Synopsis
Intrinsically disordered proteins participate in important protein-protein and protein-nucleic acid interactions and control cellular phenotypes through their prominence as dynamic organizers of transcriptional, post-transcriptional, and signaling networks. These proteins challenge the tenets of the structure-function paradigm and their functional mechanisms remain a mystery given that they fail to fold autonomously into specific structures. Solving this mystery requires a first principles understanding of the quantitative relationships between information encoded in the sequences of disordered proteins and the ensemble of conformations they sample. Advances in quantifying sequence-ensemble relationships have been facilitated through a four-way synergy between bioinformatics, biophysical experiments, computer simulations, and polymer physics theories. Here, we review these advances and the resultant insights that allow us to develop a concise quantitative framework for describing sequence-ensemble relationships of intrinsically disordered proteins.
Introduction
Emil Fischer proposed that enzyme specificity could be explained by shape complementarity [1, 2]. He used the metaphor of a lock and key to illustrate how the three-dimensional arrangements of atoms comprising an enzyme and its substrate could enable them to fit together and prevent non-specific catalysis. Interpreted literally, this metaphor suggests that proteins possess rigid structures that determine their functions. Fischer, however, felt that popular interpretations of his lock-and-key metaphor exceeded its scope and experimental justification [3]. Indeed, protein rigidity has proven to be unsatisfactory at explaining noncompetitive inhibition and cannot account for enzymes where binding of one reactive group increases the exposure of another [4]. The concepts of allosteric linkage [5] and induced fit require the invocation of protein conformational changes in response to the binding of an interaction partner [6]. The structure-function paradigm, nuanced to accommodate proteins that switch between discrete conformations with different shape complementarities for execution of specific functions provides visual clarity and mathematical simplicity.
Going beyond structure-function relationships
Advancements on scientific and technological fronts have demonstrated unequivocally that proteins can exhibit significant conformational heterogeneity. Intrinsically disordered proteins (IDPs) are at the extreme end of the heterogeneity spectrum [7-9]. They adopt ensembles of conformations in aqueous solutions for which no single structure or self-similar collection of structures provides an adequate description. By all accounts the conformational heterogeneity exhibited by IDPs is relevant for biological function [8-14]. The phrase intrinsically disordered proteins is used to imply that the amino acid sequences for this class of proteins encode a preference for heterogeneous ensembles of conformations as the thermodynamic ground state under standard physiological conditions (aqueous solutions, 150 mM monovalent salt, low concentrations of divalent ions, pH 7.0, and temperature in the 25°C – 37°C range) [9, 15].
For many IDPs, folding can be coupled to binding and they can adopt ordered structures in specific bound complexes [16-20]. The intrinsic heterogeneity in their unbound forms is reflected in their ability to adopt different folds in the context of different complexes [10]. Transcription factors represent striking examples of molecules that undergo disorder-to-order transitions in complex with their cognate DNA partners [21-24]. Highly stable complexes with DNA can make transcription factor dissociation become “unreasonably slow” when compared to the turnover time of downstream regulatory processes. Disorder in the unbound forms is proposed to be important for lowering the overall affinity, which in turn increases the off-rates of protein-DNA complexes [25].
There are a growing number of reports of “fuzzy complexes” whereby conformational heterogeneity prevails in binary and multimolecular complexes [26-28]. IDPs can also self-assemble to form ordered, supramolecular assemblies, although the degree of order within these assemblies is variable and the intermediates that seem to be obligatory for self-assembly are characterized by significant conformational heterogeneity that can be modulated to alter the mechanisms of self-assembly and the stabilities of supramolecular structures [29-38].
Connecting sequence to conformational heterogeneity
Sequence-structure relationships are well documented for proteins whose individual amino acid sequences fold autonomously into specific three-dimensional structures [39, 40]. Specificity for a well-defined fold is the result of information encoded in the amino acid sequence [41, 42]. In direct analogy, information encoded at the sequence level keeps IDPs from autonomously folding into singular, well-defined three-dimensional structures [15, 43]. The information content of IDP sequences is such that acquisition of a folded conformation (if this happens) is deferred by coupling the folding process to either binding or self-assembly providing the heterotypic or homotypic interactions in trans can stabilize the IDP in a specific fold. From a thermodynamic standpoint, the stabilities of complexes and mechanisms of binding / assembly are linked to the conformational properties of IDPs in their unbound forms. Hence, sequence-ensemble relationships are central to understanding how disorder is used in IDP function.
Quantitative descriptions regarding sequence-ensemble relationships require biophysical characterization although IDPs present significant challenges for such studies. The signals are often highly averaged and by definition these systems resist crystallization unless they can be forced into specific folded structures. IDPs also present biochemical challenges because they can be difficult to isolate from tissue or cell systems because the process of homogenization exposes them to proteases that rapidly and preferentially degrade disordered proteins [44-46]. Efforts to characterize and quantify conformational heterogeneity and understand its role in protein function have required a systematic integration of biophysical, biochemical, and bioinformatics methods. Here, we focus on advances made in describing sequence-ensemble relationships in terms of quantitative connections between IDP sequence characteristics and their coarse grain conformational descriptors such as average shapes, sizes, and amplitudes of conformational fluctuations.
The major biophysical methodologies include nuclear magnetic resonance (NMR) spectroscopy [47-54], steady state and time-resolved fluorescence (single molecule and ensemble) spectroscopies [55-58], electron paramagnetic resonance spectroscopy (EPR)[59], small angle X-ray scattering (SAXS)[60-62], and molecular simulations that are used either de novo [63-71] or in synergy with data collected from spectroscopic investigations of IDPs [54, 62, 72-80]. Methodological advances are maturing and evolving to enable comparative assessments of sequence-ensemble relationships for IDPs.
The challenge of describing conformational heterogeneity
A single set of position coordinates (and uncertainties in these coordinates) helps relate sequence-to-structure for a protein that folds autonomously into a distinct three-dimensional structure. Such coordinate sets are generated as models that fit either the electron density data from X-ray diffraction through ordered protein crystals or NMR data that report on the chemical environments of backbone and sidechain protons and nuclei in solution. The protein data bank [81] provides a comprehensive archive of coordinate sets for a range of crystallizable proteins and proteins that are amenable for studies by NMR. This rich data set has lead to systematic classification of folds and fold families [40, 41, 82, 83] thus yielding an improved understanding of sequence-structure relationships and insights regarding the evolution of protein folds.
IDPs are not amenable to descriptions by single or even small number distinct of coordinate sets. Instead statistical descriptors are required to provide a concise classification of conformational ensembles and this is the language of polymer physics [84]. We focus on these concepts [84] to provide a unifying framework for quantitative analysis and descriptions of sequence-ensemble relationships of IDPs. This framework is useful for analyzing and interpreting data obtained either from experiments or from molecular simulations.
Limiting models as descriptors of conformational heterogeneity
The two most popular statistical descriptions based on polymer physics are the Flory random coil [85] and worm like chain models [86]. In the rotational isomeric approximation [85] to the Flory random coil model, the conformational partition function for the polypeptide is written as a product of partition functions of independent interaction units. All interactions between non-nearest neighbor units or so-called Kuhn segments are explicitly ignored although the intrinsic conformational preferences of individual units are captured in terms of weights for each of the possible rotational isomers. The unit either spans the degrees of freedom of an individual residue or can take local effects into account to expand the unit to span multiple residues. In either situation case, each conformation for unit i is annotated by an intrinsic energy value that is calculated using an empirical potential function of one’s choosing. The conformations are binned into rotational isomeric states based on the similarities of the backbone and sidechain dihedral angles (see Figure 1). Residue / unit i, might have m rotational isomers whereas residue j might have n rotational isomers. For a given residue, each rotational isomer is assigned a weight that is calculated using the Boltzmann weights of energies of individual conformations that make up the rotational isomer.
Figure 1. Illustration of how the rotational isomeric approximation of the Flory random coil model is constructed.
(A) This process begins with a detailed calculation of the free energy landscape (with free energies increasing from red to blue) for an individual amino acid (or Kuhn segment), shown here for alanine. The tiles represent a coarse graining of conformational space into discrete rotational isomers, and each isomer has a label and a statistical weight that is calculated using the energies associated with conformations that make up a rotational isomer. The assumption of independence / additivity allows the statistical weights for each combination of rotational isomers to be written as a product of individual weights. Panel (B) shows this procedure, whereby there are M conformations for a polypeptide of N residues and the statistical weight for each conformation z is a product of the weights for individual residues. The result is a weighted ensemble of all conformational possibilities where each “conformation” is denoted using a combination of the coarse grain rotational isomers. Panel (C) shows a schematic conformation for one of the conformations z.
Given an amino acid sequence of N residues, one can calculate, a priori, the probabilities associated with all combinations rotational isomers. For the sequence of interest the number of rotational isomers per residue, their statistical weights, and the sequence composition dictates the total number of conformational possibilities and the likelihoods associated with each conformation. These likelihoods make up the predicted conformational distribution function and can be used to calculate a variety of conformational properties including the average end-to-end distance, the average radius of gyration, the average hydrodynamic size, the average distance between residues i and j, and any observable that can be cast as a function of a moment of the conformational distribution function.
An alternative approach is to analyze experimental data, specifically data from fluorescence [87-89] or force spectroscopy [90-92] that are functions of end-to-end distances using variants of the worm like chain model. The persistence length lp is the length scale over which the chain behaves like a continuously deformable entity. For a rod-like chain lp equals the contour length and for a freely-joined chain, lp equals the bond length, so this model ostensibly allows interpolation between two extremes. Fluctuations are highly correlated for spatial separations that are smaller than lp. For spatial separations longer than lp, the worm-like segments become uncorrelated and the model reverts to the Flory random coil limit. Therefore, if lp is found to be small, the worm like chain model does not yield any insights that go beyond the Flory random coil.
Estimates of lp values for different sequences, studied under similar solution conditions yield comparative assessments of sequence-ensemble relationships through comparative measures of “chain stiffness” although Yamakawa [93] has highlighted the limitations of lp as a measure of stiffness. The assumption of continuous deformation for spatial separations less than lP is questionable because this assumption breaks down if the chain can form heterogeneous ensembles of compact globules. Despite its inherent weaknesses, the worm like chain model retains appeal for its ease of use in interpreting experimental data for IDPs and denatured proteins [87-89].
The value of limiting models
The preceding discussion focuses on limiting models, which are analogous to limiting models / laws in other branches of physics that include the Debye-Hückel equation for calculating activity coefficients of electrolytes [94], and the Hildebrand [95] / Flory-Huggins expressions for the free energies of ideal mixtures [96, 97]. Limiting laws or models provide a route for interpreting experimental data as deviations from ideal behavior. As a limiting model, the Flory random coil model is often used to calibrate measured observables such as NMR chemical shifts [98, 99] and NMR paramagnetic relaxation enhancement effects [49] i.e., observables can be calibrated as deviations from the Flory random coil. This helps assess the contributions of spatial interactions between residues that are distal in the linear sequence. Such approaches are decidedly one-sided because deviations from a limiting model tell us what an ensemble is not and this is inadequate for developing a complete understanding of sequence-ensemble relationships.
Toward more realistic polymer models for IDPs
Sequences of IDPs are deficient in hydrophobic residues and enriched in polar and charged amino acids [15]. Electrostatic and polar interactions are typically quite large, even when screened by the surrounding solvent and this raises serious questions regarding the applicability of limiting models for describing IDP conformations. Furthermore, with a few exceptions such as proline- or glycine-rich sequences, the intrinsic flexibilities of all polypeptides are roughly equivalent. This implies that the intrinsic value of lp is essentially fixed for a wide range of sequences, even if it is erroneously treated as a free parameter when fitting experimental data. The inadequacy of limiting models for describing experimental data have lead to proposals that polymer physics concepts are inadequate for describing sequence-ensemble relationships for all IDPs [100]. This proposal ignores the rich possibilities afforded by explicit consideration of the effects of realistic interactions.
Conformational statistics are dictated by the interplay between chain-solvent and intrachain (intra-backbone, backbone-sidechain, and sidechain-sidechain) interactions [84]. As a result, polymers undergo continuous transitions between distinct conformational classes. These transitions are modulated by changes to solvent-mediated interactions either through the addition of ternary components or by changing the temperature and / or pressure. Importantly, the conformational classes are akin to distinct phases because they have distinct density profiles and the variation of spatial separation as a function of sequence separation follows distinct patterns. Transitions between conformational classes are hence akin to phase transitions [101].
Measures for assessing the phase behavior of polymers
Quantities such as the average radius of gyration (〈Rg〉), the average hydrodynamic radius (〈Rh〉), and the average end-to-end distance (〈Ree〉), respectively are different measures of chain size that can be used to quantify the average density, intrinsic viscosity, and concentration of one end of the chain around the other. In addition to measures of chain size, one can also calculate the average shapes of polymers. The average asphericity (δ*) quantifies the extent of deviation from a perfect sphere (δ* = 0). For ellipsoids, δ*≈0.4, and this quantity attains its maximum value of 1 for a perfect rod [102]. The average asphericity is calculated from the ensemble-averaged eigenvalues of the gyration tensor.
One can also calculate the average distances between residues i and j. The quantity 〈Rij〉 represents the ensemble-average of spatial separations calculated as averages over all pairs i and j that yield a sequence separation |j–i| [103]. Multiple pairs of residues i and j will have similar sequence separations |j–i|. The profile of 〈Rij〉 plotted against sequence separation |j–i| quantifies the local concentration of chain segments around each other and provides the most detailed information regarding the so-called link density [104], which is a formal order parameter in formalisms of polymer theories such as the Lifshitz approach [103, 105-108]. In addition to ensemble averages, one can also calculate the one- and two-parameter distribution functions such as P(Rg), P(Ree), P(Rh), P(δ), P(Rij| |j–i|), and P(Rg, δ). The latter quantifies the joint distribution of sizes and shapes.
Importantly, all of the quantities listed above are accessible to the appropriate combination of experiments and can be calculated using coordinates for simulated ensembles. This enables quantitative comparisons between simulation results and experiments thus facilitating direct approaches to either test predictions from simulations or routes to incorporate experimental data as restraints in simulations for generating ensembles that best describe experimental data. Both approaches are equally important and have enabled the development of quantitative sequence-ensemble relationships for IDPs.
Assessing solvent quality
The balance between chain-solvent interactions and intra-chain interactions is quantified using a parameter vex. This quantity has units of volume and is proportional to the integral of the Mayer f-function, i.e., where , is the potential of mean force for the thermally averaged inter-residue interaction and β=1/RT where R is Boltzmann's constant and T is temperature. If the effective inter-residue interactions are repulsive, then the Mayer-f function is negative, which leads to positive values for vex and the converse is true for inter-residue interactions that are attractive on average. The parameter vex is hence a measure of the volume excluded, per residue, for favorable interactions with the surrounding solvent that results from the competition between chain-chain and chain-solvent interactions. It provides a measure of the strengths of pairwise inter-residue interactions, on average, and can be related to the second virial coefficient that is accessible using light scattering measurements [84].
Classifying ensemble types based on vex
In a good solvent, vex > 0, and the chain expands to maximize the polymer-solvent interface. Expanded unfolded states are sampled in vitro in high concentrations of chemical denaturants such as urea and guanidinium chloride. Aqueous solutions with high concentrations (8 M) urea are presumed to be reasonable mimics of good solvents for generic polypeptides because urea, a carbonyl diamide, is chemically equivalent to polypeptide backbone amides. As a result, quantities such as 〈Rg〉 [109], 〈Rh〉 [110], and the average end-to-end distance 〈Ree〉 scale as N0.59 with chain length, N. In a good solvent the distances 〈R ij〉 scale as |j–i|0.59 as a function of sequence separation |j–i|.
Since the inter-residue interactions are repulsive on average, vex > 0 in good solvents. Indeed, the sizes of self-avoiding random walks also scale as N0.59 and conformational ensembles for polymers in good solvents and self-avoiding random walks are said to belong to the same “universality class” [111]. Accordingly, ensembles generated in atomistic detail for proteins in the excluded volume (EV) limit are useful reference states for expanded unfolded states [112-118]. In the EV limit, ensembles are generated using atomistic descriptions of proteins and all non-bonded interactions excepting steric repulsions are ignored.
The low overall hydrophobicity of IDP sequences implicitly suggests that these systems come under the same rubric as chemically denatured proteins. Hence, a popular approach is to generate EV limit ensembles and filter out those conformations that cause deviations from observables that are measured experimentally [89, 119-125]. Although this seems like a reasonable approach, it imposes the fiat that aqueous solutions are mimic of good solvents for IDP sequences and ignores the possibility that these sequences can sample compact phases.
Implications of decreasing values of vex
The parameter vex can change continuously going from positive values in a good solvent, through zero in a theta solvent, to negative values in a poor solvent [84]. If the effects of chain-solvent and intra-chain interactions exactly counterbalance, then vex=0, and the chain is said to be in a theta solvent. Under such conditions, the chain statistics are consistent with those of a Flory random coil model. It is important to note that this behavior comes about due to counterbalancing of interactions rather than explicitly ignoring non-local interactions. In a theta solvent, 〈Rg〉, 〈Rh〉, and 〈Ree〉 scale as N0.5 and 〈Rij〉 scales as |j–i|0.5.
In a poor solvent, vex < 0 and the chain prefers compact, globular conformations that minimize the polymer-solvent interface and 〈Rg〉 and 〈Rh〉 scale as N1/3 with chain length, N. The sizes of folded proteins follow N1/3 scaling [126]. The poorer the solvent, the more negative the value of vex. Statistics of inter-residue distances change fundamentally in a poor solvent. The distances 〈Rij〉 do not increase with sequence separation |j–i| according to a power law. Instead, for all values of |j–i| greater than larger than a so-called blob length (ca. 5-7 residues), the value of 〈Rij〉 is fixed by the average density of the globule.
Clearly, the consideration of the details of the balance between chain-chain and chain-solvent interactions affords a richer description of conformational statistics. The question is which of these models apply for describing IDPs? In order to answer this question, we need a systematic approach that assesses if typical physiological milieus are good, theta, or poor solvents for polypeptide backbones. This allows us to understand how sidechains in IDPs modulate the intrinsic backbone preferences. Recent studies of archetypal IDPs using a combination of spectroscopic experiments and molecular simulations have yielded clear insights regarding sequence-ensemble relationships. The following sections summarize these findings and the implications for predicting sequence-ensemble relationships for IDPs.
Aqueous solutions are poor solvents for generic polypeptide backbones
The free energy of hydration for N-methylacetamide (NMA) at 298 K is –10 kcal/mol [127], indicating that the transfer of NMA from the gas phase into water is highly favorable. Naïve extrapolation from the transfer model suggests that polyglycine – a poly-secondary-amide – should prefer structures that maximize the interface with the aqueous solvent. However, results based on molecular dynamics simulations show that polyglycine forms a heterogeneous ensemble of compact globules [128]. Similar simulations showed that polyglycine samples expanded, coil-like structures in 8 M urea and there is minimal overlap between ensembles sampled in the two milieus. These results and analysis of a series of order parameters drawn from polymer physics predict that water is a poor solvent for polyglycine, which is a mimic of polypeptide backbones. Recent fluorescence correlation spectroscopy experiments and solubility measurements of polyglycine peptides have confirmed these predictions [129].
Why do polypeptide backbones form compact globules in water?
In a polymer, each residue has reduced translational entropy when compared to free analogs diffusing in solvent. Mean-field theories show that the entropy of mixing between solute and solvent molecules is reduced by a factor of N if we compare the entropy of mixing for N freely diffusing solute molecules to the same N molecules concatenated to form a polymer [97, 130]. Because of this diminution in entropy, polymers, unlike small molecules, can undergo intramolecular phase separation to form globules or prefer the coil phase that is well mixed with solvent [105, 106]. Within globules, the effective concentration of residues around each other is independent of N, whereas in coils it decreases as N–0.77. As a result, each residue can make collective amide-amide contacts within globules whereas the coil state is characterized by a combination of negligible intra-chain interactions and additive interactions of individual residues with the solvent. Even for polyamides, where individual amides can be favorably solvated, it is the competition between collective self-interactions within a globule and additive interactions of individual residues with the solvent in the coil state that determines the stable phase. Model compounds do not account for the diminution in translational entropy or the competition between collective intra-polymer and additive polymer-solvent interactions. And this partially explains the preference of polypeptide backbones for compact globules.
Questions remain regarding the balance between chain / solvation entropy and enthalpy, the interplay between backbone hydration and self-solvation of amides, and the comparative roles of hydrogen bonding versus van der Waals interactions in giving rise to the observed preferences of polypeptide backbones in dilute and concentrated aqueous solutions. To answer these questions, we need a systematic investigation of the temperature and cosolute dependencies of the preference for collapsed states in dilute solutions and the solubility boundary in concentrated solutions. In addition, comparative studies of constructs with substituted amides such as amide to ester substitutions (to probe the effect of weakened hydrogen bond donors and stronger acceptors) and secondary amide to primary / tertiary amide substitutions (to probe the effect of hydrogen bond donors) will be necessary for quantifying the role of hydrogen bonding in driving polypeptide backbone collapse.
Archetype 1 – polar tracts form compact globules in aqueous solutions
Among the polar amino acids, IDPs are enriched in histidine (H), glutamine (Q), serine (S), and threonine (T) and are relatively deficient in asparagine (N) although Q/N-rich regions are the hallmark of prion forming domains in yeast [36]. Q-rich linkers were among the first disordered segments identified from sequence analysis [131]. They are abundant in transactivation domains of transcription factors and in RNA-binding proteins that play important roles in post-transcriptional regulation [13, 132].
Fluorescence correlation spectroscopy measurements revealed that 〈Rh 〉 scales as N1/3 for monomeric polyglutamine molecules [133]. These results confirmed predictions from molecular dynamics simulations [69, 134] and have been reproduced using steady state Förster resonance energy transfer (FRET) [135] measurements. The low sequence complexity of polyglutamine implies a lack of specificity for a single compact conformation and instead heterogeneous ensembles of compact conformations are energetically equivalent. Furthermore, the internal friction is uncharacteristically high for these molecules suggesting glassy dynamics for the conversions between distinct compact conformations [69]. Simulation results obtained using the ABSINTH implicit solvation model show evidence for continuous globule-to-coil transitions for polyglutamine [136-138]. In accord with polymer physics theories, the stabilities of globular conformations and the sharpness of the globule-to-coil transitions increase with increasing chain length (Figure 3).
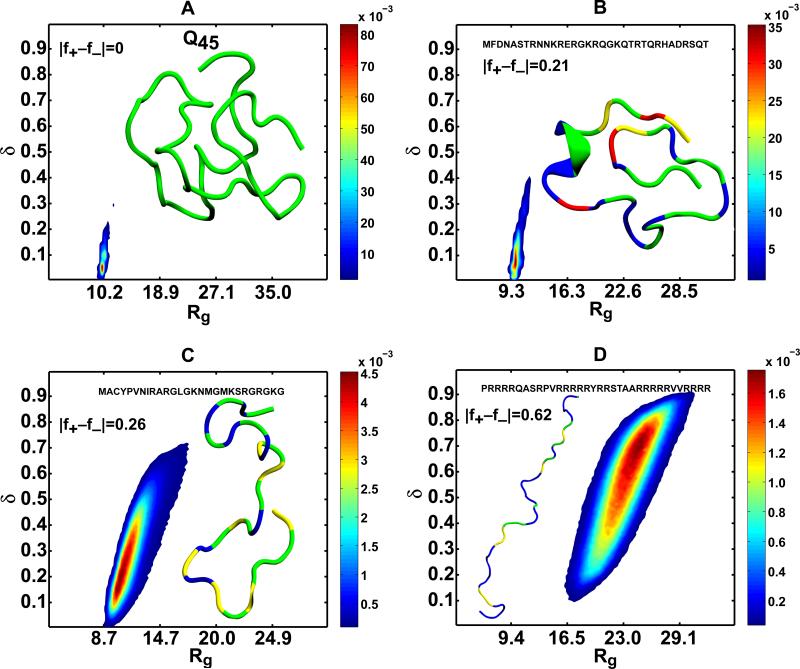
Figure 3. IDPs form globules or coils depending on the net charge per residue (|f+–f−| increases), T=298 K.
Panel (A) shows the joint distribution P(Rg,δ) and a representative conformation for polyglutamine with 45 residues. The distribution highlights the small amplitudes of conformational fluctuations and the overall compactness of the conformations that are sampled by this sequence. Panels (B), (C), and (D) show how arginine-rich sequences transition from globules (B) to theta-like conformations (C) and swollen, ellipsoidal coils (D) as the net charge per residue increases. The figures were generated using the simulation results of Mao et al. (B-D) [146] and Vitalis et al. (A) [137]. The representative conformations were drawn using the VMD package [181] and the color-coding used depicts polar residues in green, hydrophobic residues in yellow, positively charged residues in red, and negatively charged residues in blue.
Single molecule atomic force spectroscopy studies showed that polyglutamine molecules form compact globules that are mechanically resistant to forces as large as 800 pN [139]. Interestingly, the introduction of proline residues within polyglutamine tracts increases their mechanical compliance. The preference for heterogeneous ensembles of compact globular conformations has also been observed using single molecule FRET measurements for Q/N-rich tracts [55] and for glycine-serine block copolypeptides using a combination of time resolved FRET measurements [119] and molecular dynamics simulations [128]. Taken together, it is now clear that sequences enriched in polar residues form heterogeneous ensembles of compact globules as measured by a range of properties that quantify sizes, shapes, the scaling of inter-segmental distances, and the responses of these sequences to applied force. The preference for collapsed states can be traced, at least partially, to the intrinsic preferences of polypeptide backbones whose conformational properties in aqueous solutions are consistent with water being a poor solvent for polyamides. These results might seem surprising since the preference for collapsed states is realized despite the absence or deficiency of canonical hydrophobic residues in these polar tracts. The results are, however, consistent with the poor solubility profiles of IDP sequences that are enriched in polar residues and highlight the weaknesses of extrapolations based on additivity assumptions, which suggest that conformational properties and solubility profiles of polymers can be inferred exclusively from the properties of their building blocks. Indeed, the assumption of additivity has been questioned in the protein literature [140-142] and has proven to be invalid for synthetic polymers.
Archetype 2 – globules to coils as a function of increased net charge per residue
Uversky et al. [15] showed that in addition to low overall hydrophobicity, many IDP sequences also have high net charge per residue. IDP sequences populate a specific region of a two-dimensional space defined by mean hydrophobicity, 〈H〉 and mean net charge 〈q〉. In this plane, a single line, 〈q〉 = 2.785 〈H〉 – 1.151 separates IDP sequences, which lie below the line, from the sequences with well-defined folds. Given that archetypal polar IDPs form heterogeneous ensembles of collapsed structures in aqueous solutions and that the driving force for collapse originates in the intrinsic preference of polypeptide backbones for collapsed structures in water, the question is if this behavior is a generic attribute of all IDP sequences? Polymer physics theories suggest that even in poor solvents, polyelectrolytes i.e., sequences enriched in charged residues of one kind, can reverse the preference for collapsed structures [143-145]. Instead, the preference for charged residues to be solvated combined with intra-chain electrostatic repulsions leads to chain expansion. Therefore, depending on the charge content, chain sizes can go beyond values expected for self-avoiding random walks in so-called good solvents [144, 145]. This behavior results without alterations to the solvent properties, but instead is the consequence of the interplay between chain-chain interactions and chain-solvent interactions whereby the former essentially override the latter due to the large electrostatic energies involved.
Two studies on complementary systems namely, an IDP enriched in acidic residues [58], and a series of sequences enriched in basic residues (arginine) [146] showed that IDP sequences partition into globules versus coils based on their net charge per residue. This quantity, which is different from just the net charge, is calculated as |f+–f−|, where f+ and f− refer to the fraction of positive and negatively charged residues in the sequence, respectively. If |f+–f−| < 0.2, then the sequences are most likely to be globule formers and for values of |f+–f−| larger than 0.2, one observes a continuous transition into non-globular, expanded coils, where the precise power law followed by quantities such as 〈Rg〉 as a function of N or 〈Rij〉 as a function of sequence separation |j–i| depends on the value of |f+–f−|. These findings were converged upon by different groups through a combination of single molecule FRET measurements [58], synergy between molecular simulations and fluorescence spectroscopies [146], NMR measurements of hydrodynamic sizes [147], and SAXS [123].
Polymer physics theories provide a generalization of vex to include the contribution of increased charge contents [143-145, 148] for polyelectrolytes in poor solvents. For chains such as polypeptides where the interplay between linear charge density and chain thickness is important, the value of an effective excluded volume parameter Xeff determines the conformational properties of polyelectrolytes in a poor solvent. If the net charge per residue is small, then Xeff < 0, the sign and magnitude of vex dominate, and the chain collapses to a uniform distribution of compact globules. Within a narrow range of values for the net charge per residue, Xeff approaches zero. The value of the net charge per residue that yields Xeff = 0 is the one for which conformational properties are akin to those of a chain in a theta solvent – although the solvent itself is not a theta solvent. Values of Xeff > 0 are realized as the charge content increases thus increasing the net charge per residue. The chain expands and accesses coil states, the statistics of which are congruent with those of chains good solvents – although the solvent quality remains unchanged and the behavior results from increased charge content in the sequence. As Xeff becomes increasingly positive with increasing net charge per residue, theory predicts a continuous increase in chain dimensions – a feature that has been recapitulated for archetypal IDPs of increasing net charge per residue [146]. Therefore, the intrinsic solvation preferences of charged sidechains and their interactions with each other can override the intrinsic preferences of polypeptide backbones in aqueous solutions because the sidechains are akin to an effective solvent for IDP sequences that are enriched in charges.
A predictive phase diagram for sequence-ensemble relationships
Synthesis of results obtained thus far for archetypal IDPs has lead to a speculative generalization to enable the prediction of conformational classes for IDPs based on their sequence characteristics. Mao et al. introduced a schematic phase diagram shown in Figure 4 where the three axes denote mean hydrophobicity, f+ and f−, respectively and each of these parameters varies between 0 and 1. The dividing line discovered by Uversky et al. is shown as a boundary that separates folded proteins from IDPs. Below this boundary, IDPs prefer to be either globules or coils, and the net charge per residue is the determinant of this preference. The predicted phase diagram presumes that amino acid composition is a sufficient diagnostic of IDP sequence-ensemble relationships. Ongoing investigations are testing the accuracy of inferences drawn from the proposed phase diagram that is designed to quantify sequence-ensemble relationships based entirely on the composition of amino acids for the sequence of an IDP or disordered region. Although preliminary results suggest that this assumption is reasonably robust, it is also clear that the local conformational preferences such as secondary structure contents can vary considerably between sequences of fixed or similar compositions [146, 149, 150]. Indeed, recent results show that the net charge per residue is not particularly useful as a diagnostic of local conformational preferences [150]. Instead, sequence context acts as a rheostat to modulate these preferences [151].
Discussion
Going beyond polyelectrolytes
The studies that uncovered the effects of net charge per residue focused mainly on polyelectrolytes. For these systems, it appears that coarse grain sequence-ensemble relationships are robustly classifiable based on net charge per residue and hence amino acid composition alone. It is conceivable that overall charge content as well as the linear sequence patterning of oppositely charged residues alter the predictions of sequenceensemble relationships obtained on the basis of net charge per residue [152] and a systematic study of this issue is currently underway. This is especially important since a majority of IDPs are strongly polyampholytic [153] i.e., IDP sequences of high charge content that have equivalent numbers of oppositely charged residues.
Ways to modulate the net charge per residue
The pKa values of ionizable groups can shift depending on the amino acid composition, sequence context and the degree of conformational heterogeneity [154]. Changes to protonation states of ionizable residues will alter the net charge per residue and hence the predicted sequence-ensemble relationships. There is the possibility that some IDP sequences use pKa shifts to switch between conformational classes. This is relevant because IDP functions are often mediated through interactions involving linear peptide motifs that can be as short as 3-5 amino acids [155-166]. These motifs interact in trans with ordered protein domains or with motifs from other disordered regions. Shifts in pKa values of ionizable residues within disordered regions can lead to an increase in net charge per residue past threshold values causing a globule-to-coil transition to expose hidden linear motifs that mediate binding with other protein domains. Reversing these transitions from coils to globules is also feasible through shifts in pKa values that decrease the net charge per residue thus leading to sequestration of linear motifs to facilitate the dissociation of complexes.
Recent studies have also shown that disordered regions are under post-transcriptional control through tissue-specific alternative splicing [157]. These modifications might cause pKa shifts in ionizable groups by altering the sequence contexts of disordered regions in order to achieve tissue-specific control on the preferred conformational class. It is also well known that residues within disordered regions are the targets of numerous post-translational modifications [167-180]. These chemical transformations such as serine / threonine phosphorylation are reversible through the synergistic action of kinases and phosphatases. Such transformations occur often in disordered regions of hub proteins in interaction networks and afford the prospect of rewiring networks by controlling the collective exposure of linear motifs within disordered regions. Clearly, a systematic study of the impact of pH dependent conformational changes and the influence of post-translational modifications on sequence-ensemble relationships will be extremely important for understanding the expanded repertoires of functions afforded by IDPs and disordered regions. Finally, most biophysical studies of IDPs treat these systems as isolated proteins. In fact, a majority of these sequences function in cis with structured protein domains. Despite the obvious biological relevance, the influence of these naturally occurring contexts and the possible coupling between disordered regions and structured domains remains essentially unexplored – a shortcoming that needs to be addressed if we are to understand the functional implications of disordered regions within proteins.
Conclusions
A synergistic combination of computational and experimental biophysical investigations of archetypal systems is yielding important information regarding sequence-ensemble relationships of IDPs. The language of polymer physics provides a coherent framework for quantifying and hence classifying these sequence-ensemble relationships. The collection of initial results gathered from different groups has rather serendipitously been quite systematic and hence distinct archetypes within the IDP sequence space have been interrogated. The main highlights of these initial investigations are two-fold. The hydrophobic effect can – at least in spirit – be generalized to include sequences rich in polar residues and deficient in canonical hydrophobic groups. Charge content and net charge per residue appear to be the main determinants of non-globular, expanded coils. Unlike their ordered counterparts that form folded globules, the unique sequence / compositional preferences seem to afford IDPs the advantage of realizing a continuum of conformational transitions that might serve as the biophysical basis for their diverse functions. Continued investigations of sequence-ensemble relationships and their connections to function will be of utmost importance for understanding how the functional repertoires of proteins have been expanded through the use of disordered proteins and regions.
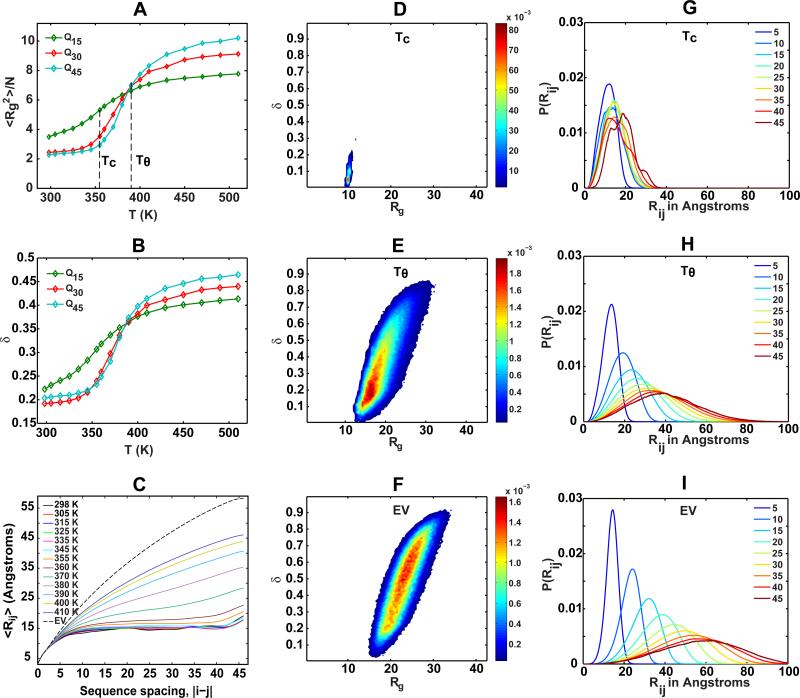
Figure 2. A partial inventory of the results and analysis afforded by combining polymer physics concepts with simulation results to describe and classify sequence-ensemble relationships.
The analyses are based on results from simulations of polyglutamine chains of different lengths. These Monte Carlo simulations utilize the ABSINTH implicit solvation and forcefield based on the OPLS-AA/L molecular mechanics parameter set. Panel (A) shows the globule-to-coil transitions plotted as the temperature dependence of the normalized mean-square radius of gyration. The sharpness of the transition increases with chain length. The increased number of favorable intra-chain interactions offset the unfavorable surface free energies with the surrounding poor solvent. The curves coincide at the theta temperature, designated as Tθ, and compact globules are preferred for all temperatures T < TC. Panel (B) depicts the globule to coil transition for different polyglutamine chains by plotting the ensemble-averaged asphericity values δ* as a function of temperature. For T < TC, spherical globules are favored by longer chains, as evidenced by the low values for δ*. This preference is reversed as chains expand to sample ellipsoidal coil-like structures (δ* > 0.4). Panel (C) shows the 〈Rij〉 profiles for a 45-residue polyglutamine chain for different temperatures. For T < TC, the profiles show little variation and have the plateauing behavior that is consistent with compact globules being preferred below TC. As T increases beyond TC toward Tθ=390 K, the profiles transition from the characteristic plateauing to the power law form characteristic of improving solvent quality i.e., weakening intra-chain interactions. Panels (D), (E), and (F) show the joint distributions P(Rg,δ ) for a 45-reisdue polyglutamine chain under three different simulation conditions, TC, Tθ, and the EV limit, which corresponds to the situation where all intra-chain interactions excepting steric repulsions are ignored (see main text). Rg is in Å and δ is unitless. These distributions underscore two points: First, the amplitudes of fluctuations viz., the range of conformations sampled increases as the chain expands and becomes more aspherical. Second, the overlap between ensembles at TC and expanded ensembles sampled at higher temperatures is minimal and decreases with increasing temperature, i.e., the EV limit ensembles bear no resemblance to the ensemble of globules sampled for T< TC and hence one should be cautious in choosing conformations from the EV limit as proxies for IDPs. Panels (G), (H), and (I) show the distributions P(Rij| |j–i|) for different sequence separations at T=TC, Tθ and the EV limit, respectively. For T=TC, the distributions overlap considerably, irrespective of sequence separation. For higher temperatures and the EV limit, the distributions shift toward larger Rij values as |j–i| increases – a feature that is consistent with the power law behavior expected for theta temperatures and beyond.
Acknowledgments
Support from the National Science Foundation and National Institutes of Health are gratefully acknowledged. We thank our colleagues, S.L. Crick, R.K. Das, and A. Vitalis for insightful discussions.
Abbreviations used
- EPR
electron paramagnetic resonance
- FRET
Förster resonance energy transfer
- IDP
intrinsically disordered protein
- NMR
nuclear magnetic resonance
- SAXS
small angle X-ray scattering
References
- 1.Fischer E. Einfuss der conguration auf die wirkung der enzyme. Berichte der deutschen chemischen Gesellschaft. 1894;27:2985–2993. [Google Scholar]
- 2.Fischer E. Einfluss der conguration auf die wirkung der enzyme. II. Berichte der deutschen chemischen Gesellschaft. 1894;27:3479–3483. [Google Scholar]
- 3.Fischer E. Bedeutung der Stereochemie fur die Physiologie. Zeitschrift fur Physiologische Chemie. 1898;26:60–87. [Google Scholar]
- 4.Koshland DE. The key-lock theory and the induced fit theory. Angewandte Chemie-International Edition in English. 1995;33:2375–2378. [Google Scholar]
- 5.Monod J, Wyman J, Changeux J-P. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1964;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 6.Yu EW, Koshland DE. Propagating conformational changes over long (and short) distances in proteins. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9517–9520. doi: 10.1073/pnas.161239298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uversky VN. Natively unfolded proteins: A point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J. Mol. Graph. Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 9.Wright PE, Dyson HJ. Intrinsically unstructured proteins: Re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 10.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21(Waf1/Cip1/Sdi1) in the free and Cdk2-bound state: Conformational disorder mediates binding diversity. Proc. Natl. Acad. Sci. U. S. A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 12.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv. Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 13.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nature Reviews in Molecular Cell Biology. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 14.Tompa P. The functional benefits of protein disorder. Journal Of Molecular Structure-Theochem. 2003;666:361–371. [Google Scholar]
- 15.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins-Structure Function And Genetics. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 16.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 17.Frankel AD, Smith CA. Induced folding in RNA-protein recognition: More than a simple molecular handshake. Cell. 1998;92:149–151. doi: 10.1016/s0092-8674(00)80908-3. [DOI] [PubMed] [Google Scholar]
- 18.Mucsi Z, Hudecz F, Hollosi M, Tompa P, Friedrich P. Binding-induced folding transitions in calpastatin subdomains A and C. Protein Sci. 2003;12:2327–2336. doi: 10.1110/ps.03138803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao LM, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat. Struct. Mol. Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 20.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins-Structure Function And Bioinformatics. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 21.Kohler JJ, Metallo SJ, Schneider TL, Schepartz A. DNA specificity enhanced by sequential binding of protein monomers. Proceedings of the National Academy of Sciences, USA. 1999;96:11735–11739. doi: 10.1073/pnas.96.21.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu JG, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuxreiter M, Tompa P, Simon I, Uversky VN, Hansen JC, Asturias FJ. Malleable machines take shape in eukaryotic transcriptional regulation. Nat. Chem. Biol. 2008;4:728–737. doi: 10.1038/nchembio.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spolar RS, Record MT. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 25.von Hippel PH. From “simple” DNA-protein interactions to the macromolecular machines of gene expression. In Annu. Rev. Biophys. Biomol. Struct. 2007:79–105. doi: 10.1146/annurev.biophys.34.040204.144521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Fuxreiter M, Tompa P. Fuzzy interactome: the limitations of models in molecular biology. Trends Biochem. Sci. 2009;34:3–3. [Google Scholar]
- 28.Mittag T, Marsh J, Orlicky S, Borg M, Tang X, Sicheri F, Chan HS, Kay LE, Tyers M, Forman-Kay JD. “Fuzzy” complexes: How much disorder can a biologically relevant complex tolerate, and can it even be beneficial? Biochem. Cell Biol. 2010;88:403–403. [Google Scholar]
- 29.Padrick SB, Miranker AD. Islet amyloid polypeptide: Identification of long-range contacts and local order on the fibrillogenesis pathway. J. Mol. Biol. 2001;308:783–794. doi: 10.1006/jmbi.2001.4608. [DOI] [PubMed] [Google Scholar]
- 30.Fandrich M, Dobson CM. The behaviour of polyamino acids reveals an inverse side chain effect in amyloid structure formation. EMBO J. 2002;21:5682–5690. doi: 10.1093/emboj/cdf573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bitan G, Kirkitadze MD, Lomakin A, Vollers SS, Benedek GB, Teplow DB. Amyloid beta-protein (Abeta) assembly: Abeta40 and Abeta42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. U. S. A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheibel T, Bloom J, Lindquist SL. The elongation of yeast prion fibers involves separable steps of association and conversion. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2287–2292. doi: 10.1073/pnas.0308754101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: The importance of being unfolded. Biochimica et Biophysica Acta - Proteins and Proteomics. 2004;1698:131. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Calamai M, Chiti F, Dobson CM. Amyloid fibril formation can proceed from different conformations of a partially unfolded protein. Biophys. J. 2005;89:4201. doi: 10.1529/biophysj.105.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krishnan R, Lindquist SL. Structural insights into a yeast prion illuminate nucleation and strain diversity. Nature. 2005;435:765–772. doi: 10.1038/nature03679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Halfmann R, Alberti S, Krishnan R, Lyle N, O'Donnell CW, King OD, Berger B, Pappu RV, Lindquist S. Opposing Effects of Glutamine and Asparagine Govern Prion Formation by Intrinsically Disordered Proteins. Mol. Cell. 2011;43:72–84. doi: 10.1016/j.molcel.2011.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heim M, Romer L, Scheibel T. Hierarchical structures made of proteins. The complex architecture of spider webs and their constituent silk proteins. Chem. Soc. Rev. 2010;39:156–164. doi: 10.1039/b813273a. [DOI] [PubMed] [Google Scholar]
- 38.Vitalis A, Pappu RV. Assessing the contribution of heterogeneous distributions of oligomers to aggregation mechanisms of polyglutamine peptides. Biophys. Chem. 2011;159:14–23. doi: 10.1016/j.bpc.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, Sillitoe I, Yeats C, Thornton JM, Orengo CA. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lo Conte L, Ailey B, Hubbard TJP, Brenner SE, Murzin AG, Chothia C. SCOP: a Structural Classification of Proteins database. Nucleic Acids Res. 2000;28:257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nagano N, Orengo CA, Thornton JM. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002;321:741–765. doi: 10.1016/s0022-2836(02)00649-6. [DOI] [PubMed] [Google Scholar]
- 42.Andreeva A, Murzin AG. Structural classification of proteins and structural genomics: new insights into protein folding and evolution. Acta Crystallographica Section F-Structural Biology and Crystallization Communications. 2010;66:1190–1197. doi: 10.1107/S1744309110007177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weathers EA, Paulaitis ME, Woolf TB, Hoh JH. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett. 2004;576:348–352. doi: 10.1016/j.febslet.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 44.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight Regulation of Unstructured Proteins: From Transcript Synthesis to Protein Degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsvetkov P, Reuven N, Shaul Y. The nanny model for IDPs. Nat. Chem. Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- 46.Tsvetkov P, Asher G, Paz A, Reuven N, Sussman JL, Silman I, Shaul Y. Operational definition of intrinsically unstructured protein sequences based on susceptibility to the 20S proteasome. Proteins-Structure Function And Bioinformatics. 2008;70:1357–1366. doi: 10.1002/prot.21614. [DOI] [PubMed] [Google Scholar]
- 47.Eliezer D, Kutluay E, Bussell R, Browne G. Conformational properties of alpha-synuclein in its free and lipid-associated states. J. Mol. Biol. 2001;307:1061–1073. doi: 10.1006/jmbi.2001.4538. [DOI] [PubMed] [Google Scholar]
- 48.Dyson HJ, Wright PE. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv. Protein Chem. 2002;62:311–340. doi: 10.1016/s0065-3233(02)62012-1. [DOI] [PubMed] [Google Scholar]
- 49.Dyson HJ, Wright PE. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 50.Barre P, Eliezer D. Folding of the repeat domain of tau upon binding to lipid surfaces. J. Mol. Biol. 2006;362:312–326. doi: 10.1016/j.jmb.2006.07.018. [DOI] [PubMed] [Google Scholar]
- 51.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr. Opin. Struct. Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 52.Bezsonova I, Forman-Kay J, Prosser RS. Molecular oxygen as a paramagnetic NMR probe of protein solvent exposure and topology. Concepts in Magnetic Resonance Part A. 2008;32A:239–253. [Google Scholar]
- 53.Jensen MR, Markwick PRL, Meier S, Griesinger C, Zweckstetter M, Grzesiek S, Bernado P, Blackledge M. Quantitative Determination of the Conformational Properties of Partially Folded and Intrinsically Disordered Proteins Using NMR Dipolar Couplings. Structure. 2009;17:1169–1185. doi: 10.1016/j.str.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 54.Salmon L, Nodet G, Ozenne V, Yin GW, Jensen MR, Zweckstetter M, Blackledge M. NMR Characterization of Long-Range Order in Intrinsically Disordered Proteins. J. Am. Chem. Soc. 2010;132:8407–8418. doi: 10.1021/ja101645g. [DOI] [PubMed] [Google Scholar]
- 55.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc. Natl. Acad. Sci. U. S. A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ferreon ACM, Gambin Y, Lemke EA, Deniz AA. Interplay of alpha-synuclein binding and conformational switching probed by single-molecule fluorescence. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5645–5650. doi: 10.1073/pnas.0809232106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nettels D, Muller-Spath S, Kuster F, Hofmann H, Haenni D, Ruegger S, Reymond L, Hoffmann A, Kubelka J, Heinz B, Gast K, Best RB, Schuler B. Single-molecule spectroscopy of the temperature-induced collapse of unfolded proteins. Proc. Natl. Acad. Sci. U. S. A. 2009;106:20740–20745. doi: 10.1073/pnas.0900622106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller-Spath S, Soranno A, Hirschfeld V, Hofmann H, Ruegger S, Reymond L, Nettels D, Schuler B. Charge interactions can dominate the dimensions of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14609–14614. doi: 10.1073/pnas.1001743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rao JN, Jao CC, Hegde BG, Langen R, Ulmer TS. A Combinatorial NMR and EPR Approach for Evaluating the Structural Ensemble of Partially Folded Proteins. J. Am. Chem. Soc. 2010;132:8657–8668. doi: 10.1021/ja100646t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paz A, Zeev-Ben-Mordehai T, Lundqvist M, Sherman E, Mylonas E, Weiner L, Haran G, Svergun DI, Mulder FAA, Sussman JL, Silman I. Biophysical characterization of the unstructured cytoplasmic domain of the human neuronal adhesion protein neuroligin 3. Biophys. J. 2008;95:1928–1944. doi: 10.1529/biophysj.107.126995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jensen MR, Salmon L, Nodet G, Blackledge M. Defining Conformational Ensembles of Intrinsically Disordered and Partially Folded Proteins Directly from Chemical Shifts. J. Am. Chem. Soc. 2010;132:1270–+. doi: 10.1021/ja909973n. [DOI] [PubMed] [Google Scholar]
- 63.Zhang WH, Ganguly D, Chen JH. Residual Structures, Conformational Fluctuations, and Electrostatic Interactions in the Synergistic Folding of Two Intrinsically Disordered Proteins. PLoS Comp. Biol. 2012;8 doi: 10.1371/journal.pcbi.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ganguly D, Zhang WH, Chen JH. Synergistic folding of two intrinsically disordered proteins: searching for conformational selection. Molecular BioSystems. 2012;8:198–209. doi: 10.1039/c1mb05156c. [DOI] [PubMed] [Google Scholar]
- 65.Ganguly D, Chen JH. Atomistic Details of the Disordered States of KID and pKID. Implications in Coupled Binding and Folding. J. Am. Chem. Soc. 2009;131:5214–5223. doi: 10.1021/ja808999m. [DOI] [PubMed] [Google Scholar]
- 66.De Sancho D, Best RB. Modulation of an IDP binding mechanism and rates by helix propensity and non-native interactions: association of HIF1 alpha with CBP. Molecular BioSystems. 2012;8:256–267. doi: 10.1039/c1mb05252g. [DOI] [PubMed] [Google Scholar]
- 67.Espinoza-Fonseca LM, Ilizaliturri-Flores I, Correa-Basurto J. Backbone conformational preferences of an intrinsically disordered protein in solution. Molecular BioSystems. 2012;8:1798–1805. doi: 10.1039/c2mb00004k. [DOI] [PubMed] [Google Scholar]
- 68.Moritsugu K, Terada T, Kidera A. Disorder-to-Order Transition of an Intrinsically Disordered Region of Sortase Revealed by Multiscale Enhanced Sampling. J. Am. Chem. Soc. 2012;134:7094–7101. doi: 10.1021/ja3008402. [DOI] [PubMed] [Google Scholar]
- 69.Vitalis A, Wang X, Pappu RV. Quantitative Characterization of Intrinsic Disorder in Polyglutamine: Insights from Analysis Based on Polymer Theories. Biophys. J. 2007;93:1923–1937. doi: 10.1529/biophysj.107.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vitalis A, Caflisch A. Micelle-Like Architecture of the Monomer Ensemble of Alzheimer's Amyloid-beta Peptide in Aqueous Solution and Its Implications for A beta Aggregation. J. Mol. Biol. 2010;403:148–165. doi: 10.1016/j.jmb.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 71.Wostenberg C, Kumar S, Noid WG, Showalter SA. Atomistic Simulations Reveal Structural Disorder in the RAP74-FCP1 Complex. J. Phys. Chem. B. 2011;115:13731–13739. doi: 10.1021/jp208008m. [DOI] [PubMed] [Google Scholar]
- 72.Lindorff-Larsen K, Best RB, DePristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 73.Vendruscolo M. Determination of conformationally heterogeneous states of proteins. Curr. Opin. Struct. Biol. 2007;17:15–20. doi: 10.1016/j.sbi.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 74.Allison JR, Varnai P, Dobson CM, Vendruscolo M. Determination of the Free Energy Landscape of alpha-Synuclein Using Spin Label Nuclear Magnetic Resonance Measurements. J. Am. Chem. Soc. 2009;131:18314–18326. doi: 10.1021/ja904716h. [DOI] [PubMed] [Google Scholar]
- 75.Robustelli P, Kohlhoff K, Cavalli A, Vendruscolo M. Using NMR Chemical Shifts as Structural Restraints in Molecular Dynamics Simulations of Proteins. Structure. 2010;18:923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 76.De Simone A, Montalvao RW, Vendruscolo M. Determination of Conformational Equilibria in Proteins Using Residual Dipolar Couplings. Journal Of Chemical Theory And Computation. 2011;7:4189–4195. doi: 10.1021/ct200361b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ullman O, Fisher CK, Stultz CM. Explaining the Structural Plasticity of alpha-Synuclein. J. Am. Chem. Soc. 2011;133:19536–19546. doi: 10.1021/ja208657z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Markwick PRL, Cervantes CF, Abel BL, Komives EA, Blackledge M, McCammon JA. Enhanced Conformational Space Sampling Improves the Prediction of Chemical Shifts in Proteins. J. Am. Chem. Soc. 2010;132:1220–+. doi: 10.1021/ja9093692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choy WY, Forman-Kay JD. Calculation of ensembles of structures representing the unfolded state of an SH3 domain. J. Mol. Biol. 2001;308:1011–1032. doi: 10.1006/jmbi.2001.4750. [DOI] [PubMed] [Google Scholar]
- 80.Marsh JA, Forman-Kay JD. Ensemble modeling of protein disordered states: Experimental restraint contributions and validation. Proteins-Structure Function And Bioinformatics. 2012;80:556–572. doi: 10.1002/prot.23220. [DOI] [PubMed] [Google Scholar]
- 81.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Orengo CA, Bray JE, Buchan DWA, Harrison A, Lee D, Pearl FMG, Sillitoe I, Todd AE, Thornton JM. The CATH protein family database: A resource for structural and functional annotation of genomes. Proteomics. 2002;2:11–21. [PubMed] [Google Scholar]
- 83.Rison SCG, Teichmann SA, Thornton JM. Homology, pathway distance and chromosomal localization of the small molecule metabolism enzymes in Escherichia coli. J. Mol. Biol. 2002;318:911–932. doi: 10.1016/S0022-2836(02)00140-7. [DOI] [PubMed] [Google Scholar]
- 84.Rubinstein M, Colby RH. Polymer Physics. Oxford University Press; Oxford and New York: 2003. [Google Scholar]
- 85.Flory PJ. Statistical Mechanics of Chain Molecules. Oxford University Press; New York: 1969. [Google Scholar]
- 86.Yamakawa H. Statisical mechanics of wormlike chains. Pure Appl. Chem. 1976;46:135–141. [Google Scholar]
- 87.Lapidus LJ, Steinbach PJ, Eaton WA, Szabo A, Hofrichter J. Effects of chain stiffness on the dynamics of loop formation in polypeptides. Appendix: Testing a 1-dimensional diffusion model for peptide dynamics. J. Phys. Chem. B. 2002;106:11628–11640. [Google Scholar]
- 88.Buscaglia M, Lapidus LJ, Eaton WA, Hofrichter J. Effects of denaturants on the dynamics of loop formation in polypeptides. Biophys. J. 2006;91:276–288. doi: 10.1529/biophysj.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Singh VR, Lapidus LJ. The intrinsic stiffness of polyglutamine peptides. J. Phys. Chem. B. 2008;112:13172. doi: 10.1021/jp805636p. [DOI] [PubMed] [Google Scholar]
- 90.Kellermayer MSZ, Smith SB, Granzier HL, Bustamante C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 1997;276:1112–1116. doi: 10.1126/science.276.5315.1112. [DOI] [PubMed] [Google Scholar]
- 91.Bemis JE, Akhremitchev BB, Walker GC. Single polymer chain elongation by atomic force microscopy. Langmuir. 1999;15:2799–2805. [Google Scholar]
- 92.Bright JN, Woolf TB, Hoh JH. Predicting properties of intrinsically unstructured proteins. Prog. Biophys. Mol. Biol. 2001;76:131–173. doi: 10.1016/s0079-6107(01)00012-8. [DOI] [PubMed] [Google Scholar]
- 93.Yamakawa H. Helical wormlike chains in polymer solutions. Springer; New York: 1997. [Google Scholar]
- 94.Debye P, Huckel E. Zur Theorie der Elektrolyte. Physikal. Z. 1923;24:185–206. [Google Scholar]
- 95.Hildebrand JH. Theory of solubility. PhRv. 1923;21:46–52. [Google Scholar]
- 96.Huggins M,L. Solutions of long chain compounds. J. Chem. Phys. 1941;9:440–440. [Google Scholar]
- 97.Flory PJ. Thermodynamics of high polymer solutions. J. Chem. Phys. 1942;10:51–61. [Google Scholar]
- 98.Schwarzinger S, Kroon GJA, Foss TR, Wright PE, Dyson HJ. Random coil chemical shifts in acidic 8 M urea: Implementation of random coil shift data in NMRView. J. Biomol. NMR. 2000;18:43–48. doi: 10.1023/a:1008386816521. [DOI] [PubMed] [Google Scholar]
- 99.Schwarzinger S, Kroon GJA, Foss TR, Chung J, Wright PE, Dyson HJ. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 2001;123:2970–2978. doi: 10.1021/ja003760i. [DOI] [PubMed] [Google Scholar]
- 100.Choi UB, McCann JJ, Weninger KR, Bowen ME. Beyond the Random Coil: Stochastic Conformational Switching in Intrinsically Disordered Proteins. Structure. 2011;19:566–576. doi: 10.1016/j.str.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.de Gennes P-G. Scaling Concepts in Polymer Physics. Cornell University Press; Ithaca and London: 1979. [Google Scholar]
- 102.Steinhauser MO. A molecular dynamics study on universal properties of polymer chains in different solvent qualities. Part I. A review of linear chain properties. J. Chem. Phys. 2005;122 doi: 10.1063/1.1846651. [DOI] [PubMed] [Google Scholar]
- 103.Imbert JB, Lesne A, Victor JM. Distribution of the order parameter of the coil-globule transition. PhRvE. 1997;56:5630–5647. [Google Scholar]
- 104.Lifshitz IM, Grosberg AY, Khokhlov AR. Some problems of the statistical physics of polymer chains with volume interaction. Rev. Mod. Phys. 1978;50:683–713. [Google Scholar]
- 105.Grosberg AY, Kuznetsov DV. Quantitative theory of the globule-to-coil transition .1. Link density distribution in a globule and its radius of gyration. Macromolecules. 1992;25:1970–1979. [Google Scholar]
- 106.Grosberg AY, Kuznetsov DV. Quantitative theory of the globule-to-coil transition .2. Density density correlation in a globule and the hydrodynamic radius of a macromolecule. Macromolecules. 1992;25:1980–1990. [Google Scholar]
- 107.Grosberg AY, Kuznetsov DV. Quantitative theory of the globule-to-coil transition .3. Globule globule interaction and polymer-solution binodal and spinodal curves in the globular range. Macromolecules. 1992;25:1991–1995. [Google Scholar]
- 108.Grosberg AY, Kuznetsov DV. Quantitative theory of the globule-to-coil transition .4. Comparison of theoretical results with experimental-data. Macromolecules. 1992;25:1996–2003. [Google Scholar]
- 109.Kohn JE, Millett IS, Jacob J, Zagrovic B, Dillon TM, Cingel N, Dothager RS, Seifert S, Thiyagarajan P, Sosnick TR, Hasan MZ, Pande VS, Ruzcinski I, Doniach S, Plaxco KW. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Penkett CJ, Redfield C, Dodd I, Hubbard J, McBay DL, Mossakowska DE, Smith RA, Dobson CM, Smith LJ. NMR analysis of main-chain conformational preferences in an unfolded fibronectin-binding protein. J. Mol. Biol. 1997;274:152–159. doi: 10.1006/jmbi.1997.1369. [DOI] [PubMed] [Google Scholar]
- 111.Schäfer L. Excluded Volume Effects in Polymer Solutions as Explained by the Renormalization Group. Springer; Berlin: 1999. [Google Scholar]
- 112.Zhou HX. Polymer models of protein stability, folding, and interactions. Biochemistry. 2004;43:2141–2154. doi: 10.1021/bi036269n. [DOI] [PubMed] [Google Scholar]
- 113.Tran HT, Wang X, Pappu RV. Reconciling observations of sequence-specific conformational propensities with the generic polymeric behavior of denatured proteins. Biochemistry. 2005;44:11369–11380. doi: 10.1021/bi050196l. [DOI] [PubMed] [Google Scholar]
- 114.Tran HT, Pappu RV. Toward an accurate theoretical framework for describing ensembles for proteins under strongly denaturing conditions. Biophys. J. 2006;91:1868–1886. doi: 10.1529/biophysj.106.086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jha AK, Colubri A, Freed KF, Sosnick TR. Statistical coil model of the unfolded state: Resolving the reconciliation problem. Proc. Natl. Acad. Sci. U. S. A. 2005;102:13099–13104. doi: 10.1073/pnas.0506078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ding F, Jha RK, Dokholyan NV. Scaling behavior and structure of denatured proteins. Structure. 2005;13:1047–1054. doi: 10.1016/j.str.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 117.Fitzkee NC, Rose GD. Reassessing random-coil statistics in unfolded proteins. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12497–12502. doi: 10.1073/pnas.0404236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bernado P, Blackledge M. A Self-Consistent Description of the Conformational Behavior of Chemically Denatured Proteins from NMR and Small Angle Scattering. Biophys. J. 2009;97:2839–2845. doi: 10.1016/j.bpj.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moglich A, Joder K, Kiefhaber T. End-to-end distance distributions and intrachain diffusion constants in unfolded polypeptide chains indicate intramolecular hydrogen bond formation. Proc. Natl. Acad. Sci. U. S. A. 2006;103:12394–12399. doi: 10.1073/pnas.0604748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Goldenberg DP. Computational simulation of the statistical properties of unfolded proteins. J. Mol. Biol. 2003;326:1615–1633. doi: 10.1016/s0022-2836(03)00033-0. [DOI] [PubMed] [Google Scholar]
- 121.Wang Y, Trewhelia J, Goldenberg DP. Small-angle x-ray scattering of reduced ribonuclease A: Effects of solution conditions and comparisons with a computational model of unfolded proteins. J. Mol. Biol. 2008;377:1576–1592. doi: 10.1016/j.jmb.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Johansen D, Jeffries CMJ, Hammouda B, Trewhella J, Goldenberg DP. Effects of Macromolecular Crowding on an Intrinsically Disordered Protein Characterized by Small-Angle Neutron Scattering with Contrast Matching. Biophys. J. 2011;100:1120–1128. doi: 10.1016/j.bpj.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Johansen D, Trewhella J, Goldenberg DP. Fractal dimension of an intrinsically disordered protein: Small-angle X-ray scattering and computational study of the bacteriophage lambda N protein. Protein Sci. 2011;20:1955–1970. doi: 10.1002/pro.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sziegat F, Silvers R, Hahnke M, Jensen MR, Blackledge M, Wirmer-Bartoschek J, Schwalbe H. Disentangling the Coil. Modulation of Conformational and Dynamic Properties by Site-Directed Mutation in the Non-Native State of Hen Egg White Lysozyme. Biochemistry. 2012;51:3361–3372. doi: 10.1021/bi300222f. [DOI] [PubMed] [Google Scholar]
- 125.Schneider R, Huang JR, Yao MX, Communie G, Ozenne V, Mollica L, Salmon L, Jensen MR, Blackledge M. Towards a robust description of intrinsic protein disorder using nuclear magnetic resonance spectroscopy. Molecular BioSystems. 2012;8:58–68. doi: 10.1039/c1mb05291h. [DOI] [PubMed] [Google Scholar]
- 126.Dima RI, Thirumalai D. Asymmetry in the shapes of folded and denatured states of proteins. J. Phys. Chem. B. 2004;108:6564–6570. [Google Scholar]
- 127.Wolfenden R. Interaction of the peptide bond with solvent water: a vapor phase analysis. Biochemistry. 1978;17:201–204. doi: 10.1021/bi00594a030. [DOI] [PubMed] [Google Scholar]
- 128.Tran HT, Mao A, Pappu RV. Role of backbone-solvent interactions in determining conformational equilibria of intrinsically disordered proteins. J. Am. Chem. Soc. 2008;130:7380–7392. doi: 10.1021/ja710446s. [DOI] [PubMed] [Google Scholar]
- 129.Teufel DP, Johnson CM, Lum JK, Neuweiler H. Backbone-Driven Collapse in Unfolded Protein Chains. J. Mol. Biol. 2011;409:250–262. doi: 10.1016/j.jmb.2011.03.066. [DOI] [PubMed] [Google Scholar]
- 130.Flory PJ. Principles of Polymer Chemistry. Cornell University Press; Ithaca and London: 1953. [Google Scholar]
- 131.Wootton JC, Drummond MH. The Q-linker - a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989;2:535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- 132.Romero PR, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins: Structure, Function and Genetics. 2000;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 133.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proceedings Of The National Academy of Sciences USA. 2006;103:16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang XL, Vitalis A, Wyczalkowski MA, Pappu RV. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins-Structure Function And Bioinformatics. 2006;63:297–311. doi: 10.1002/prot.20761. [DOI] [PubMed] [Google Scholar]
- 135.Walters RH, Murphy RM. Examining Polyglutamine Peptide Length: A Connection between Collapsed Conformations and Increased Aggregation. J. Mol. Biol. 2009;393:978–992. doi: 10.1016/j.jmb.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vitalis A, Pappu R. ABSINTH: A new continuum solvation model for simulations of polypeptides in aqueous solutions. J. Comput. Chem. 2009;30:673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vitalis A, Lyle N, Pappu RV. Thermodynamics of beta-Sheet Formation in Polyglutamine. Biophys. J. 2009;97:303–311. doi: 10.1016/j.bpj.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vitalis A, Wang X, Pappu RV. Atomistic Simulations of the Effects of Polyglutamine Chain Length and Solvent Quality on Conformational Equilibria and Spontaneous Homodimerization. J. Mol. Biol. 2008;384:279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Dougan L, Li J, Badilla CL, Berne BJ, Fernandez JM. Single homopolypeptide chains collapse into mechanically rigid conformations. Proceedings Of The National Academy of Sciences USA. 2009;106:12605–12610. doi: 10.1073/pnas.0900678106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Roseman MA. Hydrophilicity Of Polar Amino-Acid Side-Chains Is Markedly Reduced By Flanking Peptide-Bonds. J. Mol. Biol. 1988;200:513–522. doi: 10.1016/0022-2836(88)90540-2. [DOI] [PubMed] [Google Scholar]
- 141.Roseman MA. Hydrophobicity Of The Peptide C=O...H-N Hydrogen-Bonded Group. J. Mol. Biol. 1988;201:621–623. doi: 10.1016/0022-2836(88)90642-0. [DOI] [PubMed] [Google Scholar]
- 142.Dill KA. Additivity principles in biochemistry. J. Biol. Chem. 1997;272:701–704. doi: 10.1074/jbc.272.2.701. [DOI] [PubMed] [Google Scholar]
- 143.Ha BY, Thirumalai D. Conformations of a polyelectrolyte chain. Phys. Rev. A. 1992;46:R3012–R3015. doi: 10.1103/physreva.46.r3012. [DOI] [PubMed] [Google Scholar]
- 144.Dobrynin AV, Rubinstein M, Obukhov SP. Cascade of transitions of polyelectrolytes in poor solvents. Macromolecules. 1996;29:2974–2979. [Google Scholar]
- 145.Dobrynin AV, Colby RH, Rubinstein M. Scaling theory of polyelectrolyte solutions. Macromolecules. 1995;28:1859–1871. [Google Scholar]
- 146.Mao AH, Crick SL, Vitalis A, Chicoine CL, Pappu RV. Net charge per residue modulates conformational ensembles of intrinsically disordered proteins. Proc. Natl. Acad. Sci. U. S. A. 2010;107:8183–8188. doi: 10.1073/pnas.0911107107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Marsh JA, Forman-Kay JD. Sequence Determinants of Compaction in Intrinsically Disordered Proteins. Biophys. J. 2010;98:2383–2390. doi: 10.1016/j.bpj.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Loh P, Deen GR, Vollmer D, Fischer K, Schmidt M, Kundagrami A, Muthukumar M. Collapse of Linear Polyelectrolyte Chains in a Poor Solvent: When Does a Collapsing Polyelectrolyte Collect its Counterions? Macromolecules. 2008;41:9352–9358. [Google Scholar]
- 149.Potoyan DA, Papoian GA. Energy Landscape Analyses of Disordered Histone Tails Reveal Special Organization of Their Conformational Dynamics. J. Am. Chem. Soc. 2011;133:7405–7415. doi: 10.1021/ja1111964. [DOI] [PubMed] [Google Scholar]
- 150.Das RK, Crick SL, Pappu RV. N-terminal segments modulate the alpha-helical propensities of the intrinsically disordered basic regions of bZIP proteins. J. Mol. Biol. 2012;416:287–299. doi: 10.1016/j.jmb.2011.12.043. [DOI] [PubMed] [Google Scholar]
- 151.Babu MM, Kriwacki RW, Pappu RV. Versatility from protein disorder. Science. 2012;337:1460–1461. doi: 10.1126/science.1228775. [DOI] [PubMed] [Google Scholar]
- 152.Vuzman D, Levy Y. DNA search efficiency is modulated by charge composition and distribution in the intrinsically disordered tail. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21004–21009. doi: 10.1073/pnas.1011775107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dobrynin AV, Colby RH, Rubinstein M. Polyampholytes. J. Polym. Sci. [B] 2004;42:3513–3538. [Google Scholar]
- 154.Damjanovic A, Wu X, Garcia-Moreno E,B, Brooks BR. Backbone Relaxation Coupled to the Ionization of Internal Groups in Proteins: A Self-Guided Langevin Dynamics Study. Biophys. J. 2008;95:4091–4101. doi: 10.1529/biophysj.108.130906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Van Roey K, Gibson TJ, Davey NE. Motif switches: decision-making in cell regulation. Curr. Opin. Struct. Biol. 2012;22:378–385. doi: 10.1016/j.sbi.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 156.Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Molecular Biosystems. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 157.Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-Specific Splicing of Disordered Segments that Embed Binding Motifs Rewires Protein Interaction Networks. Mol. Cell. 2012;46:871–883. doi: 10.1016/j.molcel.2012.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kishore SP, Perkins SL, Templeton TJ, Deitsch KW. An Unusual Recent Expansion of the C-Terminal Domain of RNA Polymerase II in Primate Malaria Parasites Features a Motif Otherwise Found Only in Mammalian Polymerases. J. Mol. Evol. 2009;68:706–714. doi: 10.1007/s00239-009-9245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Cardarelli F, Serresi M, Bizzarri R, Beltram F. Tuning the transport properties of HIV-1 tat arginine-rich motif in living cells. Traffic. 2008;9:528–539. doi: 10.1111/j.1600-0854.2007.00696.x. [DOI] [PubMed] [Google Scholar]
- 160.Fuxreiter MJ, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Biophys. J. 2007:398A–398A. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 161.Zanuy D, Gunasekaran K, Lesk AM, Nussinov R. Computational study of the fibril organization of polyglutamine repeats reveals a common motif identified in beta-helices. J. Mol. Biol. 2006;358:330–345. doi: 10.1016/j.jmb.2006.01.070. [DOI] [PubMed] [Google Scholar]
- 162.Friedel M, Shea JE. Self-assembly of peptides into a beta-barrel motif. J. Chem. Phys. 2004;120:5809–5823. doi: 10.1063/1.1649934. [DOI] [PubMed] [Google Scholar]
- 163.Das C, Edgcomb SP, Peteranderl R, Chen L, Frankel AD. Evidence for conformational flexibility in the Tat-TAR recognition motif of cyclin T1. Virology. 2004;318:306–317. doi: 10.1016/j.virol.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 164.Kumaki Y, Matsushima N, Yoshida H, Nitta K, Hikichi K. Structure of the YSPTSPS repeat containing two SPXX motifs in the CTD of RNA polymerase II: NMR studies of cyclic model peptides reveal that the SPTS turn is more stable than SPSY in water. Biochimica Et Biophysica Acta-Protein Structure and Molecular Enzymology. 2001;1548:81–93. doi: 10.1016/s0167-4838(01)00216-3. [DOI] [PubMed] [Google Scholar]
- 165.Burley SK. DNA-binding motifs from eukartyotic transcription factors. Curr. Opin. Struct. Biol. 1994;4:3–11. [Google Scholar]
- 166.Reeves R, Nissen MS. The A.T-DNA-binding domain of mammalian high mobility group-i chromosomal-proteins - a novel peptide motif for recognizing DNA-structure. J. Biol. Chem. 1990;265:8573–8582. [PubMed] [Google Scholar]
- 167.Vuzman D, Levy Y. Intrinsically disordered regions as affinity tuners in protein-DNA interactions. Molecular BioSystems. 2012;8:47–57. doi: 10.1039/c1mb05273j. [DOI] [PubMed] [Google Scholar]
- 168.Peng ZL, Mizianty MJ, Xue B, Kurgan L, Uversky VN. More than just tails: intrinsic disorder in histone proteins. Molecular BioSystems. 2012;8:1886–1901. doi: 10.1039/c2mb25102g. [DOI] [PubMed] [Google Scholar]
- 169.Das RK, Mao AH, Pappu RV. Unmasking Functional Motifs Within Disordered Regions of Proteins. Science Signaling. 2012;5:e17. doi: 10.1126/scisignal.2003091. [DOI] [PubMed] [Google Scholar]
- 170.Khazanov N, Levy Y. Sliding of p53 along DNA Can Be Modulated by Its Oligomeric State and by Cross-Talks between Its Constituent Domains. J. Mol. Biol. 2011;408:335–355. doi: 10.1016/j.jmb.2011.01.059. [DOI] [PubMed] [Google Scholar]
- 171.Nishikawa I, Nakajima Y, Ito M, Fukuchi S, Homma K, Nishikawa K. Computational Prediction of O-linked Glycosylation Sites That Preferentially Map on Intrinsically Disordered Regions of Extracellular Proteins. International Journal of Molecular Sciences. 2010;11:4992–5009. doi: 10.3390/ijms11124991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Metallo SJ. Intrinsically disordered proteins are potential drug targets. Curr. Opin. Chem. Biol. 2010;14:481–488. doi: 10.1016/j.cbpa.2010.06.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kovacech B, Novak M. Tau Truncation is a Productive Posttranslational Modification of Neurofibrillary Degeneration in Alzheimer's Disease. Current Alzheimer Research. 2010;7:708–716. doi: 10.2174/156720510793611556. [DOI] [PubMed] [Google Scholar]
- 174.Hegde ML, Hazra TK, Mitra S. Functions of disordered regions in mammalian early base excision repair proteins. Cell. Mol. Life Sci. 2010;67:3573–3587. doi: 10.1007/s00018-010-0485-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Brown CJ, Johnson AK, Daughdrill GW. Comparing Models of Evolution for Ordered and Disordered Proteins. Mol. Biol. Evol. 2010;27:609–621. doi: 10.1093/molbev/msp277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.van Dieck J, Teufel DP, Jaulent AM, Fernandez-Fernandez MR, Rutherford TJ, Wyslouch-Cieszynska A, Fersht AR. Posttranslational Modifications Affect the Interaction of S100 Proteins with Tumor Suppressor p53. J. Mol. Biol. 2009;394:922–930. doi: 10.1016/j.jmb.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 177.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Frontiers in Bioscience-Landmark. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 178.Stein A, Pache RA, Bernado P, Pons M, Aloy P. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 2009;276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- 179.Ahmed MAM, Bamm VV, Shi L, Steiner-Mosonyi M, Dawson JF, Brown L, Harauz G, Ladizhansky V. Induced Secondary Structure and Polymorphism in an Intrinsically Disordered Structural Linker of the CNS: Solid-State NMR and FTIR Spectroscopy of Myelin Basic Protein Bound to Actin. Biophys. J. 2009;96:180–191. doi: 10.1016/j.bpj.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Eisenhaber B, Eisenhaber F. Posttranslational modifications and subcellular localization signals: Indicators of sequence regions without inherent 3D structure? Curr. Protein Peptide Sci. 2007;8:197–203. doi: 10.2174/138920307780363424. [DOI] [PubMed] [Google Scholar]
- 181.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics Model. 1996;14 doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]