Abstract
Murine norovirus (MNV) is a positive-sense, plus-stranded RNA virus in the Caliciviridae family. It is the most common pathogen in biomedical research colonies. MNV is also related to the human noroviruses, which cause the majority of non-bacterial gastroenteritis worldwide. Like the human noroviruses, MNV is an enteric virus that replicates in the intestine and is transmitted by the fecal-oral route. MNV replicates in murine macrophages and dendritic cells in cells in culture and in the murine host. This virus is often used to study mechanisms in norovirus biology, because the human noroviruses are refractory to growth in cell culture. MNV combines the availability of a cell culture and reverse genetics system with the ability to study infection in the native host. Herein, we describe a panel of techniques that are commonly used to study MNV biology.
Keywords: murine norovirus, purification, quantification, reverse genetics, transfection
Murine norovirus (MNV) is a positive-sense, plus-stranded RNA virus in the Caliciviridae family. It is the most common pathogen in biomedical research colonies. MNV is also related to the human noroviruses, which cause the majority of non-bacterial gastroenteritis worldwide. Like the human noroviruses, MNV is an enteric virus that replicates in the intestine and is transmitted by the fecal-oral route. MNV replicates in murine macrophages and dendritic cells in cells in culture and in the murine host. This virus is often used to study mechanisms in norovirus biology, because the human noroviruses are refractory to growth in cell culture. MNV combines the availability of a cell culture and reverse genetics system with the ability to study infection in the native host. Herein, we describe a panel of techniques that are commonly used to study MNV biology.
INTRODUCTION
Murine norovirus (MNV) is a small non-enveloped virus with a plus-sense RNA genome of ~7.5 kb in length. MNV is a member of the calicivirus family, the norovirus genus, and all strains isolated to date are exclusively found in norovirus genogroup V (Green 2007). MNV is highly abundant in research mice (e.g. (Hsu, Wobus et al. 2005, Kitajima, Oka et al. 2009, Mahler and Kohl 2009)). MNV-1 was originally isolated from immunocompromised mice (Karst, Wobus et al. 2003) but later shown to infect wild-type mice (Mumphrey, Changotra et al. 2007, Chachu, Strong et al. 2008). Many different strains of MNV have been isolated from wild-type or genetically modified mice in biomedical research colonies (e.g.,(Thackray, Wobus et al. 2007)). MNV has also been detected in wild rodents (Smith, McFadden et al. 2012, Tsunesumi, Sato et al. 2012). It is the only norovirus that efficiently grows in tissue culture (in macrophages and dendritic cells) and in a small animal host (Karst, Wobus et al. 2003, Wobus, Karst et al. 2004, Wobus, Thackray et al. 2006). Many biological features, including fecal-oral transmission, replication in the intestine, and fecal shedding are shared between murine and human noroviruses (Wobus, Thackray et al. 2006). Therefore, MNV is often used as a model to study norovirus biology.
The following protocols describe a variety of methods typically used to analyze different aspects of MNV biology. The protocols begin with a description of how to generate viral stocks and purify MNV. This is followed by a method to measure anti-MNV antibodies in sera of mice to verify whether mice in biomedical research colonies are seronegative prior to their use in experiments. Next, three different protocols to generate MNV mutants are described, followed by measuring viral titers either by detection of infectious particles or genome. The unit ends with protocols describing several methods to modulate a host gene of interest in a variety of cell lines or primary cells to study its effect on MNV infection.
CAUTION: MNV is a Biosafety Level 2 (BSL-2) pathogen in some countries (e.g., USA). Follow all appropriate guidelines and regulations for the use and handling of pathogenic microorganisms.
BASIC PROTOCOL 1
GENERATION OF MURINE NOROVIRUS-CONTAINING CELL LYSATE
This procedure outlines the making of a MNV-containing cell lysate (hereafter referred to as regular MNV stock). We describe the generation of an MNV-1 stock by infecting RAW 264.7 cells. However, this protocol can be used with other MNV strains and other cell lines that support viral replication and yield high viral titer, such as SRDC or BV-2 cell lines (Blasi, Barluzzi et al. 1990, Ruiz, Beauvillain et al. 2005). The regular MNV stock is useful for a wide range of applications, such as virus concentration and purification (See Support Protocols 1 and 2). Depending on the MNV strain, viral titers of 106 − 107 pfu/ml are routinely obtained after 2 days of infection.
Materials
175 cm2 tissue culture-treated flasks
37°C/5% CO2 tissue culture incubator
Cell scraper (e.g., Sarstedt – 39 cm)
RAW 264.7 cells (ATCC catalog no. TIB-71)
complete DMEM-10 medium (see recipe)
MNV-1 (or other strains of interest)
Sterile, disposable plastic tubes for storing the lysate and aliquots
10% bleach (e.g., Clorox)
−80°C freezer
Culturing of RAW 264.7 cells for MNV-1 expansion
Scrape RAW 264.7 cells from a confluent 175 cm2 flask.
Resuspend RAW 264.7 cells in fresh DMEM-10 medium and make a single cell suspension.
- Seed cells at a density of 4 × 107 cells/T175 flask in 30 ml of DMEM-10.This is roughly a 1:3 dilution of a confluent flask. Be sure to resuspend the RAW 264.7 cells well to obtain a single cell suspension.
RAW 264.7 cells infection
Infect cells with MNV-1 at MOI = 0.05 by pipetting virus directly into the medium using a micropipette. Carefully mix virus with media after flask has been closed.
Incubate in tissue culture incubator at 37°C and 5% CO2 for 40 – 48 h.
MNV-1 harvest and storage
- Check cells for the presence of cytopathic effect (CPE).CPE can be easily observed, since the cell monolayer will be detaching from the flask. If most cells are still attached, incubate for a few more hours.
Dislodge any remaining attached cells by hitting the side of the flask.
Pool contents from all flasks and transfer cell lysate containing virus into sterile container.
Freeze it at −80°C, then thaw lysate.
Repeat step 4.
- Aliquot lysate and store at −80°C.Frozen lysate is stable for months to years at −80°C. However, avoid repeated freeze/thaw of virus stocks (i.e., more than 5).
- Determine viral titers by TCID50 (Basic Protocol 5) or plaque assay (Alternate Protocol 3)It is best to titer MNV-1 regular stock at least three times independently before calculating the final virus titer of the stock.
SUPPORT PROTOCOL 1
GENERATION OF A CONCENTRATED MURINE NOROVIRUS STOCK
This procedure describes the process of generating a concentrated MNV stock from a virus lysate (see BASIC PROTOCOL 1). The concentrated MNV stock is used for techniques that require higher virus titers typically obtained by growing MNV in RAW 264.7 cells (i.e., 106 −107 PFU/ml). The MNV concentrated stock is particularly important for animal challenge experiments, due to the small size of the host and the intrinsic limitations on the volume that can be administered to mice by the different infection routes. Typically, one can expect a two log increase over the MNV lysate. So if the MNV regular stock titer is 107 PFU/ml, one can expect an MNV concentrated stock titer around 109 PFU/ml.
Materials
MNV stock (see BASIC PROTOCOL 1)
Refrigerated tabletop tissue culture centrifuge (e.g., Sorvall Legend RT)
Ultra-centrifuge (e.g., Sorvall WX Ultra 80)
Rotor and buckets for the appropriate volume (e.g., Sorvall AH-629)
36 ml clear ultra-centrifuge tubes (e.g., Beckman-Coulter product no. 344058)
PBS pH 7.4 (1×) sterile (e.g., Gibco ref no.10010-023) (here after referred to as tissue culture PBS)
0.22 µm sterile-filter (e.g., Millipore Stericup Express Plus system)
sterile-filtered 30% (w/v) sucrose solution in tissue culture PBS
Thaw MNV stock and break apart cell debris by vigorously vortexing the virus-containing lysate.
Pellet large cell debris by centrifugation in tabletop tissue culture centrifuge at 2000 g for 20 min at 4°C.
Pool supernatants.
Add 5 ml each of the sterile-filtered 30% sucrose solution to the bottom of 6 ultra-clear centrifuge tubes.
- Very carefully overlay lysate (~30 ml) onto sucrose cushion.It is imperative to add lysate very slowly onto the sucrose cushion to minimally disturb it. The ultra-centrifuge tubes should be full in order to minimize protocol duration and prevent cracking of the tubes (leave around 0.5 cm free to prevent spilling).
Balance all tubes and load into ultra-centrifuge rotor.
Centrifuge in AH-629 rotor at 95,000 g for 3 h at 4°C.
Discard supernatant.
- Repeat steps 4 to 8 until all lysate has been pelleted.We use the same tubes twice in order to minimize the volume and thus increase virus titers per volume.
Add 200 µl of tissue culture PBS per tube to resuspend pellet.
- Seal tubes (e.g., Parafilm) and incubate overnight at 4°C.The incubation period is necessary to allow pellet to loosen and to be easily resuspended.
Pool the contents of all tubes.
Rinse a set of 6 ultra-centrifuge tubes with 500 µl of tissue culture PBS and combine with the previously pooled concentrated MNV stock (from step no. 12)
Aliquot concentrated MNV stock into 2 ml screw cap tubes and store at −80°C.
Determine viral titer by TCID50 or plaque assay (see Basic Protocol 5 or Alternate Protocol 3).
SUPPORT PROTOCOL 2
PURIFICATION OF MNV BY CESIUM CHLORIDE DENSITY GRADIENT CENTRIFUGATION
This protocol describes the purification of MNV particles from cellular debris based on differential density of virions and cellular proteins/protein complexes. Several different protocols have been published for cesium chloride purification of caliciviruses (e.g., (Madore, Treanor et al. 1986, Parwani, Saif et al. 1990)). They mainly differ in the way the virus is released from cells, the method for concentration of virus particles prior to purification, and the number of density gradients performed. The protocol described below is a very basic one that includes relatively few steps. It is designed for the purification of approximately 180 ml of MNV-containing lysate (See BASIC PROTOCOL 1). For purification of MNV from larger volumes see alternate protocol 1. Keep virus-containing solutions cold whenever possible.
Materials
30% (w/v) sucrose solution in PBS (pH 7. 4) (filtered through a 0.22 µm filter)
Cesium chloride
PBS (pH 7.4)
Refrigerated ultracentrifuge and Beckmann rotors SW32 and SW55
Ultra-clear centrifuge tubes for SW32 (Beckman Coulter cat. No. 344058) and SW55 rotors (cat. No. 344057)
22G needles
Dialysis tubing, dialysis clips
Refractometer
Stir plate in cold room with beaker and stir bar
- Take flasks with MNV-containing cell lysate (see BASIC PROTOCOL 1).This protocol is for approx. 180 ml lysate from around 6× 175 cm2 flasks, with 30 ml medium in each flask.
Freeze/thaw flasks 3 times.
Break apart cell debris by vortexing flasks containing cell lysate. Pool all lysates.
Centrifuge lysate at 3000 rpm for 20 minutes at 4°C in a tabletop tissue culture centrifuge (e.g., Sorvall) to pellet large cellular debris. Save supernatant.
- Place 5 ml of 30% sucrose solution in bottom of a SW32 Beckman centrifuge tube. Very slowly layer 30 ml supernatant on top of 30% sucrose cushion in SW32 rotor.Do not mix the solutions. The clear sucrose solution and the phenol-red containing lysate should be clearly separated.
- Repeat step 5 for the other 5 centrifuge tubes.Prepare a total of 6 tubes, one for each bucket of the SW32 rotor.
- Centrifuge at 27,000 rpm for 3.0 h at 4°C.Make sure all tubes are balanced and have the same weight before starting the ultracentrifuge. Use remaining cell lysate or PBS to balance tubes.
- Discard the supernatant. Place tube upside down on paper towel to drain remaining solution.A slightly yellowish/brownish pellet should be visible at the bottom of each tube.
- Suspend the viral pellets from 3 tubes in 1 ml PBS. Rinse all tubes with 0.5 ml PBS. Pool all supernatants into a final volume of 2.5 ml.Make sure pellets are completely dissolved. You may have to incubate the pellet with PBS for several hours at room temperature or at 4C overnight. If chunks remain, vortex the pooled supernatant vigorously and pipette up and down to completely dissolve pellets. This will increase yields. In addition, rinse tubes with PBS to prevent excess loss of virus.
Suspend 1.9 g CsCl in 2ml PBS.
- Mix dissolved pellet with CsCl solution. Check the refractive index (RFI) and adjust to 1.365-1.367 if necessary by adding CsCl salt to increase density of the solution. Add solution to SW55 centrifuge tube.Make sure to prepare a balance tube of identical weight for the ultracentrifuge.
Centrifuge at 35,000 rpm overnight in SW55 Ti rotor (18–24hrs).
Fractionate gradient. Wipe bottom of tube with 95% ethanol, drill a hole in the bottom of the tube with a 22G needle, and take 0.5 – 1 ml fractions.
Determine RFI.
Dialyze fractions with RFI of 1.362–1.373 against 2 changes of 2L PBS overnight at 4°C.
Analyze gradient fractions by running 5 µl from each fraction on a SDS-PAGE gel and Coomassie stain to check the purity. The capsid protein should be visible around 58 kDa.
ALTERNATE PROTOCOL 1
PURIFICATION OF MNV BY CESIUM CHLORIDE DENSITY GRADIENT CENTRIFUGATION (LARGE SCALE)
This protocol is designed for use with 1 L of MNV-containing cell lysate (see Basic Protocol 1 on how to generate the virus stock). This protocol can be adjusted to accommodate even larger volumes of virus lysate. This protocol also includes a solvent extraction step to separate virus from cellular debris. Keep virus-containing solutions cold whenever possible.
Materials
NaCl
PEG 8000
PBS (pH 7.4)
Vertrel XF (DuPont)
CsCl
22G needles
refractometer
Sonicator with sonicator tip (e.g., Fisher Scientific, Sonic Dismembrator model 550)
Stir plate in cold room with beaker and stir bar
Mid-speed centrifuge with SLA-3000 rotor and buckets
Beckman Coulter Ultracentrifuge with SW32 rotor and tubes
Dialysis tubing, dialysis clips
Virus concentration with PEG 8000
Thaw 1L of MNV-containing cell lysate (see Basic Protocol 1).
Freeze-thaw MNV-containing cell lysate two times.
Clarify supernatants by centrifuging them at 6000 rpm for 30 min at 4°C in a SLA-3000 rotor. Save supernatants and discard pellets.
- Determine the total volume of supernatant to be concentrated. Bring the NaCl concentration to 1M.DMEM contains 6.8 g/liter NaCl, so add 51.6 g NaCl/liter of fluid to be concentrated.
Add PEG 8000 to a concentration of 8% weight to volume.
Stir fluid at 4°C overnight.
Centrifuge fluid at 10,000 rpm at 4°C in SLA-3000 rotor for 30 minutes.
Discard supernatant.
Solvent Extraction
Resuspend the pellet in 20 ml PBS. Split sample in half. Transfer to 50 ml conical tube.
Sonicate for 30 sec at 4.0.
Add an equal volume of Vertrel XF solvent, a non-hazardous freon substitute.
Sonicate with microtip at 4.0 for about one minute to make an emulsion.
Centrifuge 3000 rpm for 10 min. You should see a top opalescent aqueous phase, an interphase (white) and a lower organic phase.
Take the aqueous phase and repeat the solvent extraction one time.
Store on ice until gradient is ready.
CsCl gradient purification
Make a step gradient by placing 10 ml of 1.45g/cm3 CsCl in bottom of centrifuge tube. Then very gently overlay with 10ml of 1.35g/cm3 CsCl.
- Gently overlay ~18 ml virus-containing solution onto the gradient.Prepare another tube of same weight for balance.
Spin 48 hours at 27,000 rpm in the SW32 rotor.
Wipe bottom of tube with 95% ethanol, drill a hole in the bottom of the tube with a 22G needle, and take 1 ml fractions.
Determine refractive index (RFI) of each fraction.
Dialyze fractions with RFI ~1.362 – 1.373 against 2 changes of 2L PBS overnight at 4°C.
Analyze fractions by Coomassie staining. The capsid protein is ~58kDa and should be clearly visible.
BASIC PROTOCOL 2
MURINE NOROVIRUS ENZYME-LINKED IMMUNOSORBENT ASSAY
The purpose of an enzyme-linked immunosorbent assay (ELISA) is to detect and quantify the presence of proteins, antibodies and some other substances in a sample. The antigen is immobilized to the surface of an ELISA well and then complexed with an antibody. The antibody can be directly conjugated to an enzyme or a secondary antibody conjugated to an enzyme can be used. The enzyme typically catalyzes a reaction that produces a measurable product, most commonly color change, to measure enzyme activity. The purpose of the MNV enzyme-linked immunosorbent assay (ELISA) is to detect and quantify the presence of anti-MNV specific antibodies in a sample (e.g., mouse serum). Positive samples (i.e., containing MNV antibodies) will turn green during the development time, while negative samples remain clear.
Materials
Concentrated virus (see Support Protocol 1)
Immulon II HB flat-bottomed ELISA plate (Thermo Labsystems)
goat-anti-mouse-HRP, Jackson Immunoresearch, Cat. #115-035-003
ABTS (2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid)
Hydrogen peroxide
Repeater pipette (e.g., Eppendorf)
Multichannel pipette
ELISA Coating Buffer (see recipe)
ELISA III Buffer (see recipe)
ELISA Substrate Buffer (see recipe)
ELISA Wash Buffer (see recipe)
Blocking Buffer (see recipe)
Stop Buffer (see recipe)
Plate reader
NOTE: Perform Steps 1 – 8 in a biosafety hood.
-
1Calculate the amount of wells and final volume of virus needed to coat plate. For each well, dilute 2–5 µl of concentrated virus in phosphate-buffered saline (PBS) in a final volume of 100 µl.The optimal concentration of virus for coating per well should be determined in a pilot experiment. We typically coat with 2 µl of concentrated MNV/well.
-
2
Use repeater pipette to distribute 100 µl of diluted virus solution/well into a 96-well ELISA plate, and then incubate overnight at 4°C covered with aluminum foil.
-
3
On the next day, flick out virus solution into a biohazard bag and then wash ELISA plate once with 1× wash buffer by submerging plate, tapping out any air bubbles.
-
4
Flick out buffer into a biohazard bag and then blot on a paper towel.
-
5
Use a multichannel pipette to add 200 µl blocking buffer to each well, and then incubate plate at 37°C for 2 h covered with parafilm.
-
6While plate is blocking, use ELISA III buffer to make serum dilutions.For screening of sera, use a 1:100 dilution analyzed in duplicates Don’t forget to include a known negative and positive serum.
-
7Flick out blocking buffer into a biohazard bag and then wash plates 2 times with 1× ELISA wash buffer (flicking out buffer into biohazard bag between washes).Do not let plate dry.
-
8
Add 100 µl of the serum dilution to each well, and then incubate plate for 60 min at 37°C covered in parafilm.
-
9
Dilute secondary antibody (goat-anti-mouse-HRP) 1:2000 in ELISA III buffer.
-
10Flick out serum dilutions into a biohazard bag and then wash plates 4 times with 1× ELISA wash buffer (flicking out buffer into biohazard bag between washes).Do not let plate dry.
The following steps can be carried out on the bench
-
11
Use a multichannel pipette to add 100 µl of secondary antibody per well, and then incubate plate for 1 hour at 37°C covered in parafilm.
-
12
At the beginning of the incubation periods, thaw one 12 ml aliquot of ELISA Substrate Buffer per plate at room temperature in the dark.
-
13Flick out antibody dilutions and then wash plates 4 times with 1× ELISA wash buffer.Do not let plate dry.
-
14
Make the substrate (ABTS) by adding 1 µl of hydrogen peroxide to every 1 ml of ABTS solution immediately prior to adding it to well.
-
15
Use multichannel pipette to add 100 µl ul of ELISA Substrate Buffer to each well and incubate for up to 10 min at room temperature.
-
16
Stop the assay when your negative control begins to turn color or after 10 min (whichever comes first) by adding 100 µl phosphoric acid to each well.
-
17Measure absorption at 415 nm on ELISA reader.You can use 405nm if 415 nm is absent. Numbers are expressed as optical density (OD). Compare the OD value of your sample to the positive and negative control values for data interpretation.
BASIC PROTOCOL 3
DNA-BASED, POL II-DRIVEN GENERATION OF RECOMBINANT MURINE NOROVIRUS
Reverse genetics is a powerful tool used to recover a genetically defined recombinant virus from a cDNA clone and can be utilized to examine the effect of mutations on the virus life cycle in vitro and in vivo. Several reverse genetics systems have been described (see also Basic Protocol 4 and Alternate Protocol 2). This protocol is based on a publication by Ward et al. (Ward, McCormick et al. 2007). A DNA polymerase II promoter drives expression of viral cDNA after transfection of a DNA plasmid containing the full-length MNV genome into HEK293T cells. This usually yields about 103 PFU/ml of MNV. The recombinant virus is subsequently amplified in RAW264.7 cells to yield between 106and 107 PFU/ml of MNV in RAW264.7 cells.
Materials
Full-length MNV-containing plasmid (pMNV*)
EGFP-expressing plasmid
Transfection reagent (Fugene HD; Promega)
Tissue culture medium (DMEM-10, see recipe)
OptiMEM I (Invitrogen)
293T cells (low passage number works better)
RAW264.7 cells (if viruses are going to be passaged to RAW cells)
All steps are carried out at room temperature. Media should be warmed to 37°C or equilibrated to room temperature.
Transfect 293T cells with MNV plasmid
- Dilute 1.5 µg MNV plasmid in 94 µl OptiMEM in a 1.5 ml centrifuge tube.Include the EGFP-expressing plasmid as a control for transfection efficiency.
- Add 6 µl Fugene HD to the OptiMEM-plasmid mix and briefly vortex (for about 2 seconds) on high. Spin down transfection mix for 10 sec at 8,000×g in a centrifuge.The ratio of plasmid to Fugene HD is usually 1:4 but you can try other ratios to determine the best combination for your plasmid. Fugene HD sticks to plastics so avoid touching the plastic wall of the tube by pipetting directly into the solution. Vortex Fugene HD before use, take it directly from the fridge and put it back after pipetting Warming Fugene HD is not necessary.
- Incubate solutions for 15 min at room temperature.Do not to exceed 25 min If you are transfecting many plasmids, make sure you can proceed with the first sample after a 15 min incubation.
- In the meantime, harvest 293T cells. Aspirate the media and add 10 ml of fresh culture media. Dislodge the cells by hitting the flask. Centrifuge cells at 200 × g for 5 min, remove the supernatant, and resuspend cells at 1×107 cells/ml in culture medium.Healthy 293T cells are easily dislodged from the flask. If the cells have a problem dislodging then you may want to use a fresh batch. The effect of trypsin on 293T is not yet established so you should stay away from it during this step.
Add 100 µl of the cell suspension (1×106 cells) to the centrifuge tube containing transfection mix and incubate for 5–10 min.
Label T-75 flasks and add 10 ml of culture media to each flask.
- Transfer transfection/cells-mix into T-75 flask. Rinse centrifuge tube with 1 ml media from the prepared T-75 flask and add to flask. Incubate for at least 48 h at 37°C with 5% CO2.If incubation time will be 72–96 h, replace media on 293Ts with 10 ml culture media the next day. It is OK for 293T culture medium to have antibiotics at all times, however OptiMEM used for transfection reaction mix should NOT have added serum or antibiotics.
Passage onto RAW264.7 cells (liquid passage)
Freeze 293T cells at −80 ºC and thaw the frozen cells at 37 °C.
Repeat step 1.
Transfer lysate into 15 ml screw cap tube.
Spin at 2,500×g for 5 min at 4 °C.
Infect RAW264.7 cells in T175 flask (plated at 4 × 107cells/flask the previous day) with 5 ml of cleared 293T lysate and 25 ml culture media.
Incubate for at least 48 h at 37°C with 5% CO2 or until cytopathic effect is seen.
Harvest recombinant viruses
Freeze RAW264.7 cells at −80 °C and thaw the frozen cells at 37 °C.
Repeat step 1.
Transfer lysate into 50 ml Falcon tubes.
Spin at 2,500×g for 5 min at 4 °C.
Save supernatant. For short-term storage (1–2 days) store at 4 °C. Otherwise, freeze at −80°C.
Determine titer of recombinant virus
Use plaque assay (see Alternate Protocol 3) or TCID50 (see Basic Protocol 5) to confirm the presence of viruses both in the cleared lysates from 293T and RAW264.7 cells.
OPTIONAL STEP: plaque purification
- Pick a plaque with a sterilized glass Pasteur pipette by punching the pipette through the overlay into the plaque.Pick up to 5 plaques and freeze up to 4 at −80°C in case you need more plaques to sequence and get the desired virus.
Transfer the agar plug into 500 µl medium and vortex.
Infect RAW264.7 cells in T25 flask with 5 ml culture media with 200 µl of the medium containing the agar plug.
Proceed as described under: “Passage onto RAW264.7 cells (liquid passage)”.
Harvest virus.
Extract RNA from 140 µl virus lysate using the QIAamp viral RNA kit (QIAGEN) following the manufacturer’s recommendations.
- Prepare a 50 µl PCR reaction with the following components in a PCR tube for reverse transcription of viral RNA to cDNA using the QIAGEN one step RT-PCR kit.
Component Initial concentration Volume (µl) QIAGEN OneStep reaction buffer 5× 10.00 RNasin (Promega) 40 U/µl 0.50 dNTPs 10 mM 2.00 Forward primer 25 µM 2.00 Reverse primer 25 µM 2.00 QIAGEN enzyme mix 2.00 Nuclease-free water 26.50 RNA template 5.00 Total 50.00 Centrifuge the PCR tube briefly for about 5 sec to collect the reaction mixture at the bottom of the tube.
- Place the tube in a thermal cycler and set the PCR parameters to:
50°C for 30min 95°C for 15 min 
40× 94°C for 30 sec 55°C for 15 sec 72°C for 1 min 72°C for 10 min 4°C pause Run 5 µl of the PCR reaction on a 1 % agarose gel to confirm your cDNA.
Remove primers from the PCR product using QIAquick PCR purification kit (QIAGEN) following the manufacturer’s recommendations.
Determine the concentration of your cDNA with a spectrophotometer.
Send cDNA sample with primers for sequencing according to the sequencing company’s instructions.
Compare your sequencing results with published MNV sequences using the basic local alignment search tool (BLAST) on NCBI website.
ALTERNATE PROTOCOL 2
DNA-based, T7-driven generation of recombinant MNV
The recovery of murine norovirus (MNV) from transcripts generated from cDNA, in a process driven by T7 polymerase, is a robust, reproducible and rapid method that yields high recovery titers of infectious MNV (Figure 1). The protocol described below comprises seeding of transfectable cells permissive for MNV replication, infection with recombinant fowlpox virus (FPV-T7) for the expression of T7 RNA polymerase (see support protocol 3), and subsequent transfection of cDNA containing the MNV genome under the control of a T7 RNA polymerase promoter (Figure 2). After an appropriate incubation period, the infectious MNV particles can be titrated in suitably susceptible cells. A video is available at:http://www.jove.com/video/4145/reverse-genetics-mediated-recovery-of-infectious-murine-norovirus
Figure 1. Genome organization of MNV.
a. MNV encodes four open reading frames (ORFs). ORF1 is translated into a precursor polyprotein that is autocatalytically cleaved to release at least 6 non structural protein, NS1–7. ORF2 encodes the major capsid protein VP1 while ORF3 encodes the minor capsid protein VP2. ORF4 overlaps with ORF2 at a +1 frame and encodes VF1. During replication, MNV translates from a genomic and sub-genomic RNA, both of which include VPg at the 5’ end and a polyA tail at the 3’ end of varying lengths. b. Schematic of the recovery of infectious MNV from cDNA plasmid (representative example pT7: MNV-1 3’Rz). The MNV cDNA sequence is downstream of the T7 promoter sequence, which allows for the T7-driven transcription of MNV. At the 3’ extremity, MNV cDNA is fused to a 26-nucleotide long polyA tail. Downstream of MNV cDNA, the sequence for a self-cleaving ribozyme is fused to generate an authentic 3’ end.
Figure 2. Schematic of recovery procedure.
Step 1. Transfectable cells (i.e BSR-T7 or BHK) are infected with FPV-T7.
Step 2. Following cell entry, FPV-T7 transcription and translation results in the production of T7 RNA polymerase.
Step 3. After cells are left to incubate for two hours to allow for the production of T7 RNA polymerase, pT7: MNV-1 3’Rz is transfected using Lipofectamine 2000.
Step 4. pT7: MNV-1 3’Rz is transcribed in the cytoplasm, driven by the FPV-T7 encoded T7- RNA polymerase, to produce MNV RNA transcripts.
Step 5. The self-cleaving δ-ribozyme that is encoded by pT7: MNV-1 3’Rz generates an authentic 3’ end. It is possible that capping enzymes produced by FPV-T7 are able to cap MNV RNA transcripts.
Step 6. Capped MNV RNA transcripts are translated for the production of MNV proteins. VPg linked MNV RNA transcripts undergo replication.
Step 7. Nascent MNV RNA produced can be encapsidated to produce full infectious virus. Cells subjected to freeze-thaw cycles can facilitate virus release. Virus yields can be subsequently titred by means of end-point dilution assays, or plaque assays.
Materials
Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco), with 10% Fetal Calf Serum
Trypsin
Cultures of transfectable cells permissive for MNV replication, i.e., baby hamster kidney cells (BHK), BSR-T7 (BHK-21-derived cells expressing recombinant T7-polymerase), human embryonic kidney cells (HEK 293T), human hepatocellular carcinoma cells (Huh7) and African green monkey cells (Cos7)
A titred stock of FPV-T7
cDNA clone of MNV under the control of a T7 RNA polymerase promoter
Lipofectamine 2000 (Invitrogen)
OptiMEM (Invitrogen)
35 mm dish
Aspirator
37°C incubator (5–10% CO2)
- Trypsinise a confluent flask of BHK or BSR-T7 cells. For transfections the following day, seed 7.5×105 cells per 35 mm dish, using antibiotic-free DMEM supplemented with 10% FCS. Gently rock the dish to ensure even distribution of cells and incubate the plates at 37°C (5–10% CO2) to settle overnight. If you wish to perform the transfection on the same day, seed 1.5×106 cells and leave cells to adhere for a 2–3 hour period at 37°C (5–10% CO2) prior to proceeding to the next step.This protocol is optimized for the use of BHK or BHK-derived BSR-T7 cells. Further optimization is necessary if other transfectable cell lines are to be used. Parameters to consider include cell seeding density, multiplicity of infection of FPV to be used, amount of plasmid to be transfected and incubation period following transfection.
Remove the growth medium from the cells and add 700 µl of FPV-T7 at a multiplicity of infection of 0.5 pfu/cell. For optimal recoveries in BHK and BSR-T7 cells, ensure that FPV is diluted to infect at a multiplicity of infection of 0.5 pfu/cell. FPV-T7 should be prepared and titrated in primary chicken embryo fibroblast (CEF) cells, detailed in support protocol 3. The optimal amount of FPV-T7 must be determined for each cell line as the rate of infection differs.
- Leave cells for 1 hour in 37°C (5–10% CO2) to allow infection of FPV-T7. Add 2 ml of antibiotic-free DMEM supplemented with 10% FCS, and leave cells in 37°C (5–10% CO2) for a further 1 hour to allow FPV to express T7 RNA polymerase.It is important that no antibiotics are present in the growth medium, as it may interfere with the lipofection process.
To prepare the transfection mixture, combine 100µl of OptiMEM containing 1 µg of MNV cDNA clone that is under the control of a T7 RNA polymerase promoter (i.e., pT7: MNV-1 3’Rz), with 100 µl OptiMEM containing 4 µl of Lipofectamine 2000. The resultant mixture should be thoroughly mixed by gentle pipetting around 15 times. Allow 20 min at room temperature for Lipofectamine 2000 to form a complex with the cDNA.
Aspirate the growth medium containing unattached virus off the cells and gently wash once with 2 ml of antibiotic-free DMEM. Then add 3 ml of antibiotic-free DMEM supplemented with 10% FCS to cover the cells.
Add the transfection mix in a drop-wise manner to the cell monolayer. To ensure even distribution of the cDNA, gently rock the plates before placing them in 37°C (5–10% CO2). Plasmid pT7: MNV-1 3’Rz, containing the full MNV-1 cDNA genome sequence, produces optimal titers after 24h incubation, though other strains may require a longer incubation period. As a reference MNV-1 cDNA clone pT7: MNV-1 3’Rz requires only 24 hours to produce virus titers above 104 TCID50/ml (Table 1). For other strains (i.e. MNV-3) or viruses containing deleterious mutations affecting replication, longer incubation periods may be required for optimal yields (Arias, Bailey et al. 2012).
Freeze cells at −80°C. Once thawed, centrifuge cells at 1000×g for 5 min to pellet the cellular debris. The virus-containing supernatant can be aliquoted and stored indefinitely at −80°C. A frozen aliquot should be thawed and used to titrate the recoveries.
Table 1.
MNV Recovery titers from different transfectable cell lines
| Cell typea | Titersb with pT7: MNV-1 3’Rz |
|---|---|
| BHK | 3.4 × 104 TCID50/mLc |
| BSR-T7 | 3.4 × 104 TCID50/mL |
| HEK 293T | 6.0 × 104 TCID50/mL |
| Huh 7.5 | 2.4 × 104 TCID50/mL |
Cells transfected with pT7: MNV-1 3’Rz.
Virus yield determined 24 h post transfection.
Titers performed in triplicate and expressed as TCID50/mL.
SUPPORT PROTOCOL 3
PREPARATION OF A FPV-T7 STOCK REQUIRED FOR REVERSE GENETICS
Preceding the recovery of MNV from genomic cDNA, is the generation of a well-characterised FPV-T7 virus stock. Expression of recombinant T7 RNA polymerase in cells infected with FPV-T7 will drive the synthesis of replication-competent capped viral transcripts from cDNA. The FPV-T7 strain used was initially generated by Dr Mike Skinner, Imperial College London and is readily available via MTA (Britton, Green et al. 1996).
This approach will typically produce MNV-1 at a titer of >104TCID50/mL after 24h. Recovery titers from other commonly used transfectable cell lines are highlighted in Table 1. High titer MNV stocks can then be produced by infecting a susceptible cell line such as RAW264.7 or BV2 cells.
Materials
Dulbecco’s Modified Eagle’s Medium (DMEM; Gibco), supplemented with 10% Foetal Calf Serum and 1% Penicillin/Streptomycin
Trypsin
Primary chicken embryonic fibroblast cells (CEFs)
FPV-T7 inoculating stock (obtained via MTA from Dr. Mike Skinner, Imperial College London)
Large volume flasks (T-175 or T-225)
37°C incubator (5–10% CO2)
- Trypsinize a flask of chicken embryo fibroblasts (CEFs) primary cells and seed them at 1×105 cells/ml.Typically, large volume flasks (T-175 or T-225) are seeded with CEFs to facilitate the recovery of high volumes of FPV-T7. This seeding density equates approximately to 5×106 cells in a T-175 flask.
When cells reach 100% confluence, infect them with FPV-T7 at a multiplicity of infection of ~1 pfu/cell and incubate for 1 h at 37°C in the presence of 5–10% CO2.
- Remove the inoculum and add DMEM supplemented with 10% FCS to overlay the cells. Incubate the infected cell cultures at 37°C in the presence of 5–10% CO2until complete cytopathic effect is observed. A small mock-infected cell culture should be carried out in parallel to identify cytopathic effect in infected cultures.For a T175 flask, a volume of 20 ml is recommended to cover the cells to enable the recovery of higher virus titers. Complete cytopathic effect is observed within 48–72 hours post-infection. Typically, cytopathic effect results in the development of granular cell morphology (Somogyi, Frazier et al. 1993). A small mock-infected culture might be useful to distinguish cytopathic effect in the infected cultures.
Freeze-thaw the cell cultures twice to facilitate the release of virus particles to the supernatant.
- Remove the cell debris from the lysates by centrifugation at 600× g and 4°C for 5 min.Virus preparations should not be filtered to clarify them as FPV diameter is around 260–280 nm (larger than pore diameter size in 0.2 µm filters).
- Recover the supernatant and aliquot in different tubes that can be stored indefinitely at −80°C.The remnant of an FPV-T7 that has been used for recoveries should be marked before freezing again as the titer of the aliquot might have dropped. We suggest to combine several aliquots that have been thawed once and aliquot them in new aliquots. The virus titer for this stock of recycled FPV-T7 should be determined before use.
BASIC PROTOCOL 4
RNA-BASED GENERATION OF RECOMBINANT MURINE NOROVIRUS
Infectious MNV from cDNA is efficiently recovered viain vitro transcription and subsequentin vitro capping. The resulting capped RNA transcripts are then transfected into cells to recover infectious MNV. Transcripts are transfected into either BV-2 or BSRT7 cells. BSRT7 or parental BHK cells are used to test the specific infectivity of the RNA as the cells are not permissive to re-infection and virus yields represent those from a single growth cycle. BV-2 cells are used when minimal passage in cell culture is required for a cDNA clone that is known to be infectious. Typically, Neon®-mediated transfection of BV-2 cells can yield around 1×106 TCID50/ml MNV at 24 hours post-transfection and up to 1×109 TCID50/ml at 72 hours post-transfection, although this effectively constitutes the first passage of the virus. Recovery using BSR-T7 cells generally yields 1×106 TCID50/ml of MNV at 48 hours post-transfection.
A video for the generation of recombinant MNV can be found here: http://www.jove.com/video/4145/reverse-genetics-mediated-recovery-of-infectious-murine-norovirus
Materials
MNV cDNA (pT7: MNV 3’Rz)
BSR-T7 cells
BV-2 cells or RAW264.7 cells
NheI Restriction Enzyme (NEB R0131S)
GFX PCR DNA Gel Band Purification Kit (GE Healthcare 28-9034-70)
MEGAscript® T7 Kit (Life Technologies AM1334)
DNase I (RNAse free) (NEB M0303S)
Lithium chloride (LiCl) precipitation solution (Life Technologies AM9480)
RNA storage solution (Life Technologies AM7000)
ScriptCap m7G Capping System (Epicentre Biotechnologies SCCE0610)
Neon® transfection system (Life Technologies MPK5000)
Neon® transfection system kit (Life Technologies MPK1025)
Opti-MEM (Life Technologies 31985070)
Lipofectamine 2000 Transfection Reagent (Life Technologies 11668-027)
Agilent RNA 6000 Nano kit (optional) (Agilent Technologies 5067-1511)
Agilent 2100 bioanalyzer (optional) (Agilent Technologies G2939AA)
Synthesis of infectious capped MNV transcripts
- Digest 5–10 µg of the plasmid containing the wild type MNV cDNA (pT7:MNV 3'Rz) or recombinant MNV cDNA with NheI to obtain linear DNA. The following is a typical reaction set up for NheI digestion (see Table 2):NheI recognises a unique restriction site after the 3’ end polyA tail of MNV genome, thus it defines the 3’ end of transcription with minimal addition of nucleotides added to the 3’ end of genome (Figure 3).
- Incubate the reaction mixture at 37 °C for 3 hours. Purify the linearized plasmid using silica columns (e.g. GFX PCR DNA Gel Band Purification Kit from GE Healthcare) and elute in 50 µl water.The quality of linearized DNA is important for the subsequent in vitro RNA synthesis. Incomplete linearization or RNase contamination in the eluate will compromise the final RNA yield and integrity and as such lead to the failure to generate infectious virions. We recommend comparing 100 ng of digested and undigested DNA on an agarose gel to ensure complete digestion prior to purification.
- In vitro transcribe the linearized MNV cDNA using T7 RNA polymerase as described below (Yunus, Chung et al. 2010). Many commercial kits are available for this purpose and provide a reproducible method of large amounts of RNA synthesis such as MEGAScript (Life Technologies) and RiboMAX (Promega). Ensure all equipment used, such as microcentrifuge tubes (MCT), are certified RNase free and use filter tips throughout this protocol. The following is a typical reaction set up (50 µl in total, add the reagents following the same order) – see Table 3:Addition of pyrophosphatase is optional but without pyrophosphatase the reaction mixture will become cloudy due to the formation of free pyrophosphate. This can be removed prior to precipitation by centrifugation at 12,000×g for 1 minute and the supernatant collected.
- Incubate the reaction mixture at 37 °C for 2 to 5 hours. Load 0.5 µl or less of the RNA transcription reaction on an agarose gel and electrophorese to ensure that the reaction has worked efficiently and RNA is full-length. Denaturing gels provide valuable indication on the size of RNA however non-denaturing agarose gel electrophoresis provides a rapid method to analyse RNA integrity. The MNV genome as produced from the infectious cDNA clone pT7:MNV 3'Rz will appear as an approximately 3 kbp band relative to a dsDNA ladder on a non-denaturing TBE agarose gel (Figure 4). Alternative methods for the rapid analysis of RNA integrity include the Agilent bioanalyser.The amount of RNA loaded for agarose gel electrophoresis is important for gel resolution, overloading can result in poor resolution. Also it is essential to ensure the agarose gel is prepared using RNase-free reagents to avoid RNA degradation during electrophoresis which may also affect RNA resolution.
Centrifuge the reaction mixture at 12,000×g for 1 minute at 4°C, transfer the supernatant to a new RNase-free MCT. Then add 2 µl DNase I (RNase-free, 2 U/µl, NEB), DNase I buffer and water accordingly. Incubate at 37 °C for 30 minutes before further analysis.
- Purify the RNA sample to remove the unincorporated nucleotides. For precipitation using LiCl, bring the final volume to 100 µl with H2O and then add 40 µl of LiCl precipitation solution (7.5 M LiCl, 50 mM EDTA, pH 8.0, Life Technologies) and store the sample at −20 °C for 30 minutes to overnight.The sample can be stored overnight at this point at −20 °C and the purification and in vitro capping continued the following day.Many methods are available for removing unincorporated nucleotides, including silica column-based approaches.
Pellet the RNA by centrifugation at 12,000×g at 4 °C for 15 to 30 minutes.
- Remove the supernatant and wash it in 150 µl of 70% ethanol. Centrifuge at 12,000×g at 4 °C for 15 minutes.Be careful not to disturb the translucent RNA pellet when removing the supernatant.
- Remove the ethanol and air-dry the RNA.Avoid drying the pellet for more than one minute, as a completely dried pellet is difficult to resuspend.
Add 50–100 µl of RNA storage solution (Life Technologies) to resuspend the MNV transcripts. Care should be taken to ensure that all the RNA has dissolved properly. Heating the sample to 60 °C may help to resuspend it. Any insoluble material should then be removed by centrifugation prior to RNA quantification. The purified transcripts are uncapped and require a subsequent in vitro capping step to be infectious (ScriptCap m7G Capping System, Epicentre Biotechnologies).
- Quantify the RNA by spectrophotometry. Run 100–300 ng of sample on a 1% RNase-free agarose gel (Figure 4) to check the integrity of RNA before performing the capping reaction.Depending on the nature and scale of the transcription reaction, typical yields range from 50–150 µg of RNA per 100 µl transcription reaction.
-
To improve the efficiency of RNA capping, heat 60 to 70 µg of MNV RNA transcripts at 65 °C for 10 minutes and then place the tube immediately on ice. Pulse the RNA in a chilled microcentrifuge to collect droplets formed during the heating step.
Step annotation: This step may reduce any inhibitory effect of RNA structure on the capping reaction.
Prepare a capping reaction mixture using the ScriptCap m7G Capping System (Epicentre Biotechnologies). Table 4 is a typical reaction set up for in vitro capping reaction with a final reaction volume of 100 µl. However, the reaction volume can be scaled down according to the input amount of RNA and the amount of capped RNA required.
Keep RNA transcripts on ice to avoid degradation. Mix the reaction mixture thoroughly and then incubate at 37 °C for 1 hour. Purify the RNA by LiCl precipitation as described above (see Step 6–10). Dissolve the pellet in 50–100 µl of RNA storage solution (Life Technologies) and quantify the amount of RNA. It is convenient to adjust the RNA sample concentration to 1 µg/µl. Again, ensure that all the RNA has dissolved properly and remove by centrifugation any insoluble material prior to RNA quantification.
Check the integrity of the capped RNA again before proceeding with the transfection step. To do so, run 100–300 ng of sample on a 1% agarose gel (Figure 4)
Table 2.
Reaction set up for NheI linearization of MNV cDNA
| Components | Final Concentration | Per Reaction (µl) |
|---|---|---|
| MNV cDNA (pT7: MNV 3’Rz) | 100 ng/µl | Varies |
| 10× NEBuffer 2.1 | 1× | 10 |
| NheI (10 U/µl) | 0.2 U/µl | 2 |
| Water | To 100 µl | Varies (to 100 µl) |
Figure 3. Overview of the protocol for the recovery of infectious MNV from RNA transcribed and capped in vitro.
The plasmid pT7:MNV 3'Rz is linearised immediately downstream of the MNV genomic sequence using NheI restriction enzyme (step 1). After DNA purification (step 2), MNV RNA transcripts are generated in vitro using a recombinant T7 RNA polymerase (step 3). The RNA synthesis is confirmed by electrophoresis (step 4) and the template DNA is eliminated using a commercial RNAse-free DNase I (step 5). RNA is then purified by LiCl precipitation (step 6). The purified RNA product is then capped in vitro (steps 7). After purification by LiCl precipitation (step 8), the capped RNA is transfected into either BV-2 cells using a Neon® Transfection System or BSR-T7 cells by lipofection (steps 9). Once inside the cell, capped RNA transcripts will be translated into viral proteins which should then mediate subsequent rounds of RNA replication to generate VPg-linked RNA that can be packaged to generate infectious virions (step 10–11). To facilitate virus release from cells, one or several cycles of freeze and thaw are performed (step 12). Viral yields can be then determined by TCID50 or plaque assay procedures.
Table 3.
Reaction set up for in vitro transcription of linearized MNV cDNA
| Components | Final Concentration |
Per Reaction (µl) |
|---|---|---|
| 1 M HEPES pH7.5 | 100 mM | 5 |
| 320 mM Magnesium Acetate | 32 mM | 5 |
| 400 mM DTT | 40 mM | 5 |
| 20 mM Spermidine | 2 mM | 5 |
| 100 mM ATP | 7.5 mM | 3.75 |
| 100 mM CTP | 7.5 mM | 3.75 |
| 100 mM GTP | 7.5 mM | 3.75 |
| 100 mM UTP | 7.5 mM | 3.75 |
| Pyrophosphatase (0.1 U/µl) | 0.002 U/µl | 1 |
| RNAseOUT RNAse inhibitor (40 U/µl) | 1.6 U/µl | 2 |
| Water | 2 (to 50 µl) | |
| Linearized DNA | 20 ng/µl | 5 |
| Recombinant T7 RNA polymerase (0.5 mg/ml) | 50 ng/µl | 5 |
Figure 4. Analysis of MNV RNA transcript integrity by non-denaturing gel electrophoresis.
(A) Transcription products are run on a non-denaturing 1% agarose gel in parallel to 1-kb DNA ladder (M, lane 1). The relative mobility of viral transcripts under non-denaturing conditions is similar to a dsDNA product of 2.5–3 kb. (B) Analysis of RNA transcript after LiCl purification. (C) Analysis of RNA integrity after capping reaction. (D) Analysis of capped-RNA integrity after LiCl precipitation.
Table 4.
In vitro enzymatic capping reaction
| Components | Final Concentration |
Per Reaction (µl) |
|---|---|---|
| In vitro transcribed uncapped RNA | 0.6–0.7 µg/µl | Varies (total volume less than 73 µl) |
| 10× Capping buffer | 1× | 10 |
| 10 mM GTP | 1 mM | 10 |
| 20 mM S-adenosyl methionine | 0.1 mM | 0.5 |
| Scriptguard (40 U/µl) | 1 U/µl | 2.5 |
| Scriptcap enzyme (10 U/µl) | 0.4 U/µl | 4 |
| Water | Varies (to 100µl) |
Recovery by Neon®-mediated transfection of capped RNA into BV-2 cells
For the recovery of MNV infectious virus in a permissive cell line it is possible to electroporate the capped MNV transcripts directly into BV-2 cells using the Neon® transfection system (Life Technologies). BV-2 is an immortalized murine microglial cell line that is susceptible to MNV infection, supporting multiple rounds of virus replication and subsequent re-infection (Bocchini, Mazzolla et al. 1992, Cox, Cao et al. 2009). Yields typically approach 105 infectious units per ml or more at 24 hours post transfection and peak at >107 infectious units per ml after 48 hours, as determined by TCID50.
-
1a
One day before transfection, seed BV-2 cells at an estimated 50% confluency. Typically a confluent T75 flask (around 8.4 × 106 cells) is sufficient for 1 transfection reaction.
-
2a
On the day of transfection, aspirate the medium from the cells and detach cells using commercial trypsin.
-
3a
Neutralize trypsin using BV-2 medium composed of Dulbecco’s modified Eagle medium (DMEM), 10% foetal calf serum (FCS), 2 mM L-glutamine and 1% penicillin/streptomycin (P/S).
-
4a
Pellet the cells at 1,200 × g for 5 minutes and resuspend in 10 ml BV-2 medium without P/S. Generate a single cell suspension by repeated pipetting.
-
5a
Determine the density of live cells using a haemocytometer with trypan blue exclusion. Each transfection requires 7.2×106 cells.
-
6a
Transfer 7.2 × 106 cells to a sterile 1.5 ml MCT per transfection. Wash the cells with sterile PBS immediately prior to transfection only and pellet at 1,200 × g for 2 minutes in a table top centrifuge.
-
7aCarefully remove all PBS and resuspend cells in 120µl Buffer R to achieve the desired cell density (6.0 × 106 cells/100µl).Here prepare 20% extra cells and 20% extra Buffer ‘R’ to prevent bubble formation during taking cells by Neon® tip. Therefore the amount of capped RNA should be scaled accordingly. Cells should not remain in Buffer R for prolonged periods of time due to toxicity, resuspend immediately prior to use.
-
8a
Add 1.2 µg of capped RNA and mix thoroughly.
-
9a
Place the electrode of the Neon® system inside the hood and place a Neon® tube (tip holder) into the electrode. Add 3ml Solution E to the tube.
-
10aLabel 6-well plates (3 wells per sample) and 15 ml conical tubes. Add 9ml BV-2 medium (without P/S) to each conical tube.Step 9a and 10a can be done during the centrifugation in Step 6. Each tip holder with Solution E can be used for up to 10 transfections without contamination. However, when trying to recover genetically pure viruses for further studies it is advisable to replace the tip holder after each transfection.
-
11aMix the cells slowly and take 100 µl BV-2 cells and RNA mixture with the Neon® tip using the Neon® pipette and insert into the pipette station.Be careful not to include any bubbles in the cell suspension. The inclusion of bubbles will lead to sparks during the electroporation and cause cell death, which will compromise the transfection rate.
-
12a
Load the following parameters for optimized BV-2 electroporation: Voltage=1700 V, Width=10 ms and Pulse= 3 and electroporate.
-
13a
Remove the Neon® pipette from the pipette station and transfer the electroporated cells to the 9 ml BV-2 media in conical tubes.
-
14a
Repeat Steps 11a–13a for the remaining samples.
-
15a
Slowly mix transfected BV-2 cells with media and add 3ml to each well of labelled 6-well plates.
-
16a
Incubate the BV-2 cells at 37 °C incubator with 5 –10 % CO2 until cells adhere (4–5 hours) and then replace with media containing P/S.
-
17a
Incubate the cells at 37 °C and 5–10 % CO2 for 24 to 72 hours. Then, release infectious virions from cells by one (or more) freeze and thaw cycles and determine virus titre in the sample using either plaque assay (Alternate Protocol 3) or by TCID50 (Basic Protocol 5). Prior to titration lysates should be clarified by centrifugation for 1–2 minutes at maximum speed. Also, it is optional to pass the lysate through a 0.22 µm filter to exclude any cell debris.
Recovery by Transfection into BSR-T7 cells
-
1b
One day before transfection, trypsinize a monolayer of BSR-T7 cells and seed 7.5×105 cells into a 35 mm diameter dish (or 6-well plate) in antibiotic-free growth media and incubate the cells at 37 °C with 5–10% CO2 overnight. Double the amount of cells if the transfections are planned for the same day, and allow cells to adhere to the plate for 2–3 hours at 37 °C with 5–10% CO2.
-
2b
Remove the media from the cells and replace with 3 ml of fresh media without P/S to ensure the maximum efficiency of transfection.
-
3b
Prepare a mixture of 1 µg of capped MNV transcript into 100 µl of Opti-MEM (Life Technologies) and mix it with 4 µl of Lipofectamine 2000 pre-mixed in 100 µl of Opti-MEM (Life Technologies). Mix the sample thoroughly by pipetting it up and down 15 times. Incubate the mixture at room temperature for 10–15 minutes.
-
4b
Re-mix the transfection mixture, and add to the cell monolayer in a drop-wise fashion. Gently shake the plate in perpendicular directions.
-
5b
Incubate the cells at 37 °C and 5–10% CO2 for 24 to 72 hours. Afterwards, release infectious virions from cells by freeze and thawing and determine virus titre by plaque assay (see Alternate Protocol 3) or TCID50 (see Basic Protocol 5).
BASIC PROTOCOL 5
MEASURING MNV TITERS BY TCID50
The tissue culture infectious dose50(TCID50) assay is frequently used in virology to determine virus titers. Following serial dilutions, the titer of the virus is calculated based on forming cytopathic effect (CPE) in 50% of the infected cells after 3–5 days of incubation. The procedure involves 10-fold serial dilutions of each virus sample in a dilution plate, followed by transferring aliquots of each sample to the test plate containing cells, in quadruplicate. Typically, each sample is assayed as three replicates to ensure reproducibility.
In BV2 or RAW264.7 cells, MNV1 is able to achieve high titers of 107 or 108 following low MOI (0.01 TCID50 units/cell) infection and incubation for 24–48 hours. At that time, cytopathic effect is clear in both cells, with the cells lifting of the bottom of the dish, shrinking in size and becoming apoptotic.
Materials
DMEM-10 media (see recipe)
BV-2 mouse microglial cells (or RAW264.7 mouse macrophage cells)
30–300 µl Eppendorf Research plus multichannel pipette
10–100 µl Eppendorf Research plus multichannel pipette
300 µl sterile filter tips
Biological Safety Cabinet (Class II)
96-well plates
CO2incubator (at 37°C)
Microscope
Sterile (1.5ml) Microcentrifuge tubes
- Take out frozen samples for analysis (frozen at −80°C) and thaw them at 37°C, or room temperature.For an accurate titer, samples should be prepared in triplicate.
Transfer 100 µl-1ml of each sample into a sterile microcentrifuge tube.
Centrifuge at 13,000 × g for 3 minutes in a benchtop microcentrifuge to pellet cellular debris, and transfer the supernatant in a fresh tube.
Add 25 µl of each virus supernatant to the first well (row H) as shown in Figure 5A.
Using a 96-well tissue culture plate and the 30–300 µl Eppendorf Research plus manual pipette, transfer 225 µl of media to all wells in the dilution plate, adding to the wells containing undiluted virus (row H, Figure 5A) last. Pipette samples up and down (3 times) to mix.
Eject tips, and transfer 25µl from the diluted virus sample to the next row (row G) and mix thoroughly. Repeat this for each subsequent row until the dilution series is complete ensuring the tips are changed after each dilution (Figure 5A).
- Take a T75 Flask of BV2 cells that have reached 75% to 100% confluence. Change the media by adding 10 ml of DMEM containing 10% FBS. Then, scrape the cells and add 9 ml of media for each ml of cell suspension e.g. for 10 ml cell suspension add 90 ml of media.RAW264.7 cells may be substituted for BV2 cells.
Pipette cells up and down to ensure the cells form a single-cell suspension.
Dispense 100 µl of cells into each well of a labeled test 96-well plate using a multichannel pipette.
Using a multichannel pipette, transfer 50 µl of each dilution from the dilution plate into four columns of the test plate as shown in Figure 5B.
- Incubate at 37°C for 3 to 5 days. Check cytopathic effects (CPE) in each well. Score each well in a dish as either infected or uninfected.In BV-2 cells, CPE is apparent as cells dying and floating off the base of the dish, compared to a confluent monolayer in uninfected cells.
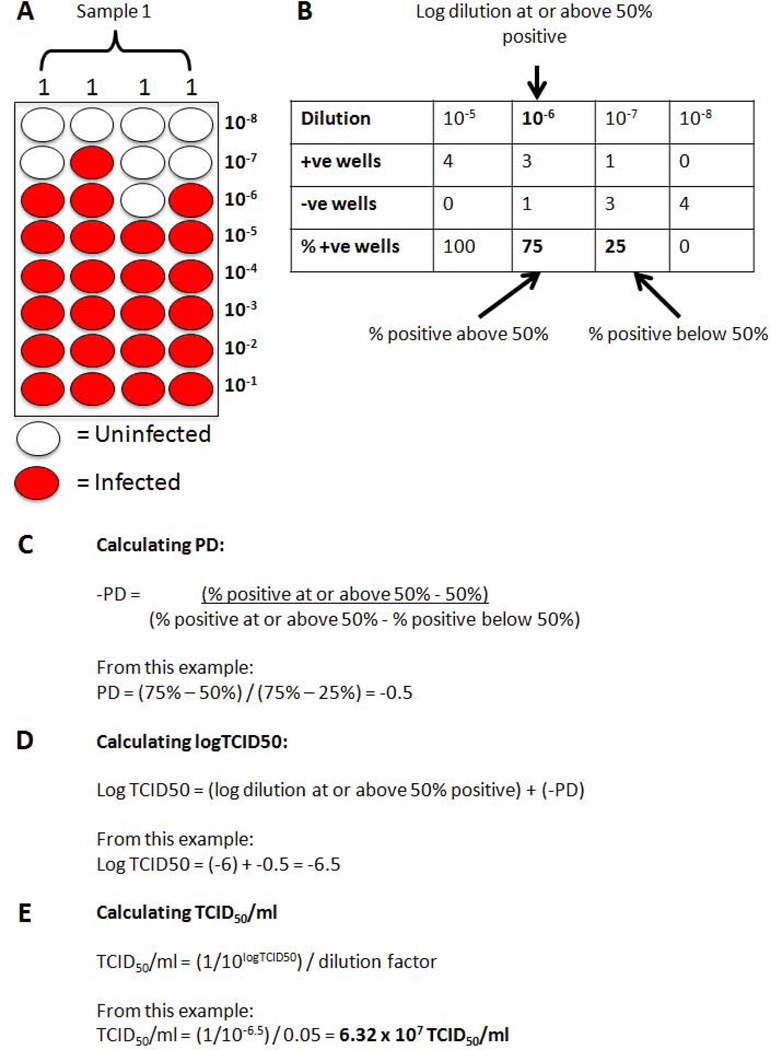
TCID50 values are calculated by the Reed & Muench method (Reed and Muench 1938, Lindenbach 2009). An example and workings for this method are given in Figure 6.
Figure 5.
Preparation of serial dilutions, and aliquotting of diluted samples. A) Serial diluations of the virus are prepared in triplicate from (row H) to (row A) in the dilution plate. B) 50ul of each dilution is transferred from the dilution plate to four columns of the test plate.
Figure 6.
Manual calculation of TCID50/ml values. A) An example TCID50 result, also given in table format in B). C) Calculation of the proportionate distance (-PD), D) Using –PD to calculate logTCID50. Calculation of TCID50/ml from the logTCID50 and dilution factor.
The dilution at which 50% of the wells are infected is calculated using the following equation:
The proportionate distance (PD) is the result of the distance between the two dilutions that gave a percentage if infection is either side of 50%. Note that the PD has a minus sign in front. The PD is then used to calculate log TCID50 as follows:
(log dilution at or above 50%) + (−PD) = log TCID50
This log TCID50 value is then used to express the virus titer as TCID50/unit volume, e.g., TCID50/ml.
TCID50/ml = (1/10logTCID50)/dilution factor
In this method, 50 μl of virus is added to each well, so the dilution factor is 0.05
For simplicity an excel spreadsheet created by Dr. Brett D. Lindenback may be used for the calculation (Lindenbach 2009). This spreadsheet is available from http://www.lindenbachlab.org/resources.html
ALTERNATE PROTOCOL 3
MEASURING MNV TITERS BY PLAQUE ASSAY
The plaque assay is used to measure infectious virions in a given sample. It is based on the published protocol by Gonzalez-Hernandez et al. (Gonzalez-Hernandez, Bragazzi Cunha et al. 2012), and is an alternative method to the TCID50 assay. Compared to TCID50, the plaque assay is shorter by 1–3 days. Herein, dilutions of a virus stock are incubated with cell monolayers, overlaid with agarose, and stained with neutral red to visualize plaques after 2 days. A video of the plaque assay can be found at: http://www.jove.com/video/4297/plaque-assay-for-murine-norovirus
Materials
DMEM-10 (see recipe)
Seaplaque agarose (Lonza, cat no. 50100)
2×MEM (Gibco, cat no. 11935)
0.33% Neutral Red (Sigma, cat no. N2889)
PBS (pH 7.4)
Repeater pipette
Multichannel pipette
Single channel pipette
Filtered tips
6-well, 48-well plates
Rocker
Tissue culture incubator
Biosafety cabinet (class II)
Plate cells at 2×106 cells per well in 2 ml (i.e., 1×106 cells/ml). Incubate cells overnight at 37°C in 5% CO2.
- Label plates and organize them based on the samples you will be testing.A suggestion for an efficient biosafety hood set-up are shown in Figure 7.
- Prepare media in 24- or 48-well plate for the number of dilutions that you are using and label them (for −1 dilutions in duplicate add 1.35 ml of media to each well and for −2 dilution add 1.5 ml of media).Use 50 ml attachment for repeater pipet for plating cells and for adding media to dilution plates. For the 50 ml repeater pipet to fit, the cell suspension and media need to be in a 250 ml stericup flask.
Pre-warming 2×MEM at 37°C and heat 3% Seaplaque in microwave, and then leave in 57°C waterbath until ready to use.
Make virus dilutions by adding 150 µl of virus to −1 dilutions for a total of 1.5 ml, and add 15 µl to −2 dilutions for a total of 1.515 ml)
Aspirate off media from 6-well plates.
Add 500 µl of a given dilution to 2 wells each for duplicate wells.
Rock for 1 hour at room temperature.
Mix pre-warmed 2×MEM and warm Seaplaque together in 1:1 ratio (add media first and then add Seaplaque from 57°C so it cools off).
Aspirate off virus and add 2 ml of SeaPlaque/2×MEM to each well.
Incubate at 37°C for 48 hours.
Figure 7. Schematic of the biosafety hood set-up.
Efficient organization of the biosafety hood minimizes spills and increases speed of the assay without compromising precision.
Overlay for Plaque Assay
Mix 3 ml of Neutral red with 100 ml PBS.
- Add 2 ml of neutral red/PBS mixture to well.Note: add the neutral red using the 50 ml repeater pipet.
Incubate at 37°C for 1–2 hours.
Aspirate off neutral red/PBS mixture.
- Count plaques and calculate titer.Note: when performing each dilution in duplicate (i.e., 1 ml of a given dilution is analyzed), add together the number of plaques from each of the two wells times the dilution factor to equal the titer.
ALTERNATE PROTOCOL 4
MEASURING MNV GENOME TITERS BY RT-qPCR
The determination of MNV levels can be achieved through amplification of viral nucleic acid (RNA) by reverse transcription quantitative polymerase chain reaction (RT-qPCR). This technique produces high enough concentrations of nucleic acid for detection and quantification by fluorescence. In general, this technique relies on purification of viral RNA, an initial synthesis of cDNA (complementary or copy DNA) by the enzyme Reverse Transcriptase using virus-specific primers and subsequent exponential amplification of that cDNA coupled to real-time detection of amplification levels via readouts of fluorescence. Reverse transcription can be performed as either a single reaction for cDNA synthesis or coupled to qPCR.
In this protocol, we discuss a one-step RT-qPCR reaction for MNV-1 amplification modified from previously published work (Taube, Perry et al. 2009), providing a reliable determination of viral RNA levels. The conditions mentioned in this protocol were optimized for use in the BioRad CFX96 Touch qPCR instrument and provide analytical sensitivity, analytical specificity, reproducibility and repeatability, thus making them suitable for publication following MIQE guidelines (Bustin, Benes et al. 2009). It is advised that conditions be optimized for alternate instruments, as variations in results from instrument to instrument can occur (technical replicate examination). If RNA quality is optimal, and no DNA contamination is present, efficient amplification of MNV-1 from a variety of starting materials (e.g. stool, tissue) is achieved.
Materials
Two sets of pipettes (one for RNA handling and one for plasmid handling)
10% bleach solution
70% ethanol
RNAse Away™ reagent (Life technologies/Ambion cat. no. 10328-011)
Viral RNA extraction kit (such as the QIAGEN QiaAmp Viral RNA Kit™ cat. no.52906)
OR
RNA extraction kit (such as QIAGEN RNeasy mini kit™ cat. no. 74104)
OR
Trizol™ reagent (Life technologies, cat. no. 15596026)
Nuclease-free 1.5 ml tubes
Nuclease-free H2O
Micro-centrifuges (one designated for RNA work and one for DNA/plasmid work)
Deoxyribonuclease (preferably Ambion Turbo DNA-free DNase™ cat. no.AM1904)
Forward Primer (5’→ 3’): GTGCGCAACACAGAGAAACG (HPLC purified)
- Reverse Primer (5’ → 3’): CGGGCTGAGCTTCCTGC (HPLC purified)Both primers bind in ORF1 of the MNV-1 genome
- Probe (5’ → 3’): [6-FAM]-CTAGTGTCTCCTTTGGAGCACCTA-[BHQ1] (HPLC purified)Probe is modified at the 5’-end with a fluorescein amidite molecule (FAM), and at the 3’-end with the non-fluorescent molecule Black Hole Quencher 1 (BHQ1).
Amplification kit (Bio-Rad iScript One-step RT-PCR kit for Probe™ cat. no.170-88940)
OR Bio-Rad iTaq Universal Probes One-Step kit™ (cat. no. 172-5140)
96-well PCR plates (Hard-Shell PCR plates 96-well white shell/clear well, Bio-Rad, cat. no. HSP9601)
Optical film (Microseal ‘B’ seal, Bio-Rad, cat. no. MSB1001)
Plasmid containing the MNV-1 genome (for standard preparation)
Viral RNA extraction
- Prepare the area for RNA extraction by thoroughly cleaning it once with a 10% bleach solution, followed by spraying it with 70% ethanol, and lastly by wiping it twice with RNAse Away™. This will help remove and/or minimize the presence of ribonucleases and DNA contamination in the area.Also wipe your gloved hands with RNase Away™ often, and change gloves every time you touch surfaces that have not been cleaned. This step is crucial for obtaining high quality RNA, which is needed for successful downstream applications. Use of an “RNA-work only” designated area is advisable.
- Carefully wipe the RNA-extraction pipettes with RNAse Away™. Make sure to also pipette a full volume of RNAse Away™ into the pipettes, without a tip, so that it can decontaminate any potential RNases and DNA inside the pipette barrel.RNA-extraction pipettes should be soaked overnight in RNAse Away™ periodically. DO NOT use RNAse Away™ on gaskets, seals, aluminum or soft metals.
- Extract viral RNA from virus-containing samples (e.g., virus-containing lysates or homogenized tissues) following the manufacturer’s protocol.If using the QIAmp Viral RNA Kit, be sure to use the “spin column method” with an “RNA-only” micro-centrifuge. A vacuum manifold method could be used only if sure the device has not been utilized to isolate plasmid DNA, as crossover contamination can occur. If you use cellular RNA (from virus-infected cells) use of the RNeasy Mini kit from QIAGEN has provided great RNA yields and quality. If isolating RNA via the TRIzol method, make sure to perform an RNA cleanup using silica membrane-based columns, to remove any phenol contaminants.
- Perform an RNA-cleanup step, in case any plasmid or DNA contamination (however minimal) has occurred in the elution step. This is done using the Ambion Turbo DNA-free DNase as follows:
- After the elution of the RNA, add sufficient amount of 10× Turbo DNA-free buffer to make it into a final 1× concentration in a volume containing 1 µl of Turbo DNAse.
- Incubate this solution at 37°C for 25–30 minutes.
- Add inactivation reagent (same volume you add of 10× buffer + enzyme and water), and vortex several times to keep it dispersed in solution.
- Incubate for 5 minutes at room temperature.
- Centrifuge at full speed to pellet inactivation reagent.
- Pipette up almost the whole volume of sample you had originally (make sure to grab about 5 µl less than what you had originally) to avoid carryover of the inactivation reagent.IMPORTANT: Inactivation reagent WILL inhibit your qPCR reactions.
- Proceed with RT-qPCR.
This step is performed because of the use of plasmid DNA containing the MNV-1 amplicon as a potential source of contamination. Cleanup could alternatively be performed as an “on-column” DNase treatment, but this protocol has not been proven successful in completely removing contaminating DNA. - To ensure RNA stability and integrity, aliquots may be made and frozen at −80°C.Addition of RNAse inhibitors are also a way of ensuring RNA stability. However, if using the commercially available kits or reagents mentioned in this protocol, and maintaining an RNAse-free environment by using and “RNA-only” work zone/gloves and cleaning with RNAse Away™,RNAses are inactivated due to highly denaturing conditions and should not affect your extraction significantly. Also, not all commercially-available RNAse inhibitors are compatible with RT-qPCR. For this protocol, it is not necessary to add an RNAse inhibitor, but the reader might do so as long as they verify that subsequent amplifications by RT-qPCR are efficient.If the RNA is isolated from infected cells, you can measure its quantity and estimate its purity/integrity via spectrophotometry, making sure you measure absorbance at 260 and 280 nm and obtain the 260/280 ratio with a value of 1.9 to 2.0. If the RNA is isolated from cell-free material with the use of a kit and carrier RNA (e.g., QIAGEN kits), spectrophotometric readings will only be useful for determining 260/280 ratios, as the amount of carrier RNA will in most cases exceed the amount of viral RNA present, and does not provide a very accurate measurement of viral RNA concentration.
Performing RT-qPCR
Note: This reaction was optimized for the BioRad CFX96Touch qPCR machine using the primer matrix, and temperature gradient methods to provide the optimal conditions for the highest primer and probe combinations. It results in the highest efficiency reaction with both optimal Cq and fluorescence intensity.
Plan the plate arrangement according to the number of samples to be assayed, always including the standards, a non-template control (NTC), a positive control, and a no-RT (–RT) control as part of the sample group. Each sample should be run in duplicate, but preferably in triplicate, to observe for pipetting error.
Thaw reaction components on ice, mix them thoroughly and briefly centrifuge them to collect the volume at the bottom of the tubes. Light-sensitive components should be protected from light (i.e. probe).
- Set-up the RT-qPCR reaction by calculating the amount of reagents needed. Reaction volume should be 20 µL final. If another volume is made, concentration of both primers should be 0.05 µM final and probe concentration should be 0.125 µM final.All steps may be performed at room temperature if a vendor-supplied mix is used, as most reaction mixes contain a Hot-start Taq polymerase and reagents that are stable at room temperature for several hours. However, reactions can be assembled on ice when degradation is suspected.Plates may be made several days in advance and stored at 4°C until the reactions are ready to run. This does not cause a substantial decline in the performance of the PCR enzyme. Advance preparation of plates is not recommended if qPCR mixes are not from commercial sources (i.e. they are home-made), as these mixes lack additives present in the commercial mixes that confer stability to the reaction components. Whenever possible, fresh reactions should be run.
- Prepare a master mix in a 1.5 mL nuclease-free tube, containing each reagent but no template, for at least 1.5 to 2 samples more than what you have to run (i.e. make an excess of master mix). This will compensate for pipetting errors. The table below can be followed as an example:
Component Initial concentration Volume/Reaction
(1× final)Master Mix (96well)× 98 samples 2× Master Mix 2× 10 µL 980 µL F primer 10 µM 0.1 µL 9.8 µL R primer 10 µM 0.1 µL 9.8 µL Probe 10 µM 0.25 µL 24.5 µL RT 50× 0.4 µL 39.2 µL Nuclease-free H2O - 4.15 µL 406.7 µL Template - 5 µL - Total 20 µL 1470 µL Mix thoroughly to ensure homogeneity and dispense 15 µL aliquots into the required wells of a 96-well plate. Use good pipetting technique to ensure precision and accuracy.
- Dispense 5 µL of each unknown sample RNA into the required well, using RNA-only pipettes. The NTC should have master mix and nuclease-free water. For the -RT, do not add RT enzyme, and compensate volume with nuclease-free water.Use equal volume for samples isolated from cell-free materials. For infected cell RNA, measure RNA concentration and load equal amounts of RNA, preferably in equal volume.
- Dispense the standards last, after performing at least 6 10-fold serial dilutions of the plasmids.It is generally not advisable to use DNA as a standard for absolute quantification of RNA because there is no control for the efficiency of the reverse transcription step. However, if it is not possible to in vitro transcribe RNA, the use of plasmid DNA with the template of interest is fine. Be sure to linearize the plasmid prior to amplification, as super-coiled DNA is not as efficiently amplified as linear DNA (template site is not as accessible to primers if DNA is super-coiled).
- To make serial dilutions of your standards, you first have to know the molar concentration of your plasmid. This way you can calculate copy numbers to be translated into genome copies later on.
- Determine the concentration of the linearized plasmid by spectrophotometry and convert the readout to grams per Liter.
- Calculate the formula weight (FW) for your plasmid as follows:
- Plasmid size (in base pairs)×662 grams/mol (average weight of a nucleotide pair).
- Calculate the molar concentration of your linearized plasmid (M):
- M=mole/L=(mass in grams/FW)/1L
- Determine copy number per microliter based on molar concentration:
- 1M is about 6.02×1023copies
- Make 10-fold serial dilutions of the plasmid in nuclease-free water and add 5 µL into the dispensed reaction mix. Make note of how many copies of plasmid each sample standard contains.Pipetting accuracy is crucial in this step for correct analyses of samples after the PCR run.
Cover the plate with the optical adhesive cover and then briefly centrifuge the plate to get contents to the bottoms of the wells, and to remove any air bubbles.
- Load the plate into the instrument and run the RT-qPCR reaction after programming the thermal cycler as follows:
50°C 10min 95°C for 5 min 94°C for 10 sec 
40× 60°C for 30 sec - Perform data analysis according to the instruments specific instructions.Make sure your standard curve has a correlation coefficient (R2) higher than 0.98, and shows a slope close to −3.3, which ensures your primers have close to 100% efficiency for amplification (you have double the original amount of template as each cycle progresses). Figure 8 shows an example of a standard curve obtained for MNV-1 standards. The most dilute standard within the linear range of the standard curve is 2 genome copies (Figure 8B). Thus, the reliable limit of detection for this particular protocol of MNV-1 amplification is 2 genome copies.
To calculate the genome copies per mL of virus in your preparation, you multiply the genome copies per well calculated in your RT-qPCR run by the dilution factor obtained after the RNA extraction experiment and according to the sample volume loaded in the reaction.
Figure 8.
RT-qPCR results for standard curve amplification of a plasmid containing the MNV-1 ORF1 region. Eight 10-fold serial dilutions were made of a plasmid containing the MNV-1 ORF1 region and were run as duplicates in an RT-qPCR experiment with Bio-Rad reagents and the Bio-Rad CFX96 Touch instrument. A) Amplification plot of MNV-1 standards. B) Automatic standard curve generated by the Bio-Rad CFX96 Touch instrument. Note the slope of the curve has a value of −3.319, which corresponds to the approximate 100.1% amplification efficiency of the primers to bind to its target template and allow for an exact doubling of nucleic acid quantities with each cycle of amplification.
BASIC PROTOCOL 6
Strand-specific quantification of MNV-1 RNA
The presence of negative strand viral RNA during any positive strand RNA virus infection is an indicator of active replication. The specific detection and quantification of the (−) strand viral RNA is therefore an important tool for the study of molecular mechanism of viral RNA replication. However, due to the fact that both most reverse transcriptase enzyme produce “self-primed” and “false-primed” cDNA, and the presence of both (+) and (−) strand during virus replication, absolute quantification of either strand is not possible by regular RT-qPCR. Several modifications can overcome these problems, including a thermo-stable reverse transcriptase and use of a non-viral tag sequence in the reverse transcription primer, which is then used for specific amplification of tagged cDNA, and make the assay strand-specific (Timofeeva and Skrypina 2001, Piche and Schernthaner 2003, Stahlberg, Kubista et al. 2004, Tuiskunen, Leparc-Goffart et al. 2010). The protocol described herein is based on a recently published method that utilizes these modifications for the detection and absolute quantification of MNV-1 positive and negative strand (Vashist, Urena et al. 2012). The basic protocol describes all the steps of strand-specific RT-qPCR (RNA-primer annealing, reverse transcription and qPCR). During reverse transcription, a strand-specific oligonucleotide primer containing sequence, complementary to the strand being detected, is annealed with the RNA and cDNA is synthesized using reverse transcriptase. This cDNA is then enzymatically amplified by SYBR based qPCR and the absolute number of RNA copies is determined by extrapolation from a standard curve. The standard curve–plotting function is available in most instrument software. If it is not, graphing software may be used instead (e.g., Microsoft Excel or GraphPad Prism). This qPCR protocol can reproducibly detect down to 50 copies per well.
Materials
Total RNA isolated from infected cells (see “Viral RNA extraction” in Alternate Protocol 4)
Control in vitro transcribed RNA (Support Protocol 4)
Nuclease-free water
10 mM dNTP mix
10 µM RT primers (Table 5)
SuperScript III (SS-III) Reverse transcriptase (Life Technologies, 18080–044)
5 × first-synthesis buffer (Life Technologies, supplied with SS-III)
0.1 M DTT (Life Technologies, supplied with SS-III)
40 U/µl RNaseOUT (Life Technologies)
2× MESA Blue Mastermix (Mesa Blue qPCR Mastermix for SYBR Assay, Eurogentec)
10 µM PCR primers (Table 5)
96-well qPCR plates
qPCR Thermal cycler (Life technologies)
Centrifuge
Thermosealer (Life technologies)
Table 5.
Primers used for RT-qPCR:
| RNA | Name | Sequence 5’ -3’ | Positiona | |
|---|---|---|---|---|
| Gpos | RT | TposGpos | CGGGAAGGCGACTGGAGTGCCCAAACATCTTTCCCTTGTTCb | 1760–1779 |
| qPCR - F | Tpos | CGGGAAGGCGACTGGAGTGCC | Non-viral | |
| qPCR -R | Gneg | TGGACAACGTGGTGAAGGAT | 1678–1697 | |
| Gneg | RT | TnegGneg | GGCCGTCATGGTGGCGAATAATGGACAACGTGGTGAAGGAT | 1678–1697 |
| qPCR - F | Tneg | GGCCGTCATGGTGGCGAATAA | Non-viral | |
| qPCR -R | Gpos | CAAACATCTTTCCCTTGTTC | 1760–1779 |
Anneal the primer to the RNA
-
1Mix the strand-specific RT primer, TposGpos or TnegGneg for positive- and negative-sense RNA respectively, RNA and dNTPs by adding the following to a microcentrifuge tube (MCT):
-
◦100 ng total RNA or serially diluted control standard RNA
-
◦0.2 µl RT primer
-
◦1 µl dNTP
-
◦H2O to 11 µl.
-
◦
-
2Heat to 65°C and snap freeze on ice.Heating the mix removes any secondary structures in the RNA and allows the primer to begin to anneal.
cDNA Synthesis
-
3
Centrifuge the sample for 10 seconds at 1000×g to collect the condensate at the bottom of the tube.
-
4
Add 9 µl of RT mix per reaction, prepared as following:
RT mix:-
◦4 µl 5× RT buffer
-
◦1 µl 0.1mM DTT
-
◦0.8 µl RNaseOUT
-
◦0.2 µl SS-III RT
-
◦H2O to 9 µl
A master mix can be prepared for more than one reaction and kept on ice. When using this approach it is advisable to include at least an extra 10–15% in the final volume of the master mix to allow for pipetting errors. Once prepared, master mix should be kept on ice. -
◦
-
5
Mix the contents of PCR tubes and incubate for 30 minutes at 55°C.
-
6
Inactivate the RT enzyme by heating the PCR tubes to 95°C for 5 minutes.
-
7Centrifuge the sample for 10 seconds at 1000×g to collect the condensate at the bottom of the tube.SS-III RT is a thermostable enzyme and has high processivity at 55°C. Use of a thermostable enzyme, a calibrated amount of RT primer and time of reaction allows minimum synthesis of false primed cDNA.It is important to centrifuge the tubes after heating to recover evaporated solution to accurately determine the RNA quantity.
Amplify the cDNA by qPCR
-
8
Mix the following:
Mix for (+) strand RNA detection-
◦0.25 µl each qPCR primer (Tpos and Gneg - Table 5)
-
◦10 µl 2× MESA Blue Mastermix
-
◦Nuclease free water to 18 µl.
Mix for (−) strand RNA detection-
◦0.25 µl each qPCR primer (Tneg and Gpos -Table 5)
-
◦10 µl 2× MESA Blue Mastermix
-
◦Nuclease free water to 18 µl.
Use of the tag primer gives specificity to the PCR reaction. Hence any false primed cDNA without the tagged RT primer would not be amplified during the PCR reaction.For each reaction, a master mix can be prepared for more than one reaction and kept on ice. When using this approach it is advisable to include at least an extra 10–15% in the final volume of the master mix to allow for pipetting errors. Once prepared, master mix should be kept on ice. -
◦
-
9
Transfer 18 µl of the PCR mix from the above step to wells of a 96 well qPCR plate.
-
10Add 2 µl cDNA standards (ranging for RNA representing 2 × 107 to 20 copies/well) to respective wells.It is important to dilute the standard RNA in presence of a carrier RNA. We typically use 20ng/µl yeast RNA.
-
11
Add 2 µl cDNA samples to respective wells.
-
12
Seal the plate with a thermosealer making sure that all the wells are sealed, particularly the corners of the plate.
-
13
Place the plate in the plate holder of the qPCR machine.
-
14
Program the machine with parameters as mentioned below. Data collection for the detection of PCR amplification is performed during the annealing and extension steps of the PCR cycle, while for the melt curve stage the data is collected at the ramp stage between annealing and dissociation.
-
15
Start the qPCR program.
qPCR Parameters:Hold Stage Heat activation: 95°C – 10 min PCR Amplification 
50 cycles Denaturation: 95°C – 15 sec Annealing: 58°C – 20 sec Extension: 72°C – 20 sec Melt curve Denaturation: 95°C – 15 sec 
Ramp rate 0.05°C/min Annealing: 58°C – 20 sec Denaturation: 95°C – 20 sec -
16
A typical layout of the plate for a growth curve using time points 0 hr, 6 hr, 12hr and 24hr, including standard curve and no template control is presented in the Figure 9.
Figure 9.
A typical layout of the qPCR plate is shown. Row 1–6 contains master mix for detection of positive sense RNA and Row 7–12 contains master mix for containing negative sense RNA. Row 1–3 and 4–6 contains samples from mentioned time-points of growth curve while row 4–6 and 10–12 contains samples for standard curve and no template control.
Analysis of the qPCR data
-
17
Analyse the data by extrapolating the standard curve.
-
18The standard curve plotting function is available on most software provided with any qPCR machine. If it is not, graphing software may be used instead by plotting Ct Values vs Log10 copy number of the control standard RNA.It is very important to analyse the melting curve before analysing the amplification data. The melting point (Tm) of all the PCR products should be identical. If the Tm deviates, it is indicative of either primer dimer formation or the generation of non-specific PCR products. Such wells should be excluded from the analysis of amplification data.
SUPPORT PROTOCOL 4
GENERATION OF STANDARD RNAs REPRESENTING MNV-1 (+) AND (−) SENSE GENOMIC RNAs
For the absolute quantification of RNA copies in a sample, it is important to make a standard curve using a known number of copies of control standard RNA representing the amplified region. This protocol describes a method for the preparation of RNA standards representing the positive and negative strands of the MNV-1 genomic RNA. The use of RNA instead of DNA for the generation of a standard curve also allows for control at the RT step. The control standard RNA can be synthesized by in vitro transcription reaction as described below. Several commercially available in vitro transcription kits could also be used to prepare the RNA.
Materials
100 ng/µl plasmid DNA template (pT7: MNV1 3’Rz, available without MTA from Dr. Ian Goodfellow, Cambridge University)
10 mM dNTP mix
KOD polymerase kit (Merck)
10 µM PCR primers (Table 6)
1% agarose TBE gels (T. Maniatis 1982)
0.5 × TBE (T. Maniatis 1982)
DNA ladder (e.g., GeneRuler 1-kb Plus, Fermentas)
PCR purification kit (GE Healthcare)
0.4 M Tris pH 8.0
320 mM Magnesium acetate
400 mM DTT
20 mM spermidine
100 mM ATP
100 mM GTP
100 mM CTP
100 mM UTP
Pyrophosphatase (Promega)
40 U/µl RNaseOUT (Life Technologies)
Nuclease-free water
T7 RNA polymerase (Promega)
37°C water bath
2 U/µl DNase I (RNase-free Life Technologies)
25:24:1 (v/v/v) phenol/chloroform/iso-amyl alcohol
24:1 (v/v) chloroform/iso-amyl alcohol
3 M sodium acetate, pH 5.2
100% (v/v) ethanol
75% (v/v) ethanol
Table 6.
Primers used for generation of standard RNAs:
| Standard RNA |
Name | Sequence 5’ −3’ | Positiona |
|---|---|---|---|
| Gpos7 | Gpos-F | GCGTAATACGACTCACTATAGGGCTTTTGGCCTCACCTCTG | 1085–1104 |
| Gpos-R | CCAAGATGAAATTGATGTGGCTGTAATCGGGCC | 1954–1986 | |
| Gneg7 | Gneg-F | GCGTAATACGACTCACTATAGGGTGCCAAGATGAAATTGATG | 1970–1990 |
| Gneg-R | GCTTTTGGCCTCACCTCTG | 1086–1104 |
The genome positions are with reference to the MNV-1.CW-1 genome RNA sequence. The standard RNAs were designed to include the neighboring RNA secondary structure complexity to imitate the actual RNA.
The 5’ end of the forward primer (underlined sequence) is T7 promoter. T7 polymerase needs this sequence to start RNA synthesis.
Generate PCR product with T7 promoter at the 5’ end6
-
1Generate Gpositive and Gnegative PCR products using pT7: MNV1 3’Rz as template and PCR primers detailed in Table 6.pT7:MNV1 3’Rz is the full length infectious clone of MNV-1 CW-1 (Chaudhry, Skinner et al. 2007).
-
2
Run the PCR product on 1% agarose gel and purify the DNA using a commercially available gel purification kit.
Generate RNA using in vitro transcription
-
3Mix the following in the order as described below:It is important to maintain the order of addition of the components in this reaction and to mix well after adding each component. Usually the master mix can be prepared for the above mix. Keep all reagents on ice once thawed. A commercially available kit (e.g., Promega, P1440) could also be used for in vitro transcription.
-
◦10 µl 0.4M Tris pH 8.0
-
◦10 µl 320mM Magnesium acetate
-
◦10 µl 400mM DTT
-
◦10 µl 20mM spermidine
-
◦7.5 µl 100mM ATP
-
◦7.5 µl 100mM GTP
-
◦7.5 µl 100mM CTP
-
◦7.5 µl 100mM UTP
-
◦2 µl Pyrophosphatase (New England Biolabs, M2403L)
-
◦5 µl 40 U/µl RNaseOUT (Life Technologies, 10777-019)
-
◦10 µl PCR product (>200ng)
-
◦10 µl T7 RNA polymerase (Promega, P2074)
-
◦Nuclease-free water to 100 µl.
-
◦
-
4
Mix well and incubate at 37°C for 2–4 hours.
-
5
Run 2 µl on denaturing 5% PAGE (Biorad mini protean gel or similar) to check the yield and purity of the RNA (T. Maniatis 1982).
-
6
Add 2 µl DNase I and continue the incubation at 37°C water bath for another30 minutes.
-
7
Add 100 µl nuclease-free water.
-
8
Add 20 µl 0.3 M sodium acetate to each sample and mix well.
-
9Extract each sample with 200 µl of 25:24:1 tube phenol/chloroform/iso-amyl alcohol. Carefully transfer only the aqueous phase from each tube to a new 1.5 ml microcentrifuge.Phenol is a neurotoxin. Local safety regulations for its use and disposal should be followed.
-
10
Add 500 µl of 100% ethanol and mix well. Microcentrifuge each sample 15 min at 13,000 × g to collect the RNA in the pellet.
-
11
Carefully remove the liquid from the small white pellets and add 400 µl of 75% ethanol to each tube. Centrifuge 5 min at 13,000×g.
-
12Carefully remove all the liquid from the pellets. Dry the pellets at room temperature and dissolve in 100 µl nuclease free water.Excessive drying would prevent the RNA from dissolving in water.
-
13
Quantitate the RNA using nano-drop (UV-spectrophotometer).
-
14
Convert RNA concentration to copy number.
Tools and scientific calculators are widely available to do this calculation (see internet resources). To calculate manually:- Calculate the concentration of RNA using formula: 1 A260 = 40 µg/ml
-
Calculate the MW weight of RNA molecule using formula: # of nucleotides × 320.5) + 159(320.5 is average MW of nucleotide and 159 is MW of a 5’triphosphate)
- Calculate copy number using formula: Copy number/µl = 6.0223×[RNA] µg/mL×10−9/[MW g/mol]
-
15
Aliquot and store the samples at −80°C at 1010 copies/µl.
BASIC PROTOCOL 7
siRNA TRANSFECTION OF BV-2 CELLS USING NEON® TRANSFECTION SYSTEM
Understanding the role of host cell genes during MNV infection is facilitated by manipulating these genes in tissue culture. siRNAs designed against specific genes are a powerful tool that can be delivered into cells to downregulate expression of their target mRNA. Transfecting nucleic acid into cells can be achieved by a variety of mechanisms, including chemical, mechanical and electrical delivery. Electroporation of nucleic acid into cells was first proposed in 1982 (Neumann, Schaefer-Ridder et al. 1982), and has since developed into a highly efficient research tool. To overcome the initial problems of low cell viability, low transfection rates and complicated protocols, in 2008 Kim et al. (Kim, Cho et al. 2008) produced a novel electroporation system using a capillary and wire-type electrode. This was initially developed as the Microporator (MP-100) by Digital Bio. This has since been optimized for, and is now marketed by Life Technologies as the Neon® transfection system. In cell culture, MNV infects cells of the macrophage lineage, such as BV-2 cells (murine microglial cell line) and RAW264.7 cells (murine leukaemic monocyte macrophage cell line). siRNA transfection of BV-2 cells via the Neon® transfection system (Life technologies) is efficient and has been successfully used to study the role of target host genes during MNV infection (Vashist, Urena et al. 2012). This protocol describes the methods used to transfect large numbers of cells with a specific siRNA, often resulting in significant reduction in the target protein within 18 hours of transfection. However, it is recommended to analyze target gene expression at various timepoints between 12 and 48 hours post transfection to determine the optimum time for maximal reduction in gene expression.
Materials
Confluent flask(s) of BV-2 cells
NEON®; pipette; cat MPP100
NEON® 100ul kit; cat MPK10096 (contains buffer E & R, gold tips, and tip holder)
Cell culture 6 well dishes
15 ml sterile conical tubes (e.g. Falcon tubes, BD Biosciences)
Autoclaved 1.5 ml micro-centrifuge tubes (MCTs)
- BV-2 growth medium pre-incubated in a humidified 37°C/5% CO2 incubator
-
◦DMEM
-
◦10% fetal calf serum (FCS)
-
◦1% NaHCO3
-
◦1% L-glutamine
-
◦
siRNAs: directed against the target and a control housekeeping gene (e.g. GAPDH)
Trypan Blue vital stain
Sterile phosphate-buffered saline (PBS)
1% penicillin/streptomycin (P/S)
- Aspirate the medium from the BV-2 cells, wash the monolayer with PBS and detach the cells using commercial trypsin. A total of 7.2×106 live cells will be required per transfection.To prepare cells for transfection, they should be passaged 24 hours prior to the start of the protocol to ensure cells are actively growing.
Neutralize trypsin using BV-2 medium, then remove the suspended cells from the culture flask and centrifuge in a table top centrifuge at 1200×g for 5 mins. Remove the supernatant and re-suspend the cells in 10 ml of BV-2 medium.
Determine the concentration of live cells in presence of trypan blue.
- Transfer 7.2 × 106 live cells to a sterile microcentrifuge tube for each transfection. Centrifuge cells at 1200×g for 2 min in table top centrifuge, remove supernatant then re-suspend in 1ml PBS.It is advisable to leave cells in growth media until immediately before washing with PBS. Typically cells can be aliquotted into sterile microcentrifuge tubes and kept at room temperature until required. The time period that cells are exposed to PBS and buffer R should be kept to an absolute minimum.
- Place the electrode of the neon system inside the tissue culture hood where the transfection is to take place and place a Neon®; tube (tip holder) in the electrode. Add 3 ml Solution E to this Neon® tube.Each Neon® tube with buffer E can be used for up to 10 transfections without contamination.
Set the instrument parameters for the following conditions; voltage 1700, width 10ms, pulse 3.
Label 6-well plates (3 wells per sample) and 15ml conical tubes. Add 9 ml BV-2 media (without P/S) to each conical tube.
- Centrifuge the 7.2×106 cells suspended in PBS from step 4 at 1200×g for 2 min in table top centrifuge. Remove the supernatant then re-suspend the cells in 120µl buffer ‘R’ to achieve the desired cell concentration of 6.6 × 106 cells/100µl for each transfection.It is advised to use 20% extra cells in 20% extra R buffer as described above to avoid bubble formation during aspiration of cells by the neon tip. The amount of siRNA should thus be altered accordingly.
Add the required amount of target siRNA to 7.2 X 106 cells cells in buffer ‘R’, mixing carefully and thoroughly. 5 µl of a 10 µM stock of siRNA (0.5 µmol) is recommended as a starting concentration.
- Gently mix the cells slowly and aspirate 100 µl of the cell-siRNA mix into the Neon® tip using the Neon® pipette. Insert this into the pipette station until a ‘click’ is heard.Note: It is essential to avoid any air bubble formation during setting up of the sample for electroporation. Formation of bubbles during mounting of the electrode is common, and the process must be carefully repeated until no bubbles are present. Air bubbles result in the generation of sparks during electroporation and hence major cell death.
Press the start button to start electroporation. The machine will confirm successful electroporation once completed.
Remove the Neon® pipette from the pipette station and transfer the electroporated cells to the 9 ml media in the conical tube (pre-prepared in step 8).
- Repeat steps (8)–(12) for the remaining samples.Note: Each Neon® tip can be reused up to a maximum of three times for transfection of cell samples with the same type of siRNA.
Mix the electroporated cells slowly in the fresh media and add 3 ml each to 3 wells of labeled 6 well dishes.
Incubate at 37°C/5% CO2 until the cells form a monolayer. This can take between 6–18 hours. When a monolayer is formed, replace the media with BV-2 cell media containing 1% P/S.
Harvest cells at pre-determined time-points (between 12 and 48 hours are suggested) and analyze protein expression of the target gene using western blotting (Figure 10).
Figure 10. siRNA mediated reduction in protein expression as demonstrated by western blotting.
siRNA targeting the host cell gene DDX3 and control siRNA were transfected into BV-2 using the Neon ™ transfection system. Cells were harvested at 18, 24 and 30 hour timepoints and protein extracted. Protein samples were then separated using SDS-PAGE and analysed using western blotting with anti-DDX3 and anti-GAPDH as a loading control.
BASIC PROTOCOL 8
LENTIVIRAL TRANSDUCTION OF PRIMARY BONE MARROW DERIVED MACROPHAGES FOR MNV INFECTION
Analysis of viral replication in primary cells is an efficient way to study the interaction of virus with its host in more physiological conditions than the study in transformed or immortalized cell lines. However, compared to established cell lines, it is difficult to obtain homogenous population of primary cells in sufficient number for experimentation and to modify the phenotype of primary cells due to the low efficiency of transfection. Bone marrow-derived macrophages obtained through culturing bone marrow progenitor cells with macrophage colony-stimulating factor (M-CSF) (Stanley, Guilbert et al. 1983) is a good solution to securing a large number of primary cells for MNV infection, and transduction of these cells with lentivirus is an efficient way to over-express or down-regulate a gene of interest. In this protocol, bone marrow cells are harvested and cultured in the medium containing M-CSF to generate bone marrow derived macrophages (Support Protocol 5) and lentiviruses are produced in 293T by transfecting lentiviral vector with packaging and pseudotyping vectors (Support Protocol 6). Prepared lentiviruses are used to transduce adherent cells from the bone marrow progenitors twice on Day 4 and 5 of culture. On Day 7, transduced bone marrow-derived macrophages are harvested for further selection or subsequent assays. This procedure optimally yields 5–10×106 macrophages, of which > 90% express the gene of interest or the cassette for down-regulating the gene of interest.
Materials
Day 4 bone marrow culture for the derivation of macrophages (Support Protocol 5)
Lentiviral vectors (Support Protocol 6)
PBS/EDTA solution (e.g., Sigma, cat no. E8008)
Sterile cell lifter (e.g., Fisher Scientific, cat. no. 08 100 240)
Macrophage complete medium (see recipe)
Additional reagents and equipment for cell counting (e.g., hemacytometer), centrifugation and cell culture (e.g., biosafety cabinet and 37°C/5% CO2 incubator)
- On Day 4 of bone marrow culture (Support Protocol 5), gently swirl the plate a couple of times to detach any loosely bound materials and remove the unbound with culture medium.Typically, 50–60% confluency is expected on Day 4.
- Add 5 ml of fresh macrophage complete media and half of the prepared lentivirus (i.e., ca. 10 ml) (support protocol 6) per each 10 cm plate.Replacing macrophage complete media help cells stay healthy.Addition of polybrene does not enhance the transduction of vesicular stomatitis virus G protein (VSV-G) pseudotyped lentiviral vectors.
On Day 5, discard the culture medium with lentivirus and add 5 ml of fresh macrophage complete media and the rest of the prepared lentivirus (i.e., ca. 10 ml) per each 10 cm plate.
Optional: Transduced cells can be selected by the expression of reporter gene (e.g., green fluorescence protein from pCDH-MCS-T2A-copGFP-MSCV) or drug resistance gene (e.g., puromycin N-acetyltransferase from pLKO.1 puro). For the drug selection of transduced cells, add drug (e.g., 1 µg/ml puromycin) for 3 days, on Days 6 through 9.
- On Day 7 (or on Day 9 for drug-selected cells), remove unbound cells and discard culture medium. Add 5 ml of cold PBS to the remaining adherent cells, swirl gently to remove loosely bound cell debris, discard the PBS, and add 5 ml of cold PBS/EDTA per each plate. Incubate at 4°C for 10 minutes.For transduced but unselected cells, a confluency of 90–100% is expected, while 70–80% confluency is expected for the transduced and drug-selected cells.
- Scrape off the cells with cell lifter, add 5 ml of cold macrophage complete media per plate, resuspend the cells by pipetting up and down, and transfer them to 50 ml conical tube.Keep macrophages cold to minimize loss due to their adherence to the tube.
Centrifuge at 300 × g, 4°C, for 5 minutes.
- Discard supernatant, resuspend the cells in 5–10 ml macrophage complete medium, and count the number of cells.The volume of the resuspension medium needs to be determined based on subsequent assays. The typical yield at this stage is 5–10×106 transduced macrophages for the unselected and 1–3×106 transduced macrophages for the drug-selected cells per 10 cm plate.
- At this time, macrophages should be replated for further experimentation. Usually, the cells are plated at 0.5–1 × 105 cells/well in 1 ml of macrophage complete medium for 24-well tissue culture plate or at 2.5–5 × 105 cells/well in 2 ml of macrophage complete medium for 6-well tissue culture plate.3 days after seeding, macrophages are ready to be used for MNV infection (for example as described in Basic Protocol 1) and subsequent assays (e.g., Western blot or quantitative PCR to measure the over-expression or down-regulation of genes of interest).The efficiency of transduction and/or expression of gene of interest in the transduced cells can vary significantly depending on the size of lentiviral vector. In general, the efficiency inversely correlates with the size of lentivirus. For example, over 90% of cells expressed green fluorescent protein upon transduction with lentiviruses from pCDH-MCS-T2A-copGFP-MSCV vector itself, but only 20–30 % of cells showed the reporter expression upon transduction with lentiviruses having an additional ca. 2 kbp gene(Hwang, Maloney et al. 2012).
SUPPORT PROTOCOL 5
PREPARATION OF MOUSE BONE MARROW SUSPENSION FOR MACROPHAGE CULTURE
This protocol describes the purification of bone marrow progenitor cells from mouse hind legs (both tibias and femurs) and differentiation into bone marrow-derived macrophages with media containing M-CSF.
Materials
Donor mice of 6 to 12 week old, specific pathogen-free
Phosphate-buffered saline (PBS)
Macrophage complete medium (see recipe)
70% Ethanol
Forceps and scissors (keep in sterile container containing 70% ethanol)
15- and 50-ml conical centrifuge tube, sterile
10- or 30-ml syringes with 26-G needles, sterile
100 × 15-mm petri dishes (non-tissue culture treated)
Additional reagents and equipment for mouse euthanasia
- Euthanize a mouse according to the guideline of home institution (e.g., CO2 asphyxiation then cervical dislocation; see Appendix 3N). Briefly sterilize the euthanized mouse either by spraying sufficient 70% ethanol to cover the whole body or by soaking the mouse in 70% ethanol. Cut out the foot, peel off skin, and cut off the hind legs at the hip joint, keeping the tibias and femurs intact.Dissection in clean environment (e.g., inside biosafety cabinet) is recommended to minimize any contamination.
Remove excess muscle from tibias and femurs, briefly dip them in the 70% ethanol in the petri dish for quick sterilization, and put them in a new clean petri dish.
- Remove the mouse carcass, clean up the biosafety cabinet, and get a new pair of gloves and sterile equipment if necessary.If the dissection equipment is going to be used again, make sure to clean and sterilize them with 70% ethanol.
- Prepare a 50 ml conical tube per mouse and fill up the tube with macrophage complete media. Load a syringe with the media in the tube and attach 26G needle.Alternatively, you can start with wash media or PBS to collect bone marrow first, spin down at 300×g, for 5 min, discard supernatant, and resuspend the cell pellet in the macrophage complete media.
- Using sterile forceps and scissors, cut the both ends of bones proximal to each joint and insert needle into bone marrow cavity. Flush bone cavity with media until the color of bone cavity changes from red to white.It takes usually 2–3 ml of media to completely flush out bone marrows from one bone.
Combine the bone marrows from a single mouse into a 50 ml conical tube.
- Mix well by pipetting and plate the cells into non-tissue culture treated petri dishes. Usually, dispense 10 ml of bone marrow suspension per petri dish and make up to 10 petri dishes per mouse.Macrophages do not come off tissue culture-treated plastic ware. Make sure to use non-tissue culture treated petri dishes to enable replating of bone marrow-derived macrophages.Each petri dish of cells is transduced with a single type of lentivirus, so the number of petri dishes of cells should match the number of lentiviruses for transduction.Typically, bone marrow progenitor cells from a single mouse can be plated in up to 10 petri dishes to produce similar number of bone marrow-derived macrophages per dish at the time of harvest (i.e., on Day 7).
- Incubate the cells in 37°C, 5% CO2 incubator for four days.All culture incubations should be performed in a humidified, 37°C, 5% CO2 incubator unless otherwise specified.
SUPPORT PROTOCOL 6
PRODUCTION OF LENTIVIRAL VECTOR
This protocol describes the preparation of VSV-G pseudotyped lentiviruses through the transfection of relevant plasmids into 293T. VSV-G is a highly stable protein with ubiquitously expressed receptors on mammalian cells, so it enhances the structural stability and tropism of the pseudotyped lentivirus.
Materials
293T (ATCC, CRL-3216)
Dulbecco’s modified Eagle medium/10% Fetal Bovine serum (DMEM-10)
Trypsin/EDTA (e.g., Cellgro, 25-053-CI)
- Plasmids:
- Pseudotyping vector encoding the G protein of vesicular stomatitis virus (VSV-G, e.g., pMD2.G, Addgene plasmid 12259)
- Lentiviral packaging vector (e.g., psPAX2, Addgene plasmid 12260)
- Lentiviral vector of choice (e.g., pCDH-MCS-T2A-copGFP-MSCV, System Biosciences CD523A-1, for the over-expression of a gene or pLKO.1 puro Addgene plasmid 8453, for the down-regulation of a gene)
Sterile water
2.5 M CaCl2, sterile
2× HeBS (see recipe), sterile
Caution: Handling of human cell line requires special caution due to possible contamination of human pathogens.
- On the same day that the bone marrow progenitor cells are harvested (Day 0), seed 4×106 293T cells in a 10-cm tissue culture treated dish with 10 ml DMEM-10. Make as many dishes as you want to produce lentivirus and set up transduction.The ideal cell density at the time of transfection is 60–70% confluency. Too few cells or too many cells can lead to the loss of cells during the harvest and reduced yield of lentiviruses.
- Day 1: For each 10 cm plate of 293T cells to be transfected, prepare the following transfection mix in a sterile 15 ml conical tube:
- 6.5 µg packaging vector
- 3.5 µg VSV-G pseudotyping vector
- 10 µg lentiviral vector
Add 100 µl of 2.5 M CaCl2 and 900 µl sterile water to the DNA mix. Mix well by pipetting up and down.
- In another 15-ml tube, add 1 ml of 2× HeBS. While vortexing the tube containing 2× HeBS, add the ca. 1 ml of DNA/CaCl2 mix dropwise to the 2× HeBS tube.Set up the speed of the vortex first to mix vigorously without spilling out the contents.
Leave the tube at room temperature for 5–30 min to allow the formation of fine precipitates of DNAs for transfection.
- While incubating, replace the medium of 293T cells with 10 ml of fresh DMEM-10.The media can be replaced as early as 3 hours before transfection.
- Add the precipitate from step 6 in a dropwise manner onto the cells (total ca. 2 ml per plate), covering the whole surface of plate. Add drops right above the surface of medium to minimize any impact that can detach 293T cells from the plate. Shake the plate gently and perpendicularly a couple of times until the medium has recovered a uniform color. Incubate the cells in 37°C, 5% CO2 incubator for 12–16 hours.Caution: 293T cells easily detach from the culture dish.
- Aspirate the medium and add 7 ml of macrophage complete media gently to the transfected cells. Incubate the cells in 37°C, 5% CO2 incubator.Biosafety Level 2 (BSL-2) practices are required for handling of lentivirus.After removing medium containing any excess transfection mix, add back the medium of target cells for transduction.
12 hours after media change, harvest the culture medium from each plate, collect them in a 50 ml conical tube, and add 7 ml of fresh macrophage complete media back to the transfected cells. Incubate the cells in 37°C, 5% CO2 incubator and store the collected medium at 4°C.
Repeat step 9 two more times every 12 hours and keep the culture medium at 4°C over the collection period. Discard the transfected 293T cells after the third harvest, according to your institutional biosafety regulations.
- Filter the pooled culture medium (ca. 20 ml final per each transfection) using a 0.45 µm filter unit.The filtered culture medium containing lentivirus can be used directly or concentrated if needed.It can be kept at 4°C for a couple of days or −80°C for a long-term storage.Avoid freeze/thaw cycles as they reduce the titer of lentivirus preparations.
ALTERNATE PROTOCOL 5
LENTIVIRAL TRANSDUCTION OF MACROPHAGE CELL LINES TO OVER-EXPRESS OR DOWN-REGULATE A GENE OF INTEREST FOR MNV INFECTION
Lentiviral vectors produced in Support Protocol 6 can be also used to transduce macrophage cell lines (e.g., RAW264.7 or BV-2) commonly used for the study of MNV to stably over-express or down-regulate genes of interest.
Materials
Lentiviral vectors (Support Protocol 6)
Macrophage cell lines: RAW264.7 (ATCC, cat. no. TIB-71) or BV-2 (Blasi, Barluzzi et al. 1990)
Sterile cell lifter (e.g., Fisher Scientific, cat. no. 08 100 240)
Dulbecco’s modified Eagle medium/10% Fetal Bovine serum (DMEM-10)
- Seed 4 × 106 RAW264.7 cells or 2×106 BV-2 cells in 10 ml DMEM-10 in a tissue culture-treated 10 cm dish approximately 24 hours before transduction.The expected cell density at the time of transduction is 30–40% confluency.
- On the day of transduction, add lentiviruses produced as in support protocol 6 onto the 10 cm dish of macrophage cell lines. Incubate for three days.In general, for the efficient transduction without titration of the produced lentivirus, match the size of plate of cells for transduction to the size of plate for the preparation of lentivirus. If necessary, scales down the size of plates for both lentivirus production and transduction proportionally. Biosafety Level 2 (BSL-2) practices are required for handling of lentivirus.
- Three days later, discard the culture medium containing lentivirus, wash once with 10 ml PBS, and add 5 ml of cold DMEM-10. Scrape off the cells with cell lifter, add additional 5 ml of cold DMEM-10, resuspend the cells by pipetting up and down, and transfer them to 50 ml conical centrifuge tube. Count the number of cells.The transduced cells can be analyzed directly at this time or they can be replated for further selection by the expression of reporter gene (e.g., green fluorescence protein from pCDH-MCS-T2A-copGFP-MSCV) or drug resistance gene (e.g., puromycin N-acetyltransferase from pLKO.1 puro).
- Optional: For the drug selection of transduced cells, cells are replated at 1×106 of RAW264.7 cells or 0.5×106 of BV-2 cells per well in 2 ml of DMEM-10 for 6-well tissue culture plates. At ca. 24 hours after seeding, add drug (e.g., 5 µg/ml puromycin) for 3 days.The selective concentration of drug for individual cell lines should be titrated before the selection in a pilot experiment. The duration and frequency of drug treatment should be adjusted depending on the degree of cell death, since too much cell death can cause false negative result.
BASIC PROTOCOL 9
TRANSFECTION OF INFECTIOUS VIRAL RNA INTO MOUSE EMBRYONIC FIBROBLAST
Macrophages and dendritic cells are the only reported cell types that can be infected by MNV (Wobus, Karst et al. 2004). However, as shown in the example of reverse genetics systems for MNV, MNV can replicate in many other cell types as long as its infectious genome is delivered into host cells (Yunus, Chung et al. 2010). For example, mouse embryonic fibroblasts (MEFs) are one cell type, which cannot be infected by MNV but can support MNV replication (Hwang, Maloney et al. 2012). MEFs isolated from genetically manipulated mouse lines are a valuable tool to study the function of mutated genes for viral replication under well-defined culture condition. In this protocol, viral RNA of MNV purified from concentrated viral stock (Support Protocol 7) is transfected into MEFs (Support Protocol 8) by lipofectamine 2000 to produce infectious MNV. The procedure optimally yields ca. 5×106 plaque forming unit of MNV per 1 µg of transfected viral RNA.
Materials
viral RNA of MNV (Support Protocol 7)
MEFs (Support Protocol 8)
Lipofectamine 2000 (Life Technologies)
Opti-MEM-I medium (Life Technologies)
- Seed 1 × 104 MEF cells in 1 ml DMEM-10 per well in tissue culture-treated 24-well plates ca. 24 hours before transfection.The expected cell density at the time of transfection is 30–40% confluency. 0.5 ml medium will be sufficient for 24-well plate, but 1 ml medium is recommended for even distribution of cells on the plate. Clumped cells significantly lower the transfection efficiency and viral yield. If necessary, drugs (e.g., interferon-gamma) can be used to treat cells at 12 hours prior to transfection.
- On the day of transfection, prepare transfection mix as follows according to the manufacturer’s instruction:
- Dilute 0.2 ~ 0.5 µg of infectious viral RNA in 50 µl of Opti-MEM-I medium. Mix gently.Handling of RNA requires extra caution to minimize any possible contamination of RNase and subsequent degradation of RNAs.
- Mix Lipofectamine 2000 gently by inverting the tube 3–4 times and dilute 2 µl in 50 µl of Opti-MEM-I medium. Mix gently by swirling and incubate for 5 minutes at room temperature.
- After the 5 minute incubation, combine the diluted RNA with the diluted Lipofectamine 2000. Mix gently by swirling and incubate for 20 minutes at room temperature.
Add the 100 µl of RNA-Lipofectamine 2000 complexes to each well of cells and mix gently by rocking the plate perpendicular a couple of times.
Incubate cells at 37°C in a CO2 incubator for 24 hours.
- To harvest MNV, freeze/thaw the entire 24-well plate by placing it at −80°C and later at room temperature. Virus-containing lysate can then be easily transferred into tubes and stored at −80°C for follow-up assays.Approximately 1×106 plaque forming units of MNV are produced per transfection (Hwang, Maloney et al. 2012). Other lipososome-mediated transfection reagents may be used, but Lipofectamine 2000 worked most reproducibly and produced the highest titer of viruses.
SUPPORT PROTOCOL 7
PREPARATION OF INFECTIOUS VIRAL RNA FROM MNV STOCK
Infectious viral RNA of MNV can be extracted directly from concentrated viral stocks (Support Protocol 1) using the standard TRIzol method. The genome-linked VPg protein, which is essential for the translation of viral RNA, stays functional after RNA preparation with TRIzol unless the RNA is treated with proteases.
Materials
TRIzol (Life Technologies) or TRI Reagent (Sigma)
Chloroform
Isopropanol
75% ethanol
RNase-free water
Water bath or heat block (55–60°C)
- Prepare concentrated MNV virus stock (Support Protocol 1) and add appropriate volume of TRIzol to homogenize, according to the manufacturer’s instruction.Concentrated viral stock is recommended as starting materials for better estimation of viral yield upon transfection of infectious viral RNA. However, a regular MNV stock can also be used as starting material.Depending on the type of TRIzol used (e.g., TRIzol vs. TRIzol LS), adjust the volume of added TRIzol. This protocol is based on the use of regular TRIzol, and thus 10 volumes (i.e., 1 ml for 0.1 ml viral stock) of TRIzol is used.
Incubate the homogenate at room temperature for 5 minutes and add 0.2 ml chloroform per 1 ml of TRIzol used.
Shake the samples vigorously for 15 seconds and incubate the resulting mixture at room temperature for 2–3 minutes.
- Centrifuge the samples at 12,000×g, 4°C for 15 minutes.Note: Centrifugation at 4°C is essential for clear phase separation.
- Transfer the aqueous phase (transparent solution on top of pink solution) to a new microcentrifuge tube.The volume of the aqueous phase will be ca. 60% of the volume of TRIzol used.
Add 0.5 ml of isopropanol, per 1 ml of TRIzol used initially, and incubate at room temperature for 10 minutes to precipitate RNA.
- Centrifuge at 12,000×g, 4°C for 10 minutes and remove the supernatant.The pellet of viral RNA may or may not be visible on the centrifugal side of the tube, depending on the amount of starting material used. Try not to touch the centrifugal side while removing the supernatant.
Add 1 ml of 75% ethanol, per 1 ml of TRIzol used, to the centripetal side of the tube. Mix the sample by inverting the tube a couple of times and centrifuge at 7,500×g, 4°C for 5 min.
- Remove supernatant, spin again the tubes briefly, and remove supernatant as much as possible.Another brief centrifugation is recommended to bring down any residual ethanol on the side of tube and thus to remove the washing ethanol more effectively.
-
9Air-dry the RNA at room temperature for 5 minutes, add the appropriate volume of RNase-free water to the centrifugal side of the tube, and incubate the samples at 60°C for 15 minutes. Keep the tubes with dissolved RNA on ice while measuring the concentration of RNA and then keep the RNA at −80°C until used.
-
9
SUPPORT PROTOCOL 8
PREPARATION OF MOUSE EMBRYONIC FIBROBLAST CELLS
MEFs are usually isolated from mouse embryos at embryonic day 13.5 (E13.5) to E15.5, but it can be prepared as early as E8.5 if the embryos have lethal mutations. It is recommended to prepare both control MEFs and mutant MEFs from mice with same backgrounds to minimize phenotypic variation due to different genetic background.
Materials
Pregnant female mouse
PBS
10-ml syringe, sterile
Dulbecco’s modified Eagle medium/10% Fetal Bovine serum (DMEM-10)
Trypsin/EDTA (e.g. Cellgro, 25-053-CI)
Prepare two 10 cm petri dishes with 20 ml of PBS.
- Euthanize a pregnant female mouse with embryos of desired genetic background according to the guideline of home institution (e.g., CO2 asphyxiation then cervical dislocation; see Appendix 3N). Briefly sterilize the euthanized mouse either by spraying sufficient 70% ethanol to cover the whole body or by soaking the mouse in 70% ethanol.Dissection in a clean environment (e.g., inside biosafety cabinet) is recommended to minimize any contamination.
- Cut out and peel off abdominal skin and open peritoneum. Dissect out the entire uterus containing all embryos without damaging the other internal organs and transfer the uterus to a 10 cm petri dish with PBS.Damaging the other internal organs, especially intestines, can lead to bacterial contamination.
- Remove the mouse carcass, clean up the biosafety cabinet, and get a new pair of gloves and sterile equipment if necessary.If the dissection equipment is going to be used again, make sure to clean and sterilize them with 70% ethanol.
- Cut out the uterine wall and take out individual embryos. Transfer all embryos into another 10 cm dish with 20 ml PBS.Remove maternal materials as much as possible, especially when the genetic background of embryos differs from their mother (e.g., embryo with homozygous genetic knock-out from heterozygous mother).
Count the number of embryos, and prepare new 10 cm dishes with 10 ml PBS each twice the number of embryos.
- Transfer each embryo individually into a 10 cm dish. Remove and discard internal organs, limbs and head of embryo. Transfer the torso of embryo to a new 10 cm dish with PBS.Keep a small piece of a discarded part of the embryo for DNA extraction and subsequent genotyping if necessary.
Using a syringe head, crush the torso as much as possible. Resuspend the crushed pieces with PBS and transfer them to a 15 ml tube. Centrifuge at 300×g for 5 min.
Discard supernatant and resuspend the pellet with 10 ml DMEM-10. Transfer the resuspended pieces of the crushed embryo into a T-175 flask and add 20 ml more media.
- Incubate the flask in 37°C, 5% CO2 incubator for 7 days.MEFs grow out from the tissue debris of the crushed embryo. Shake the flask once a day to spread out the tissue debris for maximal use of the space and even distribution of MEFs in the flask.
Discard media and loosely bound debris. Add 30 ml PBS to the flask and swirl it to detach any loosely bound debris. Discard PBS and any debris and add 5 ml of Trypsin/EDTA. Incubate the flask at 37°C for 5 minutes.
- Resuspend the cells with 25 ml more media and transfer to 50 ml conical tube.At this stage, if necessary, any remaining clumps or debris can be removed by allowing them to settle down by gravity for ca. 1 minute. Transfer the remaining cell suspension to a new tube.
Centrifuge at 300×g for 5 min. Discard supernatant and resuspend the pellet with 30 ml media. Split the resuspended cells into 3× T-175 flasks and add 20 ml more media to each flask. Incubate the flask in 37°C, 5% CO2 incubator for 2 days. These are the passage 1 (P1) MEFs.
- Upon confluency (usually, in two days), subculture the P1 MEFs for experiment or freeze them down (5×106 cells/vial in culturing media with 10% DMSO). Use the primary MEFs until P5 (based on 1:3 split).The typical yield of P1 MEFs is ca. 2–3×107 cells per embryo.
To make a stable line of MEF, immortalize the MEF either by serial passage (20–25 times) or transformation with oncogene (e.g., SV40 large T antigen).
REAGENTS AND SOLUTIONS
DMEM-10 medium
high glucose DMEM
10% (v/v) low-endotoxin fetal bovine serum (< 10 EU/mL)
10 mM HEPES
100 U/mL penicillin
100 µg/mL streptomycin
1 mM non-essential amino acids
2 mM L-glutamine
Store up to 4 weeks at 4°C
ELISA Coating Buffer
1.53 g disodiumtrioxocarbonate(IV) (Na2CO3)
2.93 g sodiumhydrogentrioxocarbonate(IV) (NaHCO3)
Check pH (pH 8.8 – 9.6), then bring to 1 l
ELISA III Buffer
0.15 M sodium chloride (NaCl)
0.001 M Ethylenediaminetetraacetic acid (EDTA)
0.05 M Tris-hydrochloric acid (Tris-HCl)
0.05 % Tween 20
pH to 7.4, bring to 1 l then add BSA to 0.1 %
ELISA Substrate Buffer
0.1 M sodium citrate + 1 mM 2,2'-azino-bis(3-ethylbenzothiazoline-6-sulphonic acid) (ABTS) + 0.016 % (v/v) H2O2
- To make 2 L:
- 25.6 g citric acid
- 25.6 g disodiumhydrogentetraoxophosphate(V) (Na2HPO4)
- pH to 4.2, bring to 2L with Milli Q
- Add 1.096 g ABTS
Aliquot 12ml/tube and store at −20°C.
Add 12 µl hydrogen peroxide (H2O2) right before adding to ELISA plate.
ELISA Wash Buffer
0.15 M NaCl
0.05 % Tween 20
ELISA Blocking Buffer
Coating buffer with 3 % BSA
ELISA Stop Buffer
0.2 N Phosphoric Acid: add 6.8 mL 85% Phosphoric Acid (14.7N) per 500 ml water
Complete 2×MEM
2× MEM
10% (v/v) low-endotoxin fetal bovine serum (< 10 EU/mL)
10 mM HEPES
100 U/mL penicillin
100 µg/mL streptomycin
4 mM L-glutamine
Equilibrate to 37°C before use.
Store at 4°C for up to 1 month.
3% Seaplaque
Add 3 g of SeaPlaque agarose in a total volume of 100 ml of distilled water (3% w/v) in a glass bottle.
Autoclave for 20–30 min.
It is important to equilibrate SeaPlaque agarose to 42°C in a water bath before use.
Store autoclaved Seaplaque at room temperature and reheat in microwave prior to use.
Seaplaque/2×MEM solution
Mix SeaPlaque agarose (at 37–42°C) and 2× MEM media (at 37°C) 1:1 in a sterile bottle immediately before use.
Neutral red/PBS solution
Add 3 ml of neutral red (0.33% w/v in DPBS; Sigma, catalog # N2889) to 100 ml of 1× PBS (tissue culture grade, Mg2+-, Ca2+-free; Gibco, catalog # 10010).
Equilibrate to 37°C before use.
Store at 4°C in dark.
10× TBE (Tris/borate/EDTA) electrophoresis buffer (1L)
Dissolve 108 g Tris base, 55 g boric acid and 40 ml 0.5 M EDTA, pH 8.0 in 700 ml water.
Add water to 1 L.
0.5 M EDTA (ethylenediamine tetraacetic acid), pH 8.0
Dissolve 408 g Na2EDTA·2H2O in 700 ml nuclease free water.
Adjust pH to 8.0 with 10 M NaOH (>45 ml).
Add water to 1 L.
3 M sodium acetate, pH 5.2
Dissolve 40.8 g sodium acetate trihydrate (NaC2H3O2·3H2O) in 80 ml nuclease-free water.
Adjust pH to 5.2 with 3 M acetic acid.
Add water to 100 ml.
Macrophage complete medium
Add the following reagents to 700 ml of Dulbecco’s modified Eagle medium (DMEM): 100 ml of Fetal Bovine Serum
50 ml of horse serum
10 ml of 200 mM L-glutamine
10 ml of 100 mM sodium pyruvate
100 ml of CMG14-12 conditioned medium (equivalent to mouse M-CSF or L-929 conditioned medium (Stanley, Guilbert et al. 1983))
Adjust the volume to 1000 ml with DMEM.
CMG14-12 conditioned medium
CMG14-12 is a mouse Ltk- cell line expressing large amount murine M-CSF (Takeshita, Kaji et al. 2000). CMG14-12 cells are cultured in α-MEM until confluent. CMG14-12 conditioned medium is then harvested every other day for five times. Culture supernatants are filtered through a 0.22 µm nitrocellulose filter, divided into ca. 50 ml aliquots and stored at −80°C until use.
HEPES-buffered saline (HeBS), 2×
Dissolve the following reagents in 800 ml of molecular biology grade water:
16.36 g NaCl (Mol. Wt. 58.44, 0.28 M final)
11.9 g HEPES (Mol. Wt. 238.3, 0.05 M final)
0.213 g anhydrous Na2HPO4 (Mol. Wt. 142, 1.5 mM final)
Adjust pH to 7.05 with 5 M NaOH and add more water to 1000 ml final
Filter sterilize through a 0.22 μm nitrocellulose filter.
Divide into 50-ml aliquots.
Store at 4°C for short-term use or at −20°C for long-term storage.
Note: An exact pH is extremely important for efficient transfection. The optimal pH range is 7.05 to 7.12. There can be wide variability in the efficiency of transfection obtained between different batches of 2× HeBS. Efficiency should be checked with each new batch.
COMMENTARY
Background Information
MNV reverse genetics
Reverse genetics serves as a cornerstone for unraveling the life cycle of many viruses, allowing the recovery of genetically defined wild type or mutant strains, making it a very important tool in modern virology. As other reverse genetics systems have been established for other members of the Caliciviridae family, efforts were made to introduce a system with MNV resulting in a total of three reverse genetics systems to date (Chaudhry, Skinner et al. 2007, Ward, McCormick et al. 2007, Yunus, Chung et al. 2010). The two DNA-based reverse genetics systems rely on either the DNA polymerase II promoter or the T7 polymerase promoter to drive expression of viral cDNA after transfection of a DNA plasmid containing the full-length MNV genome into easily transfectable cells, typically HEK293T cells. The method by Ward et al. (Ward, McCormick et al. 2007) (see Basic Protocol 3) relies on the use of a DNA polymerase II promoter to generate a capped transcript. Another method which proved successful for the recovery of FCV is the transcription of RNA from transfected cDNA, driven by the T7-RNA polymerase from vaccinia virus (MVA-T7) (Geissler, Parrish et al. 1999). However, this method failed when applied to the recovery of MNV as MVA-T7 replication interferes with the translation and replication of MNV (Chaudhry, Skinner et al. 2007). Therefore, the Alternate Protocol 2 based on FPV-T7 constitutes an improved modification on initial attempts carried out with MVA-T7. Although both FPV and MVA are members of the Poxviridae family, FPV displays a tropism only for avian cells and its replication in mammalian cells is defective. This allows the production of the T7 RNA polymerase, which drives the synthesis of the transfected MNV cDNA via the T7 promoter, without interfering with MNV translation and replication as observed with MVA-T7. Generating infectious MNV from cDNA cannot be undertaken using cell lines in which MNV is known to replicate in, such as RAW264.7 cells, due to difficulties transfecting these cells. Instead, commonly used transfectable cell lines can be used to recover the virus, mainly HEK293T cells, but HepG2, BHK-21, or COS-7 cells have also been tested (Chaudhry, Skinner et al. 2007, Ward, McCormick et al. 2007). BSR-T7 is a cell line derived from BHK-21 stably expressing T7 RNA polymerase. However, the expression of T7 RNA polymerase in these cells is insufficient to recover MNV from cDNA. This might be due to the low T7 RNA polymerase expression levels detected in these cells compared to FPV-T7 infected cells alongside the absence of capping enzymes provided by FPV-T7 that could be providing a cap to the produced MNV transcripts. Since the transfectable cell lines are not susceptible to MNV infection however, reported titers represent a single replication cycle and can provide information on the fitness of the recovered virus.
The RNA-based reverse genetic system (Basic Protocol 4) described herein represents the most efficient recovery of genetically defined MNV in cell culture compared to the DNA-based reverse genetics systems. The transfection of capped RNA, which can be recognized by the cellular translation machinery, effectively bypasses the absolute requirement for the covalent linkage of VPg to the 5' end of the viral RNA genome (Yunus, Chung et al. 2010). When transfected into BV-2 cells, virus titres recovered with the RNA-based reverse genetics systems are similar to those obtained by transfection of viral VPg-linked RNA purified from infected cell cultures (Chaudhry, Skinner et al. 2007, Yunus, Chung et al. 2010). A more direct and often more cost effective method for the recovery of infectious MNV from capped RNA transcripts is via transfection into permissive BSR-T7 cells. Given that BV-2 cells or RAW264.7 cells are difficult to transfect using lipid-based approaches, other cell lines can be used such as BSR-T7 cells, a derivative of the BHK-21 cell line. These cells are easy to transfect and are permissive to MNV replication but do not allow multiple rounds of infection. There are other cell lines also suitable to this approach including human 293T cells, human hepatocellular carcinoma Huh7 cells and African green monkey Cos7 cells.
MNV qRT-PCR
A quantitative polymerase chain reaction (qPCR) is a modification of the polymerase chain reaction (PCR, where amplification and simultaneous quantification of one or more targeted DNA sequences is achieved (Higuchi, Dollinger et al. 1992). Additionally, quantitation and amplification can be visualized in real time, because the assay is fluorescence-based which, with the proper controls, can allow for detection of absolute levels of template (Higuchi, Dollinger et al. 1992, Bustin, Benes et al. 2009). The technique originally arose in the 1990s (Higuchi, Dollinger et al. 1992), by observing that ethidium bromide had the capacity to fluoresce and this fluorescence increased in the presence of double-stranded DNA. This fluorescence could be monitored externally and could allow for determination of nucleic acid quantity. Thus, the ability to simultaneously amplify specific DNA sequences and detect the product of the amplification allowed this technique to revolutionize modern molecular biology.
Quantitative polymerase chain reaction has the capacity to detect nucleic acids in a wide range of samples from numerous sources: mammalian eukaryotic single cells and tissues, plant cells and tissues, prokaryotes, stool samples and complex fluids (e.g. blood, plasma, saliva). The quantity of amplified DNA can then be calculated as either an absolute number of copies or a relative amount when normalized to input DNA input or additional normalizing genes/parameters. Additionally, the technique has been modified to make it safer for handling by utilizing alternate fluorophores, such as fluorescein, that are minimally if at all toxic. This has made the technique easier to handle and more accessible, to the point of allowing several modifications of it to flourish.
Different kinds of qPCR have been developed in the past years. For example, multiplex qPCR allows for the simultaneous detection of more than one nucleic acid sequence in a single tube through the use of specific fluorescent probes targeting each sequence individually. If no specific probes are available, the use of the double-stranded DNA-binding dye SYBR Green can allow for amplicon detection. qPCR has also been modified to include a coupled reverse transcription step (RT-qPCR) where RNA is used as the starting template, converted to cDNA and then subsequently amplified. The technology is usually able to estimate mRNA abundance/gene expression with a remarkable degree of certainty and sensitivity. Indeed this sensitivity of the technique, for example, can be assessed when comparing it to older methods of measuring nucleic acid (e.g. Northern blot for mRNA) abundance. In contrast to older techniques, it requires minimal amounts of template to work and is reliable over a greater dynamic range of RNA concentration.
Critical Parameters and Troubleshooting
Propagation and purification of MNV
Healthy cells are a prerequisite for optimal virus growth. Cells should be split at least twice after thawing before using them for virus infections. RAW 264.7 cells do not grow to 100% confluency. Instead cells will grow on top of each other and clump. Thus, cells should be split at ~90% confluency before they pile up.
Not all MNV strains cause the clear CPE seen with MNV-1. For those strains it may be necessary to determine the optimal time of harvest by performing a viral growth curve (i.e., infect cells with MNV as described in Basic Protocol 1 and take samples every 6 – 12 hours to determine viral titers).
If the yield of purified virus is low, check the starting titer of your virus stock. Thus, is it advisable while optimizing the protocol to save a few µl of virus at each step of the protocol to check for increase in purity and/or virus titer.
MNV ELISA
The development time should not exceed 10 min as eventually all samples will turn color.
Plates should not dry out. If multiple ELISA plates are processed at one time, let plates sit in wash buffer, then flick out buffer one plate at a time and add new solution to the plate before proceeding to the next plate.
DNA-based, pol II-driven generation of recombinant MNV
Increasing DNA concentration might improve the transfection efficiency.
Incubation times are crucial. You need to find a good balance in having long transfection times and keeping the cells happy. If you change the media after 24h, keep the supernatant and test it for virus.
Make sure you do not have plasmid contamination in your analysis should you use RT-PCR on recombinant virus that does not plaque.
Make sure you have healthy 293T cells; maybe test EGFP transfection efficiency first (especially when using new serum). Mycoplasma contamination negatively affects transfection efficiency and you do not want them in your virus preps anyway.
You may want to keep 293T and RAW264.7 cell medium separate. 293T easily get contaminated with mycoplasma.
Make sure you have true 293T – if 293T’s change their behavior over time e.g. they do not come off easily anymore, this could be a HELA cell contamination (they look pretty much identical).
293T are bad cells for immunofluorescence since they come off easily and change shape during treatment.
DNA-based, T7-driven generation of recombinant MNV
Seeding density:This protocol is optimized for the use of BHK or its derivative cell line, BSR-T7. The seeding density described for these two cell lines are designed to provide a confluent cell monolayer for transfection with Lipofectamine 2000. The correct seeding density should be determined for any other cell line that is to be used. All these transfectable cell lines are permissive to MNV replication, but they are not susceptible to infection as they cannot be infected by MNV, thus the virus yields obtained represented that from a single round of replication.
cDNA transfection: The quality of the cDNA prepared can affect the recoveries. DNA preparations must be clear of any contaminating endotoxin for optimal transfection efficiency.
FPV-T7 titration: The FPV-T7 should be titrated before use and used to inoculate cells at a multiplicity of infection of 0.5 pfu/cell, based on the titer in primary CEFs. Otherwise, a higher multiplicity of infection can affect cell viability while a lower multiplicity of infection can result in insufficient number of cells infected and expressing T7-polymerase. Both scenarios can affect virus yields.
The titration of an FPV-T7 stock generated as mentioned above is performed by plaque assay in CEFs under solid overlay containing 1% agarose as previously described (Buttigieg, Laidlaw et al. 2013). Typically, a functional titration of the newly generated virus stock is performed. To this aim, we compare MNV recovery yields of a well-characterised stock of FPV-T7 with the new preparation to ensure batch-to-batch comparability. Additional options for titrations rather than plaque assay in primary CEFs include the use of an immunofocal assay for T7 RNA polymerase or the use of a reporter assay reliant on luciferase expression under the control of a T7 RNA polymerase promoter.
Low titres of infectious MNV can be due to a variety of reasons. Poor transfection efficiency or a cDNA clone that carries deleterious mutations can both culminate in low recovery yields. To determine the cause of low titers, a duplicate for each recovery should be performed. One sample would to be used to titer the recovered virus, while the other to be tested for protein extraction. NS7 detection by western blot permits the analysis of protein expression levels; a weak signal may signify a low transfection efficiency resulting in low virus titers. If this is the case, first check the integrity of the cDNA clone using restriction digestion and sequencing. Then ensure the purity of the DNA is sufficient and finally use different ratios of cDNA:Lipofectamine and/or extended incubation periods to improve the efficiency of virus recovery. Conversely, detecting a strong signal for NS7 is a sign of good transfection efficiency. Hence, lower virus yields than in wild type preparations typically suggests that the viral cDNA encodes for a lower fitness virus.
RNA-based generation of recombinant MNV
The quality of linearized MNV cDNA and subsequently synthesized genomic RNA are the key parameters for effective recovery of infectious MNV.
Ensure an RNase-free environment is maintained throughout the protocol. Degradation of RNA will compromise the final yield and may lead to failure to recover infectious viral particles.
Adhere to the step annotations in the protocol for key advice at each stage.
During the Neon®-mediated transfection of RNA into BV-2 cells, it is important to prevent the formation of bubbles by pipetting. Introduction of bubbles during the electroporation will compromise the cell viability and have negative impact on the final yield of recovery.
If infectious MNV fails to be recovered, check first whether the capped RNA used for transfections remained intact. Also, western blot analysis against the viral proteins (e.g., NS7 Pol) may be used to confirm whether the transfection is successful. The cytopathic effect (CPE) of MNV on BV-2 cells after over 24 hours post transfection is another indicator of successful transfection and production of infectious virus. It is worth noting however the possibility that mutant viruses may replicate without causing CPE. In this situation, viral RNA should be detected and can be quantified by qRT-PCR and the viral protein expression should be confirmed by western blot.
TCID50
The most common source of error in this protocol is accidental contamination and carry-over of virus from one dilution to another, or the contamination of the media or cells used for diluting the virus. To avoid this, plate out the cells in the test plates ahead of time, and be sure to exchange tips at each dilution, and between samples. Performing the mock TCID50 after the virus samples should highlight if contamination has occurred.
Plaque assay
Healthy RAW 264.7 cells are critical for the success of this assay. Do not let cells overgrow as those cells do not typically form plaques. Keep track of the passage number and use lower passage cells (we use cells below passage 30).
Do not let cell monolayers dry out. Only process up to 5 plates at a time.
Carefully count cells when plating, as no plaques form when cells are too sparse or too dense.
If no plaques are visible before staining the monolayer, incubate for an additional 4 hrs and check again. The maximum incubation time should not exceed 72 hrs.
If no plaques are visible after staining even at 72 hrs, check monolayer with light microscope. If all cells are dead, likely the agarose or staining solution was too hot when adding to the monolayer. If cells are healthy, they were likely too dense when plated and unable to form plaques. Alternatively, some MNV strains, don’t form plaques or plaques are very small and hard to visualize. In that case use TCID50 protocol (Basic Protocol 5).
MNV qRT-PCR
- For this protocol, reverse transcription can be performed as either a single reaction for cDNA synthesis or coupled to qPCR.
- If the sample of interest contains additional expressed RNA (aside from viral RNA) that is to be amplified subsequently, it is best to perform a single reaction for cDNA synthesis using non-specific primers (e.g. oligo-dT). This is done in order to avoid the worry of RNA degradation if several reactions have to be performed. However, this will only work for poly-adenylated mRNA molecules, and can skew the reaction towards the most abundant targets in the sample. If only viral RNA amplification is desired, a single-tube one-step RT-qPCR reaction, where cDNA synthesis is coupled to amplification by the use of gene-specific primers, can be performed.
- These reactions can be done using home-made reagents, as long as optimization of amplification conditions is performed. However, the use of commercially-available mixes, when possible, is recommended as they contain additives that provide stability to the reaction components and can also allow for faster amplification.Optimization of qPCR amplification should be performed using the primer/probe matrix method, temperature gradient optimization for annealing, and salt concentration modifications (Lutfalla and Uze 2006, Mikeska and Dobrovic 2009).
High-quality RNA is crucial for efficient amplification and quantification. Follow correct isolation and handling technique.
Since PCR amplifies all target nucleic acid, whether from an intact virion, defective (non-infectious) virions, or from free nucleic acids in solution, qPCR results (expressed in terms of genome copies/mL) are likely to be higher in quantity than results obtained through other methods of viral detection (e.g., TCID50 or plaque assay). This should be kept in mind when analyzing the data.
Although the use of DNA standards is effective in this situation, it is recommended that, whenever possible, in vitro transcribed RNA be used as standards for the reaction since it more accurately reflects the efficiency of reverse transcription that the unknown samples will undergo.
The probe should be protected from light at all times and not stored for long periods of time with too dilute an aliquot.
If no amplification, poor signal or no signal is seen, you have no detectable PCR product. It could be that there are inhibitors present in the reaction, that you used inadequate buffers/master mixes, that you used inadequate cycling conditions, you forgot the reverse transcription step or that the probe and/or the template degraded.
If a signal is detected in the negative controls (e.g., non-template control or no RT control), you may have contamination of your reaction components, primer dimer formation due to wrong cycling steps or contamination with DNA.
In case of low or high reaction efficiency, there might have been pipetting inaccuracy in making the standards, primer-dimer formation if conditions are not optimal (wrong temperature and master mix) or probe quality is bad.
In case the Cq values of replicate samples are not similar, the most likely cause is pipetting inaccuracy.
Strand-specific RT-qPCR
It is important to accurately measure the standard RNA concentration by UV –spectrophotometry. The OD260 of the RNA should be between 0.2–0.8 to measure the quantity accurately. If the OD260 is above this range, dilute the RNA in nuclease free water.
As the protocol deals with repetitive pipetting of small volume, care should be taken to minimize the error. Pipettes should be carefully calibrated and care should be taken to ensure no liquid either adheres to the outside of the tips during pipetting or remains within the tip after mixing of components.
It is important to analyze the melting curve before extrapolating sample copy number from the standard curve. The melting temperature of sample wells should be same as that of standard RNA wells.
It is important to run sufficient negative controls (no template control) each time, using the same aliquot of nuclease free water that has been used during the protocol.
siRNA transfection of BV-2 cells using NEON transfection system
Cells should not stay in buffer R for more than 15 minutes. It is important to keep the cells in their appropriate media until immediately prior to the first centrifugation in PBS.
- A number of protocol changes may be required if a reduction in target protein expression is not observed using the standard protocol. Excessive cell death and failure of the transfection may also occur as a result of the formation of bubbles. Extreme care should be taken to ensure this does not occur. Assuming efficient transfection has occurred, examples of changes that may be required include:
- Design of alternative siRNAs against the target gene.
- Use of a combination of pooled siRNAs targeting the same gene can be effective.
- Increasing the concentration of siRNA used to transfect the cells can increase the efficacy of siRNA.
- Multiple rounds of transfection may also be required, but care should be taken to ensure sufficient cell survival is maintained.
Preparation of primary bone marrow cells
The most common problem in preparing primary cells is contamination by bacteria and fungi. Careful sterilization of equipment and sterile operation techniques are essential to minimize the risk of contamination.
Culturing the primary cells without antibiotics (e.g., penicillin and streptomycin) helps to identify and respond to any contamination of cultures quickly.
Production of lentiviral vector
Although the 293T cell line is highly transfectable, transfection efficiency of 293T cells is the most critical parameter in the production of lentiviral vectors.
The right cell density and good health condition of the 293T cells (including low passage) and optimal pH of 2× HeBS are critical factors and need to be well controlled.
Lentiviral transduction of macrophage cell lines
Drug-mediated selection of transduced cells needs a constant monitoring of drug effect and titration of optimal drug concentration, since the screening and subsequent growth of the transduced cells is significantly affected by the death of the other cells and consequent population density.
Too much cell death due to low transduction efficiency can lead to false negative result (i.e., the death of the transduced cells).
On the contrary, drug is less effective in killing un-transduced cells if the population density of treated cells is too high and it can lead to false positive result (i.e., the survival of the un-transduced cells).
Time Considerations
Generation of MNV-containing lysate
Maximum MNV-1 titers following a low MOI infection (i.e., MOI 0.05) are reached ~42–48 hours. However, the optimal time has to be individually determined for each MNV strain.
Generation of concentrated MNV stock
The amount of MNV lysate to be concentrated determines the length of time required to concentrate virus. In one day, ~360 ml of lysate can be concentrated (i.e., 2 centrifuge runs).
MNV purification
Purification takes two - three days from start to finish. Day 1 is a long day and will be spent spinning down large cellular debris, pelleting and resuspending virus and preparing the CsCl gradients. Day 2 will be spent by fractionating the gradient and determining RFI. Alternatively, the procedure can be performed in three days. A good stopping point is to incubate the virus pellet with PBS overnight at 4oC. The gradient will then be prepared on day 2 and fractionated on day 3.
MNV ELISA
The ELISA assay takes 1 ½ days, with coating plates on day 1, while the rest is done on day 2. If needed, coating times can be extended for several days (e.g., over the weekend).
DNA-based, pol II-driven generation of recombinant MNV
Transfection of a plasmid takes ~30 min followed by an incubation time of at least 48 h to generate recombinant virus. Amplification of the recombinant virus on RAW 264.7 cells takes an additional 48 h. The optional step of plaque purification will take 2 days. To verify the mutation is contained in the virus stock, amplification of the region of interest by RT-qPCR takes another day. Sequencing time for the amplicon depends on the availability of local sequencing core. Alternatively, samples can be submitted to commercial sequencing facilities but additional shipping time will need to be considered.
DNA-based, T7-driven generation of recombinant MNV
Cell seeding as described in step 1 typically requires 30 min. They can be left overnight to settle, or for 3 hours if seeded at double the recommended amount and transfected the same day. Infecting with FPV-T7 and transfecting the cells with desired infectious clones takes a maximum of 3 h. Cells can be left to incubate from 24–72 h. After sufficient incubation time, cells can be frozen and stored in −80°C until use. After thawing, cells should be clarified from samples and the virus-containing supernatant can be aliquoted and stored at −80°C for further use.
RNA-based generation of recombinant MNV
Preparation of linearized DNA will take about 4 hours in total including 3 hours for restriction digestion and purification. In vitro transcription can be performed within 8 hours depending on how long the reaction is incubated, the amount of RNA desired and the method used to assess the RNA integrity. As mentioned above precipitation using LiCl can be performed the same day or can be left overnight. In vitro enzymatic capping can be performed within 4 hours with subsequent purification. Depending on the methods, the recovery of infectious MNV will take half a day to one day to set up and will require up to 3 days to obtain virus particles, based on how many hours post-transfection the viruses are harvested. Obtaining the titres of recovered viruses will generally take 3 to 4 days, depending which titration method is used.
TCID50
With practice, a single TCID50 can be performed in under 20 minutes. However it is common to process large numbers of samples in parallel. For example, TCID50 assays of 30–40 samples takes 3–4 hours. This can be noticeably shortened by the use of an electronic multichannel pipette (e.g. an Eppendorf Xplorer Plus) capable of serial dilutions as this represents the most time-intensive portion of this method.
MNV plaque assay
Plating cells takes approximately an hour. On the following day, the length of the infection is dependent on the number of samples being analyzed. A typical assay with 30–40 samples takes ~3 hours for virus dilutions, infection and agarose overlay. Plaques are stained 2–3 days later, which takes 30 min for applying the stain, 2–3 hour incubation for staining, and another hour for counting plaques and tabulating virus titers.
MNV qRT-PCR
Time for RNA extraction depends on the number of samples, but if using a kit it should take from 30 minutes to a couple of hours. DNAse treatment typically takes about 45 minutes (more if a large number of samples are used). qPCR plate setup should take about 15 minutes and the run in the thermal cycler takes about 40 minutes.
Strand-specific RT-qPCR
The RT step takes approx. 30 minutes and the qPCR program runs for 2 hours 40 minutes. To analyze 10 experimental samples and standard curve samples, the complete protocol can be achieved in 4–5 hours.
siRNA Transfection of BV-2 cells using NEON transfection system
Allow approximately 1 hour to complete this protocol for a single siRNA transfection and the relevant siRNA transfection control.
Lentiviral transduction of primary bone marrow derived macrophages for MNV infection
Harvesting bone marrow progenitor cells from a mouse should take less than 30 minutes. Transfection into a 10 cm dish of 293T cells takes about 40 minutes and each harvesting should take less than 1 minute per dish. Filtration of final lentiviral vector preparation takes about 5 minutes per virus. If bone marrow progenitor cells and transfectable 293T cells are prepared on Day 0, the 293T cells is transfected on Day 1 and the resulting lentiviral stocks is harvested Day 2 and 3. Bone marrow-derived macrophages are transduced on Day 4 and 5 and ready for replating on Day 7 (Day 9 in case of drug selection). The transduced cells become ready for MNV infection at 3 days after replating.
Transfection of infectious viral RNA into MEFs
Isolating embryos from a pregnant female mouse and setting up culture for MEF takes less than 2 hours. 9 additional days are required to generate passage 1 MEFs from the day of embryo isolation. Extraction of infectious viral RNA from the concentrated stocks of MNV takes about 1 hr. The transfection of the viral RNA into MEFs takes about 40 minutes and generation of infectious MNV from the transfected MEFs takes a day.
ACKNOWLEDGEMENTS
Work in the laboratory of C.E.W. was supported by National Institutes of Health (NIH) grants AI102106, AI080611, AI103961, and Defense Advanced Research Projects Agency (DARPA) Contract HR0011-11-C-0093. Work in the laboratory of I.G. was supported by Biotechnology and Biological Sciences Research Council grants BB/I012303/1 and BB/K002465/1, and Wellcome Trust grant WT097997MA. Work in the laboratory of S.H. is funded by a start-up package from the Department of Pathology at the University of Chicago.
Footnotes
The volumes and components outlined here are found in the BioRad iScript One-step RT-PCR kit for Probe mentioned in the materials section.
Internet Resources
A protocol for the generation of a neutral red-containing MNV stock to measure replicating virus can be found here: http://www.bio-protocol.org/wenzhang.aspx?id=415
A video for the generation of recombinant MNV can be found here: http://www.jove.com/video/4145/reverse-genetics-mediated-recovery-of-infectious-murinenorovirus
Excel spreadsheet for calculating TCID50 values: http://www.lindenbachlab.org/resources.html
A video for the plaque assay can be found here: http://www.jove.com/video/4297/plaque-assay-for-murine-norovirus
A link to convert RNA concentration to copy number: http://www.endmemo.com/bio/dnacopynum.php
Contributor Information
Seungmin Hwang, Email: shwang@bsd.uchicago.edu.
Armando Arias, Email: aa759@cam.ac.uk.
Sarah L Caddy, Email: slc50@cam.ac.uk.
Constantina Christodoulou, Email: cc732@cam.ac.uk.
Ed Emmott, Email: ee273@cam.ac.uk.
Marta Gonzalez-Hernandez, Email: martag@umich.edu.
Abimbola Kolawole, Email: akolaw@med.umich.edu.
Jia Lu, Email: jl766@cam.ac.uk.
Christine Rippinger, Email: cmrippinger@gmail.com.
Frédéric Sorgeloos, Email: fs391@cam.ac.uk.
Lucy Thorne, Email: lt375@cam.ac.uk.
Surender Vashist, Email: sv345@cam.ac.uk.
Ian Goodfellow, Email: ig299@cam.ac.uk.
Christiane E. Wobus, Email: cwobus@umich.edu.
LITERATURE CITED
- Arias A, Bailey D, Chaudhry Y, Goodfellow I. Development of a reverse-genetics system for murine norovirus 3: long-term persistence occurs in the caecum and colon. J Gen Virol. 2012;93(Pt 7):1432–1441. doi: 10.1099/vir.0.042176-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi E, Barluzzi R, Bocchini V, Mazzolla R, Bistoni F. Immortalization of murine microglial cells by a v-raf/v-myc carrying retrovirus. J Neuroimmunol. 1990;27(2–3):229–237. doi: 10.1016/0165-5728(90)90073-v. [DOI] [PubMed] [Google Scholar]
- Bocchini V, Mazzolla R, Barluzzi R, Blasi E, Sick P, Kettenmann H. An immortalized cell line expresses properties of activated microglial cells. J Neurosci Res. 1992;31(4):616–621. doi: 10.1002/jnr.490310405. [DOI] [PubMed] [Google Scholar]
- Britton P, Green P, Kottier S, Mawditt KL, Penzes Z, Cavanagh D, ASkinner MA. Expression of bacteriophage T7 RNA polymerase in avian and mammalian cells by a recombinant fowlpox virus. J Gen Virol. 1996;77(Pt 5):963–967. doi: 10.1099/0022-1317-77-5-963. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Buttigieg K, Laidlaw SM, Ross C, Davies M, Goodbourn S, Skinner MA. Genetic screen of a library of chimeric poxviruses identifies an ankyrin repeat protein involved in resistance to the avian type I interferon response. J Virol. 2013;87(9):5028–5040. doi: 10.1128/JVI.02738-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chachu KA, Strong DW, LoBue AD, Wobus CE, Baric RS, Virgin HW., IV Antibody is critical for the clearance of murine norovirus infection. J Virol. 2008;82(13):6610–6617. doi: 10.1128/JVI.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J Gen Virol. 2007;88(Pt 8):2091–2100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox C, Cao S, Lu Y. Enhanced detection and study of murine norovirus-1 using a more efficient microglial cell line. Virol J. 2009;6:196. doi: 10.1186/1743-422X-6-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler K, Parrish CR, Schneider K, Truyen U. Feline calicivirus capsid protein expression and self-assembly in cultured feline cells. Vet Microbiol. 1999;69(1–2):63–66. doi: 10.1016/s0378-1135(99)00089-9. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Hernandez MB, Bragazzi Cunha J, Wobus CE. Plaque assay for murine norovirus. J Vis Exp. 2012;(66):e4297. doi: 10.3791/4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KY. Caliciviridae. In: DM Knipe PH, editor. Fields Virology. Vol. 1. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 949–980. [Google Scholar]
- Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (N Y) 1992;10(4):413–417. doi: 10.1038/nbt0492-413. [DOI] [PubMed] [Google Scholar]
- Hsu CC, Wobus CE, Steffen EK, Riley LK, Livingston RS. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin Diagn Lab Immunol. 2005;12(10):1145–1151. doi: 10.1128/CDLI.12.10.1145-1151.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, Green KY, Lopez-Otin C, Xavier RJ, Thackray LB, Virgin HW. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11(4):397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Lay M, Davidson J, Virgin HW., IV STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299(5612):1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- Kim JA, Cho K, Shin MS, Lee WG, Jung N, Chung C, Chang JK. A novel electroporation method using a capillary and wire-type electrode. Biosens Bioelectron. 2008;23(9):1353–1360. doi: 10.1016/j.bios.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Kitajima M, Oka T, Tohya Y, Katayama H, Takeda N, Katayama K. Development of a broadly reactive nested reverse transcription-PCR assay to detect murine noroviruses, and investigation of the prevalence of murine noroviruses in laboratory mice in Japan. Microbiol Immunol. 2009;53(9):531–534. doi: 10.1111/j.1348-0421.2009.00152.x. [DOI] [PubMed] [Google Scholar]
- Lindenbach BD. Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol Biol. 2009;510:329–336. doi: 10.1007/978-1-59745-394-3_24. [DOI] [PubMed] [Google Scholar]
- Lutfalla G, Uze G. Performing quantitative reverse-transcribed polymerase chain reaction experiments. Methods Enzymol. 2006;410:386–400. doi: 10.1016/S0076-6879(06)10019-1. [DOI] [PubMed] [Google Scholar]
- Madore HP, Treanor JJ, Dolin R. Characterization of the Snow Mountain agent of viral gastroenteritis. J Virol. 1986;58(2):487–492. doi: 10.1128/jvi.58.2.487-492.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler M, Kohl W. A serological survey to evaluate contemporary prevalence of viral agents and Mycoplasma pulmonis in laboratory mice and rats in western Europe. Lab Anim (NY) 2009;38(5):161–165. doi: 10.1038/laban0509-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikeska T, Dobrovic A. Validation of a primer optimisation matrix to improve the performance of reverse transcription - quantitative real-time PCR assays. BMC Res Notes. 2009;2:112. doi: 10.1186/1756-0500-2-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumphrey SM, Changotra H, Moore TN, Heimann-Nichols ER, Wobus CE, Reilly MJ, Moghadamfalahi M, Shukla D, MKarst S. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J Virol. 2007;81(7):3251–3263. doi: 10.1128/JVI.02096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann E, Schaefer-Ridder M, Wang Y, Hofschneider PH. Gene transfer into mouse lyoma cells by electroporation in high electric fields. EMBO J. 1982;1(7):841–845. doi: 10.1002/j.1460-2075.1982.tb01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parwani AV, Saif LJ, Kang SY. Biochemical characterization of porcine enteric calicivirus: analysis of structural and nonstructural viral proteins. Arch Virol. 1990;112(1–2):41–53. doi: 10.1007/BF01348984. [DOI] [PubMed] [Google Scholar]
- Piche C, Schernthaner JP. Background priming during reverse transcription by oligo(dT) carried over from mRNA isolation. Biotechniques. 2003;34(4):720–722. 724. doi: 10.2144/03344bm08. [DOI] [PubMed] [Google Scholar]
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. American Journal of Hygiene. 1938;27:493–497. [Google Scholar]
- Ruiz S, Beauvillain C, Mevelec MN, Roingeard P, Breton P, Bout D, Dimier-Poisson I. A novel CD4-CD8alpha+CD205+CD11b− murine spleen dendritic cell line: establishment, characterization and functional analysis in a model of vaccination to toxoplasmosis. Cell Microbiol. 2005;7(11):1659–1671. doi: 10.1111/j.1462-5822.2005.00583.x. [DOI] [PubMed] [Google Scholar]
- Smith DB, McFadden N, Blundell RJ, Meredith A, Simmonds P. Diversity of murine norovirus in wild-rodent populations: species-specific associations suggest an ancient divergence. J Gen Virol. 2012;93(Pt 2):259–266. doi: 10.1099/vir.0.036392-0. [DOI] [PubMed] [Google Scholar]
- Somogyi P, Frazier J, ASkinner M. Fowlpox virus host range restriction: gene expression, DNA replication, and morphogenesis in nonpermissive mammalian cells. Virology. 1993;197(1):439–444. doi: 10.1006/viro.1993.1608. [DOI] [PubMed] [Google Scholar]
- Stahlberg A, Kubista M, Pfaffl M. Comparison of reverse transcriptases in gene expression analysis. Clin Chem. 2004;50(9):1678–1680. doi: 10.1373/clinchem.2004.035469. [DOI] [PubMed] [Google Scholar]
- Stanley ER, Guilbert LJ, Tushinski RJ, Bartelmez SH. CSF-1--a mononuclear phagocyte lineage-specific hemopoietic growth factor. J Cell Biochem. 1983;21(2):151–159. doi: 10.1002/jcb.240210206. [DOI] [PubMed] [Google Scholar]
- Maniatis T, F EF, Sambrook J. Molecular cloning : a laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Takeshita S, Kaji K, Kudo A. Identification and characterization of the new osteoclast progenitor with macrophage phenotypes being able to differentiate into mature osteoclasts. J Bone Miner Res. 2000;15(8):1477–1488. doi: 10.1359/jbmr.2000.15.8.1477. [DOI] [PubMed] [Google Scholar]
- Taube S, Perry JW, Yetming K, Patel SP, Auble H, Shu L, Nawar HF, Lee CH, Connell TD, Shayman JA, EWobus C. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol. 2009;83(9):4092–4101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thackray LB, Wobus CE, Chachu KA, Liu B, Alegre ER, Henderson KS, Kelley ST, Virgin IV HW. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J Virol. 2007;81(19):10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timofeeva AV, Skrypina NA. Background activity of reverse transcriptases. Biotechniques. 2001;30(1):22–24. 26–28. doi: 10.2144/01301bm02. [DOI] [PubMed] [Google Scholar]
- Tsunesumi N, Sato G, Iwasa M, Kabeya H, Maruyama S, Tohya Y. Novel Murine Norovirus-Like Genes in Wild Rodents in Japan. J Vet Med Sci. 2012 doi: 10.1292/jvms.12-0106. [DOI] [PubMed] [Google Scholar]
- Tuiskunen A, Leparc-Goffart I, Boubis L, Monteil V, Klingstrom J, Tolou HJ, Lundkvist A, Plumet S. Self-priming of reverse transcriptase impairs strand-specific detection of dengue virus RNA. J Gen Virol. 2010;91(Pt 4):1019–1027. doi: 10.1099/vir.0.016667-0. [DOI] [PubMed] [Google Scholar]
- Vashist S, Urena L, Chaudhry Y, Goodfellow I. Identification of RNA-protein interaction networks involved in the norovirus life cycle. J Virol. 2012 doi: 10.1128/JVI.00432-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vashist S, Urena L, Goodfellow I. Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J Virol Methods. 2012;184(1–2):69–76. doi: 10.1016/j.jviromet.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward VK, McCormick CJ, Clarke IN, Salim O, Wobus CE, Thackray LB, Virgin IV HW, Lambden PR. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc Natl Acad Sci U S A. 2007;104(26):11050–11055. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Krug A, Mackenzie JM, Green KY, WVirgin H. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2(12):e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobus CE, Thackray LB, Virgin HW., IV Murine norovirus: a model system to study norovirus biology and pathogenesis. J Virol. 2006;80(11):5104–5112. doi: 10.1128/JVI.02346-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus MA, Chung LM, Chaudhry Y, Bailey D, Goodfellow I. Development of an optimized RNA-based murine norovirus reverse genetics system. J Virol Methods. 2010;169(1):112–118. doi: 10.1016/j.jviromet.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]