Introduction
Abacavir is a nucleoside reverse transcriptase inhibitor (NRTI) used for treatment of HIV infection. HIV infection is usually treated with antiretroviral therapy regimens, which consist of three or more different drugs used in combination. Typical antiretrovirals used in these regimens include NRTIs, non-NRTIs, protease inhibitors, and integrase strand inhibitors [1]. Abacavir makes an ideal addition to these types of combination therapies because of its dosing flexibility. It can be administered either once or twice a day to match the dosing pattern of other drugs and can also be administered as tablets that contain other antiretroviral drugs such as lamivudine and zidovudine, allowing for a reduction in pill count [2]. Abacavir is generally well tolerated, and common side effects include nausea, headache, and diarrhea [2]. However, ~ 5–8% of patients experience a hypersensitivity reaction (HSR) within the first 6 weeks of treatment. Symptoms of an HSR include at least two of the following: fever, rash, cough, gastrointestinal symptoms, dyspnea, and fatigue [3]. These symptoms worsen with continued treatment, but typically improve within 24 h after discontinuation. However, drug rechallenge after discontinuation of abacavir because of an HSR can result in symptom recurrence within a matter of hours, and can lead to immediate and potentially fatal allergic reactions [4,5]. This hypersensitivity reaction is strongly linked to the presence of the HLA-B*57:01 allele, and testing for the allele before abacavir treatment is recommended by the US Food and Drug Administration (FDA) [6], the European Medicines Agency [7], the Clinical Pharmacogenetics Implementation Consortium [3], and the Dutch Pharmacogenetics Working Group [8]. Abacavir was also reported to be associated with a higher risk for myocardial infarction as compared with other NRTIs [9–12]. However, a meta-analysis carried out by the FDA in 2012 failed to find any such association [13].
Drug interactions
As abacavir is primarily metabolized by cytosolic alcohol dehydrogenase (ADH) and uridine diphosphate glucuronosyltransferase (UGT) enzymes, no interactions between abacavir and inducers or inhibitors of cytochrome P450 (CYP) enzymes are predicted [2]. In addition, invitro studies have shown that abacavir is unlikely to inhibit CYP enzymes at clinically relevant concentrations [14]. As non-NRTIs and protease inhibitors are primarily metabolized by CYP enzymes [15], this may eliminate the potential for drug interactions with these types of antiretrovirals. No clinically significant pharmacokinetic changes were seen when abacavir was administered with other NRTIs such as lamivudine and zidovudine [2,16]. As alcohol is also metabolized by ADH, pharmacokinetic interactions between the drug and ethanol have been analyzed, but no clinically significant changes or new adverse events have been reported [17]. Several studies have found a link between abacavir administration and virologic response in hepatitis C patients being treated with ribavirin and pegylated interferon who are also coinfected with HIV; in these patients, abacavir usage has been found to be significantly associated with early virologic failure [18] and lack of sustained virologic response [19,20]. However, subpopulation analyses from two of the studies found that the impact of abacavir on sustained virologic response was only significant in patients with baseline hepatitis C viral RNA above a certain level [20], ribavirin daily doses below a certain level [20], or ribavirin trough concentrations below a certain level [19]. In addition, a number of studies have found no association between abacavir usage and virologic response [21–24]; therefore, it is uncertain whether these two drugs have a significant and harmful interaction.
Pharmacokinetics
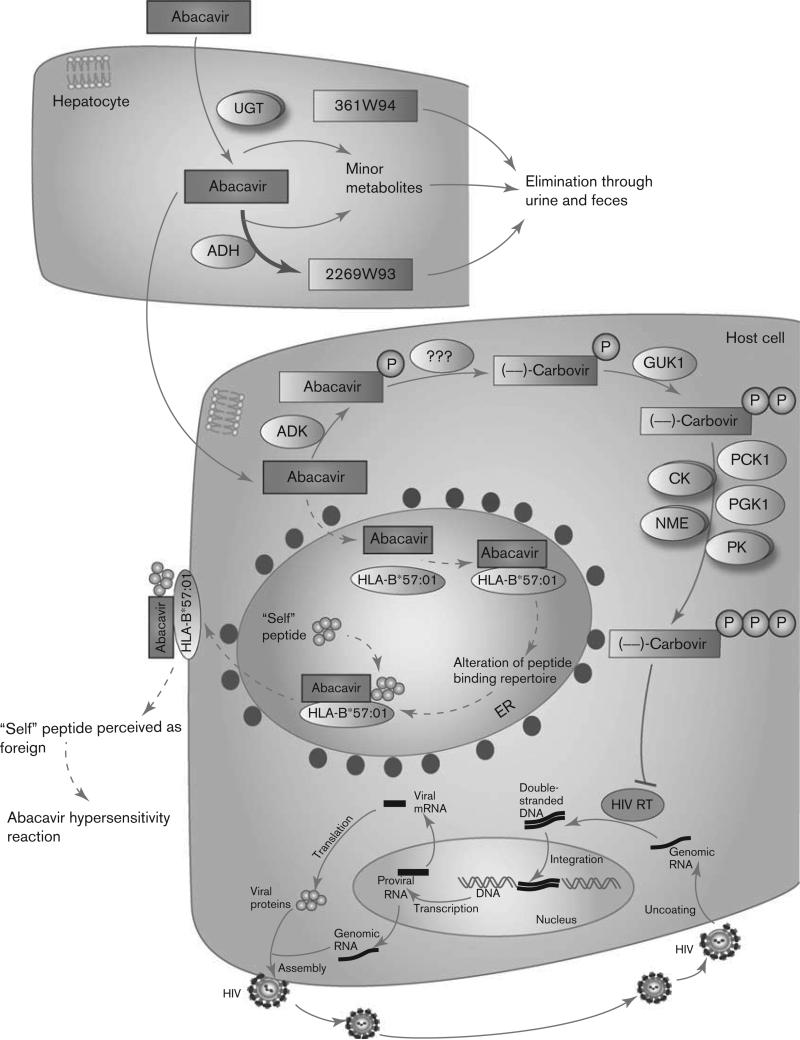
A schematic representation of abacavir disposition within the body is provided in Fig. 1. Abacavir is rapidly absorbed following oral administration, and has a mean absolute bioavailability of ~83% [2,25]. The drug is lipophilic yet also shows high water solubility, allowing it to cross cell membranes by passive diffusion alone. These properties may explain its high bioavailability, as well as its ability to easily penetrate into tissues such as the blood–brain barrier [2,26]. After absorption, abacavir is extensively metabolized within the liver, with less than 2% of the drug excreted unchanged in the urine [27]. ADH and UGT are the primary enzymes responsible for abacavir metabolism within hepatocytes. Metabolism by ADH results in the inactive carboxylate metabolite 2269W93; metabolism by the UGT enzymes results in the inactive glucuronide metabolite 361W94 [27]. A mass balance study found that 83% of the original dose was eliminated in the urine and 16% in the feces. Of the 83% eliminated through urine, 36% of the dose recovered was the glucuronide metabolite, and 30% was the carboxylate metabolite. The remaining dose was either the parent drug or trace metabolites [2,27].
Fig. 1.
Schematic representation of abacavir pharmacokinetics and pharmacodynamics. The potential mechanism of an abacavir hypersensitivity reaction is also shown and is drawn using dashed lines as it is not currently well established. A fully interactive version is available online at http://www.pharmgkb.org/pathway/PA166104634.
The parent drug that is not metabolized by hepatocytes undergoes metabolism within viral-infected cells by a different set of intracellular enzymes, converting the drug into its pharmacologically active metabolite. Initially, abacavir is metabolized to abacavir 5’-monophosphate by the enzyme adenosine phosphotransferase (encoded by the ADK gene). It then undergoes deamination by an unknown cytosolic enzyme to form (–)-carbovir 5’-monophosphate; no diphosphates or triphosphates of abacavir have been detected within cells [26,28]. (–)-Carbovir 5’-monophosphate is then converted to (–)-carbovir 5’-diphosphate by the enzyme guanylate kinase (GUK1), which is stereo-selective for the (–) enantiomer of carbovir [28–30]. One study in particular found that (–)-carbovir 5’-monophosphate was 7000 times more efficient as a substrate for guanylate kinase than the (+) enantiomer [30]. (–)-Carbovir 5’-diphosphate is then converted to the active 5’-triphosphate form by various cellular kinases. These include creatine kinases (represented by the CK gene group in Fig. 1), pyruvate kinases (PK gene group), nucleoside diphosphate kinases (NME gene group), phosphoglycerate kinase (PGK1), and phosphoenolpyruvate carboxykinase (PCK1). Given the stereoselectivity of guanylate kinase, only (–)-carbovir 5’-triphosphate (CBV-TP) is formed in any significant quantity. However, the (+) enantiomer has equivalent antiviral activity [28,30].
Abacavir can also be converted to carbovir 5’-monophosphate by other minor pathways, such as transformation into carbovir, followed by phosphorylation of carbovir into carbovir 5’-monophosphate by inosine phosphotransferase. As these pathways make up less than 2% of abacavir metabolism, they are not shown in Fig. 1 [28].
Pharmacodynamics
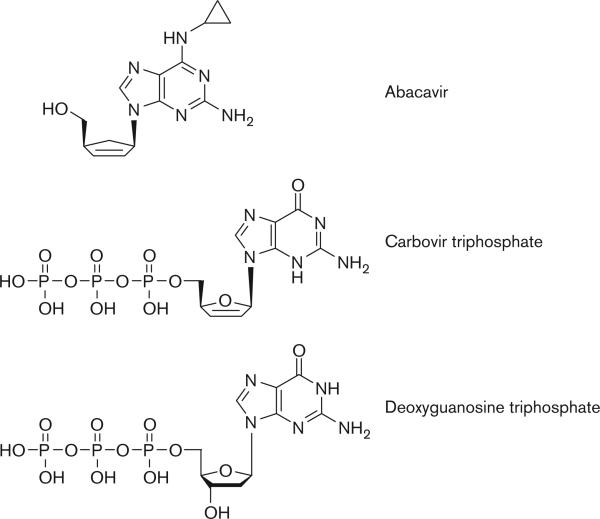
A stylized depiction of the mechanism of action of abacavir within host cells is provided in Fig. 1. CBV-TP represents the active form of abacavir and works by blocking the action of HIV reverse transcriptase (HIV-RT). HIV replicates by taking advantage of the host cell's existing genetic machinery, as well as using the viral enzyme reverse transcriptase, which is responsible for the conversion of viral RNA into double-stranded DNA. The viral DNA can then be incorporated into the host DNA, at which point the host machinery may convert the DNA into viral RNA. The viral RNA is then translated into viral proteins, which assemble to form the HIV virus [31]. CBV-TP acts as a guanosine analog, and competes for incorporation into the nucleotide chain being produced by HIV-RT from the viral RNA [26]. Other NRTIs can act as derivatives of different nucleo-sides, leading to the same type of inhibition. Examples of this are didanosine and adenosine, zalcitabine and cytidine, and zidovudine and thymidine [32]. After incorporation of CBV-TP into the nucleotide chain, its lack of a 3’-OH on the deoxyribose sugar on which subsequent nucleotides are added blocks continuing synthesis of viral DNA [28,31]. A comparison between the structures of carbovir triphosphate and deoxyguano-sine triphosphate, as well as the structure of abacavir, is shown in Fig. 2.
Fig. 2.
Structures for abacavir, CBV-TP, and deoxyguanosine triphosphate (dGTP). Note the similarities in structure between CBV-TP and dGTP, excluding the absence of the critical free 3’-OH group on the deoxyribose sugar ring in CBV-TP. This absence prevents the addition of any additional nucleotides and blocks further viral DNA synthesis. CBV-TP, (–)-carbovir 5’-triphosphate.
CBV-TP is particularly well suited for this role as it is highly selective for reverse transcriptase as compared with DNA polymerases α, β, γ, and ε. Indeed, Ki values for DNA polymerases α, β, γ, and ε were 90−, 2900−, 1200−, and 1900-fold greater, respectively, than the Ki value for HIV-RT [26]. This selectivity for reverse transcriptase prevents the potentially toxic side effects that occur when DNA polymerases are inhibited. Many antiretroviral NRTIs are associated with a range of adverse events attributed to mitochondrial dysfunction, such as lactic acidosis and hepatic steatosis. These are believed to result from the inhibition of mitochondrial DNA polymerase γ by these drugs, leading to altered mitochondrial DNA replication and resulting in mitochondrial myopathy and toxicity. Abacavir has the lowest inhibition rate for DNA polymerase γ, whereas zalcitabine, didanosine, and stavudine have the highest [32].
Pharmacogenetics
The pharmacogenetics of abacavir is well established and is almost exclusively related to the human leukocyte antigen B (HLA-B) gene and its variant allele *57:01. This particular allele has been shown to be strongly associated with abacavir HSR in a large number of studies, as discussed below. However, the positive predictive value for this allele is below 50% [33], indicating that additional factors, genetic or otherwise, may be involved in the development of an HSR. Limited research has been conducted in this area, although some potential exists for a variant in the gene HSP70-HOM (also known as HSP1AL), whose protein is hypothesized to be directly involved in the stimulation of an immune reaction to abacavir [34].
HLA-B*57:01
The HLA-B gene is a member of the major histocompatibility complex region located on chromosome 6. This genomic region encodes three groups of genes involved in the immune system. HLA-B is part of the class I group, along with HLA-A and HLA-C, all of which code for their eponymous proteins. The HLA-B protein and the other class I group members are cell-surface molecules responsible for the presentation of endogenous peptides to immune system cells and exist on almost all nucleated cells. This is in contrast to class II molecules, which display exogenous peptides and are present only on antigen presenting cells such as macrophages or dendritic cells. Briefly, class I molecules such as HLA-B are heterodimers consisting of an a-chain, encoded by the HLA-B gene, and a protein known as β2-microglobulin, which is encoded on chromosome 15. The α-chain of HLA-B has four domains: one cytoplasmic, one trans-membrane, one that binds to CD8 + cytotoxic T cells, and the last that makes up a peptide-binding groove, in which the peptide to be presented is nestled. The region of the gene encoding the peptide-binding groove is highly polymorphic, allowing for the presentation of a wide variety of peptides. Most of the peptides that HLA-B presents come from the normal breakdown of cellular proteins and are recognized by the immune system as such (i.e. ‘self’ peptides). However, when a cell becomes infected by a pathogen, the proteins presented will be from the pathogen and recognized as foreign or ‘non-self’. T-cell antigen receptors on CD8 + cytotoxic T cells are responsible for this recognition and stimulate an immune reaction and destroy the cell [35,36].
In 2002, two separate research groups published evidence that an allele known as HLA-B*57:01 was present in a significantly higher percentage of patients showing an abacavir HSR compared with patients with no reaction. One study was conducted on a North American population [37] and the other on a population known as the Western Australian HIV cohort [4]. Both included 200 patients. This was confirmed by another study that included 64 participants from the UK [38]. However, these three studies were conducted predominantly on White males, limiting their scope. Despite this limitation, several clinics began implementing prospective screening for these alleles to great success [39–41]. A later study recognized the significance of this allele in White female and Hispanic populations, but did not find any significant associations in Black populations [42]. This was likely because of the lower number of Black patients within this study (as compared with Hispanics and Whites) and the fact that Black populations have a lower carriage rate of the allele [42,43]. European populations have a *57:01 allele frequency of about 6–7%, but African populations often have allele frequencies of less than 2.5%. In addition, some Asian populations, such as the Japanese or South Koreans, have extremely low allele frequencies of 0.5% or less; in contrast, some Indian populations have *57:01 frequencies of greater than 16% [3] (allele frequency information taken from the supplementary information). In 2007, a study known as SHAPE (which included a similar number of White and Black participants) found that Black patients did have fewer cases of abacavir HSRs. However, in patients with immuno-logically confirmed HSRs, 100% of both White and Black patients were positive for the HLA-B*57:01 allele. This suggested that, although immunologically confirmed HSRs are rare among Black populations because of the reduced carriage of the allele, HLA-B*57:01 has the same clinical implications in both populations [44]. A definitive association between this allele and abacavir HSRs came in 2008 with the results of the PREDICT-1 study, a double-blind, prospective, randomized study with 1956 patients from 19 countries. Patients were observed for 6 weeks and separated into two categories: those who underwent screening for the HLA-B*57:01 allele and were eliminated if they tested positive, and those who underwent standard care without any screening. Abacavir HSRs were immunologically confirmed by skin patch testing. The results of the study showed that screening completely eliminated HSRs – 0% of the patients screened had an HSR, whereas 2.7% of the control population had an HSR. This gave screening a negative predictive value of 100%. However, the study had a positive predictive value of 47.9%, indicating that about half of all the patients who are HLA-B*57:01 positive will not develop an abacavir HSR [33]. This indicates that additional factors, genetic or nongenetic, are likely involved in the development of an HSR. This paper, along with the large amount of other existing evidence, led the FDA to implement a boxed warning in 2008, detailing the risk for HSRs in abacavir-treated patients with the HLA-B*57:01 allele [6]. The FDA [6], along with the European Medicines Agency [7], Clinical Pharmacogenetics Implementation Consortium [3], and Dutch Pharmacogenetics Working Group [8], also recommend that all patients be screened before being treated and that abacavir not be used in HLA-B*57:01-positive individuals.
The HLA-B protein has no direct effect on abacavir pharmacokinetics or pharmacodynamics, and it is still unclear how the HLA-B*57:01 allele affects susceptibility to drug hypersensitivity. Several hypotheses exist. One theory is the hapten concept, which suggests that small compounds such as drugs (called ‘haptens’) bind to the peptides bound to immune receptors such as HLA-B, causing T cells to react and stimulate an immune reaction [45]. Another theory is the p-i concept (pharmacological interactions with immune receptors), which suggests that drugs bind directly and reversibly to immune receptors, stimulating an immune reaction [45]. Recent evidence seems to support an alternative hypothesis. Two studies, both published in 2012, found that abacavir can bind noncovalently and with specificity to the F pocket of the peptide-binding groove of HLA-B*57:01 [46,47]. Because of the amino acid residues unique to the *57:01 allele, abacavir can bind only to this particular form of HLA-B. The binding of abacavir to HLA-B*57:01 is believed to change the shape and chemistry of the antigen-binding cleft, and consequently the repertoire of peptides that can bind the molecule. Indeed, both of these papers, as well as an additional paper by Norcross and colleagues, identified particular changes in the peptides presented by HLA-B*57:01 in the presence of abacavir, as compared with those when the drug was absent [46–48]. Conventional HLA-B*57:01 epitopes frequently possess large hydrophobic amino acids such as tryptophan or phenylalanine at their C-terminus; however, Illing and colleagues [46–48] found that peptides eluted in the presence of abacavir showed a preference for isoleucine or leucine at this position. This binding and subsequent peptide alteration is shown in the pathway figure – a dashed line is used, as this mechanism is not currently well established. The typical cycle of peptide loading and transport to the cell-surface plasma membrane [49] is also shown. As T cells are trained to be tolerant to a particular repertoire of peptides during their development in the thymus, an alteration in the peptides that are presented may mean that these new peptides are perceived as foreign. This change would stimulate CD8 + T-cell production and response, and would manifest as an abacavir HSR [46,47]. Indeed, CD8 + T cells are abundant in skin biopsies of patients who present with a rash during an abacavir HSR [50].
HSP70-HOM
The positive predictive value of ~50% for the HLA-B*57:01 allele and abacavir HSRs indicates the need for further studies to elucidate whether other genes affect the development of an HSR. Research in this area has been scarce, but several studies have suggested a member of the 70 kDa heat shock protein (HSP70) family to be a potential factor. The HSP70 proteins are responsible for protecting cells from stress, as well as assist in other cellular activities, such as protein folding [51]. Three genes within the human major histocompatibility complex region encode members of the HSP70 family: HSP70-1, HSP70-2, and HSP70-HOM [52]. HSP70-1 and HSP70-2 encode identical heat-inducible protein products, whereas HSP70-HOM (also known as HSP1AL) encodes a similar but non-heat-inducible protein [52]. A study using the Western Australian HIV cohort found that the reference C allele at rs2227956 in the HSP70-HOM gene (which results in a threonine at residue 493 as opposed to a methionine) was found in combination with the HLA-B*57:01 allele in 94.4% of immunologically confirmed hypersensitive cases and 0.4% of controls, whereas the HLA-B*57:01 allele appeared on its own in 94.4% of hypersensitive cases and 1.7% of controls. The authors suggested that consideration of the HSP70-HOM allele in addition to the HLA-B*57:01 allele may therefore increase the ability to discriminate between patients who would develop an HSR and tolerant controls. The population consisted of 230 controls and 18 patients with an HSR, and the alleles were found to be in strong linkage disequilibrium [53]. Further studies in larger populations are needed to verify this association. However, a later study did find that the HSP70-HOM protein colocalized with both the HLA-B*57:01 protein and abacavir within the endoplasmic reticulum. This implies that the HSP70-HOM 493T variant may lead to a protein that somehow facilitates the presentation of abacavir antigens to CD8 + T cells, perhaps by chaperoning the drug in antigen processing [34].
Conclusion
The implementation of HLA-B*57:01 testing before abacavir treatment is one of the best examples of pharmacogenetic research being used in the clinic. Genotyping for this allele is widely available in the western world. Despite this, further research should still be conducted on additional factors that lead to a propensity for an abacavir HSR. This could increase the positive predictive value, allowing more patients to be administered abacavir who could benefit from treatment. Currently there is very little evidence for the involvement of other genes, and only HSP70-HOM has emerged as a potential factor. Further advancement of our understanding in this area could prevent inappropriate denial of abacavir to patients who would tolerate it and hopefully help further elucidate the mechanism by which abacavir elicits its hypersensitivity reaction.
Acknowledgements
This work is supported by the NIH/NIGMS (R24 GM61374) and NIH grant U01 GM061390. The authors thank Li Gong for critical reading of this manuscript.
Footnotes
Conflicts of interest
There are no conflicts of interest.
References
- 1.Thompson MA, Aberg JA, Hoy JF, Telenti A, Benson C, Cahn P, et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. JAMA 2012. 308:387–402. doi: 10.1001/jama.2012.7961. [DOI] [PubMed] [Google Scholar]
- 2.Yuen GJ, Weller S, Pakes GE. A review of the pharmacokinetics of abacavir. Clin Pharmacokinet. 2008;47:351–371. doi: 10.2165/00003088-200847060-00001. [DOI] [PubMed] [Google Scholar]
- 3.Martin MA, Klein TE, Dong BJ, Pirmohamed M, Haas DW, Kroetz DL, et al. Clinical pharmacogenetics implementation consortium guidelines for HLA-B genotype and abacavir dosing. Clin Pharmacol Ther. 2012;91:734–738. doi: 10.1038/clpt.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, et al. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727–732. doi: 10.1016/s0140-6736(02)07873-x. [DOI] [PubMed] [Google Scholar]
- 5.Escaut L, Liotier JY, Albengres E, Cheminot N, Vittecoq D. Abacavir rechallenge has to be avoided in case of hypersensitivity reaction. AIDS. 1999;13:1419–1420. doi: 10.1097/00002030-199907300-00026. [DOI] [PubMed] [Google Scholar]
- 6.Ma JD, Lee KC, Kuo GM. HLA-B*57:01 testing to predict abacavir hypersensitivity. PLoS Curr. 2010;2:RRN1203. doi: 10.1371/currents.RRN1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stocchi L, Cascella R, Zampatti S, Pirazzoli A, Novelli G, Giardina E. The pharmacogenomic HLA biomarker associated to adverse abacavir reactions: comparative analysis of different genotyping methods. Curr Genomics. 2012;13:314–320. doi: 10.2174/138920212800793311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, et al. Pharmacogenetics: from bench to byte – an update of guidelines. Clin Pharmacol Ther. 2011;89:662–673. doi: 10.1038/clpt.2011.34. [DOI] [PubMed] [Google Scholar]
- 9.D:A:D Study Group. Sabin CA, Worm SW, Weber R, Reiss P, El-Sadr W, et al. Use of nucleoside reverse transcriptase inhibitors and risk of myocardial infarction in HIV-infected patients enrolled in the D:A:D study: a multi-cohort collaboration. Lancet. 2008;371:1417–1426. doi: 10.1016/S0140-6736(08)60423-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang S, Mary-Krause M, Cotte L, Gilquin J, Partisani M, Simon A, et al. Impact of individual antiretroviral drugs on the risk of myocardial infarction in human immunodeficiency virus-infected patients: a case-control study nested within the French Hospital Database on HIV ANRS cohort CO4. Arch Intern Med. 2010;170:1228–1238. doi: 10.1001/archinternmed.2010.197. [DOI] [PubMed] [Google Scholar]
- 11.Obel N, Farkas DK, Kronborg G, Larsen CS, Pedersen G, Riis A, et al. Abacavir and risk of myocardial infarction in HIV-infected patients on highly active antiretroviral therapy: a population-based nationwide cohort study. HIV Med. 2010;11:130–136. doi: 10.1111/j.1468-1293.2009.00751.x. [DOI] [PubMed] [Google Scholar]
- 12.Strategies for Management of Anti-Retroviral Therapy/INSIGHT, DAD Study Groups Use of nucleoside reverse transcriptase inhibitors and risk of myocardial infarction in HIV-infected patients. AIDS. 2008;22:F17–F24. doi: 10.1097/QAD.0b013e32830fe35e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding X, Andraca-Carrera E, Cooper C, Miele P, Kornegay C, Soukup M, et al. No association of abacavir use with myocardial infarction: findings of an FDA meta-analysis. J Acquir Immune Defic Syndr. 2012;61:441–447. doi: 10.1097/QAI.0b013e31826f993c. [DOI] [PubMed] [Google Scholar]
- 14.McDowell JA, Lou Y, Symonds WS, Stein DS. Multiple-dose pharmacokinetics and pharmacodynamics of abacavir alone and in combination with zidovudine in human immunodeficiency virus-infected adults. Antimicrob Agents Chemother. 2000;44:2061–2067. doi: 10.1128/aac.44.8.2061-2067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joly V, Descamps D, Yeni P. NNRTI plus PI combinations in the perspective of nucleoside-sparing or nucleoside-failing antiretroviral regimens. AIDS Rev. 2002;4:128–139. [PubMed] [Google Scholar]
- 16.Barry M, Mulcahy F, Merry C, Gibbons S, Back D. Pharmacokinetics and potential interactions amongst antiretroviral agents used to treat patients with HIV infection. Clin Pharmacokinet. 1999;36:289–304. doi: 10.2165/00003088-199936040-00004. [DOI] [PubMed] [Google Scholar]
- 17.McDowell JA, Chittick GE, Stevens CP, Edwards KD, Stein DS. Pharmacokinetic interaction of abacavir (1592U89) and ethanol in human immunodeficiency virus-infected adults. Antimicrob Agents Chemother. 2000;44:1686–1690. doi: 10.1128/aac.44.6.1686-1690.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bani-Sadr F, Denoeud L, Morand P, Lunel-Fabiani F, Pol S, Cacoub P, et al. Early virologic failure in HIV-coinfected hepatitis C patients treated with the peginterferon-ribavirin combination: does abacavir play a role? J Acquir Immune Defic Syndr. 2007;45:123–125. doi: 10.1097/QAI.0b013e318040b2b6. [DOI] [PubMed] [Google Scholar]
- 19.Vispo E, Barreiro P, Pineda JA, Mira JA, Maida I, Martin-Carbonero L, et al. Low response to pegylated interferon plus ribavirin in HIV-infected patients with chronic hepatitis C treated with abacavir. Antivir Ther. 2008;13:429–437. [PubMed] [Google Scholar]
- 20.Mira JA, Lopez-Cortes LF, Barreiro P, Tural C, Torres-Tortosa M, de Los Santos Gil I, et al. Efficacy of pegylated interferon plus ribavirin treatment in HIV/hepatitis C virus co-infected patients receiving abacavir plus lamivudine or tenofovir plus either lamivudine or emtricitabine as nucleoside analogue backbone. J Antimicrob Chemother. 2008;62:1365–1373. doi: 10.1093/jac/dkn420. [DOI] [PubMed] [Google Scholar]
- 21.Amorosa VK, Slim J, Mounzer K, Bruno C, Hoffman-Terry M, Dorey-Stein Z, et al. The influence of abacavir and other antiretroviral agents on virological response to HCV therapy among antiretroviral-treated HIV-infected patients. Antivir Ther. 2010;15:91–99. doi: 10.3851/IMP1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pineda JA, Mira JA, Gil Ide L, Valera-Bestard B, Rivero A, Merino D, et al. Influence of concomitant antiretroviral therapy on the rate of sustained virological response to pegylated interferon plus ribavirin in hepatitis C virus/HIV-coinfected patients. J Antimicrob Chemother. 2007;60:1347–1354. doi: 10.1093/jac/dkm373. [DOI] [PubMed] [Google Scholar]
- 23.Laufer N, Laguno M, Perez I, Cifuentes C, Murillas J, Vidal F, et al. Abacavir does not influence the rate of virological response in HIV-HCV-coinfected patients treated with pegylated interferon and weight-adjusted ribavirin. Antivir Ther. 2008;13:953–957. [PMC free article] [PubMed] [Google Scholar]
- 24.Solas C, Pambrun E, Winnock M, Salmon D, Poizot-Martin I, Dominguez S, et al. Ribavirin and abacavir drug interaction in HIV–HCV coinfected patients: fact or fiction? AIDS. 2012;26:2193–2199. doi: 10.1097/QAD.0b013e32835763a4. [DOI] [PubMed] [Google Scholar]
- 25.Chittick GE, Gillotin C, McDowell JA, Lou Y, Edwards KD, Prince WT, et al. Abacavir: absolute bioavailability, bioequivalence of three oral formulations, and effect of food. Pharmacotherapy. 1999;19:932–942. doi: 10.1592/phco.19.11.932.31568. [DOI] [PubMed] [Google Scholar]
- 26.Daluge SM, Good SS, Faletto MB, Miller WH St, Clair MH, Boone LR, et al. 1592U89, a novel carbocyclic nucleoside analog with potent, selective anti-human immunodeficiency virus activity. Antimicrob Agents Chemother. 1997;41:1082–1093. doi: 10.1128/aac.41.5.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDowell JA, Chittick GE, Ravitch JR, Polk RE, Kerkering TM, Stein DS. Pharmacokinetics of [(14)C]abacavir, a human immunodeficiency virus type 1 (HIV-1) reverse transcriptase inhibitor, administered in a single oral dose to HIV-1-infected adults: a mass balance study. Antimicrob Agents Chemother. 1999;43:2855–2861. doi: 10.1128/aac.43.12.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faletto MB, Miller WH, Garvey EP St, Clair MH, Daluge SM, Good SS. Unique intracellular activation of the potent anti-human immunodeficiency virus agent 1592U89. Antimicrob Agents Chemother. 1997;41:1099–1107. doi: 10.1128/aac.41.5.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balzarini J, Naesens L, De Clercq E. New antivirals – mechanism of action and resistance development. Curr Opin Microbiol. 1998;1:535–546. doi: 10.1016/s1369-5274(98)80086-6. [DOI] [PubMed] [Google Scholar]
- 30.Miller WH, Daluge SM, Garvey EP, Hopkins S, Reardon JE, Boyd FL, et al. Phosphorylation of carbovir enantiomers by cellular enzymes determines the stereoselectivity of antiviral activity. J Biol Chem. 1992;267:21220–21224. [PubMed] [Google Scholar]
- 31.Richman DD. HIV chemotherapy. Nature. 2001;410:995–1001. doi: 10.1038/35073673. [DOI] [PubMed] [Google Scholar]
- 32.Kakuda TN. Pharmacology of nucleoside and nucleotide reverse transcriptase inhibitor-induced mitochondrial toxicity. Clin Ther. 2000;22:685–708. doi: 10.1016/S0149-2918(00)90004-3. [DOI] [PubMed] [Google Scholar]
- 33.Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, et al. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568–579. doi: 10.1056/NEJMoa0706135. [DOI] [PubMed] [Google Scholar]
- 34.Martin AM, Almeida CA, Cameron P, Purcell AW, Nolan D, James I, et al. Immune responses to abacavir in antigen-presenting cells from hypersensitive patients. AIDS. 2007;21:1233–1244. doi: 10.1097/QAD.0b013e3280119579. [DOI] [PubMed] [Google Scholar]
- 35.Janeway CAJ, Travers P, Walport M, Shlomchik MJ. Antigen recognition by T cells. In: Janeway CAJ, editor. Immunobiology: the immune system in health and disease. Garland Science; New York: 2001. [Google Scholar]
- 36.Janeway CAJ, Travers P, Walport M, Shlomchik MJ. The major histocompatibility complex and its functions. In: Janeway CAJ, editor. Immunobiology: the immune system in health and disease. Garland Science; New York: 2001. [Google Scholar]
- 37.Hetherington S, Hughes AR, Mosteller M, Shortino D, Baker KL, Spreen W, et al. Genetic variations in HLA-B region and hypersensitivity reactions to abacavir. Lancet. 2002;359:1121–1122. doi: 10.1016/S0140-6736(02)08158-8. [DOI] [PubMed] [Google Scholar]
- 38.Hughes DA, Vilar FJ, Ward CC, Alfirevic A, Park BK, Pirmohamed M. Cost-effectiveness analysis of HLA B*5701 genotyping in preventing abacavir hypersensitivity. Pharmacogenetics. 2004;14:335–342. doi: 10.1097/00008571-200406000-00002. [DOI] [PubMed] [Google Scholar]
- 39.Waters LJ, Mandalia S, Gazzard B, Nelson M. Prospective HLA-B*5701 screening and abacavir hypersensitivity: a single centre experience. AIDS. 2007;21:2533–2534. doi: 10.1097/QAD.0b013e328273bc07. [DOI] [PubMed] [Google Scholar]
- 40.Zucman D, Truchis P, Majerholc C, Stegman S, Caillat-Zucman S. Prospective screening for human leukocyte antigen-B*5701 avoids abacavir hypersensitivity reaction in the ethnically mixed French HIV population. J Acquir Immune Defic Syndr. 2007;45:1–3. doi: 10.1097/QAI.0b013e318046ea31. [DOI] [PubMed] [Google Scholar]
- 41.Rauch A, Nolan D, Martin A, McKinnon E, Almeida C, Mallal S. Prospective genetic screening decreases the incidence of abacavir hypersensitivity reactions in the Western Australian HIV cohort study. Clin Infect Dis. 2006;43:99–102. doi: 10.1086/504874. [DOI] [PubMed] [Google Scholar]
- 42.Hughes AR, Mosteller M, Bansal AT, Davies K, Haneline SA, Lai EH, et al. Association of genetic variations in HLA-B region with hypersensitivity to abacavir in some, but not all, populations. Pharmacogenomics. 2004;5:203–211. doi: 10.1517/phgs.5.2.203.27481. [DOI] [PubMed] [Google Scholar]
- 43.Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernandez-Vina MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Hum Immunol. 2001;62:1009–1030. doi: 10.1016/s0198-8859(01)00298-1. [DOI] [PubMed] [Google Scholar]
- 44.Saag M, Balu R, Phillips E, Brachman P, Martorell C, Burman W, et al. High sensitivity of human leukocyte antigen-b*5701 as a marker for immunologically confirmed abacavir hypersensitivity in white and black patients. Clin Infect Dis. 2008;46:1111–1118. doi: 10.1086/529382. [DOI] [PubMed] [Google Scholar]
- 45.Pichler WJ, Beeler A, Keller M, Lerch M, Posadas S, Schmid D, et al. Pharmacological interaction of drugs with immune receptors: the p-i concept. Allergol Int. 2006;55:17–25. doi: 10.2332/allergolint.55.17. [DOI] [PubMed] [Google Scholar]
- 46.Ostrov DA, Grant BJ, Pompeu YA, Sidney J, Harndahl M, Southwood S, et al. Drug hypersensitivity caused by alteration of the MHC-presented self-peptide repertoire. Proc Natl Acad Sci USA. 2012;109:9959–9964. doi: 10.1073/pnas.1207934109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, Bharadwaj M, et al. Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature. 2012;486:554–558. doi: 10.1038/nature11147. [DOI] [PubMed] [Google Scholar]
- 48.Norcross MA, Luo S, Lu L, Boyne MT, Gomarteli M, Rennels AD, et al. Abacavir induces loading of novel self-peptides into HLA-B*57: 01: an autoimmune model for HLA-associated drug hypersensitivity. AIDS. 2012;26:F21–F29. doi: 10.1097/QAD.0b013e328355fe8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hewitt EW. The MHC class I antigen presentation pathway: strategies for viral immune evasion. Immunology. 2003;110:163–169. doi: 10.1046/j.1365-2567.2003.01738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chessman D, Kostenko L, Lethborg T, Purcell AW, Williamson NA, Chen Z, et al. Human leukocyte antigen class I-restricted activation of CD8 + T cells provides the immunogenetic basis of a systemic drug hypersensitivity. Immunity. 2008;28:822–832. doi: 10.1016/j.immuni.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 51.Tavaria M, Gabriele T, Kola I, Anderson RL. A hitchhiker's guide to the human Hsp70 family. Cell Stress Chaperones. 1996;1:23–28. doi: 10.1379/1466-1268(1996)001<0023:ahsgtt>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Milner CM, Campbell RD. Polymorphic analysis of the three MHC-linked HSP70 genes. Immunogenetics. 1992;36:357–362. doi: 10.1007/BF00218042. [DOI] [PubMed] [Google Scholar]
- 53.Martin AM, Nolan D, Gaudieri S, Almeida CA, Nolan R, James I, et al. Predisposition to abacavir hypersensitivity conferred by HLA-B*5701 and a haplotypic Hsp70-HOM variant. Proc Natl Acad Sci USA. 2004;101:4180–4185. doi: 10.1073/pnas.0307067101. [DOI] [PMC free article] [PubMed] [Google Scholar]