Abstract
A team of students, educators, and researchers has developed new materials to teach cell signaling within its cellular context. Two non-traditional modalities are employed: physical models, to explore the atomic details of several of the proteins in the angiogenesis signaling cascade, and illustrations of the proteins in their cellular environment, to give an intuitive understanding of the cellular context of the pathway. The experiences of the team underscore the utility of these types of materials as an effective mode for fostering students’ understanding of the molecular world, and the scientific method used to define it.
Keywords: visual learning, VEGF signaling, protein structure, angiogenesis
He said science was going to discover the basic secret of life someday, the bartender put in.
He scratched his head and frowned. Didn't I read in the paper the other day where they'd finally found out what it was?
I missed that, I murmured.
I saw that, said Sandra. About two days ago.
That's right, said the bartender.
What is the secret of life? I asked.
I forget, said Sandra.
Protein, the bartender declared. They found out something about protein.
—Kurt Vonnegut, Cat's Cradle
To appreciate the complexity and beauty of the inner workings of a living cell, students must have a solid understanding of what proteins are: what they look like, what they do, and how the former shapes the latter. Unfortunately, the beginning student rarely understands protein as anything more than something that belongs on a dinner plate. Therefore, a major goal of undergraduate educators in the molecular biosciences is to help their students develop a robust mental model of proteins and their biological activities, which is accurate at both the molecular and cellular levels. At the molecular level, students need to understand the basic principles of chemistry and physics that drive a specific sequence of amino acids into their final functional form. Just as importantly, students must understand how individual proteins interact at a cellular level, to perform the many processes needed for life.
What is the best way to ensure that our students go beyond simply memorizing amino acid structures and learning the basic connections between protein structure and function? How can we help our students truly understand how proteins function at levels ranging from atomic to cellular? What would it take for students to appreciate their world as a manifestation of the molecular world? These were the questions considered by a team of researchers, educators and students participating in the CREST project at the MSOE (Milwaukee School of Engineering) Center for BioMolecular Modeling. The premise of the CREST project (Connecting Researchers, Educators and STudents) is that the collaborative efforts of these three stakeholders will result in innovative instructional tools that effectively translate the results of active research projects into the classroom. An important corollary to this outcome is that students will understand that the knowledge in textbooks is the direct product of series of experiments performed by researchers. Speaking as a group of educators, students, and researchers, we believe that it is possible to provide students with a holistic view of biochemistry that is simultaneously accurate at the molecular level and scalable to the cellular and even macroscopic levels. The approach we describe here relies on the combined use of two new instructional tools: physical models of proteins and molecular landscapes. The goal of the latter is to put the former in proper cellular context.
Proteins in the classroom—a problem of perception
Since proteins are not directly observable, the challenge of teaching protein structure and function is largely a problem of perception. Representations of proteins must be created that capture and present the features of interest without engendering misconceptions about function in the cellular context. Historically, concepts in biochemistry have been simplified and abstracted in order to make them accessible to students. Hence, double-stranded DNA is often represented as two parallel lines drawn on the white board or illustrated in a textbook, and proteins such as RNA polymerase are represented by brightly colored ovals. This is still the case today, as evidenced by the popularity of cartoon illustrations in biochemistry textbooks [1]. While it has long been common practice to conceptually simplify and abstract proteins in this way - and, even more so, complex molecular cascades of proteins - the approach can be justifiably criticized for several reasons. Perhaps most importantly, it omits much of the information about the molecule, so important properties, such as the atomic basis of base pairing in DNA or the molecular driving forces for protein-ligand interactions, must be taken on faith instead of observed directly. Such abstracted representations of proteins are far removed from "real" life; it is therefore conceivable that the prevalence of this educational approach has contributed to public notions about science as an overly-abstract field—a peculiar conclusion to be reached about a discipline with such a firm basis in the material world. One must conclude that we as educators are not adequately bridging the gap between the abstractions we use, and the molecular reality that proteins operate in, within the cell. While there is likely to be no simple solution to this problem, we hypothesize that an important contributing factor is that we do not recognize that our biochemistry students come into our classrooms equipped with diverse learning style preferences, meaning that a successful tool for one type of learner may have limited success with other learning types. Basic two-dimensional (2-D) representations of a protein usually require students to have strong visual-spatial skills that enable them to synthesize the protein’s intricate features in 3-D. Students who are not visual learners may have difficulty with this task, and stand to benefit from the use of educational tools such as physical models and molecular landscapes.
Two-dimensional representations of proteins, based on their atomic coordinates (from the Protein Data Bank (PDB) [2]) or other data (e.g. NMR chemical shifts from Biological Magnetic Resonance Bank (BMRB) [3]), are also widely used, and computer software for 3-D visualization of proteins has become more popular in the classroom in recent years [1]. The trend toward increasing sophistication of our visual tools has great potential for the teaching of protein structure-function, with an eye towards providing a holistic molecular point of view. As observed by biomedical illustrator Linda Nye, “Humanity's ability to visualize objects and their transformations in space is a critical component of our intelligence. It makes possible the planning of actions and the anticipation of outcomes, which is the basis of science and the scientific method” [4]—this is indeed the ability required for comprehending an enzyme mechanism or protein signaling event. Visualizing the intricate details of biomolecular phenomena, such as interactions among residues in an enzyme's catalytic site, requires similarly intricate illustrations or models. However, with every new application of realism to a visual model, at this molecular level of detail, the subject becomes more complex and thus more difficult to teach (for the educator) and to comprehend (for the student). Moreover, studies have shown that for students with lower visual-spatial skills, a 3-D software program or high-resolution 2-D drawing does not make a protein more realistic, but instead makes it a more complex abstraction [1,5,6].
Recently, the use of hand-held, physical models of proteins has emerged as a new modality to help students better grasp protein structure-function in contemporary biochemistry courses. Accurately rendered physical models of proteins have been made possible by recent developments in rapid prototyping technology, but the concept of 3-D models in science is nothing new: Watson and Crick used paper models to discover the details of base pairing in DNA, and Linus Pauling discovered the basic secondary structures of proteins by experimenting with models. Assessments suggest that protein models facilitate mastery of basic protein structure-function, and students report feeling more connected to the subject when they use a tactile model [1,5,6]. It is also thought that students with low visual-spatial skills may learn more easily via kinesthetic interaction with the subject matter [5,7]. Our own experience indicates that the biochemistry classroom is comprised of students with a diversity of learning styles: we have quantified this observation using the VARK learning assessment, a survey that measures students’ preferences among Visual, Aural, Read/write, and Kinesthetic learning styles [7]. This being so, our VARK data do not indicate that any particular learning style preference puts students at an advantage for overall exam scores (see supplemental Fig. 1); although, it appears that students who are weaker visual learners may improve their molecular visualization and reasoning skills, after taking a course that employs multiple teaching modalities, including use of protein physical models to complement traditional lectures (based on the molecular visualization assessment in supplemental Fig. 2).
Studying trees—without seeing the forest
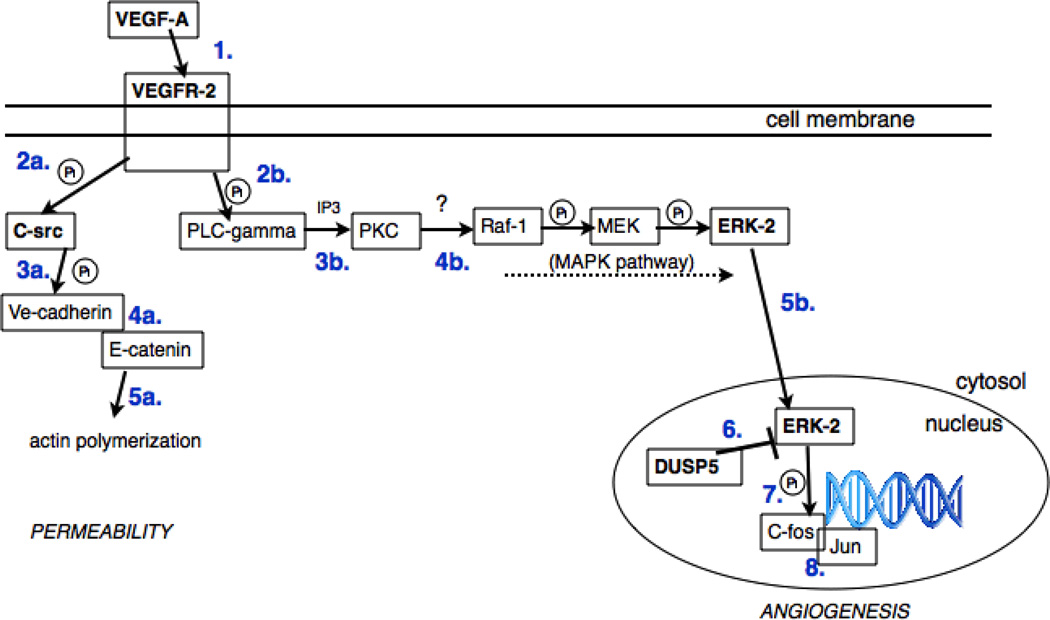
Building on our experiences, and previously reported successes with using physical models in teaching [1,5], we incorporated them into an upper-level biochemistry course that is tailored to chemistry majors. Lectures and homework assignments were centered on protein biochemistry and metabolic pathways with emphasis on mechanism and the nature of intermolecular interactions that lead to ligand binding (e.g. substrate, drug or protein partner). To complement the teaching of traditional biochemistry content, students worked in small groups (three to five students per group) on a semester-long, structure-based drug design project focused on vascular endothelial growth factor (VEGF) receptor signaling, a signaling cascade that leads to angiogenesis and is an important target for development of anticancer drugs (Fig. 1, see supplemental material for detailed pathway information).
Figure 1.
Schematic of the VEGF the pathways to be depicted in the landscape. The DUSP5/ERK-2 interaction that occurs in the nucleus (bottom right), is represented using physical models in Fig. 2.
Each group was assigned a different protein in this signaling pathway, and then visited an academic research lab that was investigating that protein. After learning about the research project involving their protein, each group then designed on the computer (using DS Visualizer then Jmol) and built a 3-D physical model of their protein with features emphasizing its structure and function, such as identifying and highlighting residues involved in ligand binding. Each group then designed a set of small molecules with shape and electronic complementarity to the active site pocket that in theory might inhibit the protein’s activity. They then prepared and presented posters along with their physical models, showing their work and demonstrating their understanding of their protein’s function, acquired based on discussions with their research mentors and their reading of the primary literature. Through its drug design component, the final project encouraged students to think at the level of molecular and atomic interactions. While these projects challenged students to think about structure-function at an advanced level, they focused on individual proteins in a cascade, even though - as a class - students had learned about multiple proteins in the cascade.
To help students expand their limited view of interacting proteins, to complicated cascades of interacting proteins (with associated structure-function implications), instruction began with a representative pair of interacting proteins involved in an important step in VEGF-mediated signaling—that between phosphorylated ERK (pERK) and DUSP-5, a phosphatase that dephosphorylates pERK (Fig. 1). Pairwise interactions such as this one were explored via molecular animations (e.g. using Pymol software) and physical models. The physical models we employed were constructed using the rapid-prototyping technology at the MSOE Center for BioMolecular Modeling (Fig. 2).
Figure 2.
Physical models of DUSP-5 and ERK-2, demonstrating the binding interaction known to occur between these proteins. DUSP-5 in comprised of two domains, an ERK binding domain and a phosphatase domain, connected by an unstructured linker region (plastic tubing in the model).
When students presented slides and posters of their work at an end-of-the-semester symposium, they demonstrated overall mastery of protein structure and function with regard to their protein member of the angiogenesis signaling cascade; however, their explanations of the signaling cascade itself tended to be weak. Despite our best efforts to use all the learning tools available to us (computer visualization/animation, lectures, physical models, active learning via presentations), students still did not demonstrate a satisfactory appreciation of the network of protein-protein interactions that are behind signaling cascades, nor of the cellular context of these interactions. While the physical models appeared to promote students’ awareness of atomic-level function of the individual proteins, the “big picture” was lost. As a result, we decided that subsequent work on the CREST project should focus on identifying and correcting this shortfall, by reinforcing the connection between all the proteins in the pathway.
How can students come to appreciate the forest?
To identify ways to help students better appreciate the cellular context in which individual proteins function in the VEGF-angiogenesis signaling pathway, we investigated the process by which most students learned about the pathway. A popular starting place for students trying to learn the pathway was an online pathway database, such as the Kyoto Encyclopedia of Genes and Genomes (KEGG) [8], which describes the networks of interacting proteins in signaling cascades and metabolic pathways. We found that students had trouble with two aspects of these diagrams. First, the diagrams have been developed primarily for the research community, and the conventions of the display may be difficult to comprehend by novices. They are often linear, glossing over details such as multi-protein complexes and subcellular compartmentalization; they often ignore the scale and population of the different protein players; and they often use coded descriptions for protein activities such as phosphorylation that may not be obvious to those outside the field. Second, and perhaps a greater problem, students were discouraged by the inconsistency among pathway diagrams from different sites, which is a natural outcome of research that is current and ongoing in developing fields, such as angiogenesis. Specifically, it is often difficult to discern between a pathway event that is strongly supported by the primary literature and an event that is less well-defined, perhaps because of ongoing scientific disputes. In short, the clarity and certainty with which textbooks presents biochemistry leaves students ill-equipped to deal with the sometimes uncertain state of our understanding of many cellular processes, which the scientific process seeks to clarify; to borrow the phraseology of Donald Rumsfeld (Feb. 12th, 2002), science textbooks teach students that science is full of “known knowns,” leaving them unprepared for the “known unknowns” and “unknown unknowns” that are pursued in active research projects.
While the first problem may be addressed with improved diagrams, such as those described below, the second problem poses a greater conceptual challenge. We have found that the best way for students to appreciate the process of scientific discovery is to have a dialog with experts in the field. Exposure to active researchers in the first phase of the CREST project (before preparing posters) was a step in that direction, but that single conversation appears to have been insufficient to empower students to interpret the sometimes conflicting literature. The need for students to have ongoing interactions with primary researchers must be balanced, of course, with the many other demands on the researchers' time. One of the researchers involved in the CREST project, and a coauthor of this paper (RR), suggested that meetings held between students and researchers approximately every two weeks might strike a good balance between teaching the scientific process, and respecting the time constraints of an active research program.
To help students integrate these molecular and cellular concepts, we have developed a more "realistic" rendering of what is happening in the cell, using what we refer to as molecular landscapes. The goal of the approach is to create an illustration that accurately simulates a portion of a cell, using visualization methods that produce an intuitively comprehensible image. The illustration and its educational goals might draw a comparison with the Harvard BioVisions project, which has developed highly effective multimedia presentations of cellular scenes that beautifully convey concepts of dynamics and scale as they apply to cellular physiology [9]. According to the project’s illustrator, David Bolinsky, the animations were designed so that Harvard students “would have an internalized view of what a cell really is in all of its truth and beauty and be able to study with this view in mind” [10]. While static two-dimensional molecular landscapes cannot convey molecular motions to the same extent as molecular animations, they have the advantage of accurately portraying cellular crowding, something that in its entirety “would prevent the vista from happening” [10] and limit the intended effect of a three-dimensional animation. In this sense, molecular landscapes and animations are complementary. Molecular landscapes attempt simulate the environment of molecules inside living cells, with illustrations based on atomic structures of individual biomolecules, electron micrographs of cells, and biochemical studies of molecular concentrations, locations, and interactions [11–14]. These molecular landscapes provide a framework for intuitive understanding that prepares the student's mind to receive the onslaught of facts we provide about pathways, interactions, cellular context, protein dynamics, and so on.
As part of one of the CREST team projects, we have created a molecular landscape of VEGF-receptor signaling at two million times magnification. In the landscape, all of the proteins are present at approximately accurate relative biological concentrations, with shapes that attempt to represent their observed domain architectures.
Construction of the molecular landscape
To integrate the cellular context effectively, it was necessary to go beyond the proteins connected with the class project in order to ensure that we had identified all the key protein players in the VEGF-angiogenesis pathway. To do so, we performed an initial KEGG inquiry into the VEGF-angiogenesis pathway, followed by primary literature searches. This work was coupled with meetings with the same researchers who participated in the in-class phase of the project. The meetings helped to sort out inconsistencies in the literature and to provide a realistic reflection of researchers’ current understanding of the pathway. From the information gathered, we generated a pathway diagram (Fig. 1).
We then compiled characteristic information about each of the proteins involved in the pathway, including size, domain arrangement/shape, biological assembly, modifications, and their interactions/functions. These characteristics, along with sequence information from UniProt [15] and PDB ID numbers (where available), were used to generate a protein description table (Fig. 3). Based on the compiled information, a series of sketches were produced by the molecular artist (Fig. 4). Several revisions were made, based on our pedagogical goals and the constraints of producing the painting. During this process, our team met with researchers actively studying angiogenesis and VEGF pathways to iteratively refine the sketch of the signaling cascade. Meetings with the researchers over the course of the CREST project played a key role, serving the dual purpose of: (a) ensuring an accurate rendering of the proteins in the cascade, with proper relative scale, subcellular location, and interacting partners, and (b) teaching students the scientific process, including how much is unknown and where the gaps in our knowledge exist—in short, teaching why science is exciting, relevant, and dynamic in a way that is not well-conveyed in textbooks.
Figure 3.
Summary of the proteins in the VEGF-mediated angiogenesis signaling cascade (Fig. 1), including relevant structural information needed for drawing the molecular landscape – such as relative size and quaternary structure.
Figure 4.
(a) An initial sketch for the molecular landscape painting depicting angiogenesis signaling, and (b) a sketch after three rounds of revisions, with markings suggesting further alterations. Relative protein sizes and shapes are based on the data compiled from Fig. 3.
Some of the features that developed with successive sketches, literature searches, and discussions included the following:
In the pathway, molecules are shown in several states to give an idea of the process in action. This includes the VEGF receptor in monomeric unbound states and dimeric active states, various combinations of signaling proteins, and monomeric and heterodimeric versions of C-fos and Jun. Initial sketches of the pathway had many of the proteins assembled into a large complex; but, discussions with active researchers suggested that these complexes should be broken into smaller units.
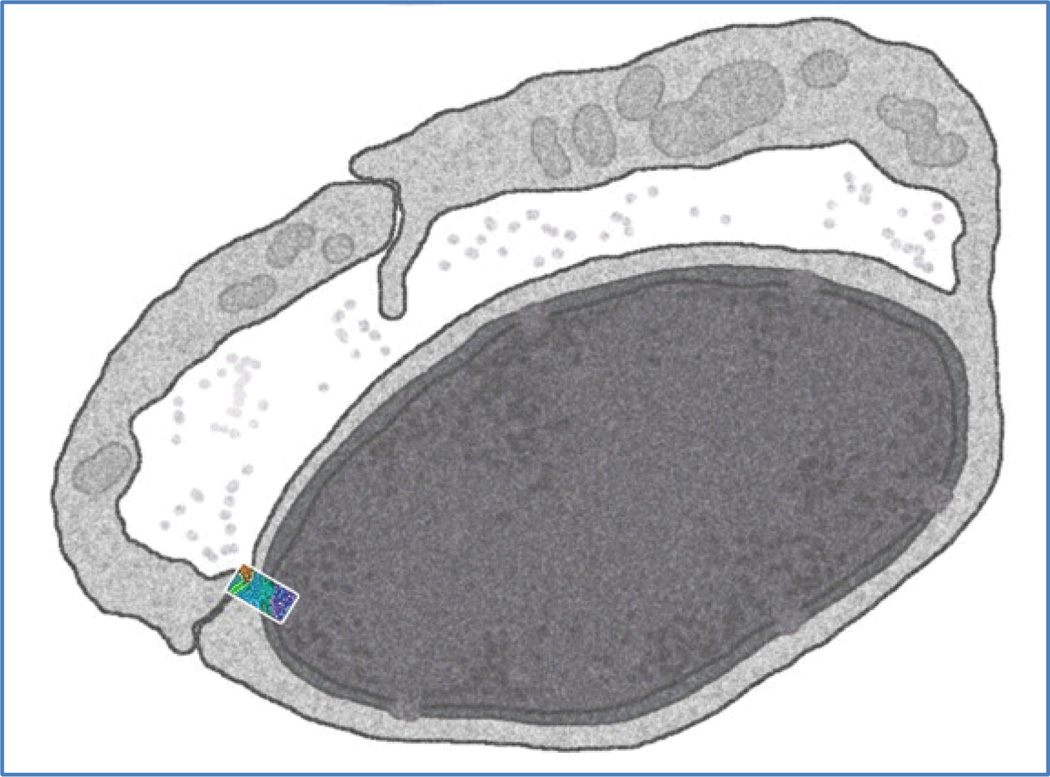
The initial concept was to produce two panels, one of the cell surface and one of the nucleus, since the portion of the cell shown in a typical painting is not sufficient to show both. Fortunately, we found an electron micrograph of capillary endothelial cells, and these cells often have the nucleus tightly pressed to the cell surface near the junctions between cells. So, it was possible to develop a scientifically accurate sketch that showed the adherens junction, cell surface, and nucleus all in one panel (Fig. 5).
The initial sketches focused on a process where Hsp27 modifies the polymerization of actin. Discussion with those actively pursuing angiogenesis research, however, determined that this is a minor biological effect, so the focus was then changed to show two effects: (a) the disassembly of tight junctions through the Src pathway and the role of the released alpha-catenin in actin reorganization, and (b) the control of transcription through the MAPK pathway and phosphorylation of C-fos by pERK.
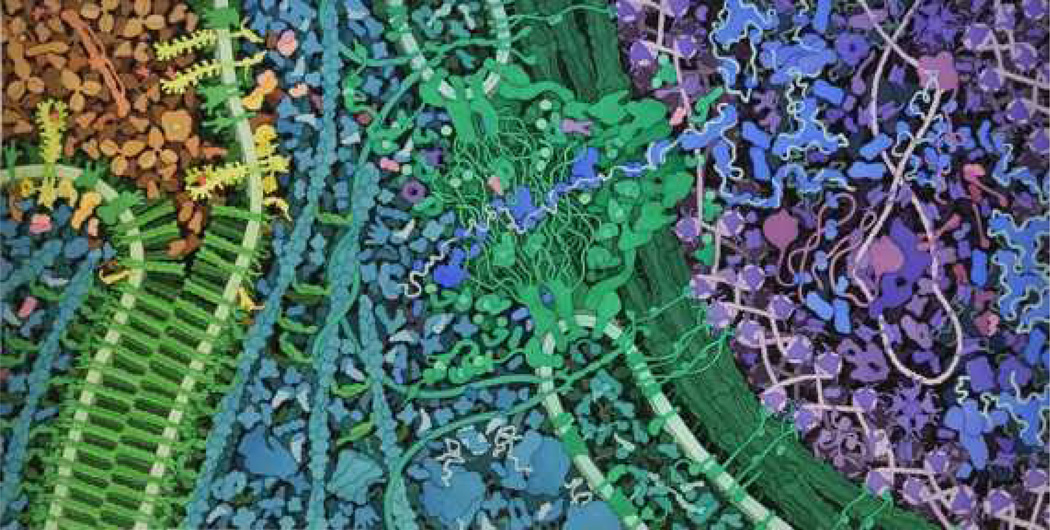
Once these details were finalized, a full sketch was created, filling in all of the other molecules (Fig. 6). This drew largely from the cellular panorama illustration from the Machinery of Life [16]. The cadherin/catenin structure of the adherens junction was adapted from an earlier paper in the Oncologist on the same subject [17] and a recent paper on the process of VEGF signaling [18]. The color design of the painting was one of the challenging aspects of the project, as there were two conflicting goals that needed to be addressed with color: 1) to highlight the molecules in the two pathways, showing the order of signaling, and 2) to make the various compartments of the cell apparent. The final color scheme uses a rainbow sequence of bright red-yellow-orange-magenta to convey the sequential action of molecules in the signaling cascade; blues, purples, and greens were used for the other molecules in the cell. This scheme may also be observed in the landscape's legends (Fig. 7).
Figure 5.
Simulated electron micrograph showing two cells from the vascular endothelium. The size and location of the portion of the vascular endothelium that is to be depicted in the molecular landscape painting is shown in color. This is the cellular context within which the angiogenesis signaling cascade is to be presented.
Figure 6.
The final molecular landscape depicting VEGF signaling. Blood serum is shown in tan at the upper left. The adherens junction between two cells is in green at left, with surface VEGF receptors shown in yellow. The cytoplasmic proteins are in turquoise, and the nuclear pore is at the center in green. The nucleus is at the right, with proteins shown in blues and purples.
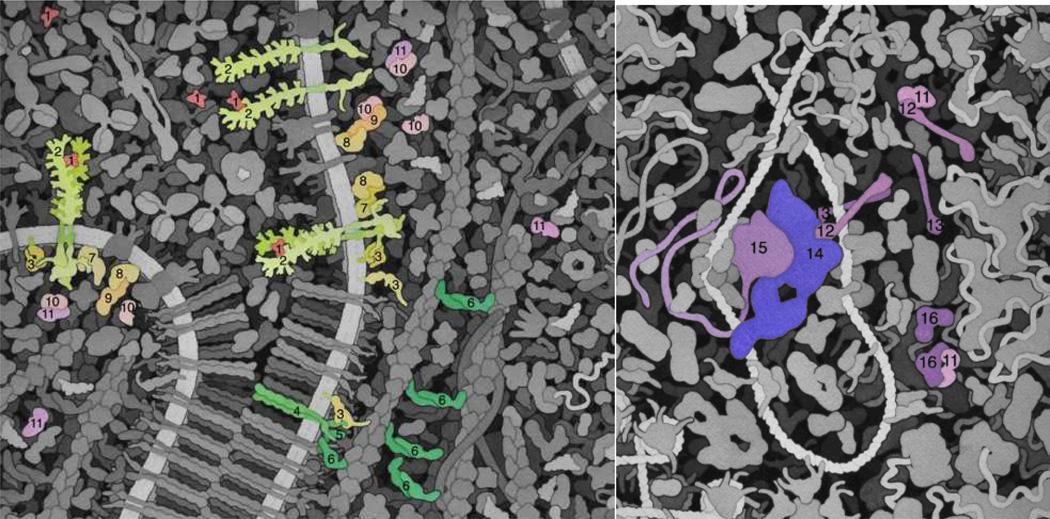
Figure 7.
Expanded and color-coded views of the molecular landscape from Fig. 6. (a) VEGF signaling, across the cell membrane into the cytosol. VEGF-A (1) in blood serum binds to VEGFR (2) and causes it to dimerize. This activates the VEGFR tyrosine kinase domains inside the cell. Two downstream pathways are shown. In one, C-src (3) is phosphorylated, causing it to open up and phosphorylate cadherins (4) in the adherens junctions, releasing alpha-catenin (6), which dimerizes and bundles actin (beta-catenin (5), which is involved in the adherens junction structure, is also shown). In the other pathway, the receptor initiates a cascade of phosphorylation reactions through PLC-gamma (7), PKC (8), Raf-1 (9), MEK (10), and ultimately ERK-2 (11). Phosphorylated ERK-2 (pERK-2) is transported through the nuclear pore (the large structure at the center of the full painting, not depicted here). (b) Continuing the signaling cascade into the nucleus. pERK-2 (11) phosphorylates C-fos (12), causing it to form a heterodimer with Jun (13) and becoming active as a transcription factor effecting transcription of proteins needed for blood vessel growth. It is shown here in an enhancer binding to the transcription mediator (14) and RNA polymerase (15). Finally, DUSP5 (16) terminates the process by dephosphorylating pERK-2. This final interaction is depicted with the physical models in Fig. 2.
Impact of the new materials on researchers, educators and students
The collaborative creation of a series of physical models of interacting proteins and the molecular landscape of VEGF signaling described here has had a significant impact on every member of the team:
For students working on the project, the physical models of a protein facilitated meaningful conversations with the researchers regarding specific details of a protein’s structure that were being investigated in the lab. At the same time, the planning of the landscape required a sophisticated synthesis of results from multiple research papers, leading to an understanding of how the destabilization of adherens junctions is a necessary prerequisite to angiogenesis, or to the transcriptional activation of a specific set of genes in the nucleus. In addition to this factual knowledge, the students were active participants in a modeling project in which the results of a wide range of experiments in different laboratories were synthesized into a conceptual model of how this signaling pathway leads to angiogenesis. This experience served as a clear example of how the concepts that are taught in a classroom are the direct product of experiments performed in a research lab. The students also had the rare experience of working closely with an educator outside the normal environment of a classroom. In the collaborative environment of this project, the “teacher” was transformed in the eyes of the students into a “scientist” who was working productively with a “researcher” to arrive at a deeper understanding of a molecular process.
The educators in this project also reported multiple beneficial outcomes, including new insights into the molecular mechanism of a specific signaling pathway, and a specific example they can use in their classrooms to demonstrate how reductionist biochemistry (details of interacting protein structures involved in ligand binding and subsequent activation of phosphorylation domains) combines with cell biology to explain the molecular process of how angiogenesis is activated in tumors. In addition, the educators reported a new appreciation for the talents and skills of the students involved in this project. In this and other modeling projects in the CREST program, educators report that the instructional materials that result from these collaborations simply would not have been developed without the efforts of students who readily embraced the use of new technologies in exploring the topic.
The researchers benefit directly from the final product of the project—both a physical model of a protein and a molecular landscape they can use to communicate their research to others. The landscape can be used to introduce new students to the “big picture” of the lab’s work, while the physical model of a protein can be used to discuss the specific details of the protein’s structure that are believed to play a critical role in the process. These same tools can also be used to explain their work to colleagues visiting from other institutions. Researchers also report satisfaction from working with undergraduate students and contributing directly in the training of a future generation of researchers. A researcher’s experience in working with a CREST modeling team is often the focus of the “broader impact” statement that is required of some research proposals.
The process of constructing a molecular landscape arguably has even more value than using an existing landscape, and the process can take multiple forms. The artistic quality of the final painting is of secondary importance—the salient matter is the process of assembling the information needed to construct the image. For example, the landscape could be prepared as a crude sketch with pencil and paper; alternatively, established graphics design software could be used to build protein shapes approximating relative scale, cellular concentration, and location. One goal of the ongoing CREST project is to create an online Landscape Creator software package with which students can construct a landscape using a palette of images of commonly encountered proteins, membranes, and nucleic acids. Regardless of how the landscape is constructed, students will be confronted with design decisions that require them to learn not only specific information about the protein players and their interacting partners, but also the skill of critically engaging with scientific literature and databases. Besides learning difficult content, they will begin to develop the skills set that will make them strong scientific thinkers. This dual instructional mode endows molecular landscape projects with high educational value and makes them decidedly promising for further classroom use.
Supplementary Material
Acknowledgments
We thank Dr. Qing (Robert) Miao and Dr. George Wilkinson at the Medical College of Wisconsin and Dr. Magdalena Chrzanowska-Wodnicka at the Blood Research Institute in Milwaukee, WI, for mentoring students and for helpful discussions on the VEGF-angiogenesis pathway, as well as Dr. Terrence Neumann at Concordia University for creative input.
This work was supported in part by NIH research grants HL102745-01A1 and HL112639-01, as well as by NSF CREST grant CCLI #1022793.
References
- 1.Harris MA, et al. A Combination of Hand-held Models and Computer Imaging Programs Helps Students Answer Oral Questions about Molecular Structure and Function: A Controlled Investigation of Student Learning. CBE—Life Sciences Ed. 2009;8:29–43. doi: 10.1187/cbe.08-07-0039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. http://www.rcsb.org/pdb/ [Google Scholar]
- 3. http://www.bmrb.wisc.edu/ [Google Scholar]
- 4.Nye LS. The Mind's Eye—Biomedical Visualization: The Most Powerful Tool in Science. Biochem. Mol. Biol. Ed. 2004;32:123–131. doi: 10.1002/bmb.2004.494032020337. [DOI] [PubMed] [Google Scholar]
- 5.Herman T, et al. Tactile Teaching—Exploring Protein Structure/Function Using Physical Models. Biochem. Mol. Biol. Ed. 2006;34:247–254. doi: 10.1002/bmb.2006.494034042649. [DOI] [PubMed] [Google Scholar]
- 6.Roberts JR, et al. Physical Models Enhance Molecular Three-dimensional Literacy in an Introductory Biochemistry Course. Biochem. Mol. Biol. Ed. 2004;33:105–110. doi: 10.1002/bmb.2005.494033022426. [DOI] [PubMed] [Google Scholar]
- 7.Fleming ND, Mills C. Not Another Inventory, Rather a Catalyst for Reflection. To Improve the Academy. 1992;11:137. [Google Scholar]
- 8. http://www.genome.jp/kegg/ [Google Scholar]
- 9.About BioVisions, BioVisions at Harvard University. [August 24, 2012];2007 http://multimedia.mcb.harvard.edu/ [Google Scholar]
- 10.Bolinsky D. Visualizing the wonder of a living cell [Video file] 2007 Retrieved from http://www.ted.com/talks/david_bolinsky_animates_a_cell.html. [Google Scholar]
- 11.Goodsell DS. Inside a Living Cell. Trends Biochem. SCi. 1991;16:203–206. doi: 10.1016/0968-0004(91)90083-8. [DOI] [PubMed] [Google Scholar]
- 12.Goodsell DS. The Machinery of Life. 2nd ed. New York: Springer; 2009. [Google Scholar]
- 13.Goodsell DS. Illustrating the Machinery of Life: Escherichia coli. Biochem. Mol. Biol. Ed. 2009;37:325–332. doi: 10.1002/bmb.20345. [DOI] [PubMed] [Google Scholar]
- 14.Goodsell DS. Putting Proteins in Context. Bioessays. 2012;34:718–720. doi: 10.1002/bies.201200072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. http://www.uniprot.org/ [Google Scholar]
- 16.Goodsell DS. Eukaryotic Cell Panorama. Biochem. Mol. Biol. Ed. 2011;39:91–101. doi: 10.1002/bmb.20494. [DOI] [PubMed] [Google Scholar]
- 17.Goodsell DS. The Molecular Perspective: cadherin. Oncologist. 2002;7:467–468. doi: 10.1634/theoncologist.7-5-467. [DOI] [PubMed] [Google Scholar]
- 18.Dejana E, Orsenigo F, Lampugnani MG. The Role of Adherens Junctions and VE-cadherin in the Control of Vascular Permeability. J. Cell. Sci. 2008;121:2115–2122. doi: 10.1242/jcs.017897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.