SUMMARY
Control of translation is a fundamental source of regulation in gene expression. The induction of protein synthesis by brain-derived neurotrophic factor (BDNF) critically contributes to enduring modifications of synaptic function, but how BDNF selectively affects only a minority of expressed mRNAs is poorly understood. We report that BDNF rapidly elevates Dicer, increasing mature miRNA levels and inducing RNA processing bodies in neurons. BDNF also rapidly induces Lin28, causing selective loss of Lin28-regulated miRNAs and a corresponding upregulation in translation of their target mRNAs. Binding sites for Lin28-regulated miRNAs are necessary and sufficient to confer BDNF responsiveness to a transcript. Lin28 deficiency, or expression of a Lin28-resistant Let-7 precursor miRNA, inhibits BDNF translation specificity and BDNF-dependent dendrite arborization. Our data establish that specificity in BDNF-regulated translation depends upon a two-part posttranscriptional control of miRNA biogenesis that generally enhances mRNA repression in association with GW182 while selectively derepressing and increasing translation of specific mRNAs.
INTRODUCTION
The control of gene expression at the level of translation is vital to neuronal function and synaptic plasticity. Dysregulated translation has been linked to cognitive disorders, including Fragile X syndrome, autism, and Parkinson’s disease. The regulation of translation plays a key role in the neuronal response to multiple stimuli, including synaptic activity (Huber et al., 2000; Raab-Graham et al., 2006; Wang et al., 2009), depolarization (Schratt et al., 2004), retinoic acid (Aoto et al., 2008), and neurotrophins (Aakalu et al., 2001; Jaworski et al., 2005; Schratt et al., 2004). Though most of these stimuli enhance total cellular protein synthesis, their responses demonstrate marked transcript specificity. This has been best defined for the brain-derived neurotrophic factor (BDNF), which is broadly expressed in the mammalian brain, and plays pivotal roles in neuronal survival, tructure, and synapse function. The effects of BDNF on protein synthesis, though physiologically important, are quite selective with an estimated 4% or less of expressed mRNAs undergoing enhanced translation (Schratt et al., 2004; Yin et al., 2002) despite a general enhancement of cap-dependent initiation and elongation by BDNF (Takei et al., 2009). Mechanisms conferring specificity to posttranscriptional control of gene expression are incompletely defined. mRNA regulatory elements and binding proteins provide significant examples of control for specific transcripts, but explanations for concerted changes in groups of mRNAs are largely lacking. Although subcellular restriction of stimulus-dependent signals in neurons likely imparts some transcript selectivity, target specificity remains inadequately explained as hundreds of mRNAs populate discrete cellular compartments such as dendrites.
We suspected that global regulatory mechanisms for mRNA translation, storage, or degradation might be enlisted to impart specificity to BDNF control of protein synthesis. RNA processing bodies (P bodies or GW bodies) are RNA granules that depend upon RNA for their formation (Teixeira et al., 2005), and harbor translationally repressed mRNAs that may be degraded or stored and released for subsequent translation (Brengues et al., 2005). In this work, we demonstrate that BDNF induces the rapid appearance of P bodies in neurons and determine that the function of miRNA biogenesis pathways is required for BDNF-mediated regulation of translation as well as the induction of P bodies. Remarkably, BDNF induces widespread changes in miRNA biosynthesis through enhancement of the general miRNA processing enzyme, Dicer, and elevation of levels of Lin28a, a protein that prevents the processing of a subset of miRNAs. The combined action of BDNF on Dicer and Lin28a mediates target specificity of BDNF-induced translation by dictating the profile of neuronal miRNAs that target mRNAs for translational repression.
RESULTS
BDNF Increases Neuronal P Body Number
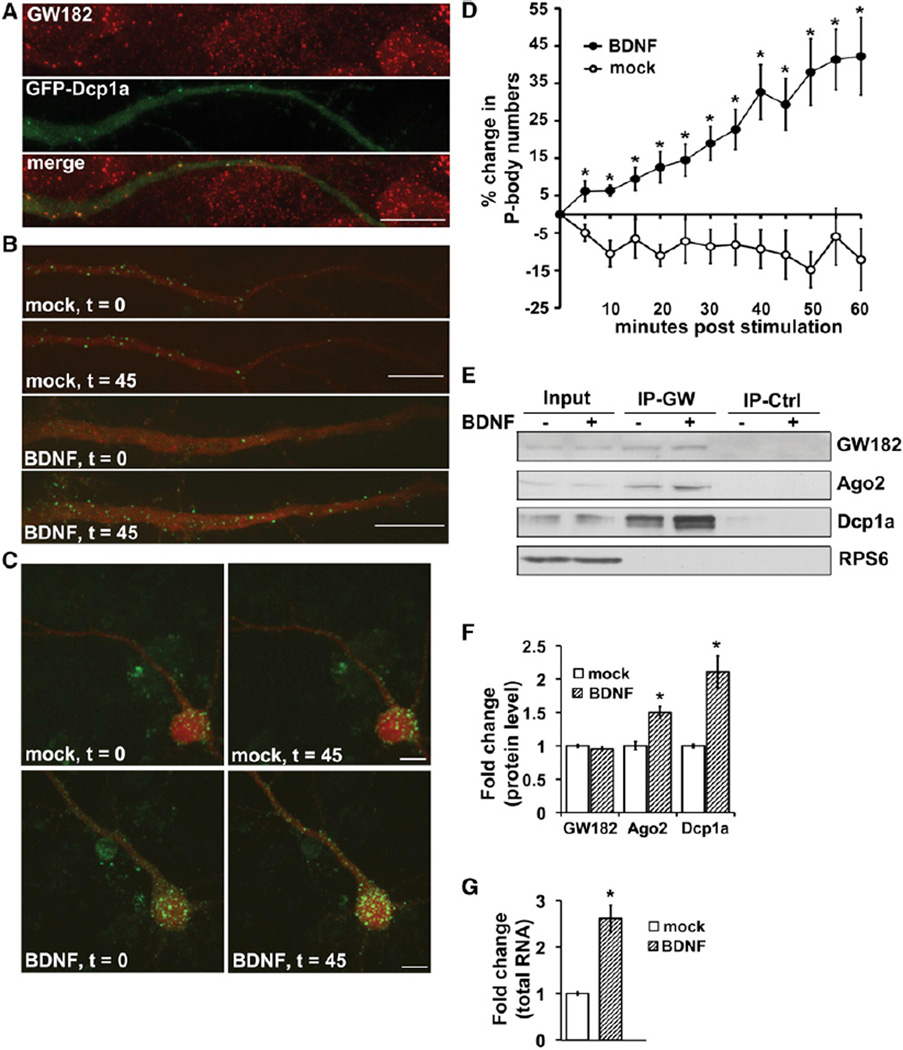
To investigate whether changes in RNA processing might be induced by BDNF, we first used live cell imaging to examine BDNF effects on neuronal P body abundance as a readout of potential broad effects on RNA regulatory mechanisms. P bodies were monitored by expression of GFP-tagged Dcp1a (GFP-Dcp1a), a decapping enzyme and specific P body marker (Anderson and Kedersha, 2006) that colocalized with endogenous Dcp1a (Figure S1A available online) and other P body components, including the RNA-binding protein GW182 (neuronal dendrites, Figures 1A and S1A–S1F). BDNF-stimulated hippocampal pyramidal neurons responded with a rapid and robust increase in the number of both dendritic and somatic P bodies, compared to mock-stimulated neurons, as assessed by live imaging of GFP-Dcp1a (Figures 1B–1D; Movies S1–S3) or endogenous staining (Figure S1G). Neurons were preincubated and imaged in the presence of the transcription inhibitor, Actinomycin-D, indicating that the rapid increase in P bodies can be mediated posttranscriptionally. BDNF induces P body complex formation rather than synthesis of components because protein levels of endogenous Dcp1a or GW182, or GFP-Dcp1a were not altered by BDNF (Figure S1H), and BDNF enhanced the total colocalization of two tagged P body components, Dcp1a and Pat1b, without altering their expression (Figures S1F and S1I). Immunoprecipitation of GW182 demonstrated that BDNF increased the association of P body components Argonaute 2 (Ago2) and Dcp1a with GW182 (Figures 1E and 1F) and, as anticipated because P bodies require RNA for formation, BDNF induced a more than 2-fold increase in the total coimmunoprecipitated RNA (Figure 1G). Exclusion of ribosomal protein S6 (RPS6) was used to corroborate immunopurification purity (Figure 1E). Collectively, these data show that the formation of P bodies, containing nontranslating RNA targeted for repression or degradation, is increased by BDNF, a stimulus known to enhance the activity of general translation factors and total cellular translation.
Figure 1. BDNF Increases P Body Formation in Soma and Dendrites of Hippocampal Neurons.
(A) Endogenous GW182 (top, red) colocalizes with GFP-Dcp1a (middle, green) in neuronal dendrites; overlay (bottom).
(B) P body formation in dendrites of hippocampal pyramidal neurons following mock (top) or BDNF stimulation (bottom, 100 ng/ml).
(C) P body formation in cell somas following mock (top) or BDNF stimulation (bottom). t = min poststimulation.
(D) Quantification and time course of percent change in GFP-Dcp1a P body numbers in neuronal dendrites following mock (open circles) or BDNF stimulation (closed circles) in the presence of Actinomycin D (0.5 µg/ml) to isolate changes due to translation.
(E) Lysates from mock (−) or BDNF (+, 1 hr) stimulated neuronal cultures immunoprecipitated (IP) with GW182 antiserum (IP-GW) or isotype-control serum (IP-Ctrl). Input is 20% of IP’d protein.
(F) Densitometric quantification from nine independent experiments, as in (E); mock condition (open bars) set as 1.0.
(G) Total RNA, measured by A260, recovered by GW182 IP from equal lysate inputs; mock (open bar) set as 1.0. BDNF increased GW182-associated RNA 2.62 ± 0.29-fold.
All error bars represent SEM. *p < 0.05 by unpaired Student’s t test. Scale bars, 10 µm. See also Figure S1 and Movies S1–S3.
Loss of GW182 Prevents BDNF Regulation of Target Protein Synthesis
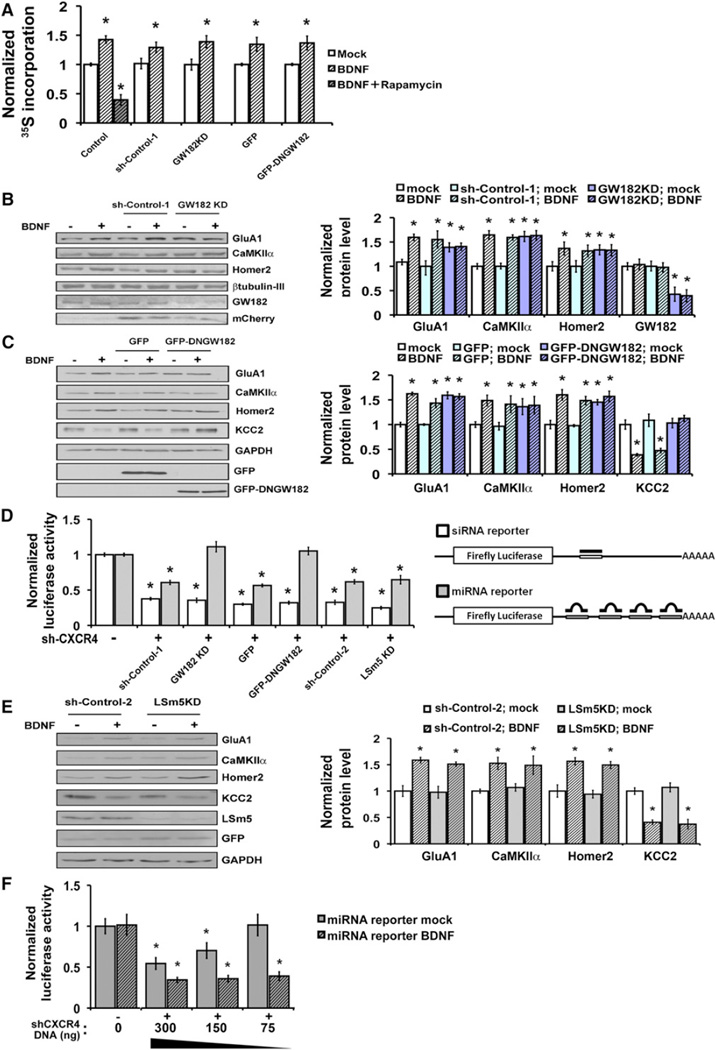
Despite modestly enhancing total cellular translation, BDNF is a highly selective modulator of protein synthesis that increases or decreases the levels of only specific target proteins. To examine whether RNA processing or repression might factor in this target specificity, we tested the effect of loss of GW182 function by either knockdown of GW182 or by expression of a GFP-tagged dominant negative GW182 (GFP-DNGW182) (Jakymiw et al., 2005). Both manipulations resulted in the loss of visible P bodies (Figures S2A–S2C), as previously reported. Loss of GW182 function did not alter the modest enhancement of total translation mediated by BDNF (Figure 2A), and also did not interfere with BDNF-regulation of another pathway, CREB-dependent transcription (Figure S2D).
Figure 2. miRNA-Mediated Repression Is Enhanced by BDNF and Associated with BDNF Target Specificity.
(A) Loss of GW182 function by shRNA targeting GW182 (GW182KD) or GFP-DNGW182 expression does not alter BDNF-enhancement of aggregate protein synthesis relative to control (uninfected) cells, or cells expressing scrambled GW182 shRNA or GFP alone. Total protein synthesis was monitored under mock (open bars) or BDNF (hatched bars, 100 ng/ml, 2 hr) stimulated conditions, and plotted relative to the control mock condition set as 1.0 (first open bar). A translation inhibitor (rapamycin, 20 µg/ml) demonstrates that observed changes are due to translation.
(B) (Left) Immunoblotting for BDNF target proteins in neurons either uninfected or infected with lentivirus expressing GW182 shRNA (GW182KD) or a mismatched control shRNA (sh-Control-1). mCherry is coexpressed from the virus. (Right) Protein levels, normalized to β-tubulin, of representative BDNF-upregulated targets under mock (open bars) or BDNF (hatched bars, 100 ng/ml, 2 hr) stimulation in the presence or absence of GW182KD (control mock, white bars, set as 1.0); n = 6 independent experiments.
(C) (Left) Immunoblotting for BDNF target proteins in neurons either uninfected or infected with lentivirus expressing GFP-DNGW182 or GFP. (Right) Protein levels, normalized to β-tubulin, of representative BDNF up- or downregulated targets under mock (open bars) and BDNF (hatched bars) stimulated conditions in cells expressing GFP-DNGW182, GFP, or control uninfected cells (control mock, white bars, set as 1.0); n = 6 independent experiments.
(D) miRNA function is inhibited by GW182KD and GFP-DNGW182, but not by knockdown of LSm5 (LSm5KD). (Left) Luciferase activities of siRNA- or miRNA-reporter constructs in cells expressing reporter alone (−sh-CXCR4), or coexpressing reporter and CXCR4 shRNA (+sh-CXCR4), with or without GW182KD, GFP-DNGW182, or LSm5KD. Normalized luciferase values are shown relative to levels without sh-CXCR4 (set as 1.0). (Right) Diagram of reporter constructs.
(E) LSm5 knockdown did not alter protein synthesis of representative BDNF targets. (Left) Immunoblotting for BDNF targets in neurons expressing control shRNA (sh-Control-2) or shRNA against LSm5 (LSm5KD) following mock (−) or BDNF (+) stimulation (100 ng/ml, 2 hr). (Right) Densitometric quantification of three independent immunoblots, normalized to GAPDH and plotted relative to mock-stimulated controls (sh-Control-2 -mock).
(F) BDNF enhances repression of a miRNA-reporter by a small RNA hairpin (sh-CXCR4). Normalized luciferase values are shown for mock (open bars) or BDNF-stimulated (hatched bars) neurons coexpressing the miRNA reporter and either sh-Control-2 or a dose titration of sh-CXCR4. Low-dose sh-CXCR4 repressed the miRNA-reporter in BDNF-stimulated, but not mock-stimulated, conditions.
All experiments done in the presence of Actinomycin-D. Error bars represent SEM. *p < 0.05 in comparison to reporter alone condition (− sh-CXCR4, D and F) or mock (open bars) by unpaired Student’s t test. See also Figures S2, S3, and S4.
In contrast, GW182 knockdown or GFP-DNGW182 expression both strikingly eliminated the mRNA target specificity of BDNF-regulated protein synthesis. The AMPA glutamate receptor subunit GluA1, calcium calmodulin-dependent protein kinase II (CaMKIIα), and Homer2 normally undergo enhanced protein synthesis in response to BDNF (Narisawa-Saito et al., 1999; Schratt et al., 2004), whereas synthesis of the potassium-chloride cotransporter, KCC2, is decreased by BDNF (Rivera et al., 2002). GW182 knockdown (Figure 2B) or GFP-DNGW182 expression (Figure 2C) in hippocampal neurons elevated the basal levels of proteins normally upregulated by BDNF (GluA1, CaMKIIα, and Homer2) and prevented their further induction by BDNF. In contrast, the basal protein level of BDNF-downregulated target (KCC2) was unchanged by loss of GW182 function, but KCC2 protein level was no longer reduced by BDNF. β-tubulin III was unchanged by BDNF and used for normalization. These experiments were performed in the presence of Actinomycin-D; similar effects were seen without Actinomycin-D (Figure S3). Effective GW182 knockdown was achieved using lentiviral transduction and verified by immunoblotting for GW182 (serum 18033, M. Fritzler, Figure 2B, and Abcam Figure S2B).
Quantitative real-time PCR (qRT-PCR) showed that mRNA levels of BDNF-upregulated targets were unchanged by loss of GW182 function (Figure S2E), suggesting that the observed changes in basal protein levels could result from altered target translation. As previously reported, BDNF stimulation reduced mRNA levels of the gene (Slc12a5) encoding the downregulated target, KCC2, in control neurons (Rivera et al., 2002). In neurons deficient in GW182, however, BDNF no longer significantly reduced the level of mRNA for KCC2 (Figure S2E). These results indicated that GW182 function is required for both baseline translational repression of BDNF-upregulated targets and for BDNF-induced mRNA degradation of a downregulated target. The composite effects implied a role for GW182 in the process that allows BDNF to differentially regulate specific mRNA targets.
The Role of miRNA-Mediated Repression in BDNF Regulation of Target Protein Synthesis
Several RNA processing events are associated with P body formation, including multiple RNA decay mechanisms, mRNA suppression by RNA binding proteins, and RNA-induced silencing complex (RISC)-mediated repressive functions. Previous reports demonstrated that P body disruption through targeting of discrete P body protein components, such as GW182, can differentially block distinct RNA processing events (Liu et al., 2005a). To test whether functions associated with GW182 in particular were required for translational specificity of BDNF, we compared the effects of loss of GW182 with loss of another P body component, LSm5. We focused initially on assessing RISC-mediated functions, in contrast to decay pathways, because transcript levels of BDNF-upregulated targets appeared unchanged by loss of GW182 (Figure S2E). Function of miRNA and siRNA pathways were tested by a reporter assay consisting of coexpression of a hairpin precursor shRNA (shCXCR4) and a luciferase reporter containing 3′ untranslated region (UTR) binding sites with either perfect (siRNA reporter) or mismatched (miRNA reporter) complementarity for the CXCR4 shRNA (Doench et al., 2003; Wang et al., 2006). Expression of either reporter without the shRNA exhibited full luciferase activity (Figure 2D); a control reporter lacking CXCR4 binding sites was unaffected by shCXCR4 coexpression (Figure S4).
P body disruption by loss of GW182 function produced a preferential miRNA pathway deficit, as shown by failure of coexpressed shRNA to repress the miRNA reporter, with no effect on siRNA-dependent inhibition (Figure 2D). Loss of GW182 has been previously reported to impair miRNA-mediated translational repression (Jakymiw et al., 2005; Liu et al., 2005a). In contrast, P body disruption by LSm5 knockdown (Figures S2F and S2G) did not significantly alter reporter suppression through siRNA or miRNA pathways in comparison to controls (shRNA-1, shRNA-2, or GFP; Figure 2D), indicating that these pathways remain intact.
The finding that loss of GW182, but not LSm5, disrupted miRNA-mediated repression, presented the opportunity to probe the importance of miRNA function in determining the specificity of BDNF-regulated translation. In contrast to the loss of specificity in BDNF-regulated protein synthesis produced by GW182 deficiency (Figures 2B and 2C), loss of LSm5 did not alter translation specificity (Figure 2E) even though LSM5 knockdown also disrupted P bodies (Figures S2F and S2G). LSm5 knockdown, like loss of GW182, also did not affect BDNF-enhancement of total cellular translation (Figure S2H). Comparing the effects of loss of GW182 or LSm5 function suggested the involvement of miRNA-mediated functions in conferring target specificity to BDNF-regulated protein synthesis.
Rapid Enhancement of Mature miRNA Biogenesis by BDNF
To further investigate the role of miRNA in the specificity of BDNF-regulated translation, we asked whether BDNF itself might affect the miRNA pathway. Intriguingly, BDNF stimulation of cells coexpressing the shCXCR4 and miRNA reporter greatly enhanced miRNA-mediated suppression. Titration of shCXCR4 in this assay revealed that a low dose that did not suppress the reporter in the absence of BDNF generated maximally effective suppression after cellular stimulation with BDNF (Figure 2F). This effect was independent of new transcription as stimulation was carried out in the presence of Actinomycin-D. CXCR4 shRNA resembles endogenous pre-miRNA and requires Dicer cleavage to generate mature duplex RNA. We reasoned that BDNF could enhance miRNA-mediated repression in the reporter assay by two potential general mechanisms: first, BDNF might increase RISC efficacy or, second, BDNF might increase the generation of functional mature duplex miRNA from transfected CXCR4 shRNA. A mechanism invoking BDNF-enhanced mature miRNA biogenesis was congruent with our earlier findings because elevated levels of miRNA can deliver additional mRNAs targeted for repression to P bodies and increase P body number (Liu et al., 2005b).
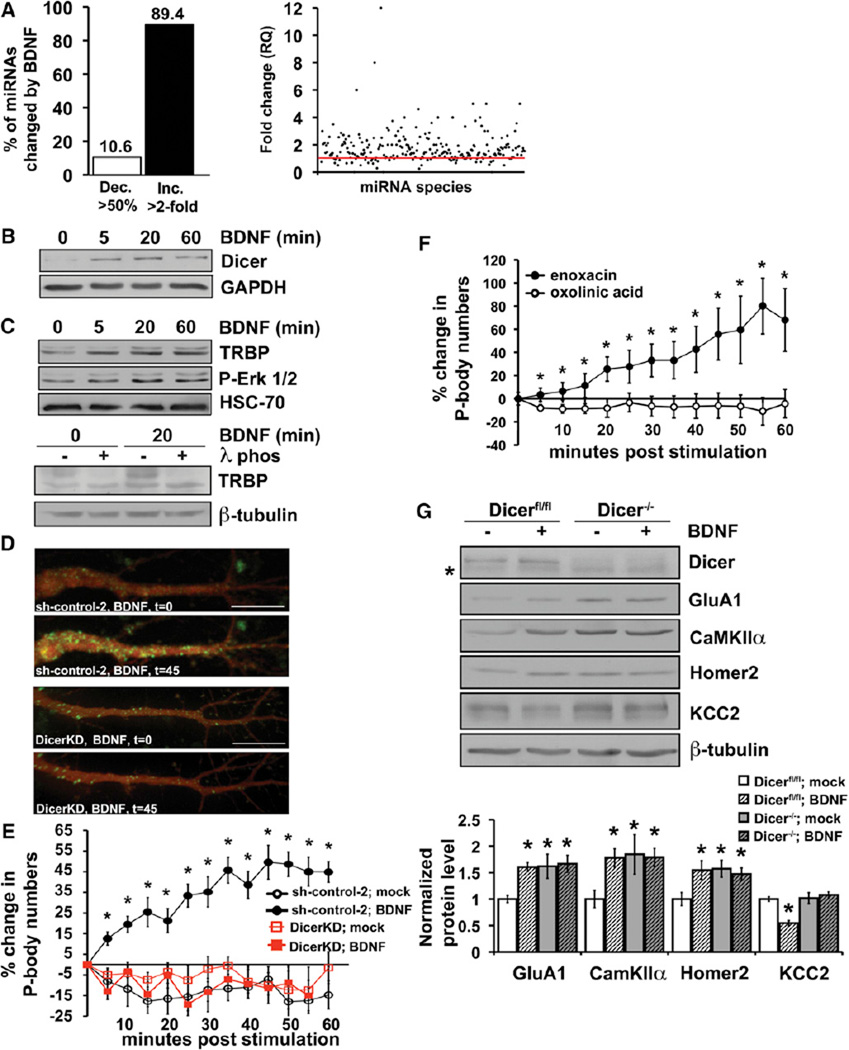
We addressed the potential for global regulation of miRNA biogenesis by BDNF using miRNA arrays that selectively measure mature miRNA, as opposed to pre-miRNA. Hippocampal neurons were treated with BDNF for 30 min in the presence of Actinomycin-D to assess changes due to processing of existing pre-miRNAs rather than new pre-miRNA production. Each array (TaqMan) contained 375 rodent miRNA targets of which 195 were detectable in hippocampus in three independent paired experiments. Remarkably, of detectable endogenous miRNAs with levels significantly altered by BDNF, 89.4% were increased more than 2-fold by BDNF, whereas only 10.6% were decreased to <50% (Figure 3A, left). Many more miRNA species showed smaller, less than 2-fold, posttranscriptional changes in abundance in response to BDNF. Though absolute changes in individual miRNAs were not reproducible between paired array experiments, the qualitative effect of a predominantly increased abundance of many miRNA species in response to BDNF was reproducible on this platform as well as an initial analysis using Geniom miRNA biochips (Febit Inc., data not included). An expression analysis of fold change for each detectable miRNA species from the arrays illustrates an overall trend toward higher miRNA quantities in BDNF compared to mock-treated primary neurons (Figure 3A, right).
Figure 3. BDNF Increases Dicer Levels and the Biogenesis of Mature miRNAs.
(A) (Left) Percentage of miRNAs from TaqMan miRNA array with levels decreased over 50% (open bar) or increased over 2-fold (black bar) by BDNF (30 min, plus Actinomycin-D). (Right) Scatter plot of relative quantities (RQ) of individual miRNA species (solid circles) following BDNF relative to mock-stimulation. Red line = 1.0 or no change; each dot above the line represents a miRNA species increased by BDNF, each dot below the line represents a miRNA species decreased by BDNF. Normalization is to averaged reference RNAs U6snRNA, and snoRNA202, which are unchanged by BDNF; n = 3 separate miRNA array pairs for mock and BDNF conditions.
(B) Immunoblot of cultured hippocampal neurons stimulated with BDNF for indicated min in the presence of Actinomycin-D. Dicer peaks near 20 and declines by 60 min.
(C) (Top) BDNF enhances TRBP and ERK phosphorylation as shown by immunoblot for TRBP and phospho-Erk. Cultured hippocampal neurons were stimulated with BDNF for indicated min in the presence of Actinomycin-D. (Bottom) Lysates incubated with λ-phosphatase (λ-phos) as indicated demonstrate loss of phosphorylated TRBP (upper band).
(D) P body appearance in dendrites of hippocampal pyramidal neurons expressing control (sh-control-2, top) or Dicer-targeting shRNA (DicerKD, bottom) following BDNF, t = min poststimulation.
(E) Quantification and time course of P body numbers in Dicer-deficient (DicerKD, boxes) or control (sh-control-2, circles) expressing hippocampal neurons following mock (open shapes) or BDNF stimulation (closed shapes).
(F) Quantification and time course of P body numbers in hippocampal pyramidal neurons treated with enoxacin (15 µM, closed circles) or oxolinic acid (15 µM, open circles).
(G) The effect of Dicer loss on BDNF-regulated protein synthesis. (Top) Immunoblotting for BDNF target proteins in Dicer-wild-type (Dicerflox/flox) or Dicer-deficient (Dicer−/−) neurons. CreERT2-expressing cells were treated with 4-hydroxy tamoxifen (800 nM) to induce recombination for 2.5 days before BDNF stimulation.
Asterisk indicates nonspecific band. (Bottom) Densitometric quantification of immunoblots.
All error bars represent SEM. *p < 0.05 by unpaired Student’s t test. See also Figure S4.
Widespread posttranscriptional upregulation of mature miRNA production suggested that BDNF might regulate an essential component of miRNA biogenesis, such as the Dicer processing complex. To assess this, we examined Dicer protein levels in BDNF-stimulated neurons. BDNF elicited a marked transcription-independent increase in Dicer levels that peaked between 5 and 20 min after stimulation (Figure 3B). The binding of BDNF to TrkB receptors triggers signaling pathways promoting growth and survival, including activation of PI3K/AKT and MAPK/ERK pathways. Previous work in tumor cell lines revealed that a component of the Dicer complex, HIV-1 TAR RNA-binding protein (TRBP), could undergo Erk-dependent phosphorylation and that phosphomimetic TRBP stabilized and enhanced Dicer levels (Paroo et al., 2009); mutations resulting in decreased TRBP protein also destabilize Dicer (Melo et al., 2009). Accordingly, we evaluated the effect of BDNF on TRBP levels and phosphorylation status in neurons. BDNF rapidly induced phospho-ERK and a multiple-banding pattern of TRBP (Figure 3C, top) that was collapsed by phosphatase treatment (Figure 3C, bottom). Total TRBP protein levels were also rapidly elevated and reached significance by 5 min after BDNF (2.61-fold increase ± 0.86).
To evaluate a requirement for Dicer activity and miRNA biogenesis in BDNF-induced recruitment of nontranslating mRNA to P bodies, we depleted Dicer by RNAi. Dicer knockdown completely prevented a BDNF-induced increase in P body number (Figures 3D and 3E) in hippocampal pyramidal neurons. Neurons expressing control nontarget shRNA responded to BDNF similarly to wild-type neurons (Figures 1D and 3E). The requirement for Dicer in P body induction by BDNF indicated a role for Dicer in targeting some mRNAs to the nontranslating pool in response to BDNF.
We next asked whether Dicer activation, and by implication an increase in mature miRNAs, was sufficient to generate P bodies in neurons. The fluoroquinolone, enoxacin, was previously shown to promote pre-miRNA processing by the Dicer/TRBP complex, whereas a structurally similar derivative, oxolinic acid, did not significantly increase miRNA biogenesis (Shan et al., 2008). Enoxacin, but not equimolar oxolinic acid, rapidly and robustly increased P body numbers in neuronal soma and dendrites (Figure 3F). In comparison to BDNF, the time course of P body induction by enoxacin was slightly more rapid, consistent with a more direct signaling mechanism. In accordance with a role for miRNA in regulating mRNA target selection, but not bulk translation, enoxacin did not alter basal or BDNF-induced total protein synthesis (Figure S4B). These results defined a potential mechanism for regulation of miRNA biogenesis by BDNF and linked BDNF upregulation of Dicer activity to rapid changes in mRNA repression.
To test whether Dicer is also required for the regulation of BDNF target genes, we examined the response of representative up- and downregulated BDNF targets in the presence or absence of Dicer. Hippocampal pyramidal neurons from mice with a conditional Dicer1 allele (Dicerflox/flox, 3A8 line) (Andl et al., 2006) were infected with lentivirus expressing 4-hydroxy tamoxifen (OHT)-inducible Cre recombinase and subsequently mock or BDNF stimulated with or without OHT. Targets that are normally low at baseline and upregulated by BDNF, including GluA1, CaMKIIα, and Homer2, were each elevated at baseline in Dicer-deficient neurons, consistent with basal derepression in the absence of Dicer, and failed to be further upregulated by BDNF. A representative target normally downregulated by BDNF, KCC2, was nonresponsive to BDNF in Dicer-deficient neurons (Figure 3G). Collectively, these results demonstrate that BDNF lacks specificity for up- or downregulated targets in the absence of Dicer, consistent with a critical role for Dicer in BDNF-induced sequestration of mRNAs in P bodies and in the mechanism determining the selective regulation of target mRNAs by BDNF.
BDNF Confers Selectivity to miRNA Biogenesis through Lin28a
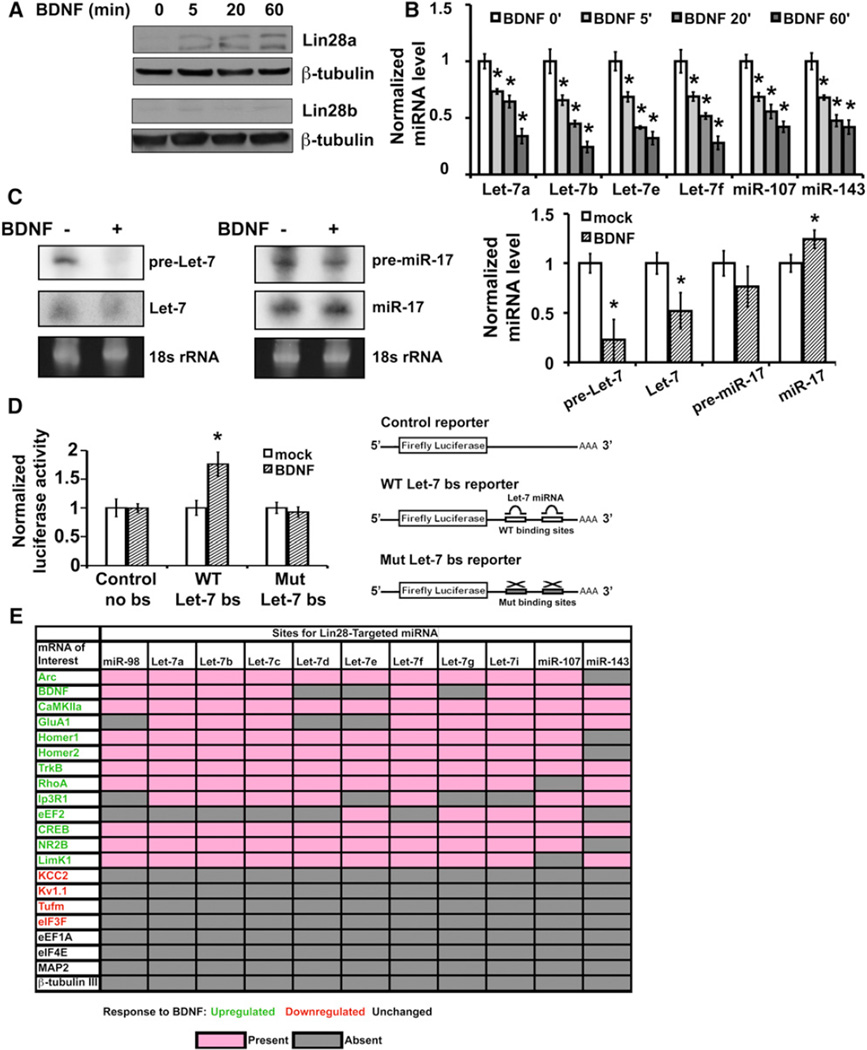
Widespread upregulation of miRNA production and consequent removal of mRNAs from the translating pool by targeted repression provides a viable negative selection mechanism to account for the low proportion of mRNAs reported to undergo BDNF-enhanced translation (Schratt et al., 2004). We next asked whether the miRNA biogenesis pathway might also be regulated to generate positive selection of BDNF-upregulated targets in protein synthesis. By miRNA array analysis, a small number of miRNAs were observed to decrease in response to BDNF; the decreases were more apparent in some individual experiments than in the collective averaged array data. Among these decreased miRNAs were several members of the Let-7 family. miRNA biogenesis can be regulated at multiple steps by trans-acting factors, including the Lin28 RNA-binding proteins (Heo et al., 2009; Newman et al., 2008; Viswanathan et al., 2008), which target Let-7 family members. Lin28 binding and subsequent pre-miRNA uridylation suppresses processing of targeted pre-miRNA to mature miRNA (Hagan et al., 2009; Heo et al., 2009), and could provide a mechanism for decreasing select mature miRNAs even in the context of Dicer elevation. Consistent with this possibility, we found a robust and rapid transcription-independent increase in Lin28a, but not Lin28b, protein in mature neurons by 5 min following BDNF exposure (Figure 4A).
Figure 4. BDNF Induces Lin28, Selectively Diminishes Lin28-Regulated miRNAs, and Specifically Upregulates a Heterologous Reporter Containing Let-7-Binding Sites.
(A) Lin28a (top) and Lin28b (bottom) immunoblots of lysates from cultured hippocampal neurons stimulated with BDNF for indicated min.
(B) Time course of BDNF-induced reductions in Lin28-regulated miRNA levels by individual TaqMan qRT-PCR reactions in mock- (BDNF 0′) or BDNF-stimulated neurons. miRNA levels were normalized to 18 s rRNA and plotted relative to each mock-stimulated condition (set as 1.0). All samples underwent equal duration Actinomycin-D incubation prior to harvest.
(C) Northern blot (left) and quantitation (right) of pre- and mature miRNA levels of a Lin28-target (Let-7a) or control miRNA (miR-17) in mock or BDNF-treated (30 min) neurons.
(D) A binding site for Let-7 miRNAs in the 3′UTR of an mRNA confers upregulation of protein synthesis in response to BDNF. Neurons expressing Let-7 reporters containing two functional (Let-7 WT) or mutated (Let-7 Mut) Let-7 miRNA binding sites in the 3′UTR of firefly luciferase, or a reporter lacking miRNA binding sites were mock or BDNF stimulated (4 hr). Luciferase activities are normalized to coexpressed constitutive β-galactosidase activity and plotted relative to mock-stimulation for each reporter. All error bars represent SEM. *p < 0.05 by unpaired Student’s t test.
(E) Predicted binding sites for Lin28-targeted miRNA. The presence of a Lin28-targeted miRNA binding site in the 3′UTR of transcripts for which translation is BDNF-upregulated (green), BDNF-downregulated (red), and BDNF-nonregulated (black) as predicted by TargetScan, PITA, Pictar, MiRanda, and miRwalk. Pink boxes denote the presence of a miRNA binding site in which the miRNA seed sequence (nucleotides 2–7) paired as a perfect or G-U wobble-containing match.
Gray boxes denote the absence of a miRNA binding site. See also Figure S5.
Analysis of the terminal loop region of the Let-7, miR-107, and miR-143 pre-miRNAs showed that each has a putative or previously functionally confirmed “GGAG” sequence motif that can permit recognition by Lin28 (Hagan et al., 2009; Heo et al., 2009). Individual qRT-PCR assays showed significant and reproducible BDNF-induced decreases in abundance of all tested members of the Let-7 family, as well as miR-107, and miR-143 (Figure 4B), even though not all were reproducibly detected through the less sensitive miRNA arrays. Significant decreases in each of these miRNAs were apparent by 5 min post-BDNF stimulation (Figure 4B). In accordance with the expected effects of Lin28, northern blotting for a member of the Let-7 family (Let-7a) showed significant decreases in both pre-Let-7 and mature Let-7 miRNA levels in BDNF-treated neurons (Figure 4C). A control mature miRNA, miR-17, is modestly increased by BDNF, consistent with enhanced Dicer processing (Figure 4C).
If Lin28 positively selects BDNF-upregulated targets by decreasing specific miRNAs, an mRNA containing functional binding sites for a Lin28-downregulated miRNA would be predicted to undergo BDNF-enhanced translation. To test this prediction, we compared the response to BDNF of luciferase reporters whose 3′UTR contained either wild-type or mutated Let-7 miRNA binding sites, or no miRNA binding sites, under conditions of transcription blockade. As predicted, the reporter containing Let-7 binding sites was significantly induced by BDNF, whereas levels of the control reporters were unchanged (Figure 4D). This result indicates that downregulation of Let-7 family members by BDNF is sufficient to relieve repression and mediate positive target selection for BDNF-enhanced translation.
To examine the extent to which this mechanism could generalize to known BDNF targets, we evaluated the presence of binding sites for Lin28-regulated miRNAs in the 3′UTRs of mRNAs known to undergo upregulated, downregulated, or unchanged translation in response to BDNF. Positive scores (pink boxes, Figure 4E) were restricted to sites in which the miRNA seed sequence (nucleotides 2–7) paired as a perfect or G-U wobble-containing match; similar miRNA seed sequence pairing was previously found important for target recognition (Guo et al., 2010). Thirteen representative BDNF-upregulated targets were all found to contain two or more sites for a Lin28-regulated miRNA (example sites in Figure S5), whereas targets known to be downregulated by BDNF (KCC2, Kv1.1; Raab-Graham et al., 2006; Rivera et al., 2002) or unregulated by BDNF (eEF1A, eIF4E, MAP2, β-tubulin III; Schratt et al., 2004; Wang et al., 2009) did not contain such sites (Figure 4E).
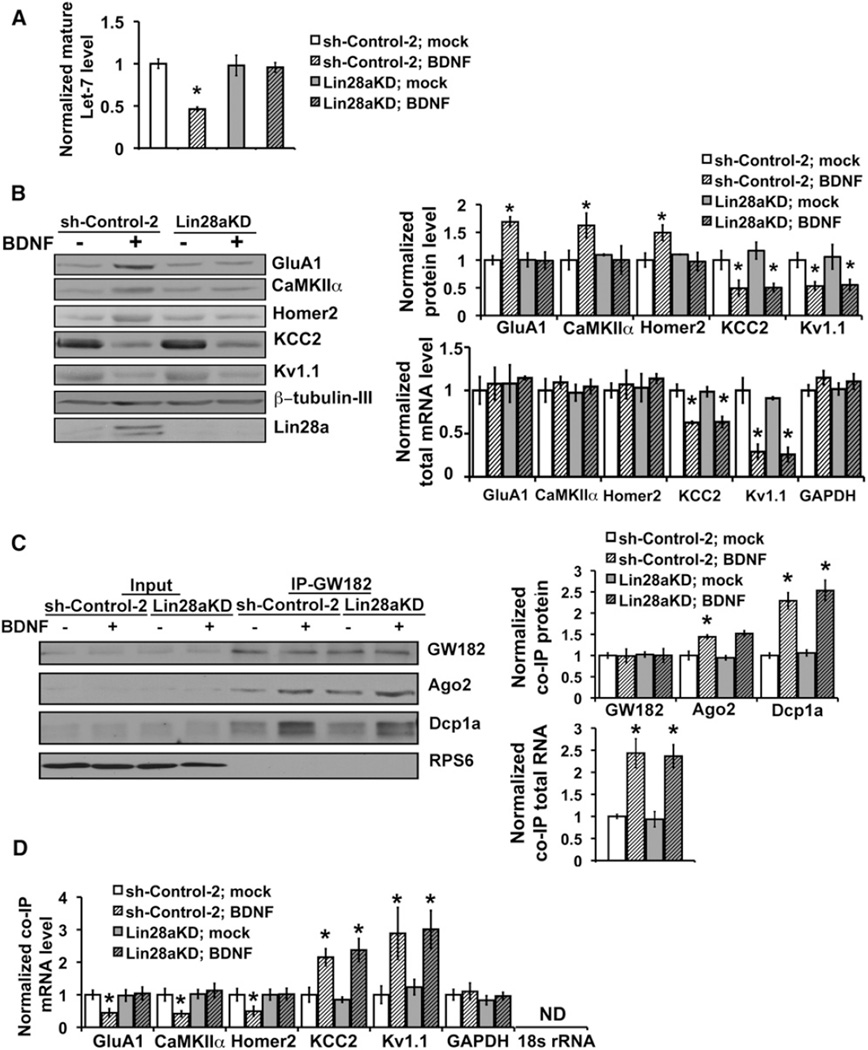
We next directly tested the role of Lin28a in BDNF target mRNA selection. Depletion of Lin28a through RNAi, but not expression of a control hairpin, prevented the decline in mature Let-7 miRNAs in hippocampal neurons responding to BDNF (Figure 5A). Lin28a knockdown also prevented the increased translation of representative mRNA targets normally upregulated by BDNF (Figure 5B). In accordance with basal mRNA repression by miRNAs that are diminished through Lin28, protein levels of these normally upregulated targets (CaMKIIα, GluA1, and Homer2) remained at low basal levels even in the presence of BDNF under Lin28a knockdown conditions (Figure 5B, left and upper-right). Lin28a knockdown did not prevent the increased association of P body protein and RNA components in response to BDNF (Figure 5C), which instead requires Dicer (Figure 3).
Figure 5. Lin28 Is Required for Relief of miRNA-Mediated Repression and Selective Induction of BDNF-Upregulated mRNA Targets.
(A) Loss of Lin28 prevents BDNF-induced decreases in mature Let-7a levels. Mature Let-7a levels were assessed by qRT-PCR from neurons infected with lentivirus expressing either control shRNA (sh-control-2) or shRNA targeting Lin28a (Lin28aKD) and mock or BDNF stimulated for 20 min (no Actinomycin-D); normalization was to control mock values (open bar, set as 1.0, n = 3).
(B) Effect of Lin28a loss on BDNF-regulated protein synthesis. Immunoblotting of BDNF targets in control or Lin28a-deficient cells, mock or BDNF stimulated (left). Densitometric quantification of protein levels (right, top, n = 6 each condition). Total mRNA levels for both BDNF-upregulated or downregulated targets (right, bottom).
(C) Effect of Lin28a KD on BDNF-induced association of protein and RNA P body components. Lysates were immunoprecipitated with anti-GW182 antibody in control (sh-Control-2) or Lin28a-deficient cells, mock or BDNF stimulated. Immunoblotting for co-IP’d Ago2 and Dcp1a (left) and densitometric quantification (right, top, n = 3). Total RNA from GW182 IP of equal lysate inputs from Lin28a knockdown (Lin28aKD) or control (sh-Control-2) neurons; mock (open bars) set as 1.0. BDNF-induced increase in GW182-associated total RNA remains intact after Lin28a knockdown (right, bottom).
(D) Abundance of BDNF mRNA targets associated with GW182 in control (sh-Control-2) or Lin28a-deficient cells. In Lin28a-deficient neurons, mRNAs for BDNF-upregulated targets remain associated with GW182 in the presence of BDNF, whereas the response of mRNAs for BDNF-downregulated targets is unchanged.
18 s rRNA is nondetectable, ND.
Error bars represent SEM. *p < 0.05 Student’s t test.
Targets normally derepressed and upregulated by BDNF remained repressed in Lin28a-deficient neurons, in contrast with the effects of loss of GW182 (Figures 2B and 2C) or Dicer (Figure 3G) which both resulted in derepression of BDNF-induced targets at baseline and occlusion of further upregulation by BDNF. Notably, targets normally downregulated by BDNF, represented by KCC2 and Kv1.1, remained responsive to BDNF in the presence of Lin28a knockdown. BDNF effects on target mRNA levels were unaffected by loss of Lin28a (Figure 5B, lower-right). These findings are consistent with translation specificity in response to BDNF being generated by a two-part regulation of the miRNA biogenesis pathway: (1) general upregulation of miRNA biogenesis that is required for repression of mRNAs whose protein products are decreased in response to BDNF, (2) downregulation of select miRNAs whose processing is blocked by BDNF-induced Lin28a, resulting in derepression and enhanced translation of mRNAs containing binding sites for Lin28-regulated miRNAs.
We further examined the role of Lin28a in BDNF-regulated translation by evaluating its effects on specific target mRNA repression in association with GW182. GW182-associated RNA was immunopurified, as in Figure 1E, under control shRNA and Lin28a knockdown conditions and the effects of BDNF on mRNA recruitment to GW182 were assessed by individual qRT-PCR assays. mRNAs undergoing regulated translation by BDNF were enriched in overall association with GW182 in comparison to a “housekeeping” Gapdh mRNA (Figure 5D). Under control shRNA conditions, BDNF reduced the GW182 association of representative mRNAs for targets whose translation is upregulated by BDNF (GluA1, CaMKIIα, and Homer2); in contrast, BDNF promoted the GW182 association of representative mRNAs for targets (KCC2 and Kv1.1) whose translation is downregulated by BDNF (Figure 5D). Translation of mRNA for β-tubulin III is unchanged by BDNF (Schratt et al., 2004) and was used for normalization; 18 s rRNA is absent from P bodies and served as a control for immunopurification purity (Figure 5D).
As expected if Lin28a regulates only selection of BDNF-upregulated targets, Lin28a knockdown altered only the GW182 enrichment profile of mRNAs for representative targets (CaMKIIα, GluA1, and Homer2) that undergo BDNF-enhanced translation (Figure 5D). mRNAs for BDNF-upregulated targets remained equivalently repressed and associated with GW182 in Lin28a knockdown neurons in the presence or absence of BDNF, and protein levels of these targets were no longer enhanced by BDNF. In contrast, mRNAs for representative BDNF-downregulated targets (KCC2 and Kv1.1) remained enriched in association with GW182 at baseline and their enrichment was equivalently increased by BDNF in both control and Lin28a knockdown neurons (Figure 5D). A target not regulated by BDNF (GAPDH) was not enriched in GW182 association at baseline, did not change in response to BDNF, and was also unaffected by Lin28a loss (Figure 5D). These findings indicate that Lin28a, induced by BDNF, is required to suppress the processing of specific pre-miRNAs and selectively decrease levels of these mature miRNAs, concomitant with a general BDNF-induced upregulation in the biogenesis of most miRNAs by enhanced Dicer levels. The negative regulation of miRNA biogenesis by Lin28a presents a mechanism for the selection of upregulated targets in BDNF-induced protein synthesis.
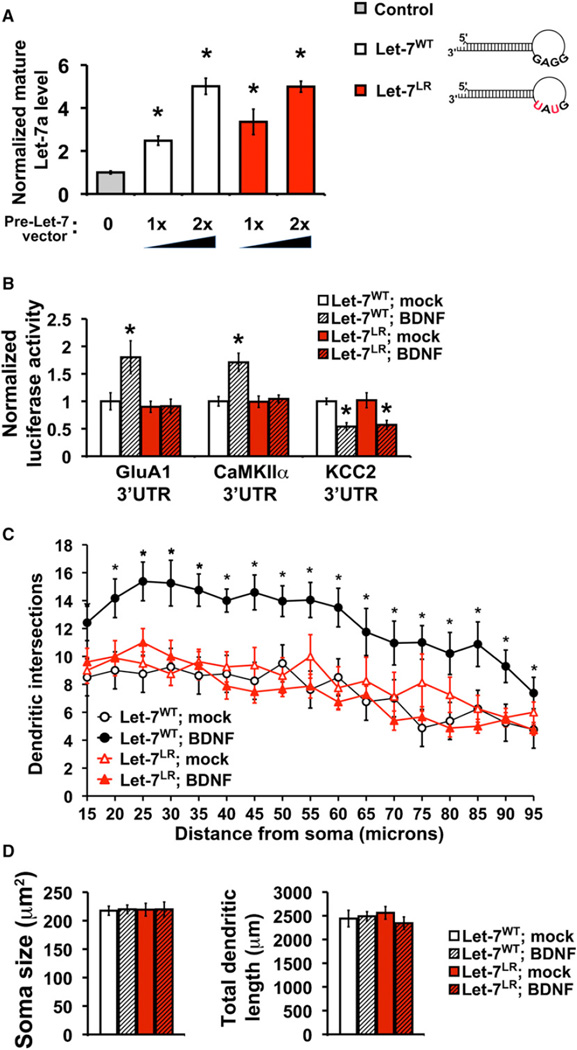
To further test the mechanism by which Lin28a mediates induced translation of BDNF-upregulated targets, we constructed a Let-7 pre-miRNA that would be resistant to Lin28-mediated degradation through mutation of the pre-miRNA terminal loop residues from GGAG to GUAU. This mutation prevents Lin28-induced uridinylation and degradation of Let-7 pre-miRNA (Heo et al., 2009), but does not alter the target specificity of the Let-7 miRNA. Lentiviral-mediated expression of either wild-type (Let-7WT) or Lin28-resistant Let-7 (Let-7LR) in hippocampal cultures enhanced mature Let-7 levels in a dose-dependent manner that could be titrated to achieve equivalent and low levels of exogenous Let-7 expression (Figure 6A). Coexpression of Let-7LR with reporters harboring the 3′UTR from either of two BDNF-upregulated targets (GluA1 or CaMKIIα) completely prevented their induction by BDNF. In contrast, coexpression of Let-7LR with a reporter harboring the 3′UTR from a BDNF-downregulated target (KCC2) had no effect on the BDNF-mediated depression of this reporter (Figure 6B). These results supported a selective role for Lin28a in mediating the specificity of BDNF for its upregulated targets.
Figure 6. Lin28-Mediated Degradation of Let-7 Precursors Is Required for Induction of BDNF-Upregulated Targets and Neuronal Outgrowth.
(A) Lentiviral-mediated expression of wild-type (Let-7WT) or Lin28-resistant (Let-7LR) Let-7 pre-miRNAs in neurons produced dose-dependent enhancement of mature Let-7a miRNA levels assessed by qRT-PCR and shown as fold change relative to infection with virus expressing GFP alone (gray bar, set as 1.0); 13 or 23 refers to viral dose.
(B) Expression of Let-7LR, but not Let-7WT, blocks specificity of BDNF for upregulated targets. Reporter assays in mock (open bars) or BDNF stimulated (hatched bars) neurons with luciferase constructs fused to the 3′UTR from BDNF-upregulated targets (GluA1 or CaMKIIα) or a downregulated target (KCC2).
(C) BDNF-induced dendrite outgrowth requires Lin28-mediated degradation of miRNA precursors. Dendrite complexity is quantitated for neurons expressing Let-7WT (black circles) or Let-7LR (red triangles) following mock (open shapes) or BDNF (25 ng/ml, closed shapes) treatment. *p < 0.05 by unpaired Student’s t test or unpaired one-way ANOVA between Let-7WT and Let-7LR in mock and BDNF conditions.
(D) Soma size (left) and total dendritic length (right) did not significantly differ between Let-7WT and Let-7LR in mock or BDNF treatment.
Error bars represent SEM. All experiments done in the presence of Actinomycin D. See also Figure S6.
Although alternative mechanisms for selectivity could coexist, our collective results strongly indicate that dual control by BDNF of the miRNA biogenesis pathway through Lin28a and Dicer critically contributes to determining both up- and downregulated target specificity in BDNF-mediated protein synthesis. These findings prompted us to investigate the effects of loss of Lin28a on a physiological response requiring BDNF-regulated protein synthesis.
Loss of miRNA-Mediated Regulation Prevents BDNF-Enhanced Dendrite Arborization
Induction of dendrite outgrowth in excitatory neurons both in culture and in vivo is a well-characterized BDNF function requiring the regulation of translation (Jaworski et al., 2005). Because inhibiting new translation blocks BDNF-induction of dendrite growth, we reasoned that BDNF-upregulated targets, selected by Lin28a, might be particularly important for this process. To test the physiological relevance of selective mRNA translation by relief of repression from Lin28-targeted miRNAs, we used low-dose BDNF to stimulate proximal dendrite growth in developing hippocampal pyramidal neurons expressing Let-7LR, or Let-7WT as a control. Based on our previous results (Figures 5, 6A, and 6B) and the distribution of sites for Lin-28-targeted miRNAs (Figure 4E; Figure S5), Let-7LR expression could be expected to function as a dominant negative to repress mRNA targets despite BDNF-mediated elevation of Lin28. Analysis of dendrite complexity (supplemental information), showed that Let-7LR expression prevented BDNF enhancement of dendrite outgrowth (Figure 6C), without significantly altering basal dendrite complexity (mock condition) in comparison to control neurons expressing either Let-7WT (Figure 6C) or GFP (Figure S6B, p = 0.78, one-way ANOVA), or cell soma size and total dendritic length, which were also unaffected by BDNF (Figure 6D). Loss of GW182 function, which would be expected to inhibit miRNA-mediated repression by both Lin28-regulated and non-Lin28-regulated miRNAs, also prevented BDNF-induced dendrite growth without altering basal dendrite complexity or total protein synthesis (Figure 2A and Figure S6). These experiments highlight the importance of miRNA-mediated target selection in a neuronal response to BDNF requiring the induction of protein synthesis. We conclude that Lin28-induced degradation of pre-miRNAs is specifically required for the appropriate specification of mRNA targets for BDNF-upregulated translation and is required for BDNF-dependent growth of neuronal dendrites.
DISCUSSION
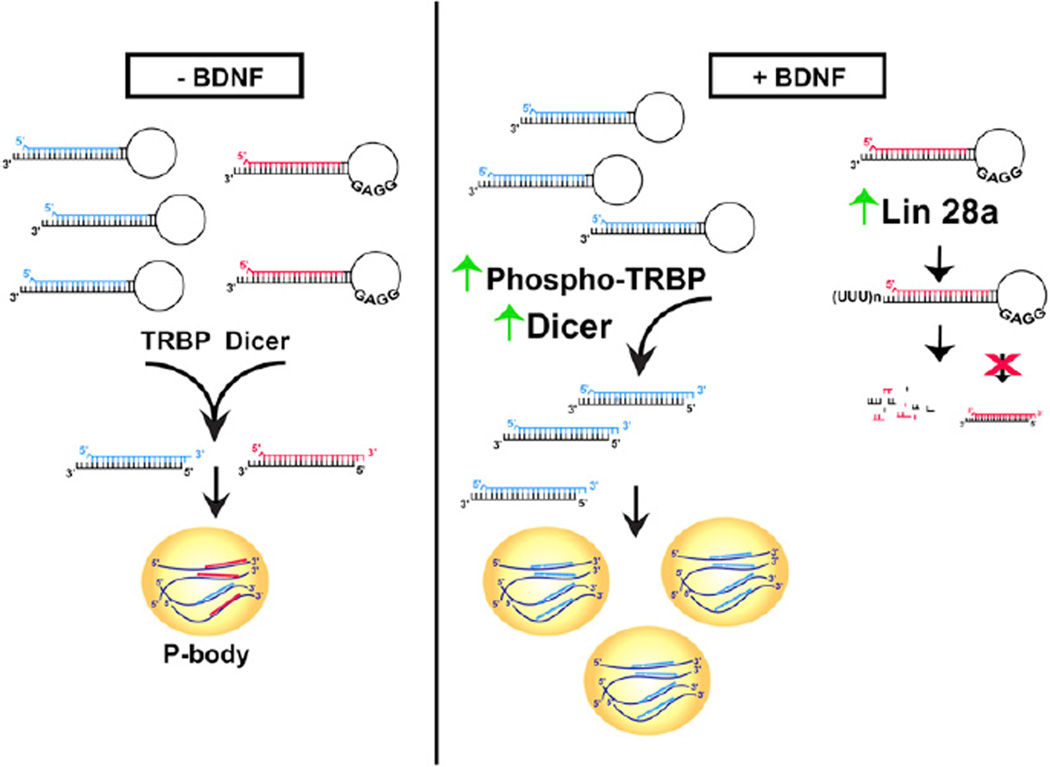
The capacity to rapidly alter the abundance of effector proteins through regulating translation is critical to the biological actions of multiple stimuli. However, the pathways that mediate stimulus-dependent selection of specific mRNAs for enhanced translation have remained poorly understood. We have defined a coordinated mechanism for genome-wide control of translation specificity that involves stimulus-dependent positive and negative regulation of miRNA biogenesis (model, Figure 7). We provide direct evidence that BDNF achieves translation target specificity by elevating levels of both Dicer and Lin28a proteins in a rapid and transcription-independent manner. The resultant action of Dicer and Lin28a on the cellular profile of miRNAs in response to BDNF effectively determines which mRNAs will participate in translation or be excluded through GW182-associated repression.
Figure 7. Proposed Model for the Determination of mRNA Target Specificity in BDNF-Mediated Translation.
(Left) In the absence of BDNF stimulation, both Lin28-targeted precursor miRNAs (GGAG, red) and non-Lin28-targeted precursor miRNAs (blue) are processed into mature miRNAs. mRNAs targeted for translational repression or degradation by these mature miRNAs accumulate in P bodies. (Right) BDNF induces both positive and negative regulation of miRNA biogenesis. In the presence of BDNF, TRBP phosphorylation and Dicer protein levels increase leading to a general enhancement of processing of precursor miRNAs (blue) into mature miRNAs. Increased abundance of mature miRNAs leads to an increase in targeting of mRNAs for repression and increases the number of P bodies in cells. However, Lin28a protein levels also increase in response to BDNF (far-right). Because Lin28a selectively prevents processing of its targeted precursor miRNAs (GGAG, red) into mature miRNAs, this population of miRNAs is diminished and mRNA targets of these miRNAs are no longer efficiently repressed and become more readily available for translation. The differential effects of BDNF on distinct miRNA populations can explain the selective increase in translation of only specific mRNAs in response to BDNF.
Our results provide the following insights into specificity for BDNF up- and downregulated protein synthesis. First, upregulation of an mRNA’s translation by BDNF requires the target mRNA to be repressed and enriched in association with P body component GW182 under basal conditions. Interference with Dicer or GW182 function prevents this basal repression and therefore occludes stimulus-dependent induction of translation. Second, interference with Dicer or GW182 blocks the downregulation of target mRNA translation by BDNF. Third, the presence of seed-matched sites for a Lin28-regulated miRNA within a 3′UTR are predictive of an upregulated BDNF-target mRNA. Interference with selective Lin28-mediated block of pre-miRNA processing prevents the induction of targets upregulated by BDNF. Fourth, the stimulus-induced association of an mRNA with GW182 is reciprocally related to its level of translation. BDNF diminishes the GW182 association of mRNA for translationally upregulated targets and enhances the GW182 association for downregulated targets.
Collective evidence indicates that GW182 interaction with miRNAs and RISC components can trigger the formation of P bodies as sites where repressed mRNAs accumulate (Eulalio et al., 2007; Liu et al., 2005b). It seems plausible that the miRNA-dependent repression induced by BDNF typically employs P bodies, consistent with the striking Dicer-dependent increase in P body number in response to BDNF. However, loss of visible P bodies by LSm5 knockdown produced no apparent interference with the specificity of BDNF-induced protein synthesis, and our data could also be consistent with a model in which the mRNA repression does not occur in P bodies per se, but elsewhere in a GW182- and miRNA-dependent manner.
BDNF-induced repression of mRNAs involves the rapid Dicer-dependent appearance of P bodies in neuronal cell soma and dendrites that can occur independently of new transcription and, as reported in other cell types (Teixeira et al., 2005) appears to result from coalescence of existing P body components. Consistent with these results, both Dicer and pre-miRNAs are present in dendrites and isolated synapses (Lugli et al., 2008), suggesting that trafficking might not be required for rapid responses; whether BDNF regulates neuronal miRNA biogenesis on a subcellular level remains to be investigated.
Recent work indicates that many miRNAs can turnover more quickly in neurons than in other cell types. miRNAs from brain or from hippocampal cultures have variable estimated half-lives of 0.5–6 hr (Krol et al., 2010; Sethi and Lukiw, 2009), compared with half-lives R24 hr in nonneuronal cells. This property might allow degradation or a block in processing of a pre-miRNA species in neurons to rapidly lower the corresponding mature miRNA level, as supported by our finding of rapid Lin28-mediated decline in mature Let-7 miRNAs. When pre-miRNA are not depleted by BDNF-induced Lin28, the available precursors (i.e., pri- and pre-miRNAs) appear sufficient to replenish mature Let-7 levels even when transcription is blocked for 1–2 hr.
miRNAs have been reported to repress target mRNA by inhibition of translation or by degradation. We observed miRNA-dependent degradation of target mRNA for representative mRNA targets that underwent decreased translation in response to BDNF; these findings are consistent with studies citing mRNA destabilization as a predominant source of miRNA-dependent reductions in protein (Guo et al., 2010; Hendrickson et al., 2009). However, our data also suggest that miRNAs can function by translation suppression in neurons under basal conditions. Specifically, BDNF-upregulated targets were repressed and associated with GW182 prior to BDNF stimulation. Disruption of this basal repression (by deficiency of GW182 or Dicer) increased protein production from BDNF-upregulated targets with no detectable elevation of their mRNA levels, consistent with reports of miRNA function by inhibition of translation (Chendrimada et al., 2007; Mathonnet et al., 2007; Petersen et al., 2006). In addition to its established role in tuning protein levels, our data highlight a role for miRNA-mediated repression in determining the specificity of stimulus-induced protein synthesis through both translation inhibition and mRNA degradation.
Mammalian Lin28 is reported to be downregulated during development with little or no expression in differentiated cells such as neurons (Moss and Tang, 2003). Our data similarly indicate low basal Lin28 expression in mature neurons, but show that BDNF induces rapid transcription-independent upregulation of Lin28a, which alters levels of Lin28-targeted miRNAs and might also perform additional functions. Lin28 expression has been associated with oncogenesis and, in conjunction with other modulators, can also induce pluripotent stem cells from differentiated tissues (Viswanathan et al., 2009; Yu et al., 2007). This underscores the concept that the reprogramming of gene expression accompanying both neoplastic transformation and induced pluripotency states may, at least in part, be additionally shared by the induction of plasticity in the adult nervous system.
Collectively, our data indicate that miRNA biogenesis undergoes dynamic posttranscriptional regulation in neurons to impart mRNA selection for BDNF-dependent protein synthesis. Our findings also reveal a role for mRNA repression in association with the P body component GW182 in conferring specificity to basal as well as stimulus-dependent translation through miRNA-dependent regulation. It is likely that other stimuli use distinct or overlapping regulatory mechanisms in the miRNA biogenesis pathway to generate specificity in the posttranscriptional regulation of gene expression.
EXPERIMENTAL PROCEDURES
Hippocampal Cultures and Stimulation
All animal procedures conformed to animal care guidelines approved by the Institutional Animal Care and Use Committee. Dissociated hippocampal cultures were prepared from postnatal day 0 (P0) mice as previously described (Meffert et al., 2003). Knockdown was by lentiviral-mediated delivery of shRNA to cultures at multiplicity of infection of 5–10, 48 hr before imaging or 4–5 days for GW182. Cultures were preincubated 10–20 min and mock or BDNF stimulated (100 ng/ml BDNF) in the presence of Actinomycin-D (0.5 µg/ml), unless indicated otherwise.
Imaging and Quantification
Confocal images of hippocampal pyramidal neurons were acquired on either a Yokogawa spinning disk (Zeiss) at 37°C (live cells), or a LSM5 Pascal system (fixed cells). Laser power and exposure time were adjusted to minimize photobleaching and avoid saturation. All experiments were from a minimum of three independent cultures and no more than three neurons per dish.
RNA Analysis
For qRT-PCR, TaqMan Gene Expression, and MicroRNA Assays (Applied Biosystems) were performed with quantitation by the standard-curve method and no preamplification, RQ was calculated as 2-ΔCtBDNF/2-ΔCtmock where ΔCt = (cycle threshold for miRNA of interest) – (cycle threshold for reference control).
Reporter Assays
The following CXCR4 siRNA/miRNA reporter assay constructs (Addgene) were used: siRNA reporter (PCD FL1P, Plasmid 12567), miRNA reporter (PCD FL4X, 12565), control luciferase reporter (PCD FL0X, 12563), and CXCR4 shRNA (pLKO.1 puro CXCR4 siRNA-2, 12272) (Wang et al., 2006). Let-7 luciferase reporters with wild-type or mutated Let-7 miRNA binding sites were gifts from G. Hannon (Liu et al., 2005b). 3′UTR reporters were constructed by inserting 3′UTRs from GluA1, CaMKIIα, or KCC2 downstream of luciferase in pGL3-Control vector (Promega). A Lin28-resistant Let-7 pre-miRNA was generated by mutation of the conserved Lin28 “GGAG” recognition motif to “GtAt” in the terminal loop of pLV-hsa-let-7a-1 (Biosettia).
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Fritzler, J. Mendell, G. Hannon, and M. Caterina for generously sharing reagents and equipment. We thank J. Pomerantz and G. Seydoux for critical reading of the manuscript and scientific suggestions, and R. Green, D. Ginty, and A. Kolodkin for helpful discussions. This work was supported by the Braude Foundation, Johns Hopkins Brain Science Institute (to M.K.M.) and Research Grant TMS-094-2-A-030 from Taiwan’s National Science Council (to Y.-W.A.H.).
Footnotes
ACCESSION NUMBERS
The miRNA microarray data has been deposited in the NIH GEO database (accession number: GSE35969).
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, six figures, and three movies and can be found with this article online at doi:10.1016/j.cell.2012.01.036.
REFERENCES
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andl T, Murchison EP, Liu F, Zhang Y, Yunta-Gonzalez M, Tobias JW, Andl CD, Seykora JT, Hannon GJ, Millar SE. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr. Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. doi: 10.1371/journal.pbio.1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- Huber KM, Kayser MS, Bear MF. Role for rapid dendritic protein synthesis in hippocampal mGluR-dependent long-term depression. Science. 2000;288:1254–1257. doi: 10.1126/science.288.5469.1254. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jaworski J, Spangler S, Seeburg DP, Hoogenraad CC, Sheng M. Control of dendritic arborization by the phosphoinositide-3′-kinase-Akt-mammalian target of rapamycin pathway. J. Neurosci. 2005;25:11300–11312. doi: 10.1523/JNEUROSCI.2270-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Busskamp V, Markiewicz I, Stadler MB, Ribi S, Richter J, Duebel J, Bicker S, Fehling HJ, Schübeler D, et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, III, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugli G, Torvik VI, Larson J, Smalheiser NR. Expression of microRNAs and their precursors in synaptic fractions of adult mouse forebrain. J. Neurochem. 2008;106:650–661. doi: 10.1111/j.1471-4159.2008.05413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat. Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, Rossi S, Fernandez AF, Carneiro F, Oliveira C, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat. Genet. 2009;41:365–370. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev. Biol. 2003;258:432–442. doi: 10.1016/s0012-1606(03)00126-x. [DOI] [PubMed] [Google Scholar]
- Narisawa-Saito M, Carnahan J, Araki K, Yamaguchi T, Nawa H. Brain-derived neurotrophic factor regulates the expression of AMPA receptor proteins in neocortical neurons. Neuroscience. 1999;88:1009–1014. doi: 10.1016/s0306-4522(98)00496-5. [DOI] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity-and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Rivera C, Li H, Thomas-Crusells J, Lahtinen H, Viitanen T, Nanobashvili A, Kokaia Z, Airaksinen MS, Voipio J, Kaila K, Saarma M. BDNF-induced TrkB activation down-regulates the K+-Cl− cotransporter KCC2 and impairs neuronal Cl− extrusion. J. Cell Biol. 2002;159:747–752. doi: 10.1083/jcb.200209011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schratt GM, Nigh EA, Chen WG, Hu L, Greenberg ME. BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development. J. Neurosci. 2004;24:7366–7377. doi: 10.1523/JNEUROSCI.1739-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ. Micro-RNA abundance and stability in human brain: specific alterations in Alzheimer’s disease temporal lobe neocortex. Neurosci. Lett. 2009;459:100–104. doi: 10.1016/j.neulet.2009.04.052. [DOI] [PubMed] [Google Scholar]
- Shan G, Li Y, Zhang J, Li W, Szulwach KE, Duan R, Faghihi MA, Khalil AM, Lu L, Paroo Z, et al. A small molecule enhances RNA interference and promotes microRNA processing. Nat. Biotechnol. 2008;26:933–940. doi: 10.1038/nbt.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei N, Kawamura M, Ishizuka Y, Kakiya N, Inamura N, Namba H, Nawa H. Brain-derived neurotrophic factor enhances the basal rate of protein synthesis by increasing active eukaryotic elongation factor 2 levels and promoting translation elongation in cortical neurons. J. Biol. Chem. 2009;284:26340–26348. doi: 10.1074/jbc.M109.023010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira D, Sheth U, Valencia-Sanchez MA, Brengues M, Parker R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng TL, Toffanin S, O’Sullivan M, Lu J, Phillips LA, Lockhart VL, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 2009;41:843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol. Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Wang DO, Kim SM, Zhao Y, Hwang H, Miura SK, Sossin WS, Martin KC. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science. 2009;324:1536–1540. doi: 10.1126/science.1173205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Edelman GM, Vanderklish PW. The brain-derived neurotrophic factor enhances synthesis of Arc in synaptoneurosomes. Proc. Natl. Acad. Sci. USA. 2002;99:2368–2373. doi: 10.1073/pnas.042693699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.