Abstract
Cell differentiation is an essential process for the development, growth, reproduction and longevity of all multicellular organisms, and its regulation has been the focus of intense investigation for the past 4 decades. The study of natural and induced stem cells has ushered an age of re-examination of what it means to be a stem or a differentiated cell. Past and recent discoveries in plants and animals, as well as novel experimental manipulations are beginning to erode many of these established concepts, and are forcing a re-evaluation of the experimental systems and paradigms presently being used to explore these and other biological process.
“The ship wherein Theseus and the youth of Athens returned had thirty oars, and was preserved by the Athenians down even to the time of Demetrius Phalereus, for they took away the old planks as they decayed, putting in new and stronger timber in their place, insomuch that this ship became a standing example among the philosophers, for the logical question of things that grow; one side holding that the ship remained the same, and the other contending that it was not the same.”
Plutarch (c. 46 – 120 AC), 75 C.E.
Introduction
The stability of the differentiated state of cells is essential for the growth, survival and perpetuation of all multicellular organisms studied to date. The generation of new cells from highly regulated, post-embryonic tissues for growth, repair, reproduction, and defense, for example, is an essential attribute of multicellular life in both Plant and Animal Kingdoms. How such capacity to generate new cells is parsed depends in great part on the evolutionary history of each species. In post-mitotic organisms such as Caenorhabditis elegans, this means that the adult animals have to live their adult lives with the fixed number of differentiated cells they were born with. Other organisms such as Drosophila melanogaster experience similar fates, except that they possess tissues such as the midgut that are renewed by resident cellular proliferation throughout adulthood (Ohlstein and Spradling, 2007). Such mixture of post-mitotic and continually renewed tissues is easily illustrated with what we know of our own biology. Tissues such as the frontal lobe of our brain is unlikely to be turning over at any appreciable rate during our adult life (Spalding et al., 2005), whereas the lining of our gut -a surface area equivalent in size to a tennis court (Heath, 2010)- is renewed approximately every three to five days (Pinto and Clevers, 2005; Pinto et al., 2003). Hence, for most known multicellular organisms their relatively constant, outward appearance is underscored by an incessant, inner transformation in which cells lost to normal physiological wear and tear (turnover) are replaced by the progeny of dividing cells (Pellettieri and Sánchez Alvarado, 2007). In other words, biological systems possess critical mechanisms driven by a balance between cell death and cell proliferation that preserve the forms and functions of developed tissues. Thus, as in the paradox of the ship of Theseus (Plutarch, 75 CE), it is through constant change that the appearance of most living organisms remains the same.
Ever since cells were first observed by Hooke in 1665, and the discovery in the early 1800’s by Treviranus (Treviranus, 1811), Moldenhawer (Moldenhawer, 1812) and Dutrochet (Dutrochet, 1824) that cells were separable units providing a fundamental element of organization to both plants and animals, their fate, functions, and behaviors have held the fascination of laypeople and biologists alike. Much research in biology has concerned itself with understanding how cell types are elaborated during embryonic development and how their functions and identities are maintained throughout life. In fact, it can be easily argued that for centuries, a significant amount of work in biology has focused on understanding the differentiation potential of cells, from Hartsoeker’s homunculus (Hartsoeker, 1694) to present day work on stem cells (Dejosez et al., 2013; Suga et al., 2011) and regeneration (King and Newmark, 2012; Sánchez Alvarado and Tsonis, 2006). Key, influential concepts have emerged from this collective and long-standing effort by biologists to understand life: potency, lineage, competence, fate, and differentiation, for example. And while these concepts have served us well, there is clear evidence that many are being eroded, while others are beginning to look more like mere suggestions rather than strict rules to be followed. Such challenges to the establishment are being ushered by a discreet, but nonetheless persistent effort to expand modern biological inquiry into novel experimental systems and paradigms, and by the wholesale embracing of the field of powerful methodologies that have increased the granularity of our studies to unprecedented levels of detail and complexity. As such, our present interrogation of cellular potency both in vivo and in vitro is leading to a re-evaluation of the explanatory system that frames our understanding of developmental processes. Here we discuss how understudied model systems and novel technologies such as induced pluripotent stem cells (iPSCs) are forcing us to question long-established concepts (Figure 1), and propose that such efforts may ultimately help marshal an age of biological discovery unconstrained by the incrustations of familiarity.
Figure 1. Potency, reprogramming and differentiation.
Discoveries and technological breakthroughs associated with the concept of cellular differentiation. The background image is plate 37 from Haeckel’s Kunstformen der Natur (Haeckel, 1904) and depicts a siphonophore.
Tissue Homeostasis, Longevity and Stem cells
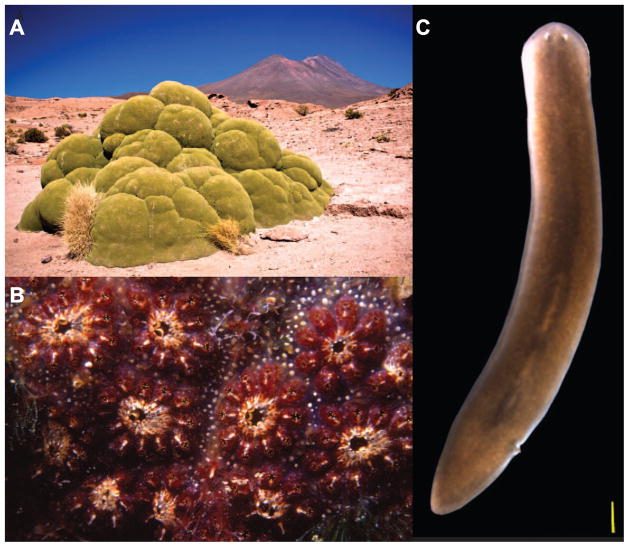
While development is normally associated with embryogenesis, this biological process does not end at birth, but continues throughout the natural lifespan of plants and animals. For many organisms this can be a remarkably long period of time during which constant cellular renewal and growth goes on for decades, sometimes centuries. In fact, the functions of many organs under normal physiological conditions depend on the continuous destruction and renewal of their cells. Therefore, understanding the mechanisms by which cell proliferation and tissue turnover are balanced in order to yield constitutive body growth, and constitutive body regeneration, should provide key insights on adult developmental processes. Consider the South American flowering plant Azorella compacta (Figure 2a), which adds about 1.5 cm of radial growth per year (Kleier and Rundel, 2004). Based of this rate of growth, it has been estimated that many specimens found in the Atacama desert likely exceed 3,000 years of age, making A. compacta one of the oldest living organisms on Earth. Or from the animal kingdom, consider the rather extreme form of tissue homeostasis that is readily found in the colonial ascidian Botryllus schlosseri, a close relative of the vertebrates (Figure 2b). This chordate is known to undergo whole-body regeneration every 7 days or so as part of its growth and reproductive cycle (Lauzon et al., 1992, 2002; Rinkevich et al., 1992). Other examples exist, such as the freshwater planarian Schmidtea mediterranea (Figure 2c), which is known to constantly, and seemingly permanently replace cells lost to physiological wear and tear with the progeny of proliferating cells, making these organisms negligibly senescent (Pellettieri and Sánchez Alvarado, 2007).
Figure 2. Biological adaptations of stem cell functions.
A) The evergreen perennial plant Azorella compacta grows constitutively through the continuous proliferation and differentiation of its meristem stem cells (Credit: Pedro Szekely (http://commons.wikimedia.org/wiki/File:3,000_Year_Old_Yareta_Plant_%282087602585%29.jpg). B) The colonial ascidian Botryllus schlosseri regenerates its whole body almost weekly as part of its sexual and asexual reproductive strategy (Credit: Parent Géry (http://commons.wikimedia.org/wiki/File:Botryllus_schlosseri_%28Pallas,_1766%29_.jpg). C) The negligibly senescent planarian Schmidtea mediterranea is a constitutive adult which constantly replaces dying differentiated cells with the freshly minted progeny of its abundant stem cell population (Credit: Erin Davies).
Because post-natal regulation of growth and homeostasis is found in both the Plant and Animal Kingdoms, it is likely that the units of selection underpinning this attribute, i.e., stem cells, and the processes regulating the perpetuation of these cells may be as ancient as the origins of multicellularity. While we have a very clear understanding of the location and number of stem cells in flowering plants (Birnbaum and Sánchez Alvarado, 2008), much remains to be resolved when it comes to animal stem cells. In plants, stem cells reside in apical and root meristems, and their cell division, determination, differentiation and patterning have been elegantly defined and characterized during embryogenesis and in mature plants in both time and space (Brand et al., 2000; Gordon et al., 2009; Reddy et al., 2004; Reddy and Meyerowitz, 2005; Sugimoto et al., 2011). The situation in animals is quite different. In the case of Botryllus, for example, while the most parsimonious explanation to whole body regeneration is the existence of stem cells, their exact anatomical location remains controversial, and the possibility of neogenesis (production of stem cells from other, differentiated cells, for example) remains a formal possibility (Laird et al., 2005; Laird and Weissman, 2004; Rinkevich et al., 2013; Voskoboynik et al., 2007; Voskoboynik et al., 2008). In planarians, the anatomical location of stem cells (neoblasts) has been known for a very long time (Elliott and Sánchez Alvarado, 2013), and while evidence exists that at least some of these cells are pluripotent (Wagner et al., 2011), it still remains to be determined whether or not there exists discrete heterogeneities of stem cells among the neoblast population and whether such heterogeneities are defined or plastic. Problematic in the field, therefore, is the significance of the extensive heterogeneity of expression of fate determinant genes in the planarian stem cells (Chen et al., 2013; Lapan and Reddien, 2012; Reddien, 2013; Scimone et al., 2014; Scimone et al., 2011), which is currently interpreted as reflecting co-existing populations of committed, yet nevertheless undifferentiated cells (Reddien, 2011, 2013).
Although one would expect our knowledge of stem cell biology in these invertebrate species to be incomplete due to the limited tools available to mechanistically interrogate them, it would not be necessarily expected that a similar state of affairs exists in better-studied animals such as mice. The biology of mammalian stem cells is usually inferred from the study of cells that were isolated based on their expression of surface markers, rather than by studying them in vivo. It is known, though, that removal of cells from their native milieu results in changes in their behavior and that the markers used to isolate them can be both transient and heterogeneous in their expression to the point where pluripotent stem cell identity can be a remarkably dynamic state (MacArthur and Lemischka, 2013). These factors have essentially conspired against us, and have prevented us from having a clear understanding of the actual number of self-renewing stem cells present in mammalian tissues, or the dynamics that govern their renewal.
Consider the mammalian small intestine as an example. For many years, the location and types of stem cells in the crypts of this organ has been hotly debated. Some placed the stem cells in specific locations of the crypt (e.g., the +4 position) (Barker et al., 2007; Clevers and Bevins, 2013), while others argued for the existence of quiescent or label retaining stem cells in other locations as being the stem cells in this organ (Tian et al., 2011; Yan et al., 2012). Genetic studies have begun to resolve these differences. We now know that some of the markers identified from isolated cells and expressed in different yet overlapping cell populations such as mTERT, Lgr5 or Hopx may represent candidate stem cell populations because all of them can give rise to long-lived clonal progeny (Barker et al., 2007; Montgomery et al., 2011; Takeda et al., 2011). We also now know that non-self-renewing secretory precursor cells in the intestine are the label retaining cells, which only under growth-promoting conditions can change their terminally differentiated fate and become stem cells. These committed secretory precursors give rise to Paneth and enteroendocrine cells, and while expressing stem cell markers, they are quiescent under homeostatic conditions (Buczacki et al., 2013). Moreover, the number of stem cells per functional intestinal crypt is remarkably small (5 to 7; (Kozar et al., 2013), begging the question of how such a minute number of equipotent, clonogenic cells can generate different numbers of cells, expressing different markers (e.g., mTERT, Lgr5, Hopx), with different degrees of overlap (Muñoz et al., 2012), and the capacity to interconvert into each other (Takeda et al., 2011).
Given the importance of knowing how many stem cells may actually exist in a given tissue, and how their population dynamics is regulated during growth, tissue homeostasis, and regeneration, it is evident that much work needs to be done to explore the fundamental nature of stemness in vivo. If the normal fate of cells in adult tissues is plastic and environmental changes such as growth-promoting conditions can switch their development from a program of terminal differentiation to one of active clonogenicity, we need to invest more effort in studying how stemness arises not only as a cell autonomous property, but rather as a property emerging from the behavior of cell populations. While stem cells in culture may help identify molecules that are associated with stem cell function, understanding the properties of stem cells will require precise lineage and fate mapping studies in vivo, and we predict that this will be accompanied by the active prospecting and identification of novel stem cell function paradigms in both currently studied and as yet to be interrogated organisms.
The Plasticity of Cell Fate
If development is a continuum from conception to death, then it is important to ask what it means for a cell to be “functionally mature” or “differentiated”. The origins of the modern concept of cellular differentiation can be traced as far back to the influential book The Theory of the Germ Plasm by August Weismann (Weismann, 1893). The concept put forward by Weismann invokes the production of two cell types early in embryogenesis: one that will pass on the individual’s genetic information to the next generation (germ cells in the gonads), and another that produces all other cell types in the organism (somatic cells). The implication being that as somatic cells develop, they become progressively and irreversibly restricted in their differentiation potency to the point that each cell can only differentiate into a single, specific cell type (e.g., muscle, neurons, epithelia, etc.). From the outset, this concept was challenged by studies of regeneration, in which new tissues are made from pre-existing, and according to Weisman’s theory, terminally differentiated cells, a difficulty Weisman himself recognized by dedicating a full, yet inconclusive chapter to this topic in his book (Weismann, 1893).
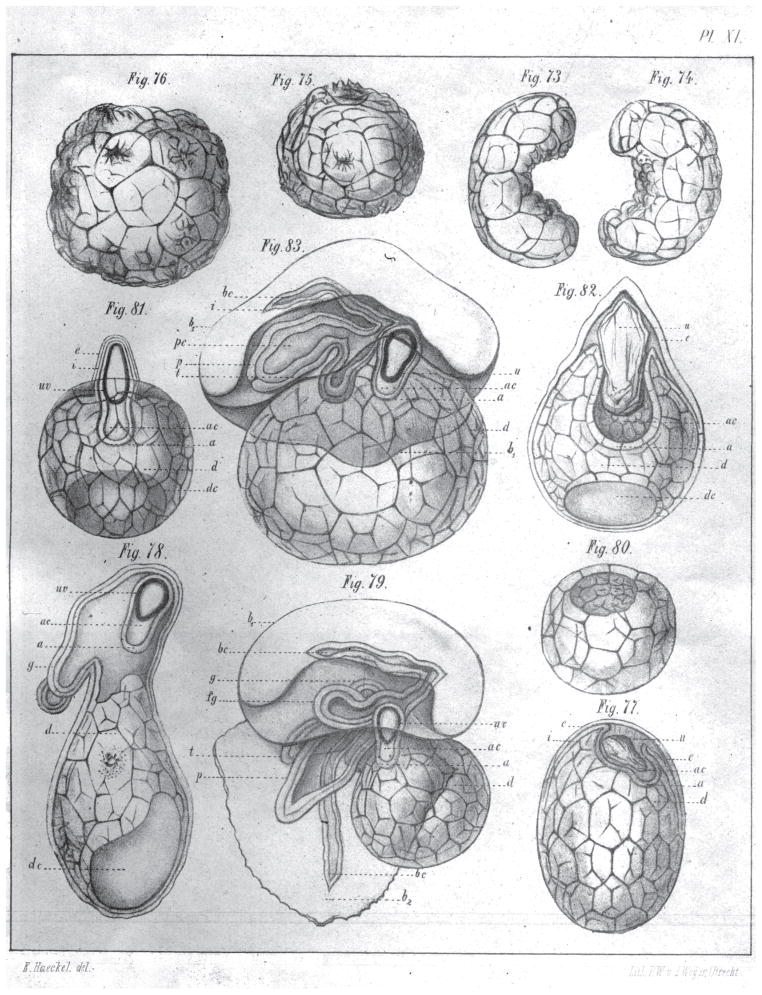
Interestingly, data was already available demonstrating that embryonic cells capable of giving rise to the soma were not restricted in their developmental potency, work that presumably should have been readily accessible to Weismann. The work involved the study of siphonophore embryos and was published in 1869 by none other than Ernst Haeckel (Haeckel, 1869). Although traditionally credited to Roux and Driesch (Gilbert, 2013), Haeckel discovered that amputated fragments of Siphonophore embryos could produce complete larvae (Haeckel, 1869) (Figure 3). Although this paper has been widely ignored by scientists and most historians of science (see (Richards, 2008) for a salient exception), it is the first clear demonstration of the pluripotency of embryonic cells, preceding the work of Roux and Driesch by almost 30 years.
Figure 3. Demonstration of pluripotency of blastomeres in siphonophores.
Dissection of siphonophore larvae in half (Fig. 73 and 74 in plate). The same larvae halves a few hours after physical separation (Figs. 75 and 76 in plate). Larvae separated into thirds 8 days after dissection (Figs. 77, 78 and 79). Figures 77 and 79 illustrate larvae fragments that developed an air sac and polyps (Fig. 77) and a normal, full larvae (Fig. 79). Finally, Figs 80, 81, 82, and 83 illustrate the results of quartering siphonophore larva with only 83 developing normally (Haeckel, 1869).
Eventually, other experimental evidence was put forward to suggest that the developmental fate of cells can indeed be quite plastic. Using hydra with different pigments (Figure 4), Ethel Browne described how the mouth (hypostomal) tissue when grafted into the body wall of another hydra produced “the necessary stimulus to call forth the development of a new hydranth” at the site of transplantation (Browne Harvey, 1909). Later, Hilde Proescholdt Mangold, a graduate student working in Hans Spemann’s laboratory, would perform similar experiments with frog embryos, in which she grafted a piece of the lip of the blastopore to the flank of another gastrula distant from the host blastopore, resulting in the induction of a secondary body axis (Mangold and Spemann, 1924). Altogether, the experiments of Haeckel, Browne, Proescholdt and others (Lewis, 1904; Spemann, 1901) showed conclusively that the fate of embryonic cells was not irreversible and that their differentiation programs could be experimentally reprogrammed.
Figure 4. Pioneering work of Ethel Browne Harvey demonstrating the ability of transplanted tissues to reprogram the fate of host cells.
Reproduced from the original (Browne Harvey, 1909), this image illustrate the specific outcomes of transplanting of various body parts between pigmented (green) and unpigmented strains of Hydra. Fig. 42: Graft of white tentacle with peristome in middle of green hydra. Fig. 43: Graft of white tentacle with peristome in foot of green hydra. Fig. 44: Graft of green tentacle without peristome in white hydra. Fig. 45: Graft of green body tissue in white hydra. Fig. 46: Graft of green hydranth in white hydra. Fig. 47: Graft of green foot in white hydra. Fig. 48: Heteromorphosis in reversed ring of green tissue grafted on white stock.
Equally interesting is the fact that such plasticity is not only confined to embryonic cells. In 1891, Colluci reported the ability of newts to regenerate their lenses after full lentectomy (Colluci, 1891), and in 1895, Wolff demonstrated that the source of the regenerated lens were the pigmented epithelial cells of the dorsal iris, providing the first example of in vivo, adult cellular reprograming (Wolff, 1895). Subsequent work demonstrated that the pigmented dorsal epithelial cells had to re-enter the cell cycle and de-differentiate in order for lens regeneration to occur, (Eguchi and Shingai, 1971; Reyer, 1977; Yamada, 1977). More recently, it has been shown in planarians that postmitotic tissues in amputated fragments devoid of stem cells can reprogram their genomic output, and both express (Reddien et al., 2007) and repress (Gurley et al., 2010) patterning signals. Such modulation is required to allow the amputated fragments to dramatically re-arrange pre-existing tissues to produce small animals with a complete complement of organ systems of appropriate allometric proportions. These observations indicate that reprograming mechanisms in differentiated cells must exist that allow a rapid change in site of expression of signaling proteins regulating planarian body patterning.
Most remarkable still is the fact that complete adult organisms can revert in their entirety to a larval state through reprogramming mechanisms that remain to be elucidated (Figure 5). Upon thermal, chemical, physical stress, or merely aging (senescence), the hydrozoan species Turritopsis dohrnii, Turritopsis nutricola, Laodicea undulate and Podocoryne carnea and possibly many more (Piraino et al., 2004) have been reported to revert their life cycle by back-transformation of the adult state into juvenile, earlier developmental stages (Carla et al., 2003; Piraino et al., 1996; Schmich et al., 2007). This exaggerated potential for dedifferentiation provides a unique experimental paradigm to understand how regulatory networks of gene expression and their attendant cell behaviors may control the directionality of ontogeny, i.e., normal vs. reverse development. Hence, it appears that in Nature, for many cells in many organisms, the differentiated state is not terminal but, instead, stable and that under varied environmental conditions such as injury and disease, or even the natural process of aging, such state can be reprogrammed.
Figure 5. Animals reported of being capable of reverse development.
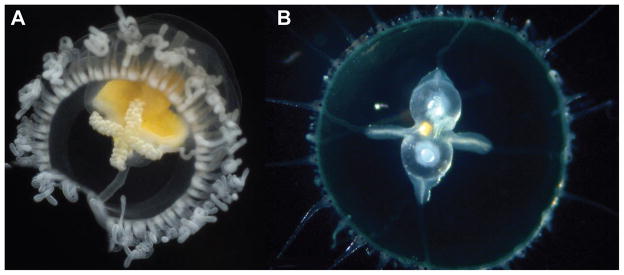
A) Turritopsis dohrnii. B) Laodicea undulate (Credit for both images: Alvaro A. Migotto http://cifonauta.cebimar.usp.br/photo/2190/ and http://cifonauta.cebimar.usp.br/photo/10792/).
Cellular reprogramming
The in vivo plasticity of the differentiated state can be induced artificially by directly manipulating cells and their environment. A notable example in which environmental perturbations were shown to drive changes in cell fate was reported in the middle of the 20th century by Holtfreter (1947). In these experiments, frog cells fated to form skin developed into brain tissues when exposed to a solution of low pH (Holtfreter, 1947). Yet, the first attempt to directly, and specifically reprogram cells involved nuclear transplantations from older, differentiated cells, into younger, undifferentiated cells. This notion was first proposed by Hans Spemann (Spemann, 1938), but was brought to fruition by the patient and groundbreaking work of Briggs and King, who developed methods to transplant the nuclei of blastula cells into enucleated frog oocytes (Briggs and King, 1952). This accomplishment was soon followed by the work of John Gurdon who extended this method to transplant nuclei from frog intestine into oocytes and produce clonal, somatically derived adult frogs (Gurdon et al., 1958). As our understanding of the molecular processes leading to the commitment and differentiation of cells increased, it became possible to reprogram cells not by transplanting their nuclei but by introducing gene functions into cells. This was brilliantly accomplished by Harold Weintraub (1989) when he and his group elegantly demonstrated that transforming fibroblast, pigment, nerve, fat and liver cells with a single transcription factor, in this case MyoD, was sufficient to push these cells into a myogenic developmental pathway (Tapscott et al., 1988; Weintraub et al., 1989).
This body of work paved the way for the generation of induced pluripotent stem cells (iPSCs) in 2006 (Takahashi and Yamanaka, 2006). By simultaneously introducing four transcription factors, namely Oct3/4, Sox2, Klf4 and c-Myc, mouse fibroblasts were transformed into cells that were nearly identical to embryonic stem (ES) cells. By culturing an inner cell mass of blastocysts, mouse ES cells were first generated in 1981 by Martin Evans (Evans and Kaufman, 1981) and Gail Martin (Martin, 1981). Such ES cells proliferate infinitely while maintaining pluripotency. The unexpected finding that somatic cells can revert all the way back to the embryonic state by a handful of transcription factors soon led to the chemical manipulation of signaling pathways to reprogram cells (Hou et al., 2013; Li et al., 2009), and to the discovery of factor-mediated conversion of fibroblasts into other cell lineages, such as neurons (Vierbuchen et al., 2010), hepatocytes (Sekiya and Suzuki, 2011) and cardiac myocytes (Ieda et al., 2010).
The fact that it is possible to reprogram cells by either perturbing their environment or changing their genomic output artificially, indicates that that the plasticity of the differentiated state may not be restricted to simple animals, and that programs for dedifferentiation may be more prevalent and much more broadly distributed among all animals (including humans) than most care to contemplate at the present time. Recent work in mice suggests that this may be the case. When white blood cells are exposed to a solution of low pH, the cells dedifferentiated and began to express gene markers typical of early embryos, a phenomenon termed stimulus-triggered acquisition of pluripotency (STAP) (Obokata et al., 2014b). Equally remarkable, when injected into embryos STAP cells produced chimaeras a property that was previously thought to be the exclusive realm of embryonic stem cells and iPSCs (Obokata et al., 2014a).
The STAP phenomenon may explain a series of controversial findings regarding the existence of pluripotent stem cells in the adult mammalian body. In 2001, Verfaillie and colleagues reported the isolation of pluripotent stem cells from adult bone marrow by a combination of selecting for specific surface markers and expanding the obtained cells in defined culture conditions. They designated these cells MAPCs for multipotent adult progenitor cells (Reyes and Verfaillie, 2001). Subsequently, these findings were followed by the report that chimeric mice could be generated by transplanting mouse MAPCs into blastocysts (Jiang et al., 2002). This body of work, however, was met with skepticism because it could only be reproduced in a few laboratories. More recently, other groups have also reported on the isolation of pluripotent stem cells from adult human tissues (Heneidi et al., 2013; Kuroda et al., 2010; Obokata et al., 2011; Roy et al., 2013). Of note, the Desawa and Chazenbalk laboratories showed that pluripotent stem cells, which they designated Muse (Multi-lineage differentiating Stress Enduring) cells, could be derived from human somatic tissues exposed to long-term exposure to collagenase, serum deprivation, low temperatures and hypoxia, and argued that Muse cells already existed in normal adult bodies but were preferentially expanded under conditions of severe cellular stress (Heneidi et al., 2013; Kuroda et al., 2010). Although the Muse and STAP procedures are remarkably similar, the STAP papers have provided a different interpretation. In essence, that the adult pluripotent stem cells of mammalian tissues that previous work regarded as pre-existing stem cells may have arisen by stress-induced reprogramming during isolation and cultivation. Hence, it appears that in Nature, for many cells, and in many organisms, the differentiated state is not terminal but, instead, stable and that under varied environmental conditions such as injury and disease, or even the natural process of aging, such state can be reprogrammed.
Technology from science: patient-derived stem cells and medical applications
The discovery in 2007 that human iPSCs could be generated opened the door to unprecedented opportunities to supply multiple functional human cells in large quantities (Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007). One potential application of iPSCs in medicine is in the field of cell therapy. The transplantation of differentiated cells derived from iPSCs has been shown to successfully induce functional recoveries in rodent models of sickle cell anemia (Hanna et al., 2007), platelet deficiency (Takayama et al., 2010), Parkinson’s disease (Wernig et al., 2008), diabetes (Alipio et al., 2010) and spinal cord injury (Tsuji et al., 2010). The first clinical trial using iPSCs will start in Japan in 2014 for patients suffering from age-related macular degeneration, a disease affecting at least 25–30 million people worldwide (WHO, 2012).
Other equally important medical application of iPSCs is drug development. Patient-derived iPSCs have proven effective in providing models of disease with which to obtain a better understanding of various disease mechanisms and the screening of chemicals and natural derivatives for the development of effective drugs (Inoue et al., 2014). To our surprise, disease phenotypes have been recapitulated using patient-derived iPSCs from not only early onset monogenic diseases, but also from polygenic late onset diseases, such as sporadic forms of Alzheimer diseases (Kondo et al., 2013). Furthermore, human somatic cells, such as cardiac myocytes and hepatocytes, are useful for predicting the toxicity of drug candidates (Guo et al., 2011; Medine et al., 2013). Therefore, iPSC technology is expected to revolutionize the process of drug discovery in the coming years.
More recently, the three-dimensional structures of the retina (Eiraku et al., 2011) and pituitary gland (Suga et al., 2011) were recapitulated in vitro by the self-organization of mouse ES cells. Furthermore, vascularized human livers were generated in mice by transplanting a mixture of human iPS cell-derived hepatocytes, endothelial cells and mesenchymal stem cells (Takebe et al., 2013). These new strategies further broaden the potential applicability of iPSCs cells in the emerging and rapidly evolving fields of regenerative medicine, disease modeling, and drug development.
Conclusions: On breaking rules
In biology, and particularly in evolution, rules are meant to be broken. Most, if not all evolutionary advances have arisen from the violation of pre-existing rules. The evolution of unicellular to multicellular, sessile to motile, and aquatic to terrestrial animals, for example, required breaking with established norms for new forms of life and lifestyles to emerge. Rule breakers drive the expansion of life into new territories that at first appeared inhospitable. In turn, these organisms will themselves evolve a new cohort of rule breakers that will continue to expand the reach of life into previously uninhabited domains. If breaking rules is as important to the perpetuation of life, as simply following them, Nature must have an abundance of rule breakers in its fold. Hence, it is not difficult to imagine how the small number and almost arbitrary species we have selected to study in depth, may be restricting our understanding of biology to a comparatively narrow set of attributes from which we hope to extract the rules and principles governing complex biological phenomena.
The study of stem cells and potency in the past decade and a half has had important repercussions in our understanding of biology in general and development in particular, and has forced us to rethink what it means for cells and tissues to be differentiated. Not only has this effort ushered new studies of key principles of developmental biology such as the regulation of genomic and epigenomic output during both embryogenesis and adult physiological functions, but it has also provided novel paradigms for the development of therapeutic strategies aimed at addressing a wide gamut of human ailments. It is becoming increasingly clear that the concept of terminal differentiation, while conceptually useful, is not biologically accurate. The accepted rules of differentiation are being broken daily by Nature and it is quite likely that many more such rules may simply be suggestions, and that the concepts of stemness, determination, biological and chronological time among others will ultimately end up being re-examined and redefined.
The concept of stable rather than terminal differentiation should, for example, lead to the re-examination of current strategies aimed at differentiating human pluripotent stem cells. Although the generation of fully functional mature cell types from human pluripotent stem cells has advanced significantly in recent years, this process still remains a major challenge. The idea that the differentiated state of a cell may not be terminal but stable underscores the importance of identifying environmental factors that maintain the stable differentiation cell state, an aspect that has not been widely explored and might offer novel solutions for producing fully functional mature cell types from human pluripotent stem cells. It is our belief, that many more conceptual dogmas will be challenged in the years to come, and that progress in our knowledge of life and the attendant understanding of complex biological systems that should emerge from it will see many more rules being broken by experimental facts. This will likely be driven by the discovery of new paradigms for the study of stem cells, gene regulation, tissue homeostasis and longevity, for example. We predict that 40 years from now biology will likely be a field hardly recognizable from what it is today.
Acknowledgments
ASA would like to acknowledge Ms. Rose Owens, the head librarian at the Stowers Institute for Medical Research for her help in procuring historical material for this article. NIH R37GM057260 to ASA funded some of the work reviewed in this article. ASA is a Howard Hughes Medical Institute and Stowers Institute for Medical Research Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alipio Z, Liao W, Roemer EJ, Waner M, Fink LM, Ward DC, Ma Y. Reversal of hyperglycemia in diabetic mouse models using induced-pluripotent stem (iPS)-derived pancreatic {beta}-like cells. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.1007884107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Birnbaum KD, Sánchez Alvarado A. Slicing across kingdoms: regeneration in plants and animals. Cell. 2008;132:697–710. doi: 10.1016/j.cell.2008.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- Briggs R, King TJ. Transplantation of Living Nuclei From Blastula Cells into Enucleated Frogs’ Eggs. Proceedings of the National Academy of Sciences of the United States of America. 1952;38:455–463. doi: 10.1073/pnas.38.5.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne Harvey EN. The production of new hydranths in Hydra by the insertion of small grafts. Journal of Experimental Zoology. 1909:7. [Google Scholar]
- Buczacki SJ, Zecchini HI, Nicholson AM, Russell R, Vermeulen L, Kemp R, Winton DJ. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- Carla EC, Pagliara P, Piraino S, Boero F, Dini L. Morphological and ultrastructural analysis of Turritopsis nutricula during life cycle reversal. Tissue & cell. 2003;35:213–222. doi: 10.1016/s0040-8166(03)00028-4. [DOI] [PubMed] [Google Scholar]
- Chen CC, Wang IE, Reddien PW. pbx is required for pole and eye regeneration in planarians. Development. 2013;140:719–729. doi: 10.1242/dev.083741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers HC, Bevins CL. Paneth cells: maestros of the small intestinal crypts. Annual review of physiology. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- Colluci VL. Sulla rigenerazione parziale dell’occhio nei Tritoni- Istogenesi e sviluppo. Studio sperimentale. Mem R Acad Sci Ist Bologna. 1891;51:593–629. [Google Scholar]
- Dejosez M, Ura H, Brandt VL, Zwaka TP. Safeguards for cell cooperation in mouse embryogenesis shown by genome-wide cheater screen. Science. 2013;341:1511–1514. doi: 10.1126/science.1241628. [DOI] [PubMed] [Google Scholar]
- Dutrochet H. Recherches anatomiques et physiologiques sur la structure intime des animaux et des végétaux, et sur leur motilité. Paris: Chez J. B. Baillière; 1824. [PMC free article] [PubMed] [Google Scholar]
- Eguchi G, Shingai R. Cellular analysis on localization of lens forming potency in the newt iris epithelium. Development, growth & differentiation. 1971;13:337–349. doi: 10.1111/j.1440-169x.1971.00337.x. [DOI] [PubMed] [Google Scholar]
- Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature. 2011;472:51–56. doi: 10.1038/nature09941. [DOI] [PubMed] [Google Scholar]
- Elliott SA, Sánchez Alvarado A. The history and enduring contributions of planarians to the study of animal regeneration. Wiley interdisciplinary reviews Developmental biology. 2013;2:301–326. doi: 10.1002/wdev.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Gilbert SF. Developmental Biology. 10. Sinauer Associates, Inc; 2013. [Google Scholar]
- Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Abrams RM, Babiarz JE, Cohen JD, Kameoka S, Sanders MJ, Chiao E, Kolaja KL. Estimating the risk of drug-induced proarrhythmia using human induced pluripotent stem cell-derived cardiomyocytes. Toxicol Sci. 2011;123:281–289. doi: 10.1093/toxsci/kfr158. [DOI] [PubMed] [Google Scholar]
- Gurdon JB, Elsdale TR, Fischberg M. Sexually mature individuals of Xenopus laevis from the transplantation of single somatic nuclei. Nature. 1958;182:64–65. doi: 10.1038/182064a0. [DOI] [PubMed] [Google Scholar]
- Gurley KA, Elliott SA, Simakov O, Schmidt HA, Holstein TW, Sánchez Alvarado A. Expression of secreted Wnt pathway components reveals unexpected complexity of the planarian amputation response. Developmental biology. 2010;347:24–39. doi: 10.1016/j.ydbio.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeckel EHP. Zur Entwicklungsgeschichte der Siphonophoren. Utrecht: C. Van Der Post Jr; 1869. [Google Scholar]
- Haeckel EHP. Kunstformen der Natur. Leipzing und Wien; 1904. [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun CW, Meissner A, Cassady JP, Beard C, Brambrink T, Wu LC, Townes TM, et al. Treatment of Sickle Cell Anemia Mouse Model with iPS Cells Generated from Autologous Skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- Hartsoeker N. Essay de Dioptrique. Paris: 1694. [Google Scholar]
- Heath JK. Transcriptional networks and signaling pathways that govern vertebrate intestinal development. Current topics in developmental biology. 2010;90:159–192. doi: 10.1016/S0070-2153(10)90004-5. [DOI] [PubMed] [Google Scholar]
- Heneidi S, Simerman AA, Keller E, Singh P, Li X, Dumesic DA, Chazenbalk G. Awakened by cellular stress: isolation and characterization of a novel population of pluripotent stem cells derived from human adipose tissue. PloS one. 2013;8:e64752. doi: 10.1371/journal.pone.0064752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtfreter J. Neural induction in explants which have passed through a sublethal cytolysis. The Journal of experimental zoology. 1947;106:197–222. doi: 10.1002/jez.1401060205. [DOI] [PubMed] [Google Scholar]
- Hou P, Li Y, Zhang X, Liu C, Guan J, Li H, Zhao T, Ye J, Yang W, Liu K, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H, Nagata N, Kurokawa H, Yamanaka S. iPS cells: a game changer for future medicine. The EMBO journal. 2014 doi: 10.1002/embj.201387098. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- King RS, Newmark PA. The cell biology of regeneration. The Journal of cell biology. 2012;196:553–562. doi: 10.1083/jcb.201105099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleier C, Rundel PW. Microsite requirements, population structure and growth of the cushion plant Azorella compacta in the tropical Chilean Andes. Austral Ecology. 2004;29:461–470. [Google Scholar]
- Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell stem cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Kozar S, Morrissey E, Nicholson AM, van der Heijden M, Zecchini HI, Kemp R, Tavare S, Vermeulen L, Winton DJ. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell stem cell. 2013;13:626–633. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Kitada M, Wakao S, Nishikawa K, Tanimura Y, Makinoshima H, Goda M, Akashi H, Inutsuka A, Niwa A, et al. Unique multipotent cells in adult human mesenchymal cell populations. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:8639–8643. doi: 10.1073/pnas.0911647107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird DJ, De Tomaso AW, Weissman IL. Stem cells are units of natural selection in a colonial ascidian. Cell. 2005;123:1351–1360. doi: 10.1016/j.cell.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Laird DJ, Weissman IL. Telomerase maintained in self-renewing tissues during serial regeneration of the urochordate Botryllus schlosseri. Developmental biology. 2004;273:185–194. doi: 10.1016/j.ydbio.2004.05.029. [DOI] [PubMed] [Google Scholar]
- Lapan SW, Reddien PW. Transcriptome analysis of the planarian eye identifies ovo as a specific regulator of eye regeneration. Cell reports. 2012;2:294–307. doi: 10.1016/j.celrep.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauzon RJ, Ishizuka KJ, Weissman IL. A cyclical, developmentally-regulated death phenomenon in a colonial urochordate. Developmental dynamics: an official publication of the American Association of Anatomists. 1992;194:71–83. doi: 10.1002/aja.1001940109. [DOI] [PubMed] [Google Scholar]
- Lauzon RJ, Ishizuka KJ, Weissman IL. Cyclical generation and degeneration of organs in a colonial urochordate involves crosstalk between old and new: a model for development and regeneration. Developmental biology. 2002;249:333–348. doi: 10.1006/dbio.2002.0772. [DOI] [PubMed] [Google Scholar]
- Lewis WH. Experimental studies on the development of the eye in amphibia. I. On the origin of the lens. Rana palustris. Am J Anat. 1904;III:13–15. [Google Scholar]
- Li W, Wei W, Zhu S, Zhu J, Shi Y, Lin T, Hao E, Hayek A, Deng H, Ding S. Generation of rat and human induced pluripotent stem cells by combining genetic reprogramming and chemical inhibitors. Cell stem cell. 2009;4:16–19. doi: 10.1016/j.stem.2008.11.014. [DOI] [PubMed] [Google Scholar]
- MacArthur BD, Lemischka IR. Statistical mechanics of pluripotency. Cell. 2013;154:484–489. doi: 10.1016/j.cell.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Mangold HP, Spemann H. Uber Induktion von Embryonalanlagen durch Implantation artfremder Organisatoren. Archiv für Mikroskopische Anatomie und Entwicklungsmechanik. 1924:599–638. [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medine CN, Lucendo-Villarin B, Storck C, Wang F, Szkolnicka D, Khan F, Pernagallo S, Black JR, Marriage HM, Ross JA, et al. Developing high-fidelity hepatotoxicity models from pluripotent stem cells. Stem cells translational medicine. 2013;2:505–509. doi: 10.5966/sctm.2012-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldenhawer JJP. Beyträge zur Anatomie der Pflanzen. Kiel: C. L. Wäser; 1812. [Google Scholar]
- Montgomery RK, Carlone DL, Richmond CA, Farilla L, Kranendonk ME, Henderson DE, Baffour-Awuah NY, Ambruzs DM, Fogli LK, Algra S, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz J, Stange DE, Schepers AG, van de Wetering M, Koo BK, Itzkovitz S, Volckmann R, Kung KS, Koster J, Radulescu S, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. The EMBO journal. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obokata H, Kojima K, Westerman K, Yamato M, Okano T, Tsuneda S, Vacanti CA. The potential of stem cells in adult tissues representative of the three germ layers. Tissue engineering Part A. 2011;17:607–615. doi: 10.1089/ten.TEA.2010.0385. [DOI] [PubMed] [Google Scholar]
- Obokata H, Sasai Y, Niwa H, Kadota M, Andrabi M, Takata N, Tokoro M, Terashita Y, Yonemura S, Vacanti CA, et al. Bidirectional developmental potential in reprogrammed cells with acquired pluripotency. Nature. 2014a;505:676–680. doi: 10.1038/nature12969. [DOI] [PubMed] [Google Scholar]
- Obokata H, Wakayama T, Sasai Y, Kojima K, Vacanti MP, Niwa H, Yamato M, Vacanti CA. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature. 2014b;505:641–647. doi: 10.1038/nature12968. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, Spradling A. Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science. 2007;315:988–992. doi: 10.1126/science.1136606. [DOI] [PubMed] [Google Scholar]
- Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- Pellettieri J, Sánchez Alvarado A. Cell turnover and adult tissue homeostasis: from humans to planarians. Annual review of genetics. 2007;41:83–105. doi: 10.1146/annurev.genet.41.110306.130244. [DOI] [PubMed] [Google Scholar]
- Pinto D, Clevers H. Wnt, stem cells and cancer in the intestine. Biology of the cell/under the auspices of the European Cell Biology Organization. 2005;97:185–196. doi: 10.1042/BC20040094. [DOI] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes & development. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piraino S, Boero F, Aeschbach B, Schmid V. Reversing the life cycle: medusae transforming into polyps and cell transdifferentiation in Turritopsis nutricula (Cnidaria, Hydrozoa) The Biological bulletin. 1996;190:303–312. doi: 10.2307/1543022. [DOI] [PubMed] [Google Scholar]
- Piraino S, De Vito D, SCHMICH J, Bouillon J, Boero F. Reverse development in Cnidaria. Can J Zool. 2004;82:1748–1754. [Google Scholar]
- Plutarch (75 CE) Plutarch’s Lives (The translation called Dryden’s) 1859. Boston, Little: Brown and Company; [Google Scholar]
- Reddien PW. Constitutive gene expression and the specification of tissue identity in adult planarian biology. Trends in genetics: TIG. 2011;27:277–285. doi: 10.1016/j.tig.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW. Specialized progenitors and regeneration. Development. 2013;140:951–957. doi: 10.1242/dev.080499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien PW, Bermange AL, Kicza AM, Sanchez Alvarado A. BMP signaling regulates the dorsal planarian midline and is needed for asymmetric regeneration. Development. 2007;134:4043–4051. doi: 10.1242/dev.007138. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131:4225–4237. doi: 10.1242/dev.01261. [DOI] [PubMed] [Google Scholar]
- Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- Reyer RW. Morphological evidence for lens differentiation from intra-ocular implants of lens epithelium in Ambystoma maculatum. Experimental eye research. 1977;24:511–522. doi: 10.1016/0014-4835(77)90272-x. [DOI] [PubMed] [Google Scholar]
- Reyes M, Verfaillie CM. Characterization of multipotent adult progenitor cells, a subpopulation of mesenchymal stem cells. Annals of the New York Academy of Sciences. 2001;938:231–233. doi: 10.1111/j.1749-6632.2001.tb03593.x. discussion 233–235. [DOI] [PubMed] [Google Scholar]
- Richards RJ. The Tragic Sense of Life: Ernst Haeckel and the Struggle over Evolutionary Thought. Chicago: The University of Chicago Press; 2008. [Google Scholar]
- Rinkevich B, Lauzon RJ, Brown BW, Weissman IL. Evidence for a programmed life span in a colonial protochordate. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:3546–3550. doi: 10.1073/pnas.89.8.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkevich Y, Voskoboynik A, Rosner A, Rabinowitz C, Paz G, Oren M, Douek J, Alfassi G, Moiseeva E, Ishizuka KJ, et al. Repeated, long-term cycling of putative stem cells between niches in a basal chordate. Developmental cell. 2013;24:76–88. doi: 10.1016/j.devcel.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Gascard P, Dumont N, Zhao J, Pan D, Petrie S, Margeta M, Tlsty TD. Rare somatic cells from human breast tissue exhibit extensive lineage plasticity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:4598–4603. doi: 10.1073/pnas.1218682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez Alvarado A, Tsonis PA. Bridging the regeneration gap: genetic insights from diverse animal models. Nature reviews Genetics. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- Schmich J, Kraus Y, De Vito D, Graziussi D, Boero F, Piraino S. Induction of reverse development in two marine Hydrozoans. The International journal of developmental biology. 2007;51:45–56. doi: 10.1387/ijdb.062152js. [DOI] [PubMed] [Google Scholar]
- Scimone ML, Lapan SW, Reddien PW. A forkhead Transcription Factor Is Wound-Induced at the Planarian Midline and Required for Anterior Pole Regeneration. PLoS genetics. 2014;10:e1003999. doi: 10.1371/journal.pgen.1003999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scimone ML, Srivastava M, Bell GW, Reddien PW. A regulatory program for excretory system regeneration in planarians. Development. 2011;138:4387–4398. doi: 10.1242/dev.068098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- Spalding KL, Bhardwaj RD, Buchholz BA, Druid H, Frisen J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Spemann H. Uber Korrelationen in der Entwicklung des Auges. Verh Anat Vers Ges. 1901;15:61–79. [Google Scholar]
- Spemann H. Embryonic Development and Induction. New Haven: Yale University Press; 1938. [Google Scholar]
- Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, Takata N, Wataya T, Muguruma K, Miyoshi H, et al. Self-formation of functional adenohypophysis in three-dimensional culture. Nature. 2011;480:57–62. doi: 10.1038/nature10637. [DOI] [PubMed] [Google Scholar]
- Sugimoto K, Gordon SP, Meyerowitz EM. Regeneration in plants and animals: dedifferentiation, transdifferentiation, or just differentiation? Trends in cell biology. 2011;21:212–218. doi: 10.1016/j.tcb.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takayama N, Nishimura S, Nakamura S, Shimizu T, Ohnishi R, Endo H, Yamaguchi T, Otsu M, Nishimura K, Nakanishi M, et al. Transient activation of c-MYC expression is critical for efficient platelet generation from human induced pluripotent stem cells. J Exp Med. 2010 doi: 10.1084/jem.20100844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang RR, Ueno Y, Zheng YW, Koike N, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng PF, Weintraub H, Lassar AB. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 1988;242:405–411. doi: 10.1126/science.3175662. [DOI] [PubMed] [Google Scholar]
- Tian H, Biehs B, Warming S, Leong KG, Rangell L, Klein OD, de Sauvage FJ. A reserve stem cell population in small intestine renders Lgr5-positive cells dispensable. Nature. 2011;478:255–259. doi: 10.1038/nature10408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treviranus LC. Beyträge zur Pflanzenphysiologie. Göttingen: Heinrich Dietrich; 1811. [Google Scholar]
- Tsuji O, Miura K, Okada Y, Fujiyoshi K, Mukaino M, Nagoshi N, Kitamura K, Kumagai G, Nishino M, Tomisato S, et al. Therapeutic potential of appropriately evaluated safe-induced pluripotent stem cells for spinal cord injury. Proc Natl Acad Sci U S A. 2010;107:12704–12709. doi: 10.1073/pnas.0910106107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskoboynik A, Simon-Blecher N, Soen Y, Rinkevich B, De Tomaso AW, Ishizuka KJ, Weissman IL. Striving for normality: whole body regeneration through a series of abnormal generations. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2007;21:1335–1344. doi: 10.1096/fj.06-7337com. [DOI] [PubMed] [Google Scholar]
- Voskoboynik A, Soen Y, Rinkevich Y, Rosner A, Ueno H, Reshef R, Ishizuka KJ, Palmeri KJ, Moiseeva E, Rinkevich B, et al. Identification of the endostyle as a stem cell niche in a colonial chordate. Cell stem cell. 2008;3:456–464. doi: 10.1016/j.stem.2008.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DE, Wang IE, Reddien PW. Clonogenic neoblasts are pluripotent adult stem cells that underlie planarian regeneration. Science. 2011;332:811–816. doi: 10.1126/science.1203983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, Miller AD. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A. The germ-plasm: a theory of heredity. New York, New York: Charles Scribne’rs Sons; 1893. [Google Scholar]
- Wernig M, Zhao JP, Pruszak J, Hedlund E, Fu D, Soldner F, Broccoli V, Constantine-Paton M, Isacson O, Jaenisch R. Neurons derived from reprogrammed fibroblasts functionally integrate into the fetal brain and improve symptoms of rats with Parkinson’s disease. Proc Natl Acad Sci U S A. 2008;105:5856–5861. doi: 10.1073/pnas.0801677105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Global data on visual impairment 2010. World Health Organization; 2012. http://www.who.int/blindness/GLOBALDATAFINALforweb.pdf. [Google Scholar]
- Wolff G. Entwicklungsphysiologische Studien. I. Die Regeneration der Urodelenlinse. Wilhelm Roux Arch Entwickl-Mech Org. 1895;1:380–390. [Google Scholar]
- Yamada T. Control mechanisms in cell-type conversion in newt lens regeneration. Monographs in developmental biology. 1977;13:1–126. [PubMed] [Google Scholar]
- Yan KS, Chia LA, Li X, Ootani A, Su J, Lee JY, Su N, Luo Y, Heilshorn SC, Amieva MR, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]