Abstract
Background
Trichobilharzia is the most species rich and widely distributed genus of schistosomes and is known throughout Europe and North America as an agent of human cercarial dermatitis. The disease is caused by an acute allergic reaction in the skin that develops as a consequence of repeated contact with water containing schistosomatid cercariae. However, despite historical outbreaks of the disease, there are no published records of accurately identified Trichobilharzia species from the UK.
Methods
Two hundred Radix auricularia (L.) were sampled from a recreational fishing lake in Hampshire and emerging schistosomatid cercariae were collected for microscopy and DNA extraction. General morphological description of the cercariae was performed, alongside sequencing and phylogenetic analysis of the 28S ribosomal DNA for accurate species identification as well as comparisons of ITS1 in order to identify evolutionary affinities with other European populations. All molecular comparisons were performed using published sequences.
Results
The phylogenetic analysis of 28S sequences identified the cercariae as Trichobilharzia franki. Two unique British ITS1 haplotypes were identified which were most closely related to haplotypes of T. franki populations from France. Haplotype network analysis indicated the mixing of T. franki populations throughout Europe. It is suggested that parasite distribution is the probable result of the movement of migratory waterfowl.
Conclusions
This is the first accurate record of T. franki in the UK. The movement of T. franki with waterfowl could pose a considerable human health risk, as in mainland Europe, and signifies T. franki-associated human cercarial dermatitis as a re-emerging disease in the UK.
Keywords: Trichobilharzia franki, Cercarial dermatitis, UK, Re-emerging disease, 28S ribosomal DNA, ITS1 haplotypes
Background
The cercariae of many species of avian schistosomatid blood flukes are known to cause human cercarial dermatitis (CD), also known as swimmer’s itch. Avian schistosomatids have a truly global distribution with agents of CD being recorded on all major continents except Antarctica. However, throughout Europe and America there has been increased incidence of reported infections and CD is now considered an emerging and re-emerging infectious disease [1-3]. Foci of infection have been identified in several European countries particularly Italy, Germany, Austria, Switzerland, the Netherlands, Czech Republic, France, Poland and Iceland with the disease impacting on local economies that depend on tourism associated with recreational water use [4].
Despite the occurrence of these parasites throughout Europe and North America [1,3], relatively little is known about the occurrence of CD and the identity and diversity of parasites that cause it in the UK, even though outbreaks of CD have been recorded in Loch Lochore in Fife in 2006, the Norfolk Broads in 2004 [5], a water sports lake in Suffolk in 1987 [6], a leisure water park in Rickmansworth, Hertfordshire in 1970 [7] and Roath Park, Cardiff between 1928–1943 [8].
Human CD presents as an acute allergic reaction in the skin that develops as a consequence of the repeated penetration of certain species of schistosomatid cercariae that emerge from freshwater snail intermediate hosts [3,9]. Maculo-papulo-vesicular eruptions occur after exposure, followed by intense itching, fever, swelling of the lymph nodes and eventually erythema and oedema [10]. Experimental studies on immunocompetent mammalian models show that although the majority of the cercariae die in the skin, some schistosomula can migrate to the lungs (visceral avian schistosome species) or nervous system (Trichobilharzia regenti) [3,9]. Severe pathologies can develop in immunodeficient mammals due to a high number of schistosomula migrating to organs. Therefore, although they are unable to complete their development and reproduce, the parasites may cause pathologies in addition to those associated with the skin (reviewed by Kolářová [9]).
Trichobilharzia[11] is considered the most species-rich and widely distributed genus of the Schistosomatidae, with species parasitizing waterfowl throughout the world, and several being leading causes of CD [1]. Schistosomatids in UK waters have been inadequately studied. Although ocellate schistosomatid cercariae have been recorded from snails in the UK [8,12,13], there is no definitive identification of a Trichobilharzia species. In the last decade, problems relating to the accurate morphological identification of Trichobilharzia species associated with CD outbreak sites in America and Europe have been overcome by using ribosomal DNA (rDNA) markers to identify cercariae to species [1,14]. Such approaches were employed in the current study to identify schistosomes released by aquatic lymnaeid snails collected from Tundry Pond, Hampshire. We report the first detailed record/description of T. franki Müller and Kimmig [15] in the UK and identify evolutionary relationships with other European populations of this schistosome. The public health implications of the findings are discussed in the context of CD being a re-emerging infectious disease in the UK.
Methods
Snail and cercariae collection
Two hundred Radix auricularia (L.) were collected and identified based on shell morphology from Tundry Pond, a recreational fishing lake in Hampshire, Southern England, UK (grid reference: SU 775 525, 51.16 N 0.53 W), in August 2011 as part of an on-going study on the diversity of digeneans and their intermediate hosts in the UK. Ocellate furcocercariae have previously been observed at this site (R. Kirk, unpublished observations). Snails were relocated to the laboratory for processing. Each snail was isolated in a 100 ml beaker filled with dechlorinated tap water that had been filtered through a Brimak carbon filter (Silverline UK) and cercarial emergence was stimulated by exposure to natural light. Three R. auricularia shed ocellate furcocercariae. These cercariae were collected for molecular work using the techniques described by Brant and Loker [1] and were pooled in Petri dishes for morphological description. Upon confirmation of species, parasite and snail reference material were deposited at the Natural History Museum, London, UK (parasite voucher NHMUK 2014.4.25.1-2; snail voucher NHMUK 20140076).
Morphological description of cercariae
Live cercariae were vitally stained with 0.5% neutral red and examined using light microscopy for initial morphological description. To obtain further morphological information, cercariae were fixed for scanning electron microscopy in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.4) for 2 h at 4°C. They were then washed in four changes of 0.1 M phosphate buffer, post-fixed in osmium tetroxide in the same buffer for 1 h, dehydrated in a graded ethanol series and dried in hexamethyldisilazane. Samples were coated with gold-palladium and examined under a Zeiss EVO50 scanning electron microscope. Morphometric parameters of cercariae were not recorded because of the unreliability of using cercarial dimensions and flame cell organization for identification of Trichobilharzia species [10]. Chaetotaxy was not performed due to known problems with misinterpretation of sensory structures [16] and staining [16,17].
Molecular identification of parasites
Live ocellate furcocercariae were morphologically identified to genus level using light microscopy and were pooled into 1.5 ml microfuge tubes (50 cercariae per snail). Each tube was centrifuged at 10000 rpm for 1 min to pellet the cercariae and DNA was extracted with the Qiagen DNeasy blood and tissue kit (Qiagen Inc.) using the manufacturer’s protocol. PCR was performed to amplify fragments of 28S ribosomal subunit (LSU) and the internal transcribed spacer region (ITS) using primers and cycling conditions specified by Olson et al. [18] and Wang et al. [19], respectively. PCR reactions were performed using 12.5 μl Thermo–StartR PCR master mix (0.625 Units of Taq DNA polymerase, 1X reaction buffer, 0.2 mM of each dNTP and 1.5 mM MgCl2) and 1–2 ng/μl of DNA. Final reactions were made up to 25 μl with PCR-grade water. Reactions were performed using a Veriti 96 well thermal cycler (Applied Biosystems™) and 5 μl of each amplicon was visualized in 1% agarose gels stained with gel red (Bioline™). The remaining 20 μl PCR products were sequenced at the DNA sequencing facility of the Natural History Museum, London, using the PCR primers with Fluorescent Dye Terminator Sequencing Kits (Applied Biosystems™); sequencing reactions were run on an Applied Biosystems™ 3730XL automated sequencer. Resultant sequences were assembled using BioEdit [20] and manually corrected for ambiguous base calls. It is important to note that despite the DNA being extracted from pooled cercariae, there were no polymorphic sites noticed in the chromatographs of both the forward and reverse sequences of LSU and ITS. Complete sequences were submitted for BLAST search using blastn (http://blast.ncbi.nlm.nih.gov/) to enable initial identification, indicating that all the sequences generated best-matched T. franki. Novel LSU sequences were aligned with previously published sequences of other species of Trichobilharzia and bird schistosomes from different genera and Schistosoma sinensium as an out group (Table 1) using MUSCLE (http://www.ebi.ac.uk), giving an alignment of comparable data of 556 bp.
Table 1.
Published sequences of schistosomatids used in the phylogenetic analysis of the LSU to identify UK specific Trichobilharzia species
| Species | Host | Accession |
|---|---|---|
|
Trichobilharzia franki |
Radix auricularia |
HM131141 |
|
T. regenti |
Anser anser |
HM439491 |
|
T. querquedulae |
Anas discors |
FJ174469 |
|
T. physellae |
Physa parkeri |
FJ174475 |
|
T. szidati |
Lymnaea stagnalis |
FJ174476 |
|
T. stagnicolae |
Stagnicola emarginata |
FJ174479 |
|
T. ocellata |
Lymnaea stagnalis |
AY157243 |
|
Anserobilharzia brantae |
Chen caerulescens |
FJ174467 |
|
Dendritobilharzia pulverulenta |
Gallus gallus (Lab host) |
AY157241 |
|
Gigantobilharzia huronensis |
Agelaius phoeniceus |
AY157242 |
|
Heterobilharzia americana |
Mesocricetus auratus |
AY157246 |
|
Schistosomatium douthitti |
Mesocricetus auratus |
AY157247 |
|
Ornithobilharzia canaliculata |
Larus delawarensis |
AY157248 |
|
Austrobilharzia terrigalensis |
Batillaria australis |
AY157249 |
|
Bilharziella polonica |
Anas platyrhynchus |
AF184265 |
| Out group |
|
|
| Schistosoma sinensium | Mus musculus (Lab host) | AY157251 |
Inter- and intra-species phylogenetic analysis
To accurately identify the novel sequences and place them into a phylogenetic context with other species of Trichobilharzia, phylogenetic analysis was performed for alignments of LSU using MEGA5 [21]. Several phylogenetic methods were employed using the neighbour joining (NJ) approach under the conditions of the Jukes-Cantor model, maximum likelihood (ML) methods using the HKY+G model of evolution also identified in MEGA5 [21]. Maximum parsimony (MP) trees were obtained using the close–neighbour–interchange algorithm. The model used for the ML tree was identified based on the lowest Bayesian information criterion scores relative to the other models tested and for all tree analysis nodal support was assessed using 1000 bootstrap replicates. The pair-wise uncorrected distances (p-distance) were also calculated for the LSU sequences between Trichobilharzia species to estimate genetic distance and to validate species identification.
In order to establish affinities between European and UK isolates of T. franki, ITS1 fragments were analysed due to the availability of comparable data from other European T. franki populations. The three novel sequences obtained were aligned with previously published sequences that represent populations of T. franki from across central and northern Europe, producing an alignment of 878 bp. Sequences were aligned and phylogenetic analysis performed as described previously, but ML trees were constructed using the K2+G model as identified by MEGA5 [21]; T. regenti was used as an out-group in all analyses. Standard population genetic analysis was performed using DnaSPv5 [22] to calculate nucleotide diversity (π) and sequence differentiation within and between sequence sets from different countries and to identify the occurrence of unique haplotypes (H) and haplotype diversity (Hd). In order to assess the association of the UK haplotypes to others from Europe, the most parsimonious haplotype network was constructed using TCS version 1.13 [23].
Results
Morphological description of Trichobilharzia franki
The morphology of the ocellate furcocercariae partly corresponded with the previous description of T. franki by Müller and Kimmig [15]. The ocelli (eye spots) were located on the dorsal part of the body, between the acetabulum and the head organ. The head organ and the acetabulum were spherical. The cercariae were apharyngeate, with a subterminal mouth and a bifurcate intestine, which were inconspicuous. The body contained five large pairs of penetration glands, two pairs circumacetabular and three pairs postacetabular, in addition to an apical gland group, with all ducts opening anteriorly. The excretory system progressed through the excretory junction at the base of the body and extended into the tail stem to the tip of the furcae. The furcae were surrounded by finfolds and ended in thorns (Figures 1 and 2A). The description of the head organ by Müller and Kimmig [15] as a fused complex of oral sucker and pharynx and the preacetabular position of two pairs of penetration glands are suggested to be errors. These authors also reported that the tegument was covered with small spines. Here, scanning electron microscopy revealed that most of the external surface of the acetabulum was devoid of spines and only the inner surface had centrally directed curved spines with rounded tips (Figure 2B). The spines on the tail stem were large and curved (Figure 2C). The spines on the body resembled those on the acetabulum (Figure 2D). All spines were posteriorly directed. Two types of uniciliate sensory endings were observed: type 1 sensory papillae (with tegumental collar) of different lengths on the body and type 2 sensory papillae (without tegumental collar) of different lengths on the tail stem, furcae and body, but not on the head organ region (Figure 2D, E), corresponding to the description for T. franki by Kock and Böckeler [16]. The tip of the head organ displayed gland duct openings surrounded by type 1 sensory papillae (Figure 2F).
Figure 1.
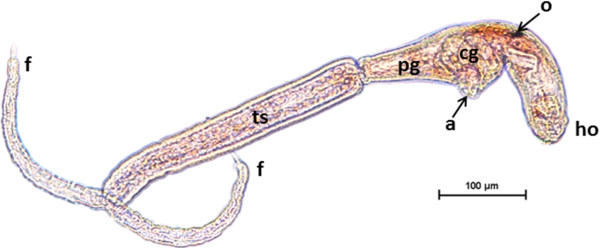
Lateral view of the cercaria of Trichobilharzia franki stained with neutral red. Acetabulum (a), head organ (ho), one of two ocelli (o) circumacetabular penetration glands (cg), postacetabular penetration glands (pg), tail stem (ts) and furcae (f).
Figure 2.
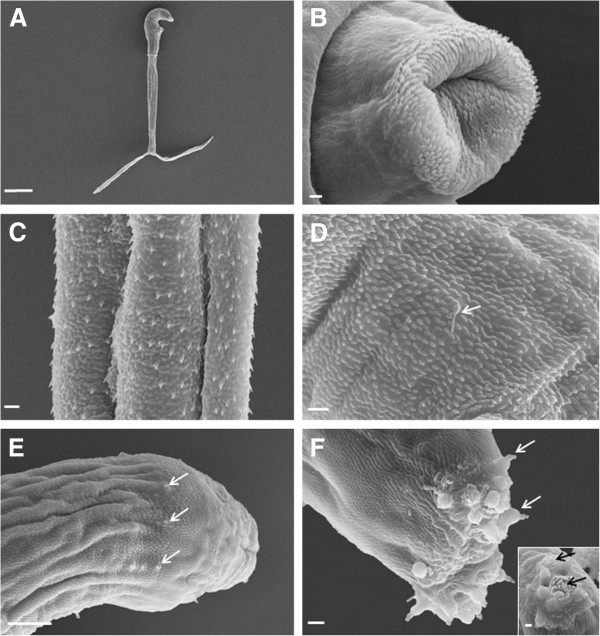
Scanning electron micrographs of the cercaria of Trichobilharzia franki. A. Entire cercaria, scale bar = 100 μm. B. Acetabulum, scale bar = 1 μm. C. Tail stem with spines, scale bar = 2 μm. D. Tegumental spines and type 2 sensory papilla on body (arrow), scale bar = 2 μm. E. Anterior of body and type 1 sensory papillae (arrows), scale bar = 10 μm. F. Apical view of body showing head organ, note type 1 sensory papillae (arrows) scale bar = 2 μm, and gland duct openings (insert, black arrows), scale bar = 1 μm.
Molecular identification of Trichobilharzia franki
Phylogenetic analysis of the T. franki LSU fragments with 15 other schistosomatids produced a well-supported Trichobilharzia clade with Dendritobilharzia pulverulenta and Gigantobilharzia huronensis forming a sister clade (Figure 3). Within the Trichobilharzia clade the sequences generated in this current study, Ham Ra6 – 8 (GenBank: KJ775865-67), clustered with T. franki only and formed a shallow, but well supported, clade separate from any of the other taxa (Figure 3). This relationship between the UK sequences and T. franki was further supported by the low p–distance values relative to comparisons with other species (Table 2). However, a noteworthy observation is that similar levels of genetic divergence seen between the UK specific sequences and T. franki, were also evident between T. franki and T. querquedulae (Table 2). Trichobilharzia querquedulae appears as a sister species to T. franki, but short branch lengths may indicate that both species are either subspecies or members of a species complex.
Figure 3.
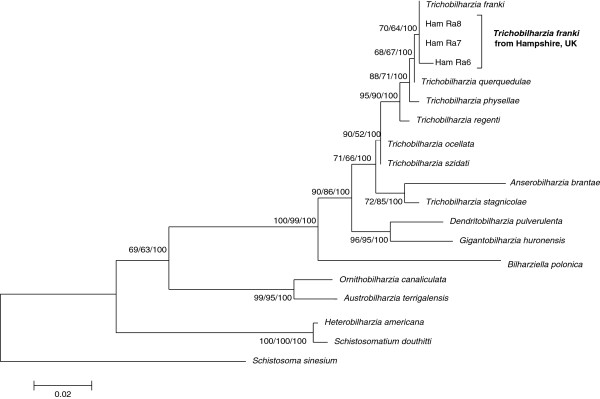
Phylogenetic tree based on partial LSU sequences for avian schistosomes in order to identify ocellate furcocercariae from Hampshire, UK. The samples sequenced from Hampshire, UK have close affinity with T. franki and do not associate with another species. The tree was constructed using the ML method in MEGA5 using the HKY + G substitution model. The scale shows the number of nucleotide substitutions per site between DNA sequences and nodal supports were generated using 1000 bootstrap replicates. The nodal support is given in NJ, ML and MP bootstraps respectively and only values >50 for at least two of the analyses are shown.
Table 2.
Uncorrected pair-wise distance (p–distance) between different species of Trichobilharzia based on number of base differences per site between sequences of LSU
| Species | T. stagnicolae | A. brantae | T. szidati | T. ocellata | T. regenti | T. physellae | T. querquedulae | T. franki | Ham Ra6 | Ham Ra7 |
|---|---|---|---|---|---|---|---|---|---|---|
|
T. stagnicolae
|
|
|
|
|
|
|
|
|
|
|
|
A. brantae
|
0.037 |
|
|
|
|
|
|
|
|
|
|
T. szidati
|
0.015 |
0.040 |
|
|
|
|
|
|
|
|
|
T. ocellata
|
0.015 |
0.040 |
0.000 |
|
|
|
|
|
|
|
|
T. regenti
|
0.024 |
0.050 |
0.009 |
0.009 |
|
|
|
|
|
|
|
T. physellae
|
0.026 |
0.051 |
0.011 |
0.011 |
0.009 |
|
|
|
|
|
|
T. querquedulae
|
0.024 |
0.050 |
0.009 |
0.009 |
0.007 |
0.006 |
|
|
|
|
|
T. franki
|
0.026 |
0.051 |
0.011 |
0.011 |
0.009 |
0.007 |
0.002 |
|
|
|
|
Ham Ra6 |
0.029 |
0.055 |
0.015 |
0.015 |
0.013 |
0.011 |
0.006 |
0.004* |
|
|
|
Ham Ra7 |
0.026 |
0.051 |
0.011 |
0.011 |
0.009 |
0.007 |
0.002 |
0.000* |
0.004* |
|
| Ham Ra8 | 0.026 | 0.051 | 0.011 | 0.011 | 0.009 | 0.007 | 0.002 | 0.000* | 0.004* | 0.000* |
Those numbers highlighted by* are indicative of the Trichobilharzia species sampled from the UK (Ham Ra6 – 8) and their confirmation as T. franki when compared to other species.
Insights into the relationships between populations of Trichobilharzia franki from the UK and Europe
Phylogenetic reconstruction showed the UK-specific sequences to have close affinity with populations of T. franki from France and clustered within a France-specific clade, but no other country specific clades were identified (Figure 4). Two unique haplotypes were identified from the three specimens with Ham Ra6 having its own individual haplotype and another being shared by Ham Ra7 and Ham Ra8 (GenBank: KJ775868-69). The molecular diversity of T. franki populations from each country appeared high with several different haplotypes being found within relatively small sample sizes (Table 3). Diversity within sequences from Switzerland showed considerable variation (S = 77; π = 0.02610) and five out of the six sequences analysed were unique haplotypes. This contrasted with sequences from France where only three haplotypes were found in the ten sequences sampled (S = 2; π = 0.00079). The high number of haplotypes within and between populations of T. franki was also illustrated in the haplotype network (Figure 5). Four distinct clades were revealed in the haplotype network, but no geographical specific lineages emerged as each clade contained a number of related haplotypes from different countries. Clade 1 contained the only haplotype shared between countries with haplotype 1 (H1) being shared by individuals from France (HM131183, HM131179, HM131180), Switzerland (AJ312041, AJ312042) and the Czech Republic (AY795573, AF356845). The UK-specific sequences were also found in clade 1 with haplotype 3 (H3) representing Ham Ra7 and 8 and haplotype 4 (H4) representing Ham Ra6. These haplotypes were unique and were not shared with other populations. However, the UK-specific sequences were most closely related to French sequences in haplotype 5 (H5) that contained HM131181, HM131177, HM131184, HM131182, HM131178, HM131176 and haplotype 2 (H2) that contained AY795572 (Figure 5). Although these data illustrate high population variation within and between T. franki populations, the sample sizes for each country are small. Further work with larger sample sizes would be required to provide in-depth insights into population differentiation and gene flow.
Figure 4.
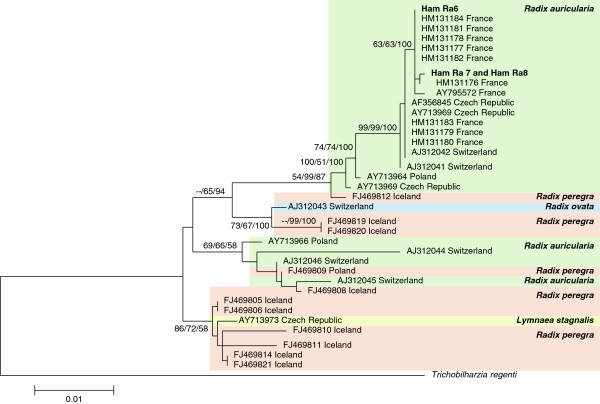
Phylogenetic tree based on the ITS1 sequences representing different populations of Trichobilharzia franki from European countries and the snail species that they infect. The samples sequenced from Hampshire, UK cluster within a France-specific clade, the only country specific clade to emerge during analysis. The tree was constructed using the ML method in MEGA5 using the K2 + G substitution model. The scale shows the number of nucleotide substitutions per site between DNA sequences and nodal supports were generated using 1000 bootstrap replicates. The nodal support is given in NJ, ML and MP bootstraps respectively and only values >50 for at least two of the analyses are shown.
Table 3.
Molecular diversity of ITS1 haplotypes of Trichobilharzia franki from several European countries
| Population | Number of sequences | S | H | Hd | K | π |
|---|---|---|---|---|---|---|
| All |
35 |
103 |
21 |
0.924 (SD ± 0.03) |
33.2546 |
0.02013 |
| UK (Hampshire) |
3 |
1 |
2 |
1 (SD ± 0.5) |
1 |
0.00118 |
| France |
10 |
2 |
3 |
0.6 (SD ± 0.131) |
0.67 |
0.00079 |
| Switzerland |
6 |
77 |
5 |
0.93 (SD ± 0.122) |
39.2 |
0.02610 |
| Czech Republic |
4 |
56 |
3 |
0.83 (SD ± 0.222) |
29 |
0.01574 |
| Poland |
3 |
58 |
3 |
1 (SD ± 0.272) |
39.67 |
0.01891 |
| Iceland | 10 | 81 | 7 | 0.93 (SD ± 0.062) | 24.07 | 0.01791 |
Where S is number of polymorphic sites including indels; H is number of haplotypes; Hd is haplotype diversity; K is average number of differences between sequences and π is nucleotide diversity.
Figure 5.
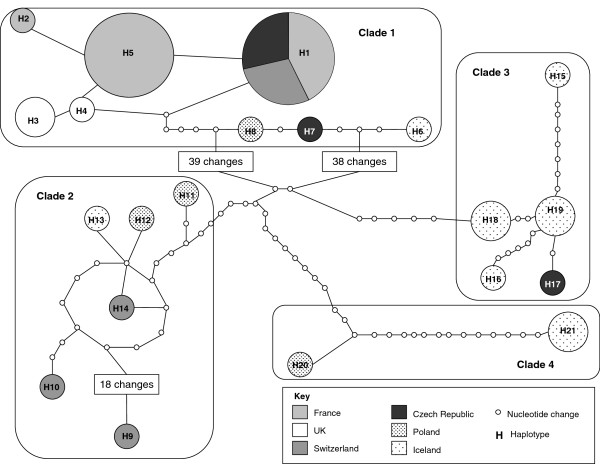
Network of ITS1 haplotypes of Trichobilharzia franki from Europe. Each circle represents a single unique haplotype and the size is proportional to the number of sequences with that specific haplotype. The ITS1 shows the UK sequences to have unique haplotypes (H3 and H4), but to be most closely related to haplotypes from France (H2 and H5) with only 1 to 2 nucleotide differences separating them. There is also some evidence of gene flow between geographical locations with the same haplotype (H1 which contains HM131183, HM131179, HM131180, AJ312041, AJ312042, AY795573, AF356845) appearing in France, Switzerland and the Czech Republic. All other haplotypes appear to be country specific, H2 contains AY795572; H3 contains Ham Ra7 and Ham Ra8; H4 contains Ham Ra 6; H5 contains HM131181, HM131177, HM131184, HM131182, HM131178, HM131176; H6 contains FJ469812; H7 contains AY713969; H8 contains AY713964; H9 contains AJ312044; H10 contains AJ312045; H11 contains AY713966; H12 contains FJ469809; H13 contains FJ469808; H14 contains AJ312046; H15 contains FJ469810; H16 contains FJ469811; H17 contains AY713973; H18 contains FJ469805 and FJ469806; H19 contains FJ469814 and FJ469821; H20 contains AJ312043; H21 contains FJ469819 and FJ469820.
Discussion
Utilising molecular techniques described by Jouet et al. [24], our study has provided the first detailed record of a Trichobilharzia parasite in the UK. The morphological description of T. franki supported by scanning electron microscopy observations, shows that the cercariae have considerable similarity to other Trichobilharzia species, particularly T. regenti. A molecular approach was therefore considered essential for identification. Phylogenetic analysis of the LSU and analysis of p–distance demonstrated that the UK-specific sequences corresponded to T. franki, showing the power of this ribosomal marker for species-specific identification and high levels of phylogenetic resolution as in other studies of avian schistosomatids [1,24]. However, there is clear evidence of a close relationship between T. franki and T. querquedulae, as also highlighted by Brant and Loker [1] and Jouet et al. [24]. Trichobilharzia querquedulae could be either a very close sister taxon, part of a species complex in which a number of Trichobilharzia species are very closely related and form the “T. franki” complex [24], or simply be a genetically distinct population of T. franki and not a species in its own right [24]. Such confounding factors on the identity of a species could be problematic for phylogenetic analysis and subsequent epidemiological surveys. Although the UK parasite was identified as T. franki, other molecular markers could be considered for identification of other species and genera of non-Schistosoma schistosomatids, to provide a comprehensive molecular phylogenetic framework for avian schistosomatid parasites [24-26].
Both the phylogenetic and haplotype network analysis of the ITS1 fragment from the UK samples show a close genetic relationship with French populations (Figures 4 and 5). The presence of Trichobilharzia spp. and occurrence of CD have been known in France for several decades [2,24] and, due to close proximity of France to the UK and the movement of migratory waterfowl between the two countries, it could be argued that although some differentiation between populations of T. franki has occurred, some gene flow is likely to take place. The occurrence of gene flow between countries was also illustrated in the haplotype network analysis due to H1 being shared between France, Switzerland and the Czech Republic (Figure 5). This is a clear indication that the distribution and prevalence of CD is intimately linked with the movement of waterfowl, and T. franki populations are moving around Europe, probably as a result of the migratory patterns of their definitive Anseriformes hosts (ducks, geese and swans) [1,9,24]. Brant and Loker [1] discussed the occurrence of genetically related individuals of T. physellae and T. querquedulae collected from snails along the major avian migratory flyways in the USA ranging from latitudes as distant as Alaska to Louisiana and Florida. This is probably also true for T. franki in the UK and the rest of Europe as some birds, such as swans and geese, migrate from Iceland to overwinter in the UK and mainland Europe from October to April, which would result in the movement of parasites within and between countries, reducing the level of genetic differentiation between T. franki populations [9]. It can be concluded from the current study that the collected parasites are closely related to populations in France, and it is highly likely that T. franki in Hampshire was brought to the UK by waterfowl migrating from France. Although the specimens used in this study were collected from a single site, it is likely that there are several different genetically unique populations of T. franki throughout the UK, particularly given the wide range of migratory waterfowl that visit the UK.
Little is known about the distribution of T. franki and other Trichobilharzia species in the UK and further investigation is required to identify the association of migratory birds and parasite distribution in order to fully understand the health risk of this parasite to the UK human population. Cercarial dermatitis is considered a global risk to human health and several authors have argued that the disease in Europe should be considered as re-emerging rather than a new threat [2,3,27]. This is partly because of changes in human activity, especially recreational use of water bodies over the previous two decades. There has been a long history and understanding of CD in mainland Europe and cases are frequently reported (reviewed by Soldánová et al. [27]). In the UK, however, the risk of infection and epidemiology of CD are relatively unknown and the disease has to be considered a potential re-emerging disease. This is mainly due to a distinct lack of knowledge of the occurrence of agents of CD and their abundance, but also to the lack of recorded cases with only a few cases being reported to physicians because symptoms can be pathologically benign and confused with other allergic reactions or insect bites [5,28]. In addition to human factors, it is important to note that recent climatic changes could also influence the epidemiology of CD [29]. All fluke infections in snails are climate/temperature sensitive with higher ambient temperatures tending to, not only result in higher cercarial emission, but also lead to higher snail numbers increasing the potential number of intermediate hosts and thus the risk of infection [29]. Climate change can also affect the distribution of definitive hosts, altering migratory routes and thus enabling infected bird species to visit the UK, which may not have done so before [29,30]. Although the UK has many native species of snail intermediate hosts of avian schistosomatids, it is possible that populations of introduced snail vectors could establish themselves in countries such as the UK that were previously too cold to sustain them; this has been observed with snail hosts of other schistosomatids. For example, Biomphalaria tenagophila, a snail vector of Schistosoma mansoni, which is a major agent of hepato-intestinal human schistosomiasis in Africa, has recently become established in Romania [31].
Conclusion
This current study describes the first specific and detailed record of T. franki in the UK, predominately based on molecular evidence, and illustrates the close relationship between the population sampled in this study and populations found throughout France. The relationship between the parasite populations is likely to be a result of the movement of parasites between countries due to migratory routes of waterfowl. Therefore, it is crucial to understand the role of waterfowl in the epidemiology of CD, and to apply appropriate tools to identify Trichobilharzia species in order to assess the risk of CD outbreaks and their impact on human health in the UK. This is particularly pertinent in view of the distinct lack of knowledge of the diversity of zoonotic flukes and their snail intermediate hosts within the UK and mainland Europe, especially when considering the potential impact to public health [32,33].
Abbreviations
CD: Cercarial dermatitis; G: Gamma distribution; H: Haplotype; Hd: Haplotype diversity; HKY: Hasegawa-Kishino-Yano substitution model; ITS1: Internal transcribed region 1; K2: Kimura 2-parameter model; LSU: Large subunit of ribosomal gene; ML: Maximum likelihood; MP: Maximum parsimony; NJ: Neighbour joining; PCR: Polymerase chain reaction; π: Nucleotide diversity.
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
SPL, JD, RC, AJW, RSK were involved in conceiving the project and wrote the manuscript. SPL, RSK, JD and RC performed the field collection and RC identified snails. RSK morphologically identified and described the furcocercariae. SPL, RML and JD performed the molecular laboratory work and phylogenetic analysis. SPL and RML performed the bioinformatics and population level analysis. All authors read and approved the final version of the manuscript.
Contributor Information
Scott P Lawton, Email: s.p.lawton@kingston.ac.uk.
Rivka M Lim, Email: rivkamlim@gmail.com.
Juliet P Dukes, Email: J.Dukes@kingston.ac.uk.
Richard T Cook, Email: R.Cook@kingston.ac.uk.
Anthony J Walker, Email: T.Walker@kingston.ac.uk.
Ruth S Kirk, Email: R.Kirk@kingston.ac.uk.
Acknowledgments
The authors would like to thank Mrs J. Llewellyn–Hughes and her staff for performing the DNA sequencing reactions at the Molecular Sequencing Facility at the Natural History Museum, London. We would also like to thank Kingston University for partial funding of the molecular work through a summer intern studentship to RML and Richard Giddens for assistance with the scanning electron microscope. The authors are grateful to the anonymous reviewers for their helpful comments.
References
- Brant SV, Loker ES. Molecular systematics of the avian schistosome genus Trichobilharzia (Trematoda: Schistosomatidae) in North America. J Parasitol. 2009;95:941–963. doi: 10.1645/GE-1870.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouet D, Ferté H, Depaquit J, Rudolfová J, Latour P, Zanella D, Kaltenbach ML, Léger N. Trichobilharzia spp. in natural conditions in Annecy Lake, France. Parasitol Res. 2008;103:51–58. doi: 10.1007/s00436-008-0926-3. [DOI] [PubMed] [Google Scholar]
- Horák P, Kolářová L. Snails, waterfowl and cercarial dermatitis. Freshwater Biol. 2011;56:779–790. doi: 10.1111/j.1365-2427.2010.02545.x. [DOI] [Google Scholar]
- Chamot E, Toscani L, Rougemont A. Public health importance and risk factors for cercarial dermatitis associated with swimming in Lake Leman at Geneva, Switzerland. Epidemiol Infect. 1998;120:305–314. doi: 10.1017/S0950268898008826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser SJ, Allan SJR, Roworth M, Smith HV, Holme SA. Cercarial dermatitis in the UK. Clin Exp Dermatol. 2008;34:344–346. doi: 10.1111/j.1365-2230.2008.02903.x. [DOI] [PubMed] [Google Scholar]
- PHLS. Report from the PHLS Communicable Disease Surveillance Centre. Br Med J. 1988;296:778–779. [PMC free article] [PubMed] [Google Scholar]
- Knight R, Worms MJ. An outbreak of cercarial dermatitis in Britain. Trans R Soc Trop Med Hyg. 1972;66:21. doi: 10.1016/0035-9203(72)90037-5. [DOI] [PubMed] [Google Scholar]
- Harding JR. Cardiff’s tropical disease: cercarial dermatitis. Med Hist. 1978;22:83–88. doi: 10.1017/S0025727300031768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolářová L. Schistosomes causing cercarial dermatitis: a mini–review of current trends in systematics and of host specificity and pathogenicity. Folia Parasitol. 2007;54:81–87. doi: 10.14411/fp.2007.010. [DOI] [PubMed] [Google Scholar]
- Horák P, Kolářová L, Adema CM. Biology of the schistosome genus Trichobilharzia. Adv Parasitol. 2002;52:156–233. doi: 10.1016/s0065-308x(02)52012-1. [DOI] [PubMed] [Google Scholar]
- Skrjabin KI, Zakharov NP. Zwei neue Trematodengattungen aus den Blutgefässen der Vögel. Izv Donsk Vet Inst. 1920;2:1–6. [Google Scholar]
- Morley NJ, Lewis JW. Anthropogenic pressure on a molluscan-trematode community over a long-term period in the Basingstoke Canal, UK, and its implications for ecosystem health. EcoHealth. 2007;3:269–280. doi: 10.1007/s10393-006-0058-0. [DOI] [Google Scholar]
- Khan D. Studies on larval trematodes infecting freshwater snails in London (UK) and some adjoining areas. Part IV. Schistosomatid cercariae. J Helminthol. 1961;35:275–284. doi: 10.1017/S0022149X0000465X. [DOI] [PubMed] [Google Scholar]
- Dvořák J, Vaňáčová Š, Hampl V, Fleger J, Horák P. Comparison of European Trichobilharzia species based on ITS1 and ITS2 sequences. Parasitology. 2002;124:307–313. doi: 10.1017/s0031182001001238. [DOI] [PubMed] [Google Scholar]
- Müller V, Kimmig P. Trichobilharzia franki n. sp. a causative agent of swimmer’s itch in south western Germany. Appl Parasitol. 1994;35:12–31. [PubMed] [Google Scholar]
- Kock S, Böckeler W. Observations on cercarial chaetotaxy as a means for the identification of European species of Trichobilharzia Skrjabin & Zakharow, 1920 (Digenea: Schistosomatidae) Syst Parasitol. 1998;43:159–166. doi: 10.1023/a:1006118619762. [DOI] [PubMed] [Google Scholar]
- Podhorsky M, Hůzova Z, Mikeš L, Horák P. Cercarial dimensions and surface structures as a tool for species determination of Trichobilharzia spp. Acta Parasitol. 2009;54:28–36. doi: 10.2478/s11686-009-0011-9. [DOI] [Google Scholar]
- Olson PD, Cribb TH, Tkach VV, Bray RA, Littlewood DT. Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda) Int J Parasitol. 2003;33:33–55. doi: 10.1016/s0020-7519(03)00049-3. [DOI] [PubMed] [Google Scholar]
- Wang CR, Li L, Ni HB, Zhai YQ, Chen AH, Chen J, Zhu XQ. Orientobilharzia turkestanicum is a member of Schistosoma genus based on phylogenetic analysis using ribosomal DNA sequences. Exp Parasitol. 2009;121:193–197. doi: 10.1016/j.exppara.2008.10.012. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user friendly biological sequence alignment program editor and analysis program for Windows 95/98/NT. Nucleic Acid Symp Ser. 1999;41:95–98. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSPv5: software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Clement M, Posada D, Crandall KA. TCS : a computer programme to estimate gene genealogies. Mol Ecol. 2000;9:1657–1660. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- Jouet D, Skírnisson K, Kolářová L, Ferté H. Molecular diversity of Trichobilharzia franki in two intermediate hosts (Radix auricularia and Radix peregra): a complex of species. Infect Genet Evol. 2010;10:1218–1227. doi: 10.1016/j.meegid.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Lockyer AE, Olsen PD, Ostergaard P, Rollinson D, Johnston DA, Attwood SW, Southgate VR, Horák P, Snyder SD, Le TH, Agatsuma T, McManus DP, Carmichael AC, Naem S, Littlewood DTJ. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology. 2003;126:203–224. doi: 10.1017/S0031182002002792. [DOI] [PubMed] [Google Scholar]
- Kolárová L, Rudolfová J, Hampl V, Skírnisson K. Allobilharzia visceralis gen. nov., sp. nov. (Schistosomatidae-Trematoda) from Cygnus cygnus (L.) (Anatidae) Parasitol Int. 2006;55:179–186. doi: 10.1016/j.parint.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Soldánová M, Selbach C, Kalbe M, Kostadinova A, Sures B. Swimmer’s itch: etiology, impact, and risk factors in Europe. Trends Parasitol. 2013;29:65–74. doi: 10.1016/j.pt.2012.12.002. [DOI] [PubMed] [Google Scholar]
- Lévesque B, Giovenazzo P, Guerrier P, Laverdière D, Prud’Homme H. Investigation of an outbreak of cercarial dermatitis. Epidemiol Infect. 2002;129:379–386. doi: 10.1017/s0950268802007379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas-Coma S, Valero MA, Bargues MD. Climate change effects on trematodiases, with emphasis on zoonotic fascioliasis and schistosomiasis. Vet Parasitol. 2009;163:264–280. doi: 10.1016/j.vetpar.2009.03.024. [DOI] [PubMed] [Google Scholar]
- Lafferty KD. The ecology of climate change and infectious diseases. Ecology. 2009;90:888–900. doi: 10.1890/08-0079.1. [DOI] [PubMed] [Google Scholar]
- Majoros G, Fehér Z, Deli T, Földvári G. Establishment of Biomphalaria tenagophila snails in Europe. Emerg Infect Diseases. 2008;14:1812–1813. doi: 10.3201/eid1411.080479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldánová M, Selbach C, Sures B, Kostadinova A, Pérez-del-Olmo A. Larval trematode communities in Radix auricularia and Lymnaea stagnalis in a reservoir system of the Ruhr River. Parasit Vectors. 2010;3:56. doi: 10.1186/1756-3305-3-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgieva S, Selbach C, Faltýnková A, Soldánová M, Sures B, Skírnisson K, Kostadinova A. A New cryptic species of the ‘revolutum’ group of Echinostoma (Digenea: Echinostomatidae) revealed by molecular and morphological data. Parasit Vectors. 2013;6:64. doi: 10.1186/1756-3305-6-64. [DOI] [PMC free article] [PubMed] [Google Scholar]


