair of DSBs.
H3K36me3 Is Required for Homologous Recombination Repair
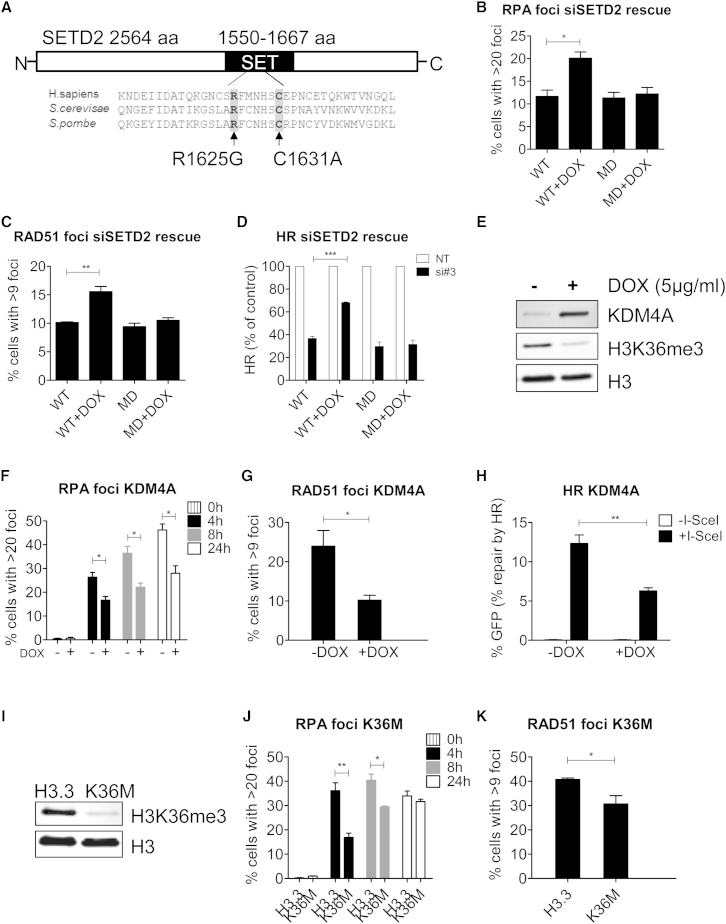
To investigate if the methyltransferase activity of SETD2 is required for its role in HR, we generated two doxycycline-inducible U2OS cell lines, where either a wild-type SETD2 cDNA or a methyltransferase-dead SETD2 cDNA was inserted behind a Tet operator in the genome of U2OS cells. Upon addition of doxycycline to the cell culture medium, the Tet operator was derepressed and the wild-type or mutant SETD2 was expressed. Both cDNA constructs were mutated so that they are refractory to SETD2 siRNA#3. The methyltransferase-dead SETD2 was mutated at two amino acid residues located within the catalytic site of the SET domain, which are conserved with yeast Set2. The corresponding mutations in yeast were shown to abolish the methyltransferase activity (Rea et al., 2000, Strahl et al., 2002) (Figure 4A). Both arginine (R) and cysteine (C) residues were mutated in the methyltransferase-dead (MD) cDNA by site-directed mutagenesis (see Supplemental Experimental Procedures). Western blot analysis confirmed that doxycycline-induced expression of wild-type SETD2 could restore the level of H3K36me3 after siSETD2 treatment, whereas expression of methyltransferase-dead SETD2 could not restore the level of H3K36me3 (Figure S4A).
Figure 4.
H3K36me3 Is Required for Efficient HR Repair
(A) Schematic map showing site-directed mutagenesis for abolishing the methyltransferase activity of SETD2, with both R and C residues mutated simultaneously. Sequence alignment of human SETD2 and Saccharomyces cerevisiae Set2 and Schizosacchromyces pombe Set2 amino acid sequences reveals evolutionarily conserved residues that reside in the SET domain.
(B) RPA32 foci formation at 2 hr after treatment with CPT (10 μM) in cells transfected with SETD2 siRNA. WT: T-REx U2OS clone (see Experimental Procedures) with wild-type SETD2 cDNA integrated but not expressing exogenous SETD2. WT+DOX: the same WT clone expressing exogenous wild-type SETD2. MD: T-REx U2OS clone with methyltransferase-dead mutant SETD2 cDNA integrated but not expressing exogenous SETD2. MD+DOX: the same MD clone expressing exogenous mutant SETD2. Error bars show SEM from three independent experiments, ∗p < 0.05.
(C) RAD51 foci formation at 4 hr after IR (5 Gy) in cells transfected with SETD2 siRNA. The clones used are the same as in (B). Error bars show SEM from three independent experiments, ∗∗p < 0.01.
(D) HR efficacy in cells transfected with NT or SETD2 siRNA. The clones used are the same as in (B); error bars show SEM from three independent experiments, ∗∗∗p < 0.001.
(E) Western blot showing levels of KDM4A, H3K36me3 and H3 following KDM4A induction with 5 μg/ml doxycycline (DOX) for 72 hr in the KDM4A T-REx U2OS cells (see Experimental Procedures).
(F) RPA32 foci formation at indicated time after 2 hr CPT (10 μM) treatment in KDM4A T-REx cells treated with or without 5 μg/ml doxycycline (DOX). Error bars show SEM from three independent experiments, ∗p < 0.05.
(G) RAD51 foci formation 8 hr after IR (5 Gy) in KDM4A T-REx cells treated with or without 5 μg/ml doxycycline (DOX). Error bars show SEM from three independent experiments. ∗p < 0.05.
(H) HR repair efficacy in KDM4A T-REx DR-GFP U2OS cells treated with or without 5 μg/ml doxycycline (DOX). Error bars show SEM for three independent experiments. ∗∗p < 0.01.
(I) Western blot showing levels of H3K36me3 and H3 in U2OS cells with stably integrated H3.3 (control) or H3.3K36M lentiviral construct (see Supplemental Experimental Procedures).
(J) RPA32 foci formation at indicated time after 2 hr CPT (10 μM) treatment in H3.3 or H3.3K36M stable expression cells. Error bars show SEM from three independent experiments.
(K) RAD51 foci formation 8 hr after IR (5 Gy) in H3.3 or H3.3K36M stable expression cells. Error bars show SEM for three independent experiments, ∗p < 0.05. See also Figure S4.
We depleted endogenous SETD2 with siRNA#3 and examined the rescue of the phenotype by the two constructs. Although the wild-type construct increased foci formation of RPA and RAD51 after DNA damage in siSETD2 cells, the methyltransferase-dead mutant did not (Figures 4B and 4C). This is consistent with the observation that the wild-type construct partially restored HR efficiency whereas the methyltransferase-dead (MD) mutant did not (Figure 4D). Five to six clones containing each construct were independently isolated and tested. All WT clones were able to restore HR efficiency, but none of the MD clones showed rescue (Figures S4B and S4C).
The observation that the methyltransferase-dead SETD2 mutant could not rescue HR efficiency suggested that H3K36me3 is required for efficient HR. To test this further, we used SETD2-independent approaches to disrupt H3K36me3 levels. First, we overexpressed KDM4A, an oncogene and a demethylase specific for tri- and dimethylated histone H3K36 and H3K9, predicting that overexpression of KDM4A would reduce the level of H3K36me3 and thus impair HR. Therefore, a doxycycline-inducible KDM4A-expression U2OS cell line was generated. Upon addition of doxycycline, KDM4A was overexpressed, which led to a global reduction in H3K36me3 levels (Figure 4E). We then compared the recruitment of the HR factors in this cell line with or without KDM4A overexpression. We found that KDM4A overexpression inhibited the formation of RPA and RAD51 foci after DNA damage (Figures 4F and 4G). Accordingly, KDM4A overexpression also reduced HR efficiency as assessed by the HR reporter (p = 0.0078) (Figure 4H). Using the same cell line without KDM4A cDNA integration, we found that doxycycline itself had no effect on HR efficiency of U2OS cells (Figure S4D). These findings indicate that overexpression of the oncogene KDM4A significantly reduces levels of H3K36me3 and HR.
Second, we utilized a dominant-negative mutation in the histone H3.3 gene (H3.3K36M) that was recently shown to reduce H3K36me3 specifically without affecting other histone methylations (Lewis et al., 2013). Accordingly, in cells expressing the mutant H3.3K36M transgene, global levels of H3K36me3 were depleted compared to the wild-type H3.3 control (Figure 4I). Cells expressing H3.3K36M also exhibited delayed RPA and RAD51 foci formation following DNA damage (Figures 4J and 4K), consistent with similar delays seen in KDM4A overexpressing cells or SETD2-depleted cells. Neither KDM4A overexpression nor H3.3K36M expression affected cell cycle progression as assessed by BrdU incorporation (Figures S4E and S4F), thus excluding the possibility that the reduction in RPA, RAD51 foci formation and HR efficacy was due to cell cycle effects. The common feature of all three different systems (SETD2 knockdown and rescue, KDM4A overexpression, and H3.3K36M expression) was the reduction in the recruitment of HR proteins and HR efficiency associated with reduced H3K36me3. Together these findings confirm a role for H3K36me3 in HR.
SETD2 Promotes DSB End Resection
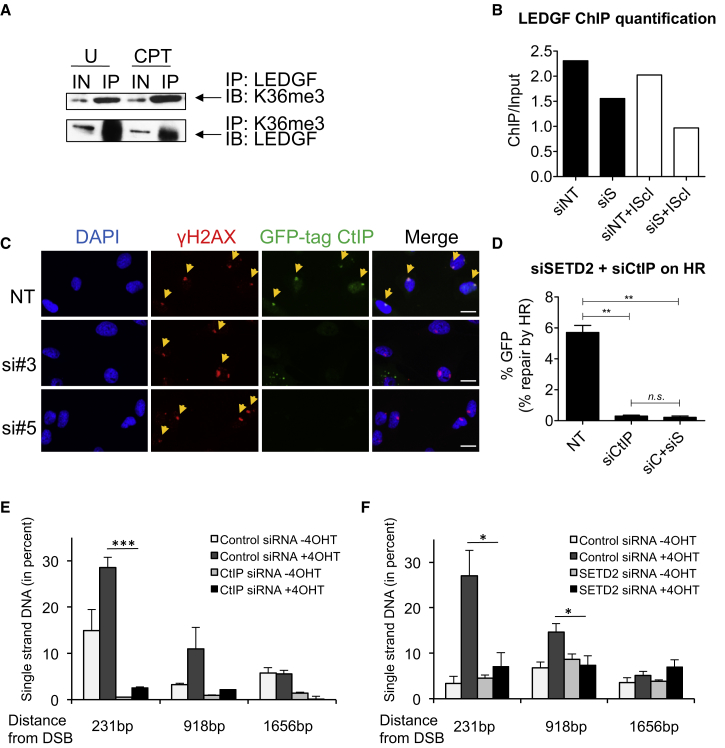
Lens epithelium-derived growth factor p75 (LEDGF) binds to H3K36me3 constitutively through its PWWP domain. Upon DNA damage LEDGF recruits CtIP, which facilitates the resection step during HR repair (Daugaard et al., 2012). Accordingly, we were able to coprecipitate LEDGF and H3K36me3, independently of DNA damage in U2OS cells (Figure 5A). LEDGF binding to chromatin was reduced upon SETD2 depletion, consistent with preferential binding of LEDGF to H3K36me3 (Eidahl et al., 2013), and was DSB independent (Figure 5B). We predicted that the reduced LEDGF binding following SETD2 depletion would reduce recruitment of CtIP upon damage. We therefore used micro-irradiation to induce localized DSBs and studied the recruitment of CtIP using a U2OS cell line expressing GFP-tagged CtIP (Sartori et al., 2007). We found CtIP to be localized to sites of DNA damage marked by γH2AX (Figure 5C). In contrast, SETD2 knockdown resulted in reduced recruitment of CtIP to the DNA damage site as marked by γH2AX (Figure 5C). If SETD2 facilitates HR through LEDGF binding and CtIP recruitment, we would expect SETD2 to be epistatic with CtIP. Indeed, codepletion of SETD2 and CtIP (siC+siS) showed the same level of reduction in HR as CtIP-depletion alone (Figures 5D and S5A). These findings are consistent with the hypothesis that SETD2-dependent H3K36me3 promotes recruitment of CtIP by LEDGF to break sites.
Figure 5.
SETD2 Promotes HR through LEDGF/CtIP-Facilitated Resection
(A) Coimmunoprecipitation of LEDGF and H3K36me3 in the presence or absence of DNA damage. U2OS cells were left untreated (U) or treated with CPT (15 μM for 4 hr). Reciprocal co-IPs are shown.
(B) ChIP analysis of LEDGF binding to the DSB site before and after I-SceI induction in NT or SETD2 knockdown DR-GFP cells. siNT, control siRNA-treated cells before cut; siS, SETD2 siRNA-treated cells before cut; siNT+IScI, control siRNA-treated cells after cut; siS+IScI: SETD2 siRNA-treated cells after cut. Numbers were quantified from one ChIP-PCR experiment and show LEDGF-ChIP over input.
(C) Microirradiation showing CtIP recruitment to the damage site in U2OS cells treated with nontargeting control siRNA (NT) or SETD2 siRNAs (si#3 and si#5). Fluorescent images were acquired using a confocal microscope (Zeiss); scale bars, 20 μm.
(D) HR efficacy (as measured by GFP reporter assay as in Figure 2D) in cells transfected with control siRNA (NT) or CtIP siRNA (siCtIP) or both CtIP and SETD2 siRNA (siC+siS). Error bars show SEM from three independent experiments, ∗∗p < 0.01, n.s. not significant.
(E) CtIP depletion impedes resection at an AsiSI induced DSB. DNA was extracted from 4OHT-treated or untreated DIvA cells, transfected with control or CtIP siRNA, as indicated. Resection was analyzed, using a protocol detailed in Zhou et al. (2014), at an AsiSI-induced DSB reported to be repaired by a RAD51-dependent pathway (DSB-II in Aymard et al., 2014). The mean and SEM (n = 4, technical replicate) of a representative experiment are presented.
(F) SETD2 depletion impedes resection at an AsiSI induced DSB. DNA was extracted from 4OHT-treated or untreated DIvA cells, transfected with control or SETD2 siRNA, as indicated. Same as in (E) except that cells were transfected with a siRNA directed against SETD2. The mean and SEM (n = 4, technical replicate) of a representative experiment are presented. See also Figure S5.
Reduced CtIP recruitment, together with the reduced RPA foci formation following SETD2 depletion, suggested that SETD2-dependent H3K36 trimethylation might promote DSB resection. To directly measure resection, we used an in vivo resection assay (Zhou et al., 2014) in which levels of ssDNA were assessed at increasing distances from a specific DSB induced by the AsiSI restriction enzyme, which was found to recruit RAD51 (Iacovoni et al., 2010, Aymard et al., 2014). Following DSB induction (+4OHT), ssDNA was readily detected 231 bp from the AsiSI break site (DSB-II in Aymard et al., 2014) and at reduced levels further from the break site. Following CtIP depletion, ssDNA levels were significantly reduced at 231 bp (p = 2.2 × 10−5) and 918 bp (p = 0.057) from the break site compared to control siRNAs (Figure 5E). Following SETD2 depletion, ssDNA levels were also significantly reduced at 231 bp (p = 0.021) and 918 bp (p = 0.037) from the break site compared to control siRNAs (Figure 5F), and SETD2 depletion did not affect DSB induction at this site as assayed by XRCC4 ChIP (Figure S5B), thus confirming a role for SETD2 in facilitating DSB resection.
Discussion
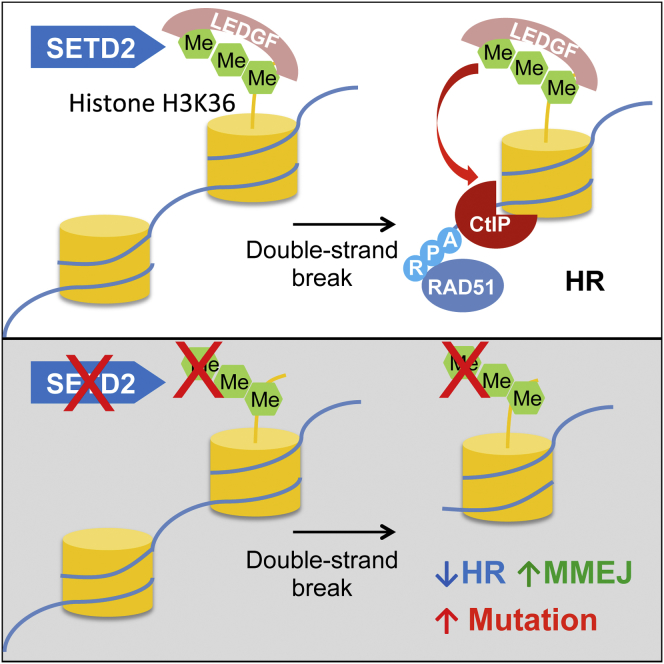
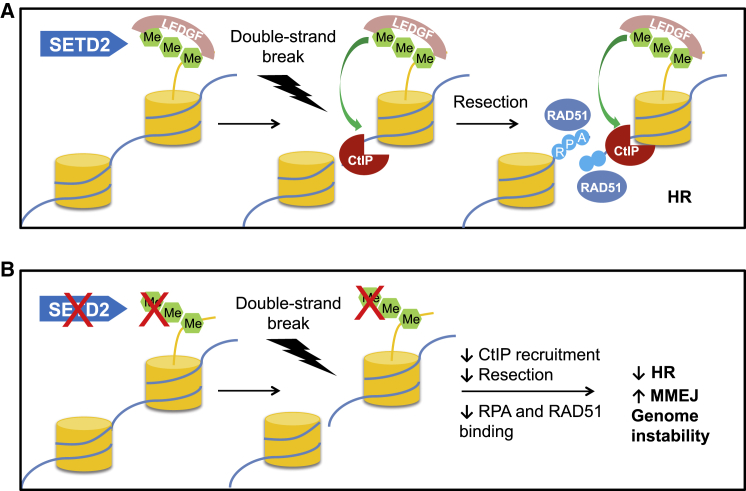
The SETD2 gene encoding the H3K36 trimethyltransferase is among the most mutated in human cancers (Lawrence et al., 2014). Yet, how SETD2 functions to suppress the genome instability frequently observed in SETD2-deficient cancer cells is unknown (Sato et al., 2013, Zhu et al., 2014). Here, we describe a role for SETD2-dependent H3K36 trimethylation in facilitating error-free DSB repair and genome stability through efficient HR. Our data support a model in which LEDGF is constitutively anchored to H3K36me3-decorated chromatin through its PWWP domain. Upon DNA damage, chromatin-bound LEDGF recruits CtIP, which promotes resection—an essential step in HR repair (Figure 6A). In the absence of SETD2-dependent H3K36me3, the chromatin association of LEDGF is compromised, and DNA damage-induced CtIP recruitment is impaired, leading to reduced resection and HR and an increase in error-prone MMEJ and subsequent genome instability (Figure 6B). Our data do not, however, exclude the possibility that other H3K36me3 binding factors may also contribute to efficient HR.
Figure 6.
Model for the Role of SETD2-Dependent H3K36me3 in HR and Genome Stability
(A) SETD2-dependent H3K36 trimethylation constitutively recruits LEDGF to chromatin. Following DNA double-strand break, LEDGF recruits CtIP, thereby promoting resection, facilitating RPA and RAD51 recruitment.
(B) In the absence of H3K36 trimethylation, lack of LEDGF chromatin association results in reduced recruitment of CtIP after double-strand breaks, impairing resection and HR, leading to elevated levels of MMEJ and genome instability.
The involvement of H3K36me3 in HR raises the question as to whether this histone mark is DSB inducible or preset to determine the choice of repair pathways. We did not detect any appreciable increase in H3K36me3 levels at the break site following I-SceI induction. These findings are consistent with previous observations indicating that H3K36me3 levels do not change following DNA damage, either globally (Fnu et al., 2011) or locally (Aymard et al., 2014, Pei et al., 2011). Given that SETD2-dependent H3K36 trimethylation strongly correlates with actively transcribed regions of the genome (Edmunds et al., 2008, Yoh et al., 2008), our findings thus support a spatial link between HR and transcriptionally active, H3K36me3 decorated regions of the genome. Indeed, because HR repair is usually error free, this would be expected to protect genome integrity following DSB induction or replication fork collapse at such sites, which would be important in maintaining cell homeostasis.
Genome-wide sequencing has suggested certain mutation signatures are associated with specific cancer types (Alexandrov et al., 2013) but lacks the power to identify driver mutations that give rise to that mutation signature. Here, we use a system to determine the impact of individual gene dysfunction on mutation patterns arising during the repair of a single DSB. We show that, by depleting SETD2 or RAD51 HR proteins, DSB induction leads to significantly increased mutation frequencies associated with deletions within six cell cycles (5 days). These findings identify a role for SETD2, like RAD51, in suppressing break-induced mutations. Further mechanistic analysis indicated that deletions arising from SETD2 or RAD51 depletion resulted from a significant increase in error-prone MMEJ. We propose that MMEJ arises from failed HR. SETD2 loss results in partial resection, short ssDNA 3′ ends and reduced RPA foci. These partially resected ends are insufficient to promote efficient HR and are no longer substrates for C-NHEJ. Instead, these partially resected ends become substrates for MMEJ. Similarly, following RAD51 depletion, whereas resection proceeds normally, HR is blocked presynaptically, resulting in the accumulation of longer unrepaired resected ends. Such ends are no longer substrates for C-NHEJ, and, because HR cannot proceed, the ends are instead rejoined by MMEJ. Extensive resection following RAD51 loss facilitates annealing between regions of microhomology further from the break site, thus leading to larger deletions than those observed for SETD2. The observations that HR was significantly reduced whereas C-NHEJ was unchanged following SETD2 or RAD51 depletion are consistent with this model. Together, these findings suggest a role for HR in maintaining genome stability through suppression of MMEJ. This “real-time” mutation study can therefore provide important insights into how loss of these genes contributes to somatic mutation and intratumor heterogeneity, and it highlights the importance of HR repair genes in maintaining genome stability.
SETD2 is a tumor suppressor and its loss is an important step in the progression of a number of cancer types (Dalgliesh et al., 2010, Fontebasso et al., 2013, Gerlinger et al., 2012). Our findings provide mechanistic insights into how SETD2 functions as a tumor suppressor. Moreover, all three Tudor domain-containing KDM4/JMJD2 proteins (A, B, and C) are frequently overexpressed in various cancers, including breast, colorectal, lung, and prostate (Berry and Janknecht, 2013). Our findings predict that overexpression of these KDM4 family members causes genome instability and tumorigenesis through loss of H3K36me3-mediated HR repair. The identification of a role for H3K36me3 in facilitating HR repair, in addition to its recently described role in mismatch repair (Li et al., 2013), establishes the H3K36me3 chromatin mark as an important functional node for maintaining genome stability.
Experimental Procedures
I-SceI-Inducible HPRT Mutation Assay
The reporter HT1080 cell line with a functional but I-SceI-cleavable HPRT gene was generated by inserting an 18 bp I-SceI recognition site into exon 6 of the HPRT gene using gene targeting. Reporter cells were transfected with the I-SceI plasmid 48 hr after siRNA knockdowns. The cells were allowed to repair in a nonselective medium for 5 days before seeding and selecting for HPRT-negative cells using 6-TG (15 μg/ml).
The mutation frequency was calculated as the number of HPRT-negative colonies divided by number of cells plated, after correcting for plating efficiency. To determine the mutation sequence, individual single-cell colonies were isolated, and genomic DNA spanning the DSB junction was PCR-amplified using the primers flanking the I-SceI site. Sequence alignment of the PCR products was conducted using the DNA Data Bank of Japan (DDBJ) ClustalW program.
In Vivo DNA End Resection Assay
DIvA cells (DSB Inducible via AsiSI [Aymard et al., 2014]) were transfected with siRNA using the Cell Line Nucleofactor kit V (Amaxa) according to the manufacturer’s instructions. Forty-eight hours after siRNA transfection, cells were treated or not with 300 nM of 4-hydroxytamoxifen (4OHT) (Sigma; H7904) for 4 hr. DNA was purified using QIAGEN DNeasy kit. Single-stranded DNA generated at an AsiSI-induced DSB was analyzed using the procedure described in Zhou et al. (2014), with the modifications described in Supplemental Experimental Procedures. The percentage of ssDNA was calculated with the following equation: ssDNA% = 1/(2(ΔCt−1) + 0.5) ×100, where ΔCt is calculated by subtracting the Ct obtained from mock-digested sample from the BanI-digested sample.
Generation of Inducible KDM4A Overexpression Cell Lines
U2OS-Flp-In/T-REx cells express the Tet repressor and contain a single Flp-In site (Gordon et al., 2009). U2OS-Flp-In/T-REx/DRGFP cells were generated by integrating the pDR-GFP expression plasmid (Pierce et al., 1999) into the U2OS-Flp-In/T-REx cells. The KDM4A cDNA (Genecopoeia) was cloned into the pDESTfrtto destination vector by a Gateway LR reaction (Life Technologies). U2OS-Flp-In/T-REx or U2OS-Flp-In/T-REx/DRGFP cells were cotransfected with the resulting KDM4A destination plasmid and pOG44 (Invitrogen). Stable clones that express the KDM4A protein under the control of the T-REx system were subjected to the treatments indicated.
Data Analysis
Data points from individual assays represent mean ± SEM. Statistical significance between two conditions was assessed by a two-tailed unpaired t test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and n.s. represents p ≥ 0.05.
Full details of all other experimental procedures are given in the Supplemental Experimental Procedures.
Author Contributions
S.X.P., S.A., L.-P.Z., S.S., and F.A. designed and performed experiments. The HPRT+:I-SceI assay was developed by A.C.G.P. The FLIP-IN cells and plasmids were developed by C.Z.B., T.C.H., A.C.G.P., T.H., N.B.L.T., and G.L. supervised the work. T.C.H. and S.X.P. wrote the manuscript with discussion and input from all authors. Experiments in Figure 1 were performed by S.A., with technology developed by A.C.G.P.; Figure 2 was performed by S.X.P. and S.A.; Figure 3 was performed by S.X.P. and L.-P.Z.; Figure 4 was performed by S.S. and S.X.P., with technology developed by C.Z.B.; Figure 5 was performed by S.S., L.-P.Z., S.X.P., and F.A.; Figure 6 was produced by S.X.P. Experiments in Figure S1 were performed by S.A. and S.S.; Figure S2 was performed by S.X.P. and S.A.; Figure S3 was performed by L.-P.Z.; Figure S4 was performed by S.X.P. and S.S.; Figure S5 was performed by F.A. and S.X.P.
Acknowledgments
We thank David Allis for the H3.3 and H3.3K36M lentiviral vectors; Catherine Millar for the U2OS-Flp-In/T-REx cell line; Atsushi Shibata and Hideaki Hogiwara for the NHEJ reporter cell line; Wojciech Niedzwiedz for the siRNAs against CtIP; and Annika Baude and Jiri Lukas for CtIP-GFP U2OS cells. We thank Mick Woodcock for helping with FACS analysis and Fereydoon Ahrabi for statistical analysis. We thank Peter McHugh and Valentine Macaulay for helpful discussions. This research was supported by the Medical Research Council (C.Z.B., T.H., S.S., T.C.H. grant R066538 and R19583), (L.-P.Z., N.B.L.T. grant ANRUBT00); Cancer Research UK (S.A. grant C5255/A15935) (L.-P.Z., N.L.T. grant 300/A13058); the Clarendon Scholarship (S.A., S.X.P.); the BBSRC (A.C.G.P. grant BB/H003371/1 and C.Z.B. grant BB/K019597/1); the Agence Nationale pour la Recherche (G.L. grant ANR-09-JCJC-0138); Association Contre le Cancer (G.L., F.A.); and the Torsten and Söderberg Foundation (T.H.).
Published: June 12, 2014
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2014.05.026.
Supplemental Information
References
- Adamson B., Smogorzewska A., Sigoillot F.D., King R.W., Elledge S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012;14:318–328. doi: 10.1038/ncb2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Sarakbi W., Sasi W., Jiang W.G., Roberts T., Newbold R.F., Mokbel K. The mRNA expression of SETD2 in human breast cancer: correlation with clinico-pathological parameters. BMC Cancer. 2009;9:290. doi: 10.1186/1471-2407-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.L., Australian Pancreatic Cancer Genome Initiative. ICGC Breast Cancer Consortium. ICGC MMML-Seq Consortium. ICGC PedBrain Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaudeau C., Lundin C., Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., Iacovoni J.S., Daburon V., Miller K.M., Jackson S.P., Legube G. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry W.L., Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res. 2013;73:2936–2942. doi: 10.1158/0008-5472.CAN-12-4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black J.C., Manning A.L., Van Rechem C., Kim J., Ladd B., Cho J., Pineda C.M., Murphy N., Daniels D.L., Montagna C. KDM4A lysine demethylase induces site-specific copy gain and rereplication of regions amplified in tumors. Cell. 2013;154:541–555. doi: 10.1016/j.cell.2013.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couture J.F., Collazo E., Ortiz-Tello P.A., Brunzelle J.S., Trievel R.C. Specificity and mechanism of JMJD2A, a trimethyllysine-specific histone demethylase. Nat. Struct. Mol. Biol. 2007;14:689–695. doi: 10.1038/nsmb1273. [DOI] [PubMed] [Google Scholar]
- Dalgliesh G.L., Furge K., Greenman C., Chen L., Bignell G., Butler A., Davies H., Edkins S., Hardy C., Latimer C. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugaard M., Baude A., Fugger K., Povlsen L.K., Beck H., Sørensen C.S., Petersen N.H., Sorensen P.H., Lukas C., Bartek J. LEDGF (p75) promotes DNA-end resection and homologous recombination. Nat. Struct. Mol. Biol. 2012;19:803–810. doi: 10.1038/nsmb.2314. [DOI] [PubMed] [Google Scholar]
- Edmunds J.W., Mahadevan L.C., Clayton A.L. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27:406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eidahl J.O., Crowe B.L., North J.A., McKee C.J., Shkriabai N., Feng L., Plumb M., Graham R.L., Gorelick R.J., Hess S. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41:3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fnu S., Williamson E.A., De Haro L.P., Brenneman M., Wray J., Shaheen M., Radhakrishnan K., Lee S.H., Nickoloff J.A., Hromas R. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl. Acad. Sci. USA. 2011;108:540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontebasso A.M., Schwartzentruber J., Khuong-Quang D.A., Liu X.Y., Sturm D., Korshunov A., Jones D.T., Witt H., Kool M., Albrecht S. Mutations in SETD2 and genes affecting histone H3K36 methylation target hemispheric high-grade gliomas. Acta Neuropathol. 2013;125:659–669. doi: 10.1007/s00401-013-1095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Rowan A.J., Horswell S., Larkin J., Endesfelder D., Gronroos E., Martinez P., Matthews N., Stewart A., Tarpey P. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon W.R., Vardar-Ulu D., L’Heureux S., Ashworth T., Malecki M.J., Sanchez-Irizarry C., McArthur D.G., Histen G., Mitchell J.L., Aster J.C., Blacklow S.C. Effects of S1 cleavage on the structure, surface export, and signaling activity of human Notch1 and Notch2. PLoS ONE. 2009;4:e6613. doi: 10.1371/journal.pone.0006613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W.D., Ehmsen K.T., Liu J. Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends Genet. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Jeggo P., Lavin M.F. Cellular radiosensitivity: how much better do we understand it? Int. J. Radiat. Biol. 2009;85:1061–1081. doi: 10.3109/09553000903261263. [DOI] [PubMed] [Google Scholar]
- Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Mermel C.H., Robinson J.T., Garraway L.A., Golub T.R., Meyerson M., Gabriel S.B., Lander E.S., Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis P.W., Müller M.M., Koletsky M.S., Cordero F., Lin S., Banaszynski L.A., Garcia B.A., Muir T.W., Becher O.J., Allis C.D. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science. 2013;340:857–861. doi: 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Mao G., Tong D., Huang J., Gu L., Yang W., Li G.M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153:590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M., Lee S.E. MMEJ repair of double-strand breaks (director’s cut): deleted sequences and alternative endings. Trends Genet. 2008;24:529–538. doi: 10.1016/j.tig.2008.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan M.E., Cui T.Y., Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- Newbold R.F., Mokbel K. Evidence for a tumour suppressor function of SETD2 in human breast cancer: a new hypothesis. Anticancer Res. 2010;30:3309–3311. [PubMed] [Google Scholar]
- Ogiwara H., Ui A., Otsuka A., Satoh H., Yokomi I., Nakajima S., Yasui A., Yokota J., Kohno T. Histone acetylation by CBP and p300 at double-strand break sites facilitates SWI/SNF chromatin remodeling and the recruitment of non-homologous end joining factors. Oncogene. 2011;30:2135–2146. doi: 10.1038/onc.2010.592. [DOI] [PubMed] [Google Scholar]
- Pei H., Zhang L., Luo K., Qin Y., Chesi M., Fei F., Bergsagel P.L., Wang L., You Z., Lou Z. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470:124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O’Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Yoshizato T., Shiraishi Y., Maekawa S., Okuno Y., Kamura T., Shimamura T., Sato-Otsubo A., Nagae G., Suzuki H. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat. Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- Shanbhag N.M., Rafalska-Metcalf I.U., Balane-Bolivar C., Janicki S.M., Greenberg R.A. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 2010;141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G., van Attikum H. The chromatin response to DNA breaks: leaving a mark on genome integrity. Annu. Rev. Biochem. 2013;82:55–80. doi: 10.1146/annurev-biochem-061809-174504. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Grant P.A., Briggs S.D., Sun Z.W., Bone J.R., Caldwell J.A., Mollah S., Cook R.G., Shabanowitz J., Hunt D.F., Allis C.D. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol. Cell. Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., New J.H., Kowalczykowski S.C. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington L.S., Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Carpenter P.B. Understanding the language of Lys36 methylation at histone H3. Nat. Rev. Mol. Cell Biol. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Yoh S.M., Lucas J.S., Jones K.A. The Iws1:Spt6:CTD complex controls cotranscriptional mRNA biosynthesis and HYPB/Setd2-mediated histone H3K36 methylation. Genes Dev. 2008;22:3422–3434. doi: 10.1101/gad.1720008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–163. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Caron P., Legube G., Paull T.T. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., He F., Zeng H., Ling S., Chen A., Wang Y., Yan X., Wei W., Pang Y., Cheng H. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 2014;46:287–293. doi: 10.1038/ng.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.