Abstract
Land plants associate with a root microbiota distinct from the complex microbial community present in surrounding soil. The microbiota colonizing therhizosphere(immediately surroundingthe root) and the endophytic compartment (within the root) contribute to plant growth, productivity, carbon sequestration and phytoremediation1-3. Colonization of the root occurs despite a sophisticated plant immune system4,5, suggesting finely tuned discrimination of mutualists and commensals from pathogens. Genetic principles governing the derivation of host-specific endophyte communities from soil communities are poorly understood. Here we report the pyrosequencing of the bacterial 16S ribosomal RNA gene of more than 600 Arabidopsis thaliana plants to test the hypotheses that the root rhizosphere and endophytic compartment microbiota of plants grown under controlled conditions in natural soils are sufficiently dependent on the host to remain consistent across different soil types and developmental stages, and sufficiently dependent on host genotype to vary between inbred Arabidopsis accessions. We describe different bacterial communities in two geochemically distinct bulk soils and in rhizosphere and endophytic compartments prepared from roots grown in these soils. The communities in each compartment are strongly influenced by soil type. Endophytic compartments from both soils feature overlapping, low-complexity communities that are markedly enriched in Actinobacteria and specific families from other phyla, notably Proteobacteria. Some bacteria vary quantitatively between plants of different developmental stage and genotype. Our rigorous definition of an endophytic compartment microbiome should facilitate controlled dissection of plantmicrobe interactions derived from complex soil communities.
Roots influence the rhizosphere by altering soil pH, soil structure, oxygen availability, antimicrobial concentration, and quorum-sensing mimicry, and by providing an energy source of dead root material and carbon-rich exudates6,7. The microbiota inhabiting this niche can both benefit and undermine plant health; shifting this balance is of agronomic interest. Mutualistic microbes may provide the plant with physiologically accessible nutrients and phytohormones that improve plant growth, may suppress phytopathogens or may help plants withstand heat, salt and drought8,9. The rhizosphere community is a subset of soil microbes that are subsequently filtered via niche utilization attributes and interactions with the host to inhabit the endophytic compartment10 (EC). Although a variety of microbes may enter and become transient endophytes, those consistently found inside roots are candidate symbionts or stealthy pathogens10,11. Notably, Arabidopsis and other Brassicaceae are not well colonized byarbuscular mycorrhizal fungi, implying that other microorganisms may fill this niche.
Microbial community structure differs across plant species12,13, and there are reports of host-genotype-dependent differences in patterns of microbial associations14,15. However, the divergent methods used in those studies relied on small sample sizes and low-resolution phylotyping techniques potentially confounded by off-target sequences and chimaeric amplicons. We developed a robust experimental system to sample repeatedly the root microbiome using high-throughput sequencing. Our results confirm many of the general conclusions from earlier studies and, because of controlled experimental design and the power of deep sequencing, provide a key step towards the definition of this microbiome’s functional capacity and the host genes that potentially contribute to microbial association phenotypes. Such plant genes would constitute major agronomic targets.
We used 454 pyrosequencing to sequence 16S ribosomal RNA (rRNA) gene amplicons for DNA prepared from eight diverse, inbred A. thaliana accessions. Plants were grown from surface-sterile seeds in climate-controlled conditions in two diverse soils, respectively termed Mason Farm and Clayton (Supplementary Table 1; detailed in Supplementary Information). For each soil, we assayed multiple individuals from each A. thaliana accession grown from sterile seeds in both soils across independent full-factorial biological replicates, in which all genotypes and bulk soils (pots without a plant) for a given soil type were grown in parallel (Supplementary Table 2). We isolated separate rhizosphere and EC fractions from individual plant root systems (Supplementary Fig. 1 and Supplementary Table 2). We established 1114F and 1392R as our primer pair (Supplementary Information and Supplementary Fig. 2). Using an otupipe-based pipeline (http://drive5.com/otupipe/), we grouped sequences into 97%-identical operational taxonomic units (OTUs), reduced noise and removed chimaeras. We determined technical reproducibility thresholds to conclude that OTUs defined by ≥25 reads in ≥5 samples (hereafter 25 × 5) are individually ‘measurable OTUs’16,17 (Supplementary Figs 2 and 10). All data reported here are from one run of our otupipe-based pipeline (Supplementary Fig. 3 and Supplementary Database 1).
Excluding additional control samples, we ribotyped 1,248 samples comprising 111 bulk soil, 613 rhizosphere and 524 EC samples, generating 9,787,070 high-quality reads (Supplementary Figs 3 and 4a-c). After removing plant-sequence-derived OTUs, we obtained a table of usable OTU read counts per sample containing 6,387,407 reads distributed across 18,783 OTUs. We normalized this table of usable reads by rarefying to 1,000 reads per sample (Supplementary Database 2a) or, alternatively, by dividing the reads per OTU in a sample by the sum of usable reads in that sample, resulting in a table of relative abundances (frequencies) (Supplementary Database 2b). Using the 25 × 5 threshold, we defined 778 measurable OTUs representing 54% (3,463,632) of the usable reads (Supplementary Fig. 4c and Supplementary Table 3). The diversity of the 778 measurable OTUs in soil, rhizosphere and EC fractions showed expected relative trends when compared with the diversity by fraction of all usable OTUs (Supplementary Fig. 4d). We display the rarefaction-normalized data; parallel analyses of frequency-normalized data are provided in Supplementary Figures.
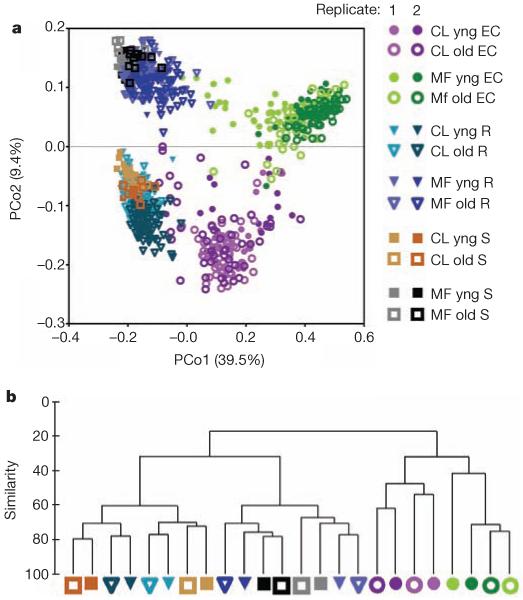
We used principal coordinate analysis on pairwise, normalized, weighted UniFrac distances between all samples, considering all usable OTUs, to identify the main factors driving community composition (Fig. 1a and Supplementary Fig. 5a). The first principal coordinate (PCo1) revealed that the two bulk soils and their associated rhizospheres were differentiated from the respective EC fractions. Soil type was the main factor in the second component (PCo2). This pattern was recapitulated by hierarchical clustering of pairwise Bray–Curtis dissimilarities considering only measurable OTUs (Fig. 1b and Supplementary Fig. 5b). Samples harvested at different developmental stages clustered together, indicating that this variable does not have a major effect on overall community composition (Fig. 1 and Supplementary Fig. 5a, b; yng versus old, where yng refers to the time of appearance of an inflorescence meristem and old refers to fruiting plants with greater than 50% senescent leaves). Additional control samples from the reference genotype Col-0 harvested from four independent digs of Mason Farm soil underscored the reproducibility of these bacterial community profiles (Supplementary Fig. 6). Together, these data demonstrate that the interaction of diverse soil communities with plants determines the assembly of the rhizosphere, leading to winnowed ECs, that the ECs from at least these two diverse soils are very different from the starting soil communities and that there is little difference in communities over host developmental time.
Figure 1. Sample fraction and soil type drive the microbial composition of root-associated endophyte communities.
a, Principal coordinate analysis of pairwise, normalized, weighted UniFrac distances between samples based on rarefaction to 1,000 reads in unthresholded, usable OTUs. CL, Clayton; MF, Mason Farm; R, rhizosphere; S, soil. b, Rarefied counts for the 25 × 5 thresholded, measurable OTUs from each of 24 soil, stage or fraction groups were log2-transformed (Methods) to make 24 representative samples (branch labels), and pairwise Bray–Curtis similarity was used to cluster these representatives hierarchically (group-average linkage).
We fitted a general linear mixed model (GLMM) to samples from each set of plant fractions (rhizosphere or EC), plus the bulk soil controls, to identify measurable OTUs whose abundances differ significantly between plant and bulk soil as a result of soil type, developmental stage, fraction and genotype (Supplementary Information and Supplementary Database 3). This approach allowed us to quantify the contribution from each variable to the community composition (Supplementary Table 4). Controlling for sequencing plate effects, plant fraction is the most important factor; its effect is strongest for the EC, consistent with our UniFrac and Bray–Curtis analyses. Soil type is less important, followed by experiment, developmental stage and, finally, genotype, which had a small but consistent effect.
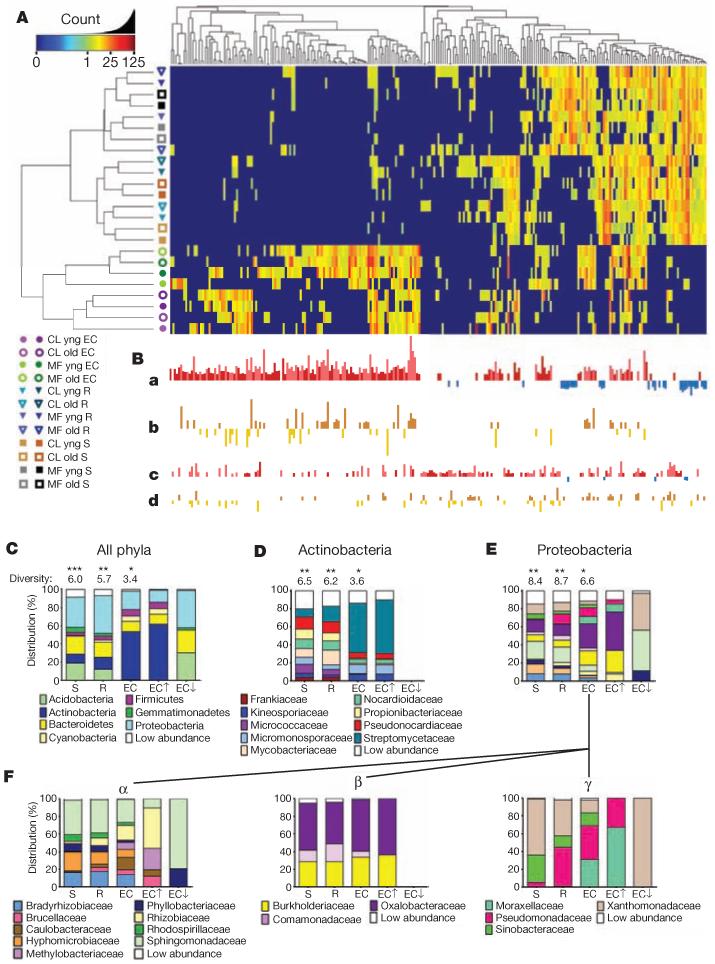
Hierarchical clustering of sample groups considering 256 OTUs identified by the GLMM to differentiate rhizosphere and EC from soil recapitulated the separation of EC from soil and rhizosphere (Fig. 2A and Supplementary Fig. 7a, left; compare with Fig. 1 and Supplementary Fig. 5). Of these, 164 OTUs were enriched in EC samples (Fig. 2B, a; dark and light red bars), defining an A. thaliana ‘EC microbiome’. Of these 164, 97 were enriched in EC samples from both soil types (Fig. 2B, a; dark red bars), potentially representing a core EC microbiome. By contrast, 67 of these 164 were enriched in EC to a greater extent in one soil than the other (Fig. 2B, a; light red bars; Fig. 2B, b)). Importantly, 32 OTUs were depleted in EC samples (Fig. 2B, a; blue bars). Some OTUs exhibited rhizosphere enrichment; these significantly overlapped the EC-enriched OTUs (P < 10−16, one-sided hypergeometric test) and also sometimes had a soil-type component (Fig. 2B, c and d). Only a few rhizosphere-specific enrichments were not also enriched in the EC (Supplementary Table 3). Hence, the A. thaliana EC microbiome is enriched for both a shared set of OTUs commonly assembled across two replicates from two diverse soils, and a set of OTUs that are assembled from each soil.
Figure 2. OTUs that differentiate the EC and rhizosphere from soil.
A, Heat map showing OTU counts from the rarefied OTU table (Supplementary Database 2a; log2-transformed) from each of the 256 rhizosphere- and EC-differentiating OTUs present across replicates. Samples and OTUs are clustered on their Bray–Curtis similarities (group-average linkage). The key relates colours to the untransformed read counts. Different hues of the same colour correspond to different replicates as in Fig. 1. B, The strength of GLMM predictions (best linear unbiased predictors) is represented by bar height. a, OTUs predicted as EC enriched (red, up) or EC depleted (blue, down). b, OTUs higher in the EC in Mason Farm soil than Clayton (brown, up) or higher in Clayton soil than Mason Farm (gold, down). OTUs in a that are not differentially affected by soil type are shown there in darker hues. c, OTUs predicted as rhizosphere enriched (as in a). d, OTUs higher in rhizosphere in one soil type (as in b). C, Histograms showing the distributions of phyla present in the 778 measurable OTUs in soil, rhizosphere and ECs compared with phyla present in the subset of EC OTUs enriched (EC↑) or depleted (EC↓) relative to soil. Shannon diversity (considering phyla as individuals) is given above each bar. A differential number of asterisks above the diversity values represents a significant difference (P < 0.05, weighted analysis of variance; Supplementary Methods and Supplementary Table 5). D, Distribution of families present among the OTUs from the phylum Actinobacteria. E, Distribution of families present among the OTUs from the phylum Proteobacteria. F, Distribution of families present among the OTUs of three classes of the phylum Proteobacteria: Alphaproteobacteria (α), Betaproteobacteria (β) and Gammaproteobacteria (γ). Statistical evidence for presence, enrichment in or depletion from EC is in Supplementary Table 6.
We assessed taxonomic distributions, first those of the 778 measurable OTUs in soil, rhizosphere and EC fractions, and then those of the 256 EC-enriched and 32 EC-depleted OTUs (Fig. 2A, Supplementary Fig. 7a and Supplementary Table 3). Measurable OTUs were distributed across seven dominant phyla (Fig. 2C and Supplementary Fig. 7c) and contained ~50–70% of the usable reads in all fractions (Supplementary Fig. 4c). Phyla distribution of the ECenriched OTUs reflected that of the entire EC. Conversely, the phyla distribution of the EC-depleted OTUs typically resembled that of the rhizosphere fraction (Fig. 2C). The lower Shannon diversity of the EC fraction is consistent with enrichment for a subset of dominant phyla. Specifically, the EC microbiome was dominated by Actinobacteria, Proteobacteria and Firmicutes, and was depleted of Acidobacteria, Gemmatimonadetes and Verrucomicrobia, when soil types were considered either together or separately (Fig. 2C, Supplementary Figs 7c and 15 and Supplementary Table 5). Lower-order taxonomic analysis (Fig. 2D and Supplementary Fig. 7d) demonstrated that enrichment of a low-diversity Actinobacteria community in the EC was driven by a subset of families, predominantly Streptomycetaceae.
Other phyla, such as Proteobacteria, were represented by both EC enrichments and EC depletions at the family level (Fig. 2E and Supplementary Fig. 7e). Strikingly, two alphaproteobacterial families, Rhizobiaceae and Methylobacteriaceae, and two gammaproteobacterial families, Pseudomonadaceae and Moraxellaceae, dominated the EC population in their respective classes (Fig. 2F, α and γ, and Supplementary Fig. 7f, α and γ). Equally striking was the EC redistribution of particular alpha- and gammaproteobacterial families that were common in soil and rhizosphere (Fig. 2F and Supplementary Fig. 7f).
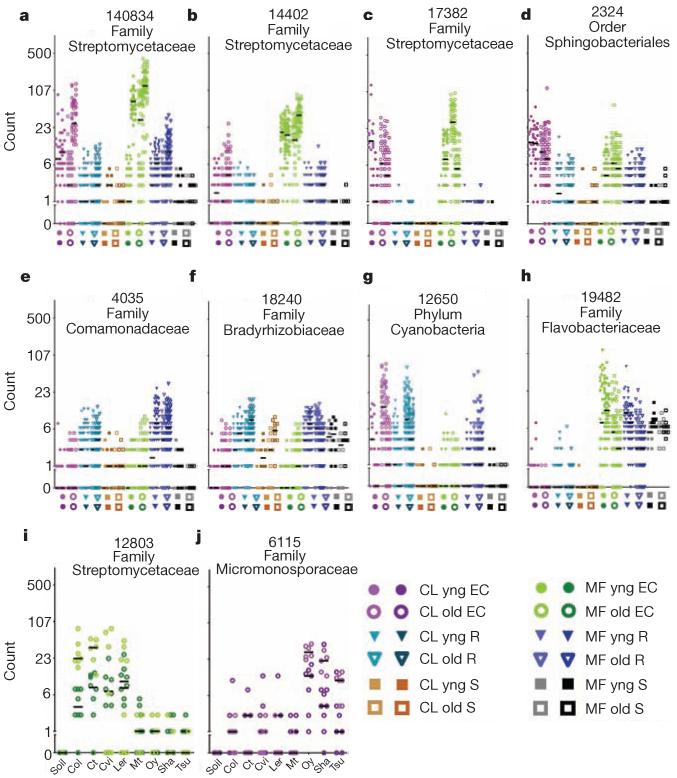
Specific OTUs, three from the family Streptomycetaceae and one from the order Sphingobacteriales, demonstrate the robustness of EC enrichments (Fig. 3a–d and Supplementary Fig. 11a– d). A few OTUs were either significantly enriched in rhizosphere but not in the EC (Fig. 3e, f, Supplementary Fig. 11e, f and Supplementary Table 3), or were associated with one of the two developmental stages (Fig. 3g, h, Supplementary Fig. 11g, h and Supplementary Table 3). Data in Fig. 2, Supplementary Fig. 7, Fig. 3, Supplementary Fig. 11 and Supplementary Table 3 demonstrate that entire taxa at various levels are enriched in or depleted from the EC microbiome. Additionally, rhizosphere taxa capable of colonizing the root vicinity are nonetheless prevented from colonizing the EC.
Figure 3. Dot plots of notable OTUs.
Counts for each OTU (number at top keyed to Supplementary Table 3) from the rarefied table were log2-transformed and the counts for each sample plotted as an individual symbol. The y axis is labelled with the actual (untransformed) counts. a–h, Each position on the x axis is labelled with a symbol to represent the sample group, and samples from that group are plotted in the column directly above. Biological replicates in the same column have different hues. The median of each replicate is shown with a horizontal black bar; some are invisible because they are at 0. i, j, Each x-axis position is labelled by Arabidopsis accession; samples from that accession are plotted above each label. Each OTU in the figure has model predictions in several categories (Supplementary Table 3).
Several OTUs differentiated inbred A. thaliana accessions. Genotype-dependent enrichments and depletions were significant but weak (Supplementary Tables 5 and 3). To identify accession-dependent effects specific to a soil type or a developmental stage, we fitted a partial GLMM that modelled each genotype against bulk soil for each experiment or developmental stage group, and tested the model’s predictions with a non-parametric Kruskal–Wallis test corrected for multiple testing (Supplementary Information). We considered only those significant accession-dependent effects that were present in the same direction in both biological replicates. We further required that these OTUs have a consistent prediction in the full GLMM, which narrowed the field to 12 OTUs (or 27 with frequency-normalized data; Supplementary Table 3). In Fig. 3, we display relative abundances of two such OTUs, one for each soil type, both Actinobacteria (Fig. 3i, j and Supplementary Fig. 11i, j). That these enrichments were detected by the full GLMM (which accounts for plate effects due to 454 sequencing), and were sequenced over several plates (Supplementary Fig. 14) supports a true genotype effect. Thus, a small subset of the EC microbiome is likely to be quantitatively influenced by host-genotype-dependent fine-tuning in specific soil environments. This could allow compensatory contributions of the EC microbiome and host genome variation to overall metagenome function.
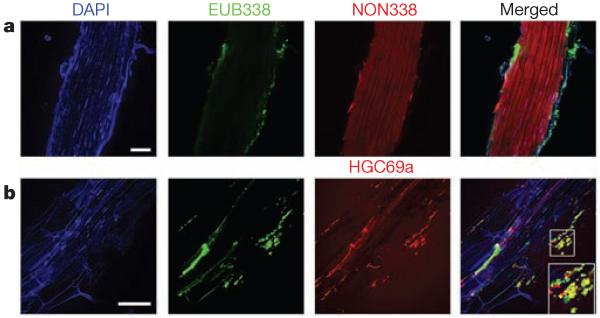
Because the rhizoplane is stripped during preparation of EC fractions, we confirmed the presence of live bacteria on roots using catalysed reporter deposition and fluorescence in situ hybridization (CARD–FISH) to whole Col-0 root segments18. Eubacteria were common on unsonicated roots (Fig. 4a). Actinobacteria detected with probe HGC69a were visible on the surface of roots grown in Mason Farm soil, and co-localized with a subset of the eubacterial signals using double CARD–FISH (Fig. 4b), suggesting that their enrichment in EC fractions either comes from, or egresses through, the rhizoplane. Similarly, we confirmed the rare presence on the rhizoplane of Bradyrhizobiaceae (Supplementary Fig. 12c), a family with members defined by the GLMM as more abundant in Mason Farm rhizosphere than Mason Farm EC (Fig. 3f and Supplementary Fig. 11f). We enumerated the relative number of CARD–FISH signals on a set of filters made from equal amounts of material harvested in the same way as were the samples processed for pyrotag sequencing (Supplementary Fig. 12a, b). We confirmed that Actinobacteria were found in higher abundance, and that Bradyrhizobiaceae were present in lower abundances, in EC samples than in the bulk soil and rhizosphere samples. We also noted that emerging lateral roots were typically heavily colonized by a variety of bacteria (Supplementary Fig. 12d) consistent with previous observations19. These results are PCR-independent support for our sequencing methods.
Figure 4. CARD–FISH confirmation of Actinobacteria on roots.
A single set of Mason Farm yng Col-0 roots were fixed and stained using CARD–FISH. DAPI, 4′,6-diamidino-2-phenylindole. Double CARD–FISH was applied using the EUB338 eubacterial probe (green) and either the NON338 probe (a), which is the nonsense negative control of EUB338, or the HGC69a Actinobacteria probe (b). Inset, twofold enlargement of boxed region. Scale bars, 50 μm.
We present a reduced-complexity, robust experimental platform with which to study root microbiota. Our data, and similar conclusions presented in a companion publication20 using a similar platform, provide the deepest analysis available regarding the principles of root microbiome assembly for any plant species. Remarkably, our conclusions are very similar to those in ref. 20 and we identify phyla and family level enrichments in the EC fraction that largely overlap with those reported in ref. 20. We note three main differences between our study and that of ref. 20: different soils from a different continent, a different primer pair and a different portion of root harvested (top 3 cm in ref. 20; whole root here).
A subset of the soil bacterial population is typically enriched in rhizosphere samples7. Thus, a diverse bacterial community can surround the root surface and thrive there, recruited by biophysical and/or host-derived metabolic cues. We demonstrate that the A. thaliana microbiome undergoes dramatic loss of diversity as the spatial level of plant–microbe ‘intimacy’ further increases from the external rhizosphere to the intercellular EC. Both common and soil-type-specific OTUs are established inside roots grown in diverse soils. A small number of bacterial taxa, particularly the Actinobacteria family Streptomycetaceae, and several Proteobacteria families, are highly enriched in the EC. Actinobacteria are well known for production of antimicrobial secondary metabolites9, and many proteobacterial families contain plant-growth-promoting members. Conversely, several taxa (Acidobacteria, Verrucomicrobia and Gemmatimonadetes, and various proteobacterial families) that are common in soil and rhizosphere are depleted from the EC. This depletion suggests that these taxa are either actively excluded by the host immune system, outcompeted by more-successful EC colonizers or metabolically unable to colonize the EC niche. Our identification of a limited-diversity EC facilitates detailed characterization of the isolates comprising the core A. thaliana microbiome, which could facilitate the design of community-based plant probiotics.
Within the EC, we identified rare cases of quantitative variation in the enrichment of specific bacteria at two developmental stages or by different host genotypes, consistent with rare genotype-dependent associations noted in ref. 20. The former result suggests that the EC microbiome is robust to the source–sink differences across these two developmental stages, which may be related to the relatively high frequency of putative saprophytes defined in ref. 20. The latter result suggests that host genetic variation can drive either differential recruitment of beneficial microbes and/or differential exclusion. A limited-diversity EC microbiome with common features suggests similar host needs across A. thaliana, potentially extending to other plant taxa. These are probably fulfilled by contributions from a limited number of bacterial taxa across diverse soils. The identification of genotype-specific endophyte associations in particular soils may signal interactions that meet environment-specific host needs, balancing contributions of EC microbiome and host genome variation to overall metagenome function. These two generalities suggest that the A. thaliana root microbiome might assemble by core ecological principles similar to those shaping the mammalian microbiome, in which core phylum level enterotypes provide broad metabolic potential combined with modest levels of host-genotype-dependent associations that individualize the metagenome21,22. Isolation and characterization of the microbes that define host-genotype-dependent associations, and characterization beyond the 16S gene, should be particularly instructive in unravelling the molecular rules contributing to endophytic colonization and persistence.
METHODS
General strategy
Seed sterility was verified by plating and deep-sequencing of homogenates from sterile seedlings (Supplementary Fig. 13). We established seedling growth, harvesting and DNA preparation pipelines as detailed in the specific sections below. We defined the bacterial community within each soil, and the community associated with plant roots across a number of controlled experimental variables: soil type, plant sample fraction, plant age and plant genotype. For plant age, we harvested roots from two developmental stages: at the formation of an inflorescence meristem (yng) and during fruiting when ≥50% of the rosette leaves were senescent (old). The former represents plants at the peak of photosynthetic conversion to carbon, whereas the latter represents a stage well after the source-sink shift has occurred, marking the change in carbon allocation from vegetal to reproductive utilization23. We prepared two microbial sample fractions from each individual plant: a rhizosphere (bacteria contained in the layer of soil covering the outer surface of the root system that could be washed from roots in a buffer/detergent solution), and EC (bacteria from within the plant root system after sonication-based removal of the rhizoplane; Supplementary Fig. 1). We also collected control soil samples (soil treated in parallel, but without a plant grown in it).
Soil collection and analysis
For each full-factorial experiment, the top 8 in of earth were collected with a shovel and transported to the lab in closed plastic containers at room temperature from two collection sites. The first collection site, Mason Farm, is managed by the North Carolina Botanical Garden and is free of pesticide use and heavy human traffic and is located in Chapel Hill, North Carolina, USA (+35° 53′ 30.40″, −79° 1′ 5.37″). The second collection site is the Central Crops Research Station in Clayton, North Carolina, USA (135° 39′ 59.22″, −78° 29′ 35.69″) and is also free of pesticide use. Visible weeds, twigs, worms, insects and so on were removed with gloves, and the soil was then crushed with an aluminium mallet to a fine consistency and sifted through a sterile 2-mm sieve. Because sieved soil from Mason Farm drained poorly and test plants grown in it suffered from hypoxia, we adopted the practice of mixing sterile (autoclaved) playground sand into both Mason Farm (MF) and Clayton (CL) soils at a soil:sand ratio of 2:1. Soil micronutrient analysis was performed on pure and 2:1 mixed soils by the University of Wisconsin soil testing labs.
Seed sterilization and germination
All seeds were surface-sterilized by a treatment of 1 min in 70% ethanol with 0.1% Triton-X100, followed by 12 min in 10% A-1 bleach with 0.1% Triton-X100, followed by three washes in sterile distilled water. Seeds were spread on 0.5% agar containing half-strength Murashige & Skoog (MS) vitamins and 1% sucrose. Seeds were stratified in the dark at 4 °C for one week, then germinated at 24 °C under 18 h of light for one week. Seed coat sterility was confirmed by lack of visible contamination on MS plates during germination, and also by absence of visible contamination after plating some of the whole seeds on KB, 1/10-strength LB and 1/10-strength ‘869’ bacterial growth media.
To address whether there were seed-borne microbes that might survive surface sterilization, one-week-old seedlings were taken from sterile MS plates and homogenized by aseptic bead beating under non-bacteriolytic conditions (three 3-mm glass balls per 2-ml tube, with 300-μl PBS, using a FastPrep from MP Bio at speed 4.0 m s2−1 for 10 s). The homogenate was streaked onto 1/10-strength LB, 1/10-strength ‘869’ and KB media. No colonies were observed. To detect potential unculturable microbes, we pyrosequenced 16S amplicons from the same homogenates using bacteriolytic DNA preps from the genotypes Col-0, Cvi-0, Sha-0 and Tsu-0 (Supplementary Fig. 13). Each accession was individually barcoded and sequenced with 1114F and 1392R, yielding 21,935, 20,747, 23,141 and 20,272 reads, respectively. A matching number of total reads was sampled from each accession using pooled data from the full experimental data set for comparative analysis. Thus, 86,095 high-quality reads were obtained from both non-sterile plants and sterile plants, the majority of which were chloroplast sequences. See Supplementary Fig. 13 for results.
Seedling growth
One-week-old healthy seedlings were aseptically transplanted from MS plates to sterile (autoclaved) 2.5-inch-square pots filled with either MF or CL soil, with one seedling per pot. Seedlings were transferred by lifting from underneath the cotyledon leaves using open tweezers; no pressure was applied to the hypocotyl. Some pots were designated ‘bulk soil’ and were not given a plant. All pots, including bulk soil controls, were always watered from the top with a shower of distilled water (non-sterile) as an accessible proxy for rain water that avoids chlorine and other tapwater additives. Pots were spatially randomized and placed in growth chambers providing short days of 8 h light (800–1,000 lx) at 21 °C and 16 h dark at 18 °C. The use of short days was to help synchronize flowering time between A. thaliana genotypes and to facilitate robust rosette and root growth. After harvesting the floral transition developmental stage, remaining plants and bulk soils were moved from the growth chamber to 16-h days in the greenhouse to promote a more synchronized flowering and senescence for the senescent developmental stage.
Harvesting
Each plant was killed and harvested at one of two developmental time points: (1) at the floral transition and (2) after fruiting when senescence is well underway. We considered the floral transition to have begun when the shoot apical meristem was first apparent in five or more plants. Cvi-0, Sha-0 and Ct-1 occasionally flowered one to two weeks earlier under our conditions than the other A. thaliana genotypes. The senescence harvest began when five or more plants showed 50% or more yellow and/or brown rosette leaves24; this occurred approximately four to five weeks after transfer to the greenhouse. Senescence occurred in the same order as bolting (flowering).
Our maximum harvesting and processing capacity was 30 plants per day, meaning that each harvesting period for each full-factorial biological replicate (90 pots) lasted between one and two weeks. On each harvest day, we strove to represent all genotypes and at least one bulk soil to avoid potential confounding harvesting artefacts with genotype effects. Because we harvested as many pots each day as time allowed, we did not always harvest in multiples of our genotype number and did not have equal representation of each genotype on each harvest day.
The aboveground plant organs were aseptically removed. Loose soil was manually removed from the roots by kneading and shaking with sterile gloves (sprayed with 70% EtOH) and by patting roots with a sterile (flamed) metal spatula—this ‘neighbouring soil’ fell to the sterile (flamed) work surface. We followed the established convention of defining rhizosphere soil as extending up to 1 mm from the root surface25 and we removed loose soil on all root surfaces until remaining aggregates were within this range. Roots were placed in a clean and sterile 50-ml tube containing 25 ml phosphate buffer (per litre: 6.33 g of NaH2PO4·H2O, 16.5g of Na2HPO4·7H2O, 200 μl Silwet L-77). Tubes were vortexed at maximum speed for 15 s, which released most of the rhizosphere soil from the roots and turned the water turbid. The turbid solution was then filtered through a 100-μm nylon mesh cell strainer into a new 50-ml tube to remove broken plant parts and large sediment. The roots were transferred from the empty tube to a new sterile 50-ml tube with 25-ml sterile phosphate buffer, and the turbid filtrate was centrifuged for 15 min at 3,200g to form a pellet containing fine sediment and microorganisms.
Most of the supernatant was removed and the loose pellets were resuspended and transferred to 1.5-ml microfuge tubes, which were then spun at 10,000g for 5 min to form tight pellets, from which all supernatant was removed. These rhizosphere pellets, averaging 250 mg, were flash-frozen in liquid nitrogen and stored at −80 °C until processing. The root systems, while in the 25 ml of new buffer, were cleaned of remaining debris with sterile tweezers and transferred to new sterile buffer tubes until the buffer was clear after vortexing (without major sediment on the tube bottom). The roots were then sonicated in a Diagenode Bioruptor at low frequency for 5 min (five 30-s bursts followed by five 30-s rests). The sonication further disrupted tiny soil aggregates and attached microbes, cleaning the root exterior. We opted for physical removal of surface microbes by sonication instead of killing them with bleach because sequencing measures DNA; at lower concentrations, bleach kills microbes without necessarily destroying the DNA. Although an extended bleach treatment would also destroy unwanted DNA, it could also enter roots and destroy DNA of interest.
After sonication, the roots were snap-frozen, freeze-dried to remove ice and then stored at −80 °C until processing. Our rhizosphere and EC fractions were collected using time-practical protocols designed to partition sequencing-quality DNA and may differ slightly from classic definitions of these fractions that rely on partitioning culturable bacteria. We note that sonication may leave some rhizoplane microbes behind, especially if they are in a microniche shielded from the ultrasound. Such artefacts may cause our collected fractions to differ from theoretical definitions.
DNA extraction
To extract DNA, the samples were resuspended in a lysis buffer and microbial cells were mechanically lysed through bead beating. For all bulk soil and rhizosphere data, bead beating and purification were performed with the MoBio PowerSoil kit (SDS/mechanical lysis) because of its unmatched ability to remove humics and other PCR inhibitors in our soil. EC DNA from Arabidopsis experiments was prepared with the MP Bio Fast DNA Spin Kit for soil (also a SDS/mechanical lysis) because the more intense bead-beating protocol and lysis matrix gave improved lysis of whole roots and higher DNA yield, and soil PCR inhibitors were less of a problem with these samples. Our procedure yielded around 1 μg of DNA per rhizosphere sample, and more total DNA for EC samples (although a significant portion of EC DNA sequenced was of host origin). Although MoBio Powersoil and MP Bio Fast DNA use highly similar bead-beating/mechanical lysis methods, we developed a custom method of sample pre-homogenization that allowed us to prepare some EC samples using the MoBio kit. A comparison of Col-0 fractions soil, rhizosphere and EC across four soil digs of MF, where EC was prepared using MoBio in two digs and MP Bio in the other two digs, shows that although we cannot rule out a slight kit effect, both kits produce highly similar clustering separating EC from rhizosphere and soil fractions (Supplementary Fig. 6, replicates 3 and 4). DNA quantity was assessed with the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen) and a plate fluorospectrometer.
PCR
For each 1114F-barcoded 1392R primer set, PCR reactions with ~10 ng of template were performed in triplicate along with a negative control to reveal contamination. The PCR program used was 95 °C for 3 min followed by 30 cycles each of 95 °C for 30 s, 55 °C for 45 s and 72 °C for 1 min, followed by 72 °C for 10 min and then cooling to 16 °C. We first verified that the no-template control did not contain DNA via gel electrophoresis, and then pooled the three replicate PCR products and quantified DNA from each pool with PicoGreen (Invitrogen). Pooled PCR products from 30–48 barcoded samples were then combined in equimolar ratios into a master DNA pool, which was cleaned with Mo-Bio UltraClean PCR Clean-Up kit before submission for standard JGI pyrosequencing using a half-plate of Roche 454-FLX with titanium reagents.
454 pyrotag sequencing
To identify organisms present in each sample, 454 sequencing of the SSU rRNA genes was performed. For 454 sequencing, the SSU rRNA genes present in each sample were amplified with the primers 1114F and 1392R containing the 454 adaptors26. Each sample was assigned a reverse primer with a unique 5-bp barcode, allowing 30–48 samples to be pooled per half-plate. In preparation for sequencing, working aliquots of the master pool were immobilized on beads and amplified by emulsion PCR, the emulsion was broken with isopropanol, DNA-carrying beads were enriched and the enriched beads were loaded on the instrument for sequencing. During the emPCR protocol, we reduced the amplification primer amount from 460 μl in the standard protocol to 58 μl per emulsion cup. This is the same amount of primer used for the paired-end emPCR protocol. One-and-three-quarter million beads were loaded in each plate region (reduced from 2,000,000 beads per region in the standard protocol). A detailed standard protocol is available on request.
Primer test and technical reproducibility
We first tested three sets of broad-specificity 16S rRNA 5′ primers4 (Supplementary Fig. 2a,b) and established technical reproducibility metrics. We used 13 samples chosen from each of the three sample fractions (soil, rhizosphere and EC) and both soil types (MF and CL) (Supplementary Fig. 2c). Each sample was amplified individually with each of the forward primers (804F, which broadly targets bacteria and archaea; 926F, a universal primer; and 1114F, which broadly targets bacteria), paired with the barcoded universal reverse primer (1392R) and sequenced twice to measure technical reproducibility. We identified bacteria by grouping highly similar (97% identity) sequences into OTUs (Supplementary Methods). We chose 1114F for our experiments, on the basis of its broad coverage of the bacterial domain27 and higher usable data yield (Supplementary Fig. 2f-i and Supplementary Fig. 10).
We identified bacteria present by grouping highly similar (97% identity) sequences into OTUs using a standard QIIME (quantitative insights into microbial ecology)-based pipeline6 with default settings; thus, this stand-alone test consists of a different set of OTUs than those described in this work. The primer test samples are included in our submitted data and are found on 454 half-plates 26b and 27a. The progressive drop-out analysis, displaying the coefficient of determination (R2) of the least-squares regression between the two technical replicates as low-abundance OTUs are sequentially discarded, was calculated using the software R with a custom script.
Primer specificity sequence
804F prokaryote: 5′-agattagatacccdrgtagt-3′. 926F universal: 5′-actcaaaggaattgacgg-3′. 1114F bacteria: 5′-gcaacgagcgcaaccc-3′. 1392R barcoded universal: 5′-XXXXXacgggcggtgtgtrc-3′.
Sequence processing pipeline and assignment of OTUs
As each 454 plate was sequenced, raw reads from individual plates were immediately run through PYROTAGGER28 to diagnose plate quality so that plates could be re-queued if necessary. Plates with a reasonable number of long, high-quality raw reads with matching barcodes were used in the final analysis of OTU picking and taxonomy assignment. Using QIIME-1.4.029, short reads were removed and the remaining reads were trimmed to 220 bp, and low-quality reads were removed from the analysis using default quality settings (http://qiime.org/scripts/split_libraries.html). These high-quality sequences were clustered into OTUs using a custom script derived from otupipe (http://drive5.com/otupipe). The three main steps used from otupipe include (1) de-replicating sequences to reduce the size of the data set and the run time of clustering analysis, (2) de-noising sequences by forming clusters of 97% identity and representing these with the consensus sequence, and (3) forming OTUs by clustering de-noised consensus sequences at 97% identity.
The consensus sequence of sequences in each OTU was used as a representative sequence. Each representative sequence was assigned a taxonomy by two methods: (1) using the RDP classifier30 trained on the 4 February 2011 Greengenes reference sequences and (2) by assigning the Greengenes31 taxonomy of the best BLAST hit within a combined database including the complete Greengenes 16S database and 18S A. thaliana sequences from NCBI. By the BLAST-based method, sequences without a hit below the E-value threshold of 0.001 are considered unclassified.
Once OTUs were assigned a taxonomy, all OTUs annotated as chloroplasts, Viridiplantae or Archaea by any of the methods were removed from the OTU table, resulting in the set of usable OTUs.
We pooled usable reads from each bulk soil and rarefied to 200,000 reads per soil; this was permuted 100 times. We observed a median of 9,709 OTUs in MF soil and 9,897 OTUs in CL soil. Rarefaction curves to 200,000 reads in each bulk soil (not shown) indicated that, even at 200,000 reads, we were not capturing the entire community in either soil. Consequently, the total number of OTUs we report for our bulk soils may be lower than that found in some reports aimed at finding the true microbial diversity in soils.
A handful of samples had been sequenced more than once, over more than one 454 half-plate (for example to increase the read depth from problematic samples). These duplicated samples were pooled into a single sample by adding the unnormalized counts in the OTU table, and the resulting column was renamed to reflect the pooling that took place. Next any sample that had fewer than 50 usable reads was discarded, resulting in the unnormalized usable OTU table. At this point, both a frequency table and a rarefied table (1,000 usable reads per sample) were created as alternative normalization techniques.
The frequency table was made from the unnormalized usable OTU table by dividing the number of reads for each OTU in a given sample by the total number of reads inthat sample and multiplying by 100, and repeating this across all samples.
We also created a rarefied table; because some samples, particularly samples from the EC, had fewer than 1,000 usable reads in the unnormalized usable OTU table, counts from independent samples sharing the same soil type, genotype, fraction, age and experiment were pooled to make groups of at least 1,000 reads, and the sample names were changed to reflect the pooling that had taken place (Rarefaction_MappingFile‖ in Supplementary Database 1). Then all samples were rarefied to 1,000 counts using the rrarefy() function in the vegan package of R32.
We present both methods because each has advantages and limitations. The advantage of the frequency table is that it keeps each individual plant separate, contains more individual samples and uses all of the data, but this comes at the cost of increased granularity in the normalized relative abundance percentages for some of the samples with fewer reads, causing problems with direct comparability. The major advantage of the rarefied table is that comparisons are not biased by sampling depth and all read counts have equal weight, but this comes at the cost of reduced sample number and samples that mix information from several replicated individuals because we needed to pool some of our samples to meet our rarefaction threshold, and also at the cost of higher overall granularity because we discarded many reads from more deeply sequenced samples.
Because the majority of OTUs were represented by a very small number of reads and these OTUs were not technically reproducible (Supplementary Fig. 2d, e), both the rarefaction-normalized and the frequency-normalized OTU tables were thresholded to generate measurable OTUs for the majority of analyses (the major exception being the UniFrac analysis in Fig. 1: weighted UniFrac distance is robust to rare OTUs). An OTU was deemed measurable if and only if there were ≥25 reads in ≥5 samples in the unnormalized usable OTU table. As described in the text and Supplementary Fig. 2, this threshold was derived from the fact that the correlation between abundance in the same OTU in technical replicates improved greatly as OTUs approached an abundance of 25 reads, and from the fact that although contamination might create an OTU at this abundance once, the probability of an OTU being spurious decreases greatly if it occurs at a measurable level in several (we chose ≥5) independent samples.
Detection of differentially enriched OTUs by the GLMM
The OTU abundances were analysed with a GLMM to estimate the effect of the different variables on each measurable OTU. The lme4 R package33 was used to fit the model. The abundance of each OTU on each sample (yij) was log2-transformed and modelled as a function of the abundance of the same OTU in bulk soil samples (std_check) as a fixed effect, and plant genotype (b1), sample type (plant or bulk soil, b2), plant developmental stage (b3), soil type (b4), sequencing half-plate (b5) and biological replicate (b6) were modelled as random effects. The full model is specified by
where eij is the residual error and std_check was calculated as the mean abundance of each OTU in all the bulk soil samples from each combination of experiment and developmental stage.
There were not enough paired samples of rhizosphere and EC from the same individual plant to model the effect of both fractions directly. Instead, the abundance table was split into EC and rhizosphere samples, and the effect of each fraction with respect to bulk soil controls was estimated. The same model specification was used independently on both fractions, and for both the frequency and the rarefied tables (see Supplementary Methods on sequence processing pipeline). The percentage of total variance explained by each random variable on the OTU abundances is reported in Supplementary Table 5.
For each level of the random effects, the conditional mode and 95% prediction interval were estimated by Markov chain Monte Carlo sampling from the fitted model. A specific level is considered to have an effect on an OTU if the prediction interval of its conditional mode does not include zero. OTUs detected this way are reported in Supplementary Database 3.
Partial GLMM
There were not enough samples to estimate all the interaction effect between all variables without drastically reducing the size of the data set and our statistical power (Supplementary Table 2). To assess specific interactions of the genotype effect with other variables, a constrained version of the previously defined GLMM was used that employed only the fixed effect (std_check) and the random effects for plant genotype (b1) and sample type (b2). Samples were split into groups of the same experiment, developmental stage and fraction (thus, all the other variables from the full model are tested within each group), and the model was fitted and analysed in the same way as the full GLMM. A non-parametric Kruskal–Wallis test was used to verify independently the predictions of the partial GLMM for significance, where P values were corrected to Q values using the Benjimani–Hochberg FDR method; predictions from each partial GLMM with a Q value >0.05 were discarded as insignificant. The intersection of the significant genotype predictions between both biological replicates of each condition was calculated. The intersection analysis from the partial GLMM is displayed in Supplementary Table 3.
Scanning electron microscopy sample preparation
Arabidopsis roots were fixed in 2% paraformaldehyde, 2.5% glutaraldehyde and 0.15 M sodium phosphate buffer, pH 7.4. The samples were dehydrated using a gradual ethanol series (30%, 50%, 75%, 100%, 100%) and dried in a Samdri-795 supercritical dryer using carbon dioxide as the transitional solvent (Tousimis Research Corporation). Roots were mounted on aluminium planchets with double-sided carbon adhesive and coated with 10 nm of gold–palladium alloy (60:40 Au:Pd, Hummer X Sputter Coater, Anatech USA). Images were made using a Zeiss Supra 25 FESEM operating at 5 kV and a working distance of 5 mm, and with a 10-μm aperture (Carl Zeiss SMT Inc.), at the Microscopy Services Laboratory, Pathology and Laboratory Medicine, UNC at Chapel Hill.
Log2 transformation
All log2 transformations on OTU tables followed the formula log2(1000x + 1), where x is the rarefied read counts (or frequency) per OTU.
Heat maps
Heat maps were constructed using custom scripts and the function heatmap.2 from the R package gplots34. For better visualization, all data was log2-transformed. Hierarchical clustering of rows and columns in the heat maps is based on Bray–Curtis similarities and uses group-average linkage.
Diversity
The Shannon diversity index and the non-parametric Chao1 diversity were calculated with the vegan package in R32. The exponential function was applied to the Shannon diversity index to calculate the true Shannon diversity (effective number of species).
Rarefaction curves
Rarefaction curves were made with custom scripts that sampled each sample fraction only once at each read depth. To reveal the variance in sampling, no attempt was made to smooth the curves by taking the average of repeated samplings.
Taxonomy histograms and statistics
Taxonomy histograms were created using custom scripts and visualized in GraphPad PRISM version 5.0 for Windows 35 (GraphPad Software, Inc.; http://www.graphpad.com). The ‘low-abundance’ category was created to help remove visual clutter, and contained any taxonomic group that did not reach at least 5% in any one fraction. The Shannon diversity index was calculated as described above. Differences in distribution at varying taxonomic levels, and differences in Shannon diversity between soil, rhizosphere and EC fractions, were tested by weighted analysis of variance (to account for differing numbers of soil, rhizosphere and EC samples), invoking the central limit theorem (>60 samples in each group in all tests for both frequency-normalized and rarefaction-normalized tests). For more details about tests, see additional notation in Supplementary Table 5.
Sample clustering using UniFrac
A phylogenetic tree was built with the representative sequence for each OTU and the pairwise, normalized, weighted UniFrac distance36. For UniFrac, representative sequences from all non-plant OTUs, including those that did not meet the 25 × 5 sample threshold, were considered. UniFrac distances between samples are based on the fraction of branch length that is unique to each sample in a shared phylogenetic tree composed of OTU representative sequences from all samples. Thus, samples containing OTUs of highly divergent sequences will be more distant from each other, because the OTUs comprising each sample will occupy different major branches on the shared phylogenetic tree of OTUs, whereas samples containing highly similar OTUs will share these major branches. In weighted UniFrac, the branch length unique to each sample is multiplied by the frequency at which that OTU occurs in the sample. Thus, weighted UniFrac can detect differences between two samples that have the same set of OTUs that differ quantitatively between the samples.
Principal coordinate analysis was performed using pairwise, normalized, weighted UniFrac distances between all samples on the unthresholded but normalized OTU tables, and the first two principal coordinates of UniFrac were visualized with GraphPad PRISM version 5.0 for Windows.
CARD–FISH application to roots
We applied a modified protocol described previously37. Briefly, several root systems from a bolting Col-0 grown in MF were fixed using 4% formaldehyde in PBS at 4 °C for 3 h, washed twice in PBS and stored in 1:1 PBS:molecular-grade ethanol at −20 °C. Treatments with lysozyme solution (1 h at 37 °C, 10 mg ml−1; Fluka) and achromopeptidase (30 min at 37 °C, 60 U ml−1; Sigma) were sequentially used for prokaryotic cell-wall permeabiliza-tion. Endogenous peroxidases were inactivated with methanol treatment amended by 0.15% H2O2 at room temperature for 30 min and washed again. Probes targeting eitherthe16S or the 23S rRNA (EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′, 35% formamide), NON338 (5′-ACTCCTACGGGAGGCAGC-3′, 30% formamide), HGC69a (5′-TATAGTTACCACCGCCGT-3′, 25% formamide) and Brady4 (5′-CGTCATTATCTTCCCGCACA-3′, 30% formamide)) were defined using probeBase38 (http://www.microbial-ecology.net/default.asp), labelledwith enzyme horseradish peroxidase on the 5′ end (Invitrogen), diluted in hybridization buffer (final concentration of 0.19 ng ml−1) with each probe’s optimum formamide concentration, and hybridized at 35 °C for 2 h. Unbound probes were washed away from samples in wash buffer (NaCl content adjusted according to the formamide concentration in the hybridization buffer) at 37 °C for 30 min. Fluorescently labelled tyramide was used for signal amplification, and samples were washed before mounting on glass slides.
For double CARD–FISH, a subset of samples went through a second round of the protocol, starting at the peroxidase inhibition with a second variety of fluorescently labelled tyramide used to be able to distinguish the signals from each probe. Roots were mounted on glass slides using Vectashield with DAPI (Vector Laboratories, catalogue no. H-1200) for mounting solution, and sealed with nail polish for storage. All microscopy images were made on a confocal laser scanning microscope (Zeiss LSM 710 META) located in the Biology Department at UNC. The Brady4 probe, which has not been used for this application previously, was tested on filters of cultured Bradyrhizobiaceae and three negative control cultured strains to determine the most specific formamide concentration in the hybridization buffer.
For application of samples onto filters, bulk MF soil, rhizosphere and EC samples from four sets of Col-0 roots were pooled and harvested in the way described above before DNA extraction. Samples were then fixed as described above and passed through a 10-μm filter. The concentrations of plant material were made equal and samples were sonicated in a water bath for 5 min. The sample suspension was further diluted to 1:500 in water and applied to a 25-mm polycarbonate filter with a pore size of 0.2 μm (Millipore) using a vacuum microfiltration assembly. Filters were embedded in 0.2%, low-melting-point agarose and dried, and CARD–FISH was applied as described above. For quantification of bacteria, filters were visualized on a Nikon Eclipse E800 epifluorescence microscope. Positive EUB338 probe signals that co-localized with a DAPI signal were counted as Eubacteria. Positive Actinobacteria or Bradyrhizobiaceae signals were counted as positive when the HGC69a or Brady4 probe co-localized with both EUB338 and the DAPI signal.
Sample naming in OTU tables
All sample names in OTU tables are in the following form: [soil type].[genotype].[sample number][fraction].[age].[experiment]_[plate]. For example, M21.Col.6E.old.M1_2b should be interpreted as [soil type] = M21 = MasonFarm2:1,[genotype] = Col = Col-0,[samplenumber] = 6, [fraction] = E = endophyte compartment, [age] = old, [experiment] = M1 = Mason Farm replicate 1, [plate] = 2b.
Supplementary Material
Acknowledgements
We thank A. Smithlund, M. Gonek, V. Madden, H. Schmidt, M. Rott and N. Zvenigorodsky for technical assistance. We thank A. Spor, J. Pfeiffer and J. Rawls for discussions and C. Herring for research field soil. This work was supported by US NSF grant IOS-0958245 (J.L.D.), the JGI Director’s Discretionary Grand Challenge Program and the HHMI-GBMF Plant Science Program. J.L.D. is an HHMI-GBMF Plant Science Investigator. Work conducted by the US Department of Energy Joint Genome Institute is supported by the Office of Science of the US Department of Energy under contract no. DE-AC02-05CH11231. S.L.L. was supported by the National Institutes of Health, Minority Opportunities in Research division of the National Institute of General Medical Sciences grant K12GM000678.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions D.S.L., S.L.L. and J.L.D. designed the project, D.S.L. and S.L.L. set up the experiments and organized construction of the sequencing libraries. D.S.L., S.L.L., S.H.P. and J.G. harvested samples and prepared DNA for sequencing. A.E., V.K., T.G.d.R., S.M., P.H. and S.G.T. worked together at the JGI to perform, optimize and quality-control the sequencing. S.H.P. applied the GLMM to the data. D.S.L., S.H.P., S.Y. and R.E. created and managed the data analysis pipeline. S.Y. oversaw the data deposition. D.S.L., S.L.L., S.H.P., S.Y., J.G. and J.L.D. analysed the data and created figures. S.L.L. performed the CARD–FISH microscopy in the laboratory of T.E., at the Max-Planck-Institute for Plant Breeding in Cologne, and at UNC. R.E.L. and P.H. advised on primer design and appropriate statistical methods. D.S.L., S.L.L., S.H.P. and J.L.D. wrote the manuscript with significant input from S.Y., R.E.L., P.H. and S.G.T.
Author Information 454 pyrosequencing data are deposited in the ENA Sequence Read Archive under study number ERP001384. Custom R scripts are available with documentation at http://labs.bio.unc.edu/dangl/resources/scripts_Lundberg_et_al_2012.htm and additional code is available on request. Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Rodriguez RJ, et al. Stress tolerance in plants via habitat-adapted symbiosis. ISME J. 2008;2:404–416. doi: 10.1038/ismej.2007.106. [DOI] [PubMed] [Google Scholar]
- 2.De Deyn GB, Cornelissen JHC, Bardgett RD. Plant functional traits and soil carbon sequestration in contrasting biomes. Ecol. Lett. 2008;11:516–531. doi: 10.1111/j.1461-0248.2008.01164.x. [DOI] [PubMed] [Google Scholar]
- 3.van der Lelie D, et al. Poplar and its bacterial endophytes: coexistence and harmony. Crit. Rev. Plant Sci. 2009;28:346–358. [Google Scholar]
- 4.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 5.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nature Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 6.Marschner H, Römheld V, Horst WJ, Martin P. Root-induced changes in the rhizosphere: importance for the mineral nutrition of plants. Z. Pflanz. Boden. 1986;149:441–456. [Google Scholar]
- 7.Dennis PG, Miller AJ, Hirsch PR. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol. Ecol. 2010;72:313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- 8.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 9.Firáková S, šturdíková M, Múčková M. Bioactive secondary metabolites produced by microorganisms associated with plants. Biologia. 2007;62:251–257. [Google Scholar]
- 10.Schulz BJE, Boyle CJC, Sieber TN, Schulz B, Boyle C. Microbial Root Endophytes. Vol. 9. Springer; 2006. pp. 1–13. [Google Scholar]
- 11.Hallmann J, Quadt-Hallmann A, Mahaffee WF, Kloepper JW. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997;43:895–914. [Google Scholar]
- 12.Redford AJ, Bowers RM, Knight R, Linhart Y, Fierer N. The ecology of the phyllosphere: geographic and phylogenetic variability in the distribution of bacteria on tree leaves. Environ. Microbiol. 2010;12:2885–2893. doi: 10.1111/j.1462-2920.2010.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hardoim PR, van Overbeek LS, Elsas JD. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 14.Inceoglu O, Salles JF, van Overbeek L, van Elsas JD. Effect of plant genotype and growth stage on the β-proteobacterial community associated with different potato cultivars in two fields. Appl. Environ. Microbiol. 2010;76:3675–3684. doi: 10.1128/AEM.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inceoğlu O, Al-Soud WA, Salles JF, Semenov AV, van Elsas JD. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE. 2011;6:e23321. doi: 10.1371/journal.pone.0023321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson AK, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc. Natl Acad. Sci. USA. 2010;107:18933–18938. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottel NR, et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 2011;77:5934–5944. doi: 10.1128/AEM.05255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eickhorst T, Tippkötter R. Improved detection of soil microorganisms using fluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH) Soil Biol. Biochem. 2008;40:1883–1891. [Google Scholar]
- 19.Chi F, et al. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 2005;71:7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulgarelli D, et al. Structure of and assembly cues for the Arabidopsis root-inhabiting bacterial microbiota. Nature. doi: 10.1038/nature11336. doi:10.1038/nature11336 (this issue) [DOI] [PubMed] [Google Scholar]
- 21.Arumugam M, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature Rev. Microbiol. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 23.Masclaux C, Valadier M, Brugière N, Morot-Gaudry J, Hirel B. Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta. 2000;211:510–518. doi: 10.1007/s004250000310. [DOI] [PubMed] [Google Scholar]
- 24.Levey SAW. Natural variation in the regulation of leaf senescence and relation to other traits in Arabidopsis. Plant Cell Environ. 2005;28:223–231. [Google Scholar]
- 25.van Elsas JD, Trevors JT, Starodub ME. Bacterial conjugation between pseudomonads in the rhizosphere of wheat. FEMS Microbiol. Lett. 1988;53:299–306. [Google Scholar]
- 26.Engelbrektson A, et al. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- 27.Lane DJ. In: Nucleic Acid Techniques in Bacterial Systematics. Stackebrandt MGE, editor. Wiley; 1991. pp. 115–175. [Google Scholar]
- 28.Kunin V, Hugenholtz P. PyroTagger: a fast, accurate pipeline for analysis of rRNA amplicon pyrosequence data. Open J. 2010:1–8. [Google Scholar]
- 29.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sul WJ, et al. Bacterial community comparisons by taxonomy-supervised analysis independent of sequence alignment and clustering. Proc. Natl Acad. Sci. USA. 2011;108:14637–14642. doi: 10.1073/pnas.1111435108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oksanen J, et al. Vegan: R Functions for Vegetation Ecologists. 2011 http://cc.oulu.fi/~jarioksa/softhelp/vegan.html.
- 33.Bates D, Maechler M, Bolker B. Lme4: Linear Mixed-Effects Models using S4 Classes (R package version 0.999375-42) 2011 http://CRAN.R-project.org/package5lme4.
- 34.Warnes GR. Gplots: Various R Programming Tools for Plotting Data. 2011 http://cran.r-project.org/web/packages/gplots/index.html.
- 35.Motulsky HJ. Prism 4 Statistics Guide: Statistical Analyses for Laboratory and Clinical Researchers. GraphPad Software, Inc.; 2003. [Google Scholar]
- 36.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eickhorst T, Tippkötter R. Improved detection of soil microorganisms using fluorescence in situ hybridization (FISH) and catalyzed reporter deposition (CARD-FISH) Soil Biol. Biochem. 2008;40:1883–1891. [Google Scholar]
- 38.Loy A, Maixner F, Wagner M, Horn M. probeBase–an online resource for rRNA-targeted oligonucleotide probes: new features 2007. Nucleic Acids Res. 2007;35:D800–D804. doi: 10.1093/nar/gkl856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.