Abstract
The p53 tumor suppressor plays a central role in dictating cell survival and death as a cellular sensor for a myriad of stresses including DNA damage, oxidative and nutritional stress, ischemia and disruption of nucleolar function. Activation of p53-dependent apoptosis leads to mitochondrial apoptotic changes via the intrinsic and extrinsic pathways triggering cell death execution most notably by release of cytochrome c and activation of the caspase cascade. Although it was previously believed that p53 induces apoptotic mitochondrial changes exclusively through transcription-dependent mechanisms, recent studies suggest that p53 also regulates apoptosis via a transcription-independent action at the mitochondria. Recent evidence further suggests that p53 can regulate necrotic cell death and autophagic activity including mitophagy. An increasing number of cytosolic and mitochondrial proteins involved in mitochondrial metabolism and respiration are regulated by p53, which influences mitochondrial ROS production as well. Cellular redox homeostasis is also directly regulated by p53 through modified expression of pro- and anti-oxidant proteins. Proper regulation of mitochondrial size and shape through fission and fusion assures optimal mitochondrial bioenergetic function while enabling adequate mitochondrial transport to accommodate local energy demands unique to neuronal architecture. Abnormal regulation of mitochondrial dynamics has been increasingly implicated in neurodegeneration, where elevated levels of p53 may have a direct contribution as the expression of some fission/fusion proteins are directly regulated by p53. Thus, p53 may have a much wider influence on mitochondrial integrity and function than one would expect from its well-established ability to transcriptionally induce mitochondrial apoptosis. However, much of the evidence demonstrating that p53 can influence mitochondria through nuclear, cytosolic or intra-mitochondrial sites of action has yet to be confirmed in neurons. Nonetheless, as mitochondria are essential for supporting normal neuronal functions and in initiating/propagating cell death signaling, it appears certain that the mitochondria-related functions of p53 will have broader implications than previously thought in acute and progressive neurological conditions, providing new therapeutic targets for treatment.
p53 Functions centered around the mitochondria
p53 is a transcription factor that activates or represses the expression of multiple genes [1], but it is also found in the cytosol and mitochondria eliciting an increasing repertoire of extra-nuclear, non-transcriptional functions. p53 expression is upregulated in response to a diverse array of cellular stresses, including DNA damage, hypoxia, oxidative and nutritional stress, ribonucleotide depletion, disruption of nucleolar function and oncogene activation [2], [3], regulating DNA repair, metabolism, cell cycle progression, senescence and apoptosis and thus playing a key role in tumor suppression, aging and neurodegeneration [4], [5], [6], [7]. This review is focused upon p53 functions that directly or indirectly regulate mitochondrial physiology and its immediate up- and down-stream events (Figure 1) and provides current, still very limited assessment of those functions in neurons.
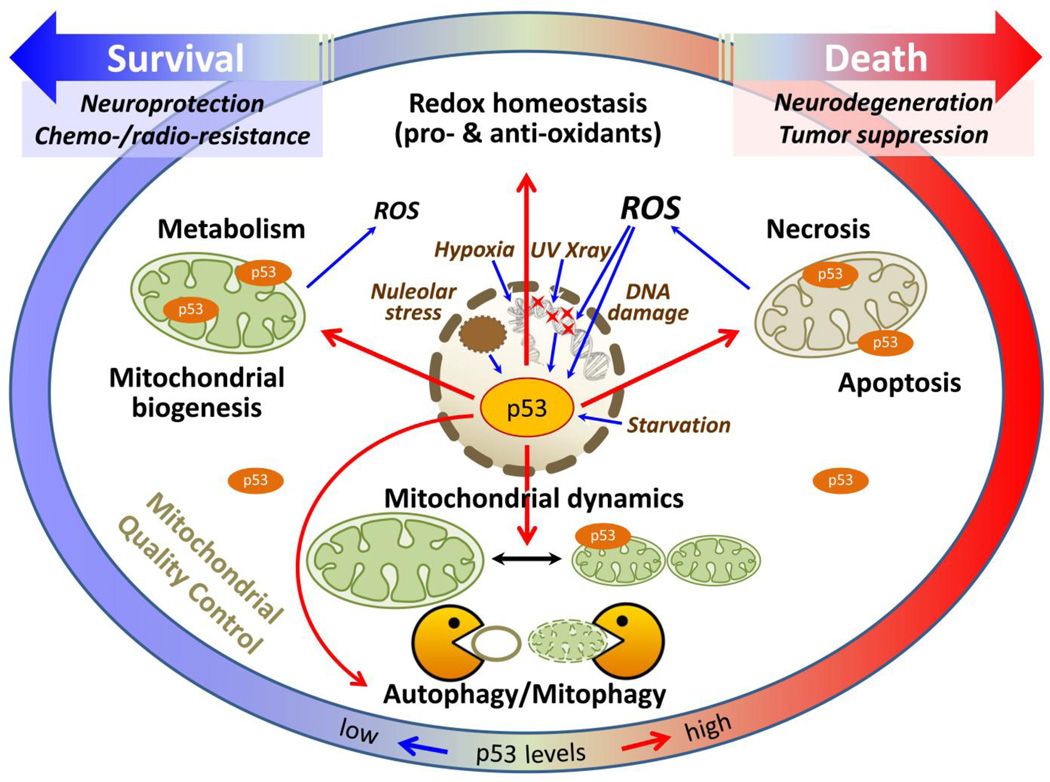
Figure 1.
p53 plays numerous distinct roles in mitochondria-related processes, such as apoptosis/necrosis, autophagy/mitophagy, mitochondrial quality control and cellular redox regulation, depending on its expression levels, subcellular localization, availability of cell-specific binding partners, and cellular state (i.e. resting versus stressed). Nuclear p53 is activated (upregulated and/or stabilized) by cellular stress including DNA damage, hypoxia, oxidative stress (ROS), nucleolar stress and starvation, and transcriptionally activates or represses p53 target genes, leading to a variety of downstream effects. p53 can also translocate to the cytoplasm and to mitochondria, where it can directly bind to and activate or inhibit proteins and pathways related to mitochondrial function. p53 actions related to mitochondrial quality control (to the left) are largely functional at basal (physiological) levels of p53 expression, while its pro-death function (to the right) requires higher levels of p53. The same is generally true at the individual gene/protein level within any specific category of p53 function where pro-survival actions are seen with physiological levels of p53 expression, while pro-death actions are induced by upregulated levels of p53.
p53-mediated apoptosis (Figure 2A)
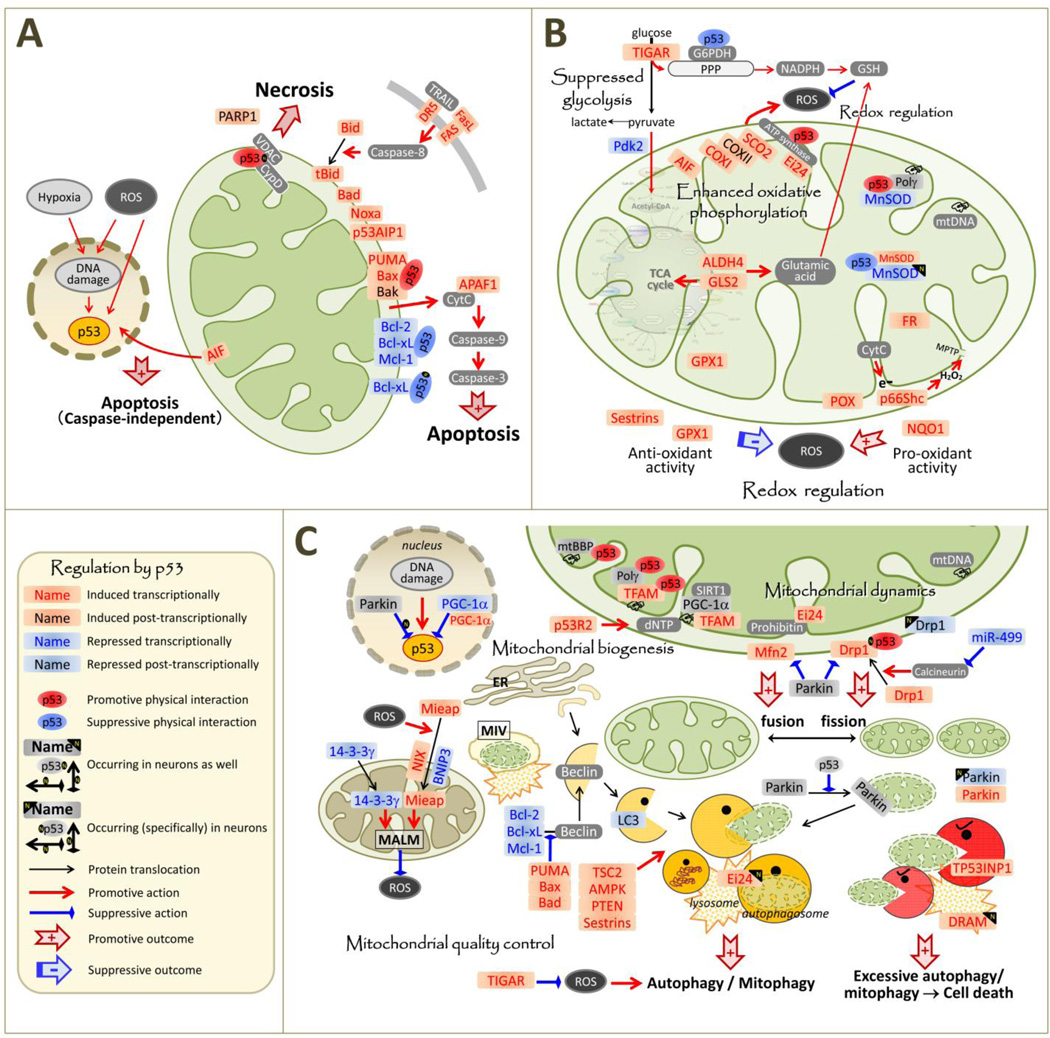
Figure 2.
p53-regulated proteins and their interacting proteins and pathways in relation to mitochondrial function. Note that p53-mediated regulation of gene expression is most likely cell and stress type-specific and the proteins listed as “induced/repressed by p53” may not be regulated that way in neurons. Proteins are listed as “transcriptionally induced/repressed by p53” only when the transcriptional regulation has been demonstrated at the gene structure level (promoter reporter assay, CHIP analysis). Not all proteins/genes mentioned in the text are presented in these figures. Note that essentially all the apoptosis-related proteins shown in (A) are transcriptionally regulated in similar ways in neurons at least under certain specific apoptotic conditions, but this is not reflected in the figure for the purpose of simplicity. Abbreviations: AIF: apoptosis-inducing factor, mitochondrion-associated, 1; ALDH4; aldehyde dehydrogenase 4 family, member A1: AMPK: 5' adenosine monophosphate-activated protein kinase; APAF1: Apoptotic peptidase activating factor 1; Bad: Bcl-2-associated death promoter; Bak: Bcl-2-homologous antagonist/killer; Bax: Bcl-2-associated X protein; Bcl-2: B-cell lymphoma 2; Bcl-xL: B-cell lymphoma-extra-large (BCL2-like 1); Bid/tBid: (truncated) BH3 interacting domain death agonist; BNIP3: BCL2/adenovirus E1B 19 kDa protein-interacting protein 3; COX: cytochrome c oxidase; CypD: cyclophilin D; CytC: cytochrome c; dNTPs: deoxyribonucleotide triphosphates; DR5: tumor necrosis factor receptor superfamily, member 10b; DRAM: DNA damage-regulated autophagy modulator protein; Drp1: dynamin-related protein 1; Ei24: Etoposide-induced protein 2.4 homolog (PIG8); ER: endoplasmic reticulum; FAS: Fas cell surface death receptor; FASL: FAS ligand; FR: ferredoxin reductase; G6PDH: glucose-6-phosphate dehydrogenase; GLS2: Phosphate-activated mitochondrial glutaminase; GPX1: glutathione peroxidase 1; GSH: reduced glutathione; LC3: microtubule-associated protein light chain 3; MALM: Mieap-induced accumulation of lysosome-like organelles within mitochondria; Mcl-1: myeloid cell leukemia-1; Mfn2: mitofusin 2; Mieap: mitochondria eating protein; MIV: Mieap-induced vacuole; MnSOD: manganese superoxide dismutase; MPTP: mitochondrial permeability transition pore; mtSSB; mitochondrial single-stranded DNA-binding protein: mtDNA: mitochondrial DNA; Nix: NIP-like protein X (a.k.a., BNIP3-like protein); Noxa: phorbol-12-myristate-13-acetate-induced protein 1 (PMAIP1); NQO1: quinone oxidoreductase or NAD(P)H dehydrogenase, quinone 1; p53: tumor protein 53; p53AIP1: tumor protein p53 regulated apoptosis inducing protein 1; p53R2: ribonucleotide reductase M2 B (RRM2B); p66Shc: p66 isoform of Src homology 2 domain-containing-transforming protein C1; PARP1: poly(ADP-ribose) polymerase 1; Pdk2: Pyruvate dehydrogenase kinase isoform 2; PGC-1α: Peroxisome proliferator-activated receptor gamma coactivator 1-alpha; POX: proline oxidase 1; PPP: pentose phosphate pathway; Polγ: DNA polymerase subunit gamma; PTEN: Phosphatase and tensin homolog; PUMA: p53 upregulated modulator of apoptosis; ROS: reactive oxygen species; SCO2: cytochrome c oxidase assembly; SIRT1: sirtuin 1; TSC2: Tuberous sclerosis protein 2; TFAM: mitochondrial transcription factor A; TIGAR: TP53-inducible glycolysis and apoptosis regulator; TP53AIP1: tumor protein p53 regulated apoptosis inducing protein 1; p53INP1: VDAC: voltage-dependent anion channel.
Numerous studies have established that p53 promotes apoptosis by transcriptionally activating or repressing the expression of a panel of pro- and anti-apoptotic proteins. For apoptotic processes involving mitochondria, p53 transcriptionally activates Fas/Fas ligand and DR5/KILLER for the extrinsic apoptotic pathway. For the intrinsic pathway p53 induces expression of PUMA, Noxa, Bid, Bad, p53AIP1, Bax and APAF1 among others [1], [8], [9], maintains basal expression of apoptosis-inducing factor (AIF) [10] and represses expression of Bcl-2 [11], Bcl-xL [12] and Mcl-1 [13] for the intrinsic pathway, consequently triggering release of apoptogenic proteins including cytochrome c and AIF from the mitochondrial intermembrane space. These pathways contribute to neuronal cell death and neurodegeneration but the critical players mediating the pathway may vary depending upon the nature of the apoptotic stimulus [14–18].
The last decade of research, however, has revealed a role for p53 as a non-transcriptional inducer of apoptosis, which involves its direct action at the mitochondria [19], [20]. In this model, a cytoplasmic pool of p53 rapidly translocates to the surface of the outer mitochondrial membrane in response to stress, where it behaves like a BH3-only protein physically interacting with anti-apoptotic (Bcl-xL, Bcl-2, Mcl-1) and/or pro-apoptotic (PUMA, Bax, Bak) members of the Bcl-2 family. These interactions eventually facilitate Bax/Bak-mediated permeabilization of the outer mitochondrial membrane leading to release of cytochrome c and the activation of the caspase cascade. Consistent with p53 acting directly at mitochondria, p53 protein has been localized in mitochondrial fractions after stress [21], [22], [23] and directly targeting p53 to the mitochondrial outer membrane is sufficient to promote apoptosis in the absence of any external stress [21], [24]. While the evidence for a direct action of p53 at the mitochondria is compelling, it was derived from studies using normal and tumor cells of non-neuronal origin.
Evidence for a direct mitochondrial action of p53 in neurons is not as abundant and remains inconclusive. Although initial studies appeared to have established that p53 immunoreactivity is confined to the nucleus in neurons destined to die [15], [25], [26], [27], in agreement with nuclear p53 function being essential for apoptosis induction, there have been several reports describing cytoplasmic accumulation of p53 in neurons, but with no demonstration that this had functional consequences for viability [28], [29], [30]. Subsequent to the demonstration of extra-nuclear apoptotic activity of p53 in non-neuronal cells, several studies have since demonstrated a potential role for mitochondrial p53 in mediating neuronal apoptosis using various in vitro and in vivo models [31], [32], [33], [34], [35], [36], [37]. However, none of these studies has provided definitive proof for a mitochondrial site of p53 action in neurons. Definitive evidence in support of a mitochondrial site of action for p53 would include its physical association with mitochondria and its molecular interaction with either members of the Bcl-2 family of proteins or other intrinsic mitochondrial proteins. The evidence available from these studies is still not conclusive in that a mitochondrial association for p53 has principally been demonstrated by immunofluorescence colocalization. When data is presented in support of p53’s physical association with mitochondria and/or its molecular interaction with an appropriate protein in mitochondrial fractions, the results cannot be strictly associated with neurons because the analysis has been done using whole brain tissue. In this regard, Endo et al. [31] used micro-dissected CA1 pyramidal cell layer tissue to demonstrate a p53-Bcl-xL interaction in mitochondrial fractions and, therefore, the result is more likely to represent a neuronal event. A functional contribution of mitochondrial p53 to apoptosis has been demonstrated by using pifithrin compounds. Pifithrin-α [38] and pifithrin-μ [39] specifically inhibit the nuclear (transcriptional) and mitochondrial (non-transcriptional) action of p53, respectively, although their specificities have not been critically tested in neurons. The results obtained with these inhibitors are not consistent and suggest context-dependent, variably proportionate contributions of nuclear and mitochondrial p53 to neuronal apoptosis [31], [32], [35], [36], [37]. A shift in sensitivity from pifithrin-α to pifithrin-μ is also observed as neurons mature in culture [34]. Collectively, these studies appear to suggest that mitochondrial p53 action contributes to neuronal apoptosis but to a variable extent. Results from some of these studies [31], [32], [36], however, suggest that the action of pifithrin-α and pifithrin-μ may not be exclusively confined to their expected sites of action, nucleus and mitochondria, respectively, although some “non-specific” effects of pifithrin-μ on nuclear p53 activity may be explained by a potential increase in nuclear p53 concentration when p53 translocation to mitochondria is blocked by pifithrin-μ. Pifithrin-α has been widely used to inhibit the nuclear, transcriptional activity of p53, but it may possess some nonspecific actions [40], [41], [42]. Caution must be taken when assessing mitochondrial p53 function solely on the basis of the sensitivity to pifithrin-μ because the specificity of this compound has recently been challenged. Leu et al. [43] demonstrate that the direct target of pifithrin-μ action is actually heat shock 70 kDa protein (HSP70). Pifithrin-μ disrupts the association between HSP70 and its cofactors and client proteins, the latter of which include p53, APAF1 and autophagy-related proteins (p62, LAMP2). This results in compromised activation of both the caspase cascade and autophagy as well as reduced localization of p53 to mitochondria. Also, pifithrin-μ inhibits NF-κB signaling [43]. Thus, pifithrin-μ is an HSP70 inhibitor and its multi-faceted action is not exclusively restricted to the inhibition of mitochondrial p53 action.
We evaluated postnatal mouse cortical neurons in culture to determine if there was evidence for a significant apoptogenic contribution of the cytosolic/mitochondrial p53 pathway, but the results instead corroborated the sole requirement for the transcriptional activity of p53 [17]. The p53 protein did not accumulate at detectable levels in the mitochondria during p53-and Bax-dependent apoptosis caused by camptothecin (CPT; induces DNA strand breaks via topoisomerase I inhibition) or nutlin-3 (upregulates p53 by inhibiting Mdm2-mediated degradation). Expression of a dominant-negative p53 mutant, p53R273H, nearly completely rescued neurons from these forms of apoptosis while suppressing the induction of p53 target genes, PUMA and Fas, a finding that rules out the presence of apoptogenic mitochondrial p53 action since p53R273H possesses little dominant-negative activity against mitochondrial wild-type p53 action [44]. In addition, exogenously-expressed p53 lost its apoptotic activity when directly targeted to the mitochondrial outer membrane in neurons but not in non-neuronal cells [17].
The uniqueness of p53-mediated apoptotic signaling in postnatal cortical neurons was further illustrated by the distinct function the proline-rich (PP) domain of p53 plays in neurons and non-neuronal cells [17]. In non-neuronal cells, the PP domain of p53 is required for its overall apoptosis-inducing activity [45], [46], [47] and specifically for the transcription-independent “BH3-only protein-like” death-promoting action of p53 in the mitochondria as well [48], [49]. In contrast, when expressed in p53-deficient neurons, a deletion mutant lacking the PP domain (amino acids 62–91: p53ΔPP) exhibited the same degree of apoptotic activity as wild-type p53 even in the absence of DNA damage [17]. The naturally occurring polymorphism at codon 72 of human p53 (Pro vs. Arg) occurs in this PP domain, affecting nuclear export of p53 in favor of p53Arg72 and, consequently, p53Arg72 exhibits greater mitochondrial translocation and transcription-independent apoptotic activity than p53Pro72 [23]. In mouse postnatal cortical neurons, however, both human p53Pro72 and p53Arg72 showed exclusive nuclear localization and indistinguishable apoptotic activity [17]. Thus, our results point to the possibility that the cellular context in postnatal cortical neurons recognizes the p53 molecule and mediates its apoptotic activity differently compared to non-neuronal cells and, consequently, is not conducive to p53 promoting apoptosis through the mitochondria. Cellular contexts may vary, however, even among neurons depending upon the neuronal population being examined, the age of neurons (embryonic, postnatal, adult), culture conditions, in vitro vs. in vivo, the type of apoptotic stress employed, etc. Indeed, when expressed in rat embryonic cortical neurons, p53Arg72, but not p53Pro72, accumulates in the cytosol and activates the intrinsic apoptotic pathway, increasing vulnerability to excitotoxic and ischemic apoptotic death [36].
The occurrence and significance of p53 acting directly at the mitochondria in neurons remains to be further explored. The data accumulated thus far suggest that a mitochondrial site of p53 action in neurons may occur under certain conditions, but biochemical analyses using whole brain tissue and functional analyses based on pifithrin sensitivity make it difficult to draw a firm conclusion at this point. Neurons may utilize the transcription-independent mitochondrial p53 pathway to a variable extent depending on parameters not fully understood.
p53 regulates necrosis and autophagy (Figure 2A, C)
The cell fate-determining action of p53 on mitochondria may not be restricted solely to the activation of apoptotic cell death. p53 has recently been suggested to regulate programmed necrotic death, an irreversible form of cell destruction that results from an acute failure in energy production due to the opening of the mitochondrial permeability transition pore (MPTP). Oxidative stress has been shown to stabilize and promote p53 accumulation in the mitochondrial matrix [50], [51]. A recent study demonstrates that, in response to oxidative stress, p53 localized in the mitochondrial matrix can promote the opening of the MPTP via its direct interaction with the MPTP regulator, cyclophilin D [51]. A complex between p53 and cyclophilin D is also detected in necrotic brain tissue in a model of transient ischemic stroke. Intriguingly, a more prominent reduction in infarct size was observed in mice lacking one p53 allele (p53+/−) than in p53-null (p53−/−) mice, which is, in fact, consistent with a previous observation [52] but requires further clarification for the underlying molecular mechanism. This finding is provocative as it ties p53 to necrotic cell death in the brain for the first time, but it remains to be demonstrated if this necrotic mode of mitochondrial p53 action indeed occurs in neurons and is it associated with the loss of neurons only in the ischemic core (necrotic) or in the penumbra (apoptotic) as well. An interaction between p53 and the voltage-dependent anion channel (VDAC), another key structural component of the mitochondrial permeability transition pore, was also found by co-immunoprecipitation in PC12 cells and, interestingly, the interaction was attenuated upon etoposide treatment [53]. p53-mediated formation of large VDAC-containing protein complexes has also been reported [51]. Thus, p53 may interact with VDAC under normal conditions and then shift the interaction toward cyclophilin D under conditions that precipitate necrosis. A more recent study demonstrates another mode of p53-dependent “non-apoptotic” cell death in mouse embryonic fibroblasts (MEFs) and human tumor cell lines [54]. This mode of cell death is induced by H2O2 at a concentration that exceeds the levels that trigger the p53-cyclophilin D interaction mentioned above [51] and occurs in wild-type and even Bax/Bak double knockout (DKO) MEFs but not in p53−/− MEFs, thus making it p53-dependent and Bax/Bak-independent (or non-apoptotic, necrotic) cell death. DNA damage is involved with the resulting activation of poly(ADP-ribose) polymerase 1 (PARP-1) and p53 deficiency causes a decrease in PARP-1 activity consistent with the observation that inhibition of PARP-1 activity is protective. This p53-dependent, PARP-1-mediated non-apoptotic (or necrotic) cell death is not inhibited by cyclosporine A, an MPTP inhibitor, and is largely unchanged in cyclophilin D KO cells and, therefore, is unlikely to be mediated by the opening of the PTP [54].
An increasing number of proteins involved in autophagy are now known to be regulated by p53. Autophagy is pro-survival, for example, under conditions of mild nutritional stress, while dysregulated and excessive autophagy can precipitate cell death. Autophagic activity is also critical for the execution of mitophagy, a selective form of autophagy responsible for the degradation of damaged mitochondria. Under conditions of stress, p53 can promote autophagy via the transcriptional activation of autophagic genes such as TSC2 (tuberous sclerosis 2), the beta 1 and beta 2 subunits of AMPK, sestrin 1 and 2, and PTEN [55]. Additionally, p53 can promote autophagy by facilitating the release of the autophagic protein beclin 1 from sequestration by Bcl-2, Bcl-xL and Mcl-1 by repressing transcription of these sequestering proteins (Bcl-2, Bcl-xL and Mcl-1) [11], [12], [13], or by increasing expression of Bax, Bad and PUMA [56], [57]. Tumor protein p53 inducible nuclear protein 1 (TP53INP1), originally identified as a p53 target gene and designated as p53DINP1, encodes a nuclear protein that works to further enhance p53 function [58]. Upon induction of autophagy this protein translocates to autophagosomes and purportedly promotes autophagy by binding to LC3 to displace p62. TP53INP1 overexpression can induce caspase-dependent autophagic cell death [59]. TP53INP1 mRNA is upregulated in response to copper-induced apoptosis in NTERA-2-N neurons, but its autophagic contribution to cell death has not been demonstrated [60]. p53 also activates the autophagy pathway by transcriptionally upregulating a lysosomal protein, damage-regulated autophagy modulator (DRAM), which causes little cell death by itself when overexpressed, but is required for p53-mediated cell death [61]. DRAM is expressed in multiple spliced forms and their additional localization at multiple sites other than lysosomes is indispensable for the whole range of DRAM functions [62]. p53-mediated induction of DRAM has been reported in a rat model of Huntington’s disease where striatal neuron death is induced by striatal administration of a mitochondrial complex II inhibitor, 3-nitropropionic acid. Induction of DRAM together with LC3 upregulation is blocked by pifithrin-α administration and striatal neuron death is strongly attenuated by autophagy inhibitors [63]. A similar relationship between p53-mediated induction of DRAM and autophagic activity is observed when excitotoxic death of striatal neurons is induced by quinolinic acid [64]. In kainic acid-induced excitotoxic cell death in striatal neurons in vitro, autophagic proteins and activity are induced in a p53-dependent manner based on their sensitivity to pifithrin-α and, to a lesser extent, pifithrin-μ [37].
Conversely, in contrast to the transcriptional activation of autophagy by nuclear p53 under conditions of stress as described above, cytoplasmic p53 can suppress autophagy [65]. Deletion, depletion or inhibition of p53 in a variety of normal and transformed human cell lines is sufficient to stimulate autophagy, while various inducers of autophagy (starvation, rapamycin and ER stress) enhance p53 degradation. Restoring cytoplasmic, but not nuclear, p53 in p53-deficient HCT116 human colon carcinoma cells can inhibit autophagy induction [65]. Transcription-independent effects of p53 on the AMPK/mTOR axis, a key pathway for autophagy regulation, is suspected of mediating the autophagy-suppressing action of cytoplasmic p53 [65], but p53-mediated post-transcriptional downregulation of LC3 protein as demonstrated in the same HCT116 cells in response to prolonged starvation [66] may also have some mechanistic implication in this cytoplasmic p53 action. The autophagy-suppressing action of cytoplasmic p53 has not yet been demonstrated for neurons.
A p53-regulated increase in autophagic activity may be broadly associated with different forms of neuronal cell death, but its overall contribution to the demise of neurons requires further clarification since autophagy can be protective when stress levels are sub-lethal or when autophagy is activated early on in the process of cell death. For example, nucleolar stress-induced p53 in striatal neurons in a model of Huntington’s disease transiently upregulates PTEN and the resulting induction of autophagy exerts a neuroprotective effect [67]. Since both impaired autophagy and excessive activation of autophagy can be pathogenic in nervous system injury and neurodegenerative diseases [68], autophagy-related actions of p53 may also variably affect viability in neurons. The autophagy-modulating action of p53 is also relevant to mitochondrial quality control through autophagic clearance of damaged mitochondria (see below).
p53 and mitochondrial quality control
Maintaining a pool of viable mitochondria is essential for sustaining normal cellular function, especially for neurons, a cell type with high energy demands that can only be met through the process of oxidative phosphorylation. The quality of mitochondrial function is assured “structurally” by the production of healthy mitochondria (mitochondrial biogenesis) and removal of non-functional mitochondria (mitophagy) and “functionally” by the health of the systems involved in metabolism (glycolysis, TCA cycle, oxidative phosphorylation) and the associated production of reactive oxygen species (ROS), a critical component that determines cellular redox homeostasis that is kept in balance with the anti-oxidant system. Recent studies have revealed a significant contribution of p53 to each of these aspects of mitochondrial function as detailed below.
p53 and mitophagy (Figure 2C)
Mitophagy is a selective form of autophagy by which damaged mitochondria are eliminated in autolysosomes [69]. Damaged or non-functional mitochondria produce excess ROS, potentially activating cell death signaling and, therefore, their timely removal is critical in maintaining the cellular redox homeostasis. When this degradative process becomes dysregulated and excessive, however, mitophagy may be detrimental, culminating in a bioenergetic crisis. Mitophagy is regulated by an increasing number of proteins that reside in the mitochondria or become associated with mitochondria when required, such as PINK1, parkin, BNIP3, NIX and FUNDC1 [69], [70], [71]. The expression and function of some of these proteins is regulated by p53.
The PINK1/parkin pathway plays a major role in mitochondrial quality control by marking damaged (loss of membrane potential) mitochondria for mitophagy [69], [70], [71], although this remains to be established for neurons [72]. Parkin, a gene associated with early onset forms of Parkinson’s disease, was recently identified as a p53 target gene, increasing its expression upon p53 upregulation both in human and mouse cells [73]. In contrast, we found that p53 suppresses parkin protein expression in postnatal mouse cortical neurons with little suppression at the mRNA level [74], suggesting that p53 can regulate parkin expression both transcriptionally and post-transcriptionally and distinctly between non-neuronal cells and at least some neuronal populations. Interestingly, parkin reciprocally acts on the p53 gene as a transcriptional repressor by binding to the p53 promoter region [75]. Parkin overexpression suppresses p53 expression and apoptosis (caspase-3 activity) in 6-hydroxydopamine-treated fibroblasts, neuronal cell lines and primary neurons [75]. Elevated levels of p53 expression are observed in fibroblasts and brain from parkin knockout mice and in human brain afflicted by autosomal recessive juvenile parkinsonism caused by parkin mutations, which abolish the protective action of wild-type parkin [75]. Consistent with this finding, a recent report suggests that modification of parkin by S-nitrosylation abolishes the ability of parkin to suppress p53 expression and the levels of both S-nitrosylated parkin and p53 are simultaneously elevated in Parkinson’s disease brain [76].
The interaction between p53 and parkin can also occur through a direct protein-protein interaction. The main function of parkin in mitochondrial quality control is its ability to mark damaged mitochondria for mitophagy through its ubiquitin ligase activity. In a model of doxorubicin-induced cardiotoxicity and aging-associated heart failure in mice cytosolic p53 directly binds to parkin, blocking its translocation to damaged mitochondria and thereby their subsequent clearance by mitophagy [77]. The significance of this p53-parkin interaction in neurons remains to be determined as it is highly dependent on cytosolic expression of p53. In ischemic cardiac tissue, mitophagy is also impaired due to p53-dependent transcriptional upregulation of TIGAR [77], which suppresses autophagy through its ability to counteract mitochondrial ROS production via enhanced regeneration of NADPH and, consequently, reduced glutathione (GSH) [78]. In p53−/− myocardium, therefore, elevated ROS production is followed by homodimerization and activation of BNIP3, an initiator of mitophagy (see below), that serves as a basis for the observed cardioprotection [77].
BNIP3 (Bcl2 and adenovirus E1B 19kDa-interacting protein 3) and its homolog NIX (NIP-like protein X; a.k.a., BNIP3-like protein or BNIP3L) are BH3 domain-only proteins residing in or translocating upon activation to the outer mitochondrial membrane. However, they exhibit relatively low apoptotic activity [79] and can additionally induce caspase-independent permeability transition-mediated cell death and autophagy/mitophagy [80]. They promote mitophagy by interacting with LC3, a protein critical for autophagosome formation [71], [80]. While BNIP3 and NIX are inducible transcriptional targets of HIF-1α, they are also transcriptionally regulated by p53, but in different directions in response to hypoxic conditions in non-neuronal cells such that BNIP3 is downregulated [81] while NIX is upregulated by p53 [82]. Although these regulatory pathways have not yet been established for neurons, both BNIP3 [83], [84], [85] and NIX [86], [87] are upregulated in neurons in response to stress, suggesting that BNIP3 expression may be regulated differently in neurons. In studies with neurons, however, mitigating the upregulation of BNIP3 induced by oxidative and ischemic stress is neuroprotective, suggesting that BNIP3/NIX-dependent mitophagy may be excessive and detrimental or that BNIP/NIX work as pro-death proteins.
Recent studies have implicated BNIP3 and NIX in a different form of mitochondrial quality control that involves Mieap (mitochondria-eating protein; a.k.a, spermatogenesis-associated protein 18 or SPATA18), a p53-inducible gene that encodes a novel protein found in the mitochondrial matrix [88], [89]. In response to stress, such as DNA damage, Mieap is induced and translocates to the mitochondrial matrix in a ROS-dependent manner [89], allowing lysosomal enzymes to move into the matrix thereby facilitating degradation of oxidatively damaged mitochondrial proteins (designated MALM for Mieap-induced accumulation of lysosome-like organelles within mitochondria [88]). This occurs only when BNIP3 and NIX coexist at the mitochondrial surface [90]. The interaction between Mieap, BNIP3 and NIX appears to facilitate the formation of a mitochondrial membrane pore involving both mitochondrial membranes, thus causing a drop in membrane potential. However, the drop in membrane potential is not sensitive to cyclosporine A, suggesting that the pore induced by the interaction between Mieap, NIX and BNIP3 is not the traditional mitochondrial permeability transition pore and, indeed, it does not lead to cell death in this model [90]. When the Mieap pathway is activated, 14-3-3γ also translocates from the cytosol to the mitochondrial matrix, where it interacts with Mieap and is required for Mieap-promoted degradation of oxidized proteins by lysosomal enzymes [91]. Interestingly, 14-3-3γ expression is suppressed by p53 [92], suggesting that p53 can also curtail this pathway. When NIX is not available (not yet known for BNIP3), cytosolic Mieap contributes to the formation of a vacuole-like structure called MIV (Mieap-induced vacuole). The MIV engulfs damaged mitochondria and fuses with lysosomes to promote the elimination of unhealthy mitochondria, acting as a form of mitophagy [89]. Thus, Mieap regulates the repair (within mitochondria) and elimination (in cytosol) of oxidatively damaged mitochondria. Since all of these proteins (Mieap, BNIP3, NIX, 14-3-3γ) are regulated in some manner by p53, this pathway may prove to be very important in p53-mediated mitochondrial quality control although its regulation is likely complex. High levels of p53 expression can precipitate the opening of a cyclosporine A-sensitive mitochondrial permeability transition pore associated with necrosis [51], [90]. In contrast, this Mieap-mediated mitochondrial quality control pathway may be implicated in a cytoprotective response to stress that is associated with more modest activation of p53, as observed under chronic moderate oxidative stress. Mieap indeed maintains normal mitochondrial function by suppressing ROS generation and promoting ATP production under irradiation-induced oxidative stress [88]. The expression and function of Mieap in neurons has not yet been described.
The DNA damage response gene Ei24 (etoposide-induced 2.4 kb transcript, also known as p53-induced gene 8 or PIG8) is a p53-inducible gene [93] associated with the regulation of autophagy. Mice displaying a neural-specific knockout of this gene demonstrate defects in basal autophagy in oligodendrocytes and neurons and develop age-dependent neurological abnormalities characterized by significant axonal demyelination and degeneration as well as extensive neuron loss in brain and spinal cord [94]. Thus, although it’s inducibility by p53 in neurons has not yet been determined, p53-mediated Ei24 expression may sustain the basal rate of autophagic flux in neurons, playing an important role in the clearance of aggregate-prone proteins [94]. Ei24 is known to be localized to the endoplasmic reticulum and to bind to Bcl-2 in cancer cells [95]. A proteomic analysis [96] further supports the association of Ei24 with, among others, the ATP synthase complex and prohibitin, a mitochondrial inner membrane protein that affects the processing of the inner membrane fusion protein OPA1 [97], suggesting that Ei24 may function directly in mitochondria. Its mechanism of action and possible pro-survival function together with its relationship to mitochondrial structure and function in neurons requires further exploration.
p53 and mitochondrial biogenesis (Figure 2C)
The production and maintenance of functional mitochondria constitutes another aspect of mitochondrial quality control. Maintenance of mitochondrial DNA (mtDNA) integrity and copy number is fundamental to sustaining mitochondrial function. Accumulating evidence indicates that p53 works as a guardian of the genome for mitochondria as well. Among those proteins directly involved in this function within mitochondria, nuclear DNA-encoded mitochondrial transcription factor A (TFAM), essential for mtDNA replication, repair and transcription, is the only protein thus far identified to be transcriptionally regulated by p53 [98]. However, TFAM expression is p53-dependent in fibroblasts and muscle tissue, but not in liver or heart, indicating cell type specific inducibility [98]. Having been identified in the mitochondrial matrix, p53 also protects the mitochondrial genome through direct interactions with repair enzymes. p53 interacts with mitochondrial single-stranded DNA-binding protein (mtSSB)[99] and TFAM [100] forming p53/TFAM/mtDNA complexes [98], [101], [102] and also interacts with mitochondrial DNA polymerase (Polγ) [103], thereby promoting replication and base excision repair of mtDNA [103], [104]. Consequently, mtDNA copy number and mitochondrial mass are greater in p53-expressing cells relative to p53-deficient counterparts under various conditions [98], [103], [105], [106]. Also observed in p53-deficient cells is reduced expression of p53R2 [105], [106], one of the two small subunits (R2 and p53R2) comprising ribonucleotide reductase that supplies dNTPs for DNA replication and repair. p53R2 (RRM2B gene) is transcriptionally induced by p53 upon DNA damage and, unlike the R2 subunit, translocates to the nucleus, where it is thought to constitute ribonucleotide reductase activity and supply dNTPs locally for DNA repair [107]. Although it is not known if p53R2 also resides in mitochondria, p53R2 is required for mtDNA synthesis in quiescent cells [108]. Consistently, mtDNA depletion is seen in muscle from human patients with p53R2 mutations and in muscle, liver and kidney of p53R2 knockout mice [109]. Thus, proper maintenance of mtDNA relies on the expression of p53R2. Although some neurological conditions are reported in patients with p53R2 mutations, the effects of p53R2 deficiency on mitochondria in neural tissue have not been examined [109]. Nonetheless, p53R2 is probably the only small subunit of ribonucleotide reductase available in post-mitotic cells including neurons because the other small subunit, R2, but not p53R2, is a degradation target for the APC/C-Cdh1 E3 ubiquitin ligase [110], which is highly expressed in neurons [111]. Although p53 inducibilty of p53R2 in neurons has not yet been demonstrated, it would be worthwhile to determine if p53-induced p53R2 expression can be used to protect mitochondrial function in neurons under conditions of aging and neurodegeneration. p53 also interacts with an F1F0 ATP synthase subunit to promote the assembly of the ATP synthase complex, at least partially explaining how basal levels of p53 in the mitochondrial matrix can increase oxygen consumption and decrease mitochondrial superoxide production [112]. Collectively, these studies indicate that, under physiological, mildly-stressed conditions, constitutively expressed p53 can protect the mitochondrial genome and support mitochondrial biogenesis and function. In contrast, as described above, accumulation of excess p53 in the mitochondrial matrix in response to noxious levels of oxidative stress can trigger opening of the permeability transition pore leading to programmed necrotic cell death [51].
PGC-1α (peroxisome-proliferator-activated receptor gamma co-activator-1α) is a co-transcriptional regulator that induces mitochondrial biogenesis by activating a host of different transcription factors involved in the regulation of energy metabolism [113], [114], [115]. In p53 knockout mice mitochondrial content and the levels of PGC-1α in skeletal muscle tissue are reduced compared to wild-type tissue [116], and reduced mtDNA content is associated with p53 deficiency in other studies [98], [103], [105], [106]. However, a recent study on mitochondrial dysfunction stemming from telomere damage demonstrates that telomere damage-induced p53 transcriptionally suppresses the expression of PGC-1α (and PGC-1β) and, consequently, of PGC-1α transcriptional targets including TFAM in different tissues of mice [117]. Changes in the engagement of inducible/repressive p53-binding sites in the PGC-1α promoter are confirmed to be consistent with the suppressed expression of PGC-1α [117]. This repressive regulation of PGC-1α expression by p53 is also demonstrable in p53-null fibroblasts upon adenovirus-mediated p53 overexpression, independently of telomere damage [117]. Interestingly, a more recent study conversely demonstrates that p53 can transcriptionally induce PGC-1α expression upon GSH depletion in a nitric oxide-dependent manner in various human cell types including SH-SY5Y cells and in mouse brain [118]. The discrepancy between the two studies reflects whether p53 preferentially binds to the positive or negative response elements in the PGC-1α promoter. The negative response elements are predominantly occupied by p53 in the telomere-damaged cells [117] and also in the non-GSH-depleted control cells/tissues [118], but the positive response element is predominantly occupied by p53 induced in response to GSH depletion [118]. Inhibition of nitric oxide synthase abolishes PGC-1α induction in response to GSH depletion, suggesting that nitroxidative stress caused by GSH depletion is critical for PGC-1α induction [118]. This could actually be a strictly nitric oxide-specific event because H2O2-mediated oxidative stress results in p53-mediated PGC-1α downregulation [119]. Also, the levels of p53 induced under these conditions may be an important determinant of outcome with high (pro-death) levels of p53 suppressing and low (pro-survival) levels of p53 inducing PGC-1α expression. In these studies PGC-1α downregulation is pro-death [117], [119] while its upregulation is pro-survival [118], as expected from the purported function of PGC-1α. Since p53-mediated PGC-1α upregulation is demonstrable in dopaminergic SH-SY5Y neuroblastoma cells and mouse brain [118], the PGC-1α gene in neurons may be positively regulated by p53 under certain conditions, if not as a default, and thus activation of the p53-PGC-1α pathway may be neuroprotective. A neuroprotective role for PGC-1α has been demonstrated for various neurodegenerative conditions in vivo and in vitro [113], [115].
Interestingly, PGC-1α, in turn, can regulate p53’s transcriptional activity by binding to p53 and modulating its transactivation preference toward pro-cell cycle arrest and pro-metabolic target genes under conditions of metabolic stress by blocking p53 acetylation at Lys120, which is necessary for pro-apoptotic activation of p53 [120]. As this feedback mechanism is also observed in SH-SY5Y neuroblastoma cells among other cell types [120], the mechanism may also be intact in neurons and contribute, at least in part, to the neuroprotective (anti-apoptotic) effects of PGC-1αas revealed in overexpression and knockout studies [113], [115]. PGC-1α also directly resides in mitochondria and forms complexes with TFAM and SIRT1 to associate with the D-loop region of mtDNA [121], where it may also interact with p53 recruited to mtDNA (see above). Interestingly, PGC-1α is more abundant (when normalized against total protein) in mitochondria than in the nucleus in different tissues including brain [121], suggesting that PGC-1α may contribute to mitochondrial biogenesis as a transcriptional co-activator and/or via protein-protein interactions within mitochondria.
p53 and metabolism (Figure 2B)
Accumulating evidence indicates that p53 also plays an important role in cellular metabolism by directly regulating glucose metabolism and oxidative phosphorylation [122], [123]. p53 decreases the expression of pyruvate dehydrogenase kinase isoenzyme-2 (Pdk2) by transcriptional repression [124], which facilitates the flux of pyruvate to acetyl-CoA rather than to lactate. In addition, p53 directly transactivates phosphate-activated mitochondrial glutaminase (GLS2) [125], [126], which promotes a glutamine to glutamate conversion leading to enhanced generation of α-ketoglutarate, a key TCA cycle intermediate. p53 also contributes to maintenance of the mitochondrial oxidative phosphorylation machinery by transcriptionally activating cytochrome c oxidase (COX) I subunit (COX1/MT-COI) [127] and “synthesis of cytochrome c oxidase 2” (SCO2) [128], which is required for the assembly of the COX II subunit into the COX complex (complex IV), while post-transcriptionally maintaining COX II subunit expression [129]. In addition, p53 transcriptionally induces the expression of AIF [10], which promotes the proper assembly and function of respiratory complex I [130] separately from its apoptosis-related function. Collectively, these p53 actions promote mitochondrial metabolism and oxidative phosphorylation with a natural consequence of increased production of reactive oxygen species (ROS). To counteract this oxidative stress, p53 simultaneously upregulates anti-oxidant activity through metabolic processes. p53-mediated transcriptional activation of GLS2 [125], [126] and aldehyde dehydrogenase 4 (ALDH4) [131] helps to maintain elevated levels of reduced glutathione (GSH) by supplying its constituent, glutamic acid, from glutamine and proline, respectively, for glutathione synthesis, thereby reducing ROS levels and conferring resistance to oxidative stress. Perhaps more importantly, p53 regulates the use of the pentose phosphate pathway (PPP) by transcriptionally inducing “TP53-induced glycolysis and apoptosis regulator” (TIGAR) [132]. TIGAR possesses a fructose-2,6-bisphosphatase activity and shuttles glucose to the PPP while suppressing glycolysis. The PPP is a key metabolic pathway supplying NADPH required for regeneration of reduced glutathione (GSH). However, there is another layer of regulation imposed on the PPP as cytosolic p53 can inhibit glucose-6-phosphate dehydrogenase (G6PDH) [133], which is the first, rate-limiting enzyme of the PPP. Therefore, the net effect of p53 on PPP activity may be determined by the relative levels of nuclear versus cytoplasmic p53 and is clearly cell and context dependent. In HCT116 colon cancer cells, p53’s suppressive effect on the PPP through G6PDH inhibition appears to dominate its stimulatory effect through TIGAR induction as p53 deficiency results in increased PPP flux and higher NADPH levels [133].
It is not known if p53 regulates the PPP to any significant extent in neurons. Maintaining the integrity of this pathway is a prerequisite for survival in neurons, and neurons possess a neuron-specific mechanism to favor the PPP over glycolysis to cope with inherently high ROS production. The enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) acts as a key regulator of glycolysis by synthesizing fructose-2,6-bisphosphate (the target of TIGAR) to promote glycolysis. In neurons, the levels of PFKFB3 are kept low through its continuous degradation by the highly expressed APC/C-Cdh1 E3 ubiquitin ligase [111], [134]. Because of this, the p53-TIGAR pathway may be redundant in neurons at least under resting conditions. The PPP is probably not being obstructed by the p53-G6PDH interaction under resting conditions because cytosolic p53 levels are likely to be very low in healthy neurons. Therefore, p53-mediated regulation of the PPP may not be relevant in unstressed neurons. The intriguing question then arises: how does this process change when p53 is upregulated in neurons in response to stress? TIGAR induction may serve to ensure the activity of the PPP in case stress-induced changes in intracellular signaling favors the glycolytic pathway over the PPP, which may be detrimental to neuronal viability as suggested by the finding that inhibition of the PPP can exacerbate stress-induced neuronal cell death [134]. Conversely, inhibition of G6PDH may help dampen the excessive generation of reduced glutathione (GSH) to enable mitophagy/autophagy which is known to be activated by p53 (see above). Elevated TIGAR activity, which favors the PPP, is known to mitigate mitophagy [78], [135]. A partial answer may come from the observation that, during glutamate-induced apoptosis in cortical neurons, which is at least partially dependent on p53 [136], PFKFB3 escapes degradation and its cytoplasmic accumulation promotes glycolysis. This results in oxidative stress and cell death that can be blocked by directly promoting the PPP via overexpression of G6PDH [137], a PPP enzyme that is inhibited by p53 via direct interaction [133]. Therefore, in the context of excitotoxic stress, increased PPP activity is clearly neuroprotective. Although upregulated p53 can work both protectively through TIGAR induction and adversely by inhibiting G6PDH, the outcome would heavily rely on the levels of nuclear vs. cytosolic p53 and it would be beneficial if p53 could be contained in the nucleus. Despite having been extensively discussed in the context of the Warburg effect and tumor suppression [122], [123], these metabolic effects of p53 have not been addressed for neuroprotection or neurodegeneration. A recent study implicates TIGAR expression in Alzheimer’s disease by showing that TIGAR protein expression is lower with increasing severity of dementia, raising the possibility that reduced TIGAR expression equates to a progressive loss of protection against oxidative stress [138].
p53 and redox homeostasis (Figure 2B)
Mitochondria are the major source of ROS production both in health and death of a cell. ROS production is countered by the scavenging actions of anti-oxidants. Physiological levels of ROS are important as signaling molecules affecting cellular processes through modification of the activity of kinases and transcription factors including p53 [139]. Conversely, excess levels of ROS can provoke irreparable oxidative damage and trigger cell death. p53 influences both the production and scavenging of ROS by directly inducing and suppressing the expression of anti- and pro-oxidant proteins, thereby maintaining redox homeostasis on one hand, but also actively promoting ROS production and tipping the redox balance toward cell death.
As mentioned above, the anti-oxidant function of p53 is partly mediated through the induction of ALDH4 [131], GLS2 [125], [126] and TIGAR [132], facilitating the synthesis of glutathione and the production of NADPH which collectively increase the supply of reduced glutathione (GSH). The remainder is accomplished through transcriptional induction of antioxidant proteins, sestrins (peroxiredoxin reductase) [140] and glutathione peroxidase 1 (GPX1) [141]. p53-inducible TP53INP1 also shows anti-oxidant activity, but the exact mechanism for this activity remains unknown [142]. Most notable among the other anti-oxidant proteins, including those mentioned above, is another p53-regulated anti-oxidant protein, manganese superoxide dismutase (MnSOD/SOD2). Residing in the mitochondrial matrix, MnSOD protects mitochondria from oxidative damage by converting superoxide radicals to H2O2, which are subsequently detoxified by peroxidases such as mitochondrially localized GPX1 [143] and mitochondrial glutathione [144]. Thus, intra-mitochondrial ROS detoxification significantly relies on p53 target anti-oxidant proteins. Unlike other p53-inducible anti-oxidant proteins, however, MnSOD expression is regulated by p53 in both inductive and repressive ways although most studies report a repressive action of p53 [145, 146]. This may be explained by the observation that, when p53 levels are low, NF-κB is induced and binds to an enhancer promoting MnSOD expression, whereas p53 and SP1 are recruited to the promoter overriding the enhancer activity and suppressing MnSOD expression when p53 levels are increased [147]. As found for other p53 target proteins, p53 can also affect MnSOD function via direct protein-protein interactions. Upon apoptosis induction with a tumor promoter (TPA) in JB6 skin epidermal cells p53 translocates into mitochondria, preceding its nuclear translocation, and binds to and inactivates MnSOD causing mitochondrial depolarization [50]. In mouse skin, however, UV-induced p53 translocates into the mitochondrial matrix and forms a complex with MnSOD and mitochondrial DNA polymerase (Polγ), an important enzyme for mtDNA base excision repair, enabling MnSOD-dependent protection of Polγ from nitration-induced inactivation [148]. Thus, p53’s effect on MnDOS activity through direct interactions appears to be context-dependent.
Pro-oxidant proteins are also induced by p53. p53 transcriptionally upregulates ROS-generating enzymes such as quinone oxidoreductase (NQO1/PIG3) [149], proline oxidase (POX/PIG6) [149] and ferredoxin reductase (FR/FDXR) [150]. While overexpression of NQO1 [149] and FR [150] only sensitize cells to apoptosis, POX overexpression induces apoptosis [151], [152], which is inhibited by MnSOD [152]. p66Shc (p66 isoform of Src homology 2 domain-containing-transforming protein C1) has also attracted attention as a molecule that can mediate p53-promoted ROS production by mitochondria [146], [153], [154]. Consistent with targeted mutation of the p66shc gene prolonging life span in mice [155], p66Shc−/− animals and cells show reduced levels of intracellular ROS and increased resistance to oxidative stress such as UV, H2O2 or paraquat [155], [156]. p66Shc is transcriptionally regulated by p53 in endothelial cells [157] and is also stabilized by p53 post-translationally in mouse embryo fibroblasts [156]. p66Shc induces apoptosis by locally producing H2O2 through oxidation of cytochrome c and, as a result, causes cyclosporine A-inhibitable mitochondrial permeability transition [156].
The regulation of redox status by p53 in neurons may differ from its regulation in other cell types. p53 deficiency lowers intracellular ROS levels in cultured cortical neurons and cortical tissue while it elevates ROS levels in embryonic lung fibroblasts and liver and lung tissue [158]. In normal fibroblast and carcinoma cell lines increased ROS levels are observed in response to p53 knockdown as well under non-stressed conditions [159]. Similarly, p53 maintains low ROS levels in neural progenitor cells to suppress premature neurogenesis, indicating that the mode of p53-mediated redox regulation is not yet neuronal [160]. Consistent with these contrasting effects of p53 on ROS levels, the expression of pro- and anti-oxidant genes is differentially regulated by p53 in neurons compared to fibroblasts. The overall effect of p53 is to repress expression of anti-oxidant genes including those that have been demonstrated to be p53 inducible in non-neuronal cells [158]. Although MnSOD transcript levels did not differ significantly between p53+/+ and p53−/− embryonic cortical neurons [158], another study reveals significantly increased expression of the MnSOD protein in mitochondrial fractions from p53−/− mouse brain relative to wild-type with reduced oxidative/nitrosative damage seen in mitochondrial and nuclear fractions from p53−/− brain [161]. These studies indicate that, in contrast to non-neuronal cells, constitutive p53 expression is pro-oxidant in neurons and in the nervous system, suggesting its implication in aging and neurodegenerative conditions [162]. Under conditions associated with neurodegeneration, reduced expression of MnSOD is observed in association with p53 upregulation in models of Alzheimer’s disease [163] while p53-dependent sestrin 2 upregulation occurs under conditions of Parkinson’s disease [164]. p66Shc deficiency reduces axonal injury in a mouse model of multiple sclerosis [165] and protects hippocampal neurons against cell death induced by H2O2 or NO donor treatment [166]. p66Shc also mediates β-amyloid neurotoxicity [167] and is required for successful ischemic preconditioning in mixed neuron/glia cultures [168].
p53 and mitochondrial dynamics (Figure 2C)
Maintaining proper mitochondrial length is essential for normal mitochondrial function in neurons and dysregulated mitochondrial dynamics has been associated with neuropathological conditions [169], [170], [171]. Mitochondrial fusion and fission affect bioenergetic efficiency and both excessive and impaired fission/fusion lead to mitochondrial dysfunction [172], [173], [174], [175]. Specifically, excessive mitochondrial fission or fragmentation is associated with and often causally related to neuronal cell death caused by a variety of experimental toxic stressors [176], [177], [178], [179]. Also, the process of fission is tightly coupled with and is a prerequisite for mitophagy [71], [180], while fusion is required for maintenance of mutation-free mtDNA assuring respiratory competence [181]. Mitochondrial fission and fusion are regulated by dynamin-related GTPases, and include dynamin-related protein 1 (Drp1) for fission and mitofusins (Mfn1 and Mfn2) and optic atrophy-1 (OPA1) for fusion [182].
Recent studies suggest that p53 has direct implications for the regulation of mitochondrial dynamics by altering the expression and activity of some of these fission/fusion proteins. p53 transcriptionally induces expression of Mfn2 in HepG2 hepatocellular carcinoma cells [183] and Drp1 in cardiomyocytes undergoing apoptosis [184]. p53 can also induce mitochondrial fission in HeLa cells by promoting Drp1 translocation to mitochondria through transcriptional suppression of miR-499 and its target calcineurin [185], a known Drp1 phosphatase and activator [186]. p53-dependent regulation of fission/fusion proteins in neurons is also observed for Drp1. A recent study implicates p53-Drp1 binding as a causal event to increased mitochondrial fragmentation and pathology in Huntington’s disease (HD) using immortalized mutant huntingtin (mtHtt: HdhQ111) knock-in mouse striatal cell lines, human HD-iPS cell-derived striatal neurons and R2/6 HD mice [187]. In this model, the levels of Drp1, but not p53, regulate mitochondrial co-accumulation of Drp1 and p53 although it was not demonstrated if p53/Drp1 complexes, which may also include huntingtin, are formed in the cytosol before mitochondrial translocation or on the mitochondrial surface [187]. Functionally, p53 suppression effectively mitigates disease-associated mitochondrial fragmentation and dysfunction (ROS production, loss of membrane potential) and cell death. Since there is no additional benefit observed with simultaneous direct inhibition of Drp1 activity, the findings suggest that the majority of mitochondrial pathology due to Drp-1 hyperactivation is mediated by p53’s interaction with Drp1 [187]. Studies from our laboratory demonstrate that treatment of cultured postnatal cortical neurons with the DNA-damaging agent camptothecin (CPT) results in elongated mitochondria, in contrast to fragmented mitochondria observed upon staurosporine and glutamate treatment [74]. This is due to p53-dependent, post-transcriptional suppression of the expression of Drp1 and the E3 ubiquitin ligase parkin, and overexpression of Drp1 or parkin corrects mitochondrial morphology and blocks associated neuronal cell death [74]. In contrast, CPT induces mitochondrial fragmentation in fibroblasts [74], suggesting cell type-specific regulation of Drp1 by p53. All of these effects of p53 on mitochondrial fission/fusion proteins should also influence mitophagy as mitochondrial fission is a prerequisite for this important process [71], [180].
Summary
The diverse range of functions performed by p53, even when considered in the narrow context of mitochondria-related events, is telling evidence for the multifunctional nature of the protein originally identified as a tumor suppressor. In addition to its function as a transcription factor evidence supports a role for direct mitochondria-related and cytosolic functions of p53 in non-neuronal cells. In neurons, the apoptosis/necrosis-inducing action of p53 directly at mitochondrial membranes is gradually being unraveled, but little is known about its expression levels and actions in the cytosol and the mitochondrial matrix under physiological and pathological levels of stress. The mitochondrial proteome currently contains more than 1000 proteins in humans and mice [188]. Interestingly, only about half of the proteins registered in the mitochondrial proteome may be shared between different tissue types [189], reflecting cell type-specific gene expression and protein localization for mitochondrial proteins. Given such heterogeneity, it comes as no surprise that neurons lack certain specific p53 functions demonstrated for non-neuronal cells and in turn display some neuron-specific ones. One notable example may be the overall pro-oxidant activity of p53 in neurons and brain, which imposes a constitutive oxidative stress burden on the nervous system [162]. The lack of compelling evidence for the participation of the PINK1/parkin pathway in mitophagy in neurons [72] in conjunction with p53-mediated downregulation of parkin expression [74] also suggests the presence of neuron-specific regulatory pathways for mitophagy.
It is clear that p53 exerts both pro-survival and pro-death actions through mitochondria-related pathways. This dichotomy in p53 action is clearly a product of the context-dependent action of p53 dictated, for instance, by its levels of expression and activity (chemical modifications), its subcellular localization (nuclear vs. cytosolic/mitochondria, transcriptional vs. non-transcriptional) and the availability of its interacting partners. Although knowledge regarding each of these parameters is still very limited for neurons, both actions may be simultaneously but separately targetable for neurodegenerative conditions, potentially yielding a more neuroprotective outcome.
Highlights.
The p53 tumor suppressor regulates cell survival and death as a sensor for stress
p53 induces mitochondrial changes through transcription-dependent and independent mechanisms
p53 can also regulate necrotic cell death and autophagic activity including mitophagy
p53 regulates proteins involved in mitochondrial metabolism and respiration
p53 has a much wider influence on mitochondrial integrity and function than previously expected
Acknowledgments
This work was supported by grants from the National Institutes of Health NS35533 and NS056031 to R.S.M. and by an NINDS Institutional Center Core Grant to support the viral core facility in the Neuroproteomics Center at the University of Washington (NS055088).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402–412. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 2.Hu W, Feng Z, Levine AJ. The Regulation of Multiple p53 Stress Responses is Mediated through MDM2. Genes Cancer. 2012;3:199–208. doi: 10.1177/1947601912454734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vlatkovic N, Boyd MT, Rubbi CP. Nucleolar control of p53: a cellular Achilles' heel and a target for cancer therapy. Cell Mol Life Sci. 2013 doi: 10.1007/s00018-013-1361-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: the critical roles of p53. Oncogene. 2013 doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 5.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 6.Lanni C, Racchi M, Memo M, Govoni S, Uberti D. p53 at the crossroads between cancer and neurodegeneration. Free radical biology & medicine. 2012;52:1727–1733. doi: 10.1016/j.freeradbiomed.2012.02.034. [DOI] [PubMed] [Google Scholar]
- 7.Berkers CR, Maddocks OD, Cheung EC, Mor I, Vousden KH. Metabolic Regulation by p53 Family Members. Cell metabolism. 2013;18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura Y. Isolation of p53-target genes and their functional analysis. Cancer science. 2004;95:7–11. doi: 10.1111/j.1349-7006.2004.tb03163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady CA, Attardi LD. p53 at a glance. Journal of cell science. 2010;123:2527–2532. doi: 10.1242/jcs.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stambolsky P, Weisz L, Shats I, Klein Y, Goldfinger N, Oren M, Rotter V. Regulation of AIF expression by p53. Cell death and differentiation. 2006;13:2140–2149. doi: 10.1038/sj.cdd.4401965. [DOI] [PubMed] [Google Scholar]
- 11.Miyashita T, Krajewski S, Krajewska M, Wang HG, Lin HK, Liebermann DA, Hoffman B, Reed JC. Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene. 1994;9:1799–1805. [PubMed] [Google Scholar]
- 12.Sugars KL, Budhram-Mahadeo V, Packham G, Latchman DS. A minimal Bcl-x promoter is activated by Brn-3a and repressed by p53. Nucleic acids research. 2001;29:4530–4540. doi: 10.1093/nar/29.22.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietrzak M, Puzianowska-Kuznicka M. p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol Chem. 2008;389:383–393. doi: 10.1515/BC.2008.039. [DOI] [PubMed] [Google Scholar]
- 14.Morrison RS, Kinoshita Y, Johnson MD, Guo W, Garden GA. p53-dependent cell death signaling in neurons. Neurochemical research. 2003;28:15–27. doi: 10.1023/a:1021687810103. [DOI] [PubMed] [Google Scholar]
- 15.Cregan SP, Arbour NA, Maclaurin JG, Callaghan SM, Fortin A, Cheung EC, Guberman DS, Park DS, Slack RS. p53 activation domain 1 is essential for PUMA upregulation and p53-mediated neuronal cell death. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:10003–10012. doi: 10.1523/JNEUROSCI.2114-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Culmsee C, Mattson MP. p53 in neuronal apoptosis. Biochemical and biophysical research communications. 2005;331:761–777. doi: 10.1016/j.bbrc.2005.03.149. [DOI] [PubMed] [Google Scholar]
- 17.Uo T, Kinoshita Y, Morrison RS. Apoptotic Actions of p53 Require Transcriptional Activation of PUMA and Do Not Involve a Direct Mitochondrial/Cytoplasmic Site of Action in Postnatal Cortical Neurons. The Journal of Neuroscience. 2007;27:12198–12210. doi: 10.1523/JNEUROSCI.3222-05.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Niizuma K, Endo H, Chan PH. Oxidative stress and mitochondrial dysfunction as determinants of ischemic neuronal death and survival. Journal of neurochemistry. 2009;109(Suppl 1):133–138. doi: 10.1111/j.1471-4159.2009.05897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaseva AV, Moll UM. The mitochondrial p53 pathway. Biochimica et biophysica acta. 2009;1787:414–420. doi: 10.1016/j.bbabio.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. The Journal of biological chemistry. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 22.Sansome C, Zaika A, Marchenko ND, Moll UM. Hypoxia death stimulus induces translocation of p53 protein to mitochondria. Detection by immunofluorescence on whole cells. FEBS Lett. 2001;488:110–115. doi: 10.1016/s0014-5793(00)02368-1. [DOI] [PubMed] [Google Scholar]
- 23.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–365. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 24.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. p53 has a direct apoptogenic role at the mitochondria. Molecular cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 25.Wood KA, Youle RJ. The role of free radicals and p53 in neuron apoptosis in vivo. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1995;15:5851–5857. doi: 10.1523/JNEUROSCI.15-08-05851.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hughes PE, Alexi T, Yoshida T, Schreiber SS, Knusel B. Excitotoxic lesion of rat brain with quinolinic acid induces expression of p53 messenger RNA and protein and p53-inducible genes Bax and Gadd-45 in brain areas showing DNA fragmentation. Neuroscience. 1996;74:1143–1160. doi: 10.1016/0306-4522(96)00174-1. [DOI] [PubMed] [Google Scholar]
- 27.Sakhi S, Sun N, Wing LL, Mehta P, Schreiber SS. Nuclear accumulation of p53 protein following kainic acid-induced seizures. Neuroreport. 1996;7:493–496. doi: 10.1097/00001756-199601310-00028. [DOI] [PubMed] [Google Scholar]
- 28.LaFerla FM, Hall CK, Ngo L, Jay G. Extracellular deposition of beta-amyloid upon p53-dependent neuronal cell death in transgenic mice. The Journal of clinical investigation. 1996;98:1626–1632. doi: 10.1172/JCI118957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadoul R, Quiquerez AL, Martinou I, Fernandez PA, Martinou JC. p53 protein in sympathetic neurons: cytoplasmic localization and no apparent function in apoptosis. J Neurosci Res. 1996;43:594–601. doi: 10.1002/(SICI)1097-4547(19960301)43:5<594::AID-JNR9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 30.Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 31.Endo H, Kamada H, Nito C, Nishi T, Chan PH. Mitochondrial translocation of p53 mediates release of cytochrome c and hippocampal CA1 neuronal death after transient global cerebral ischemia in rats. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:7974–7983. doi: 10.1523/JNEUROSCI.0897-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nair VD, McNaught KS, Gonzalez-Maeso J, Sealfon SC, Olanow CW. p53 mediates nontranscriptional cell death in dopaminergic cells in response to proteasome inhibition. The Journal of biological chemistry. 2006;281:39550–39560. doi: 10.1074/jbc.M603950200. [DOI] [PubMed] [Google Scholar]
- 33.Nijboer CH, Heijnen CJ, Groenendaal F, May MJ, van Bel F, Kavelaars A. Strong neuroprotection by inhibition of NF-kappaB after neonatal hypoxia-ischemia involves apoptotic mechanisms but is independent of cytokines. Stroke. 2008;39:2129–2137. doi: 10.1161/STROKEAHA.107.504175. [DOI] [PubMed] [Google Scholar]
- 34.Martin LJ, Liu Z, Pipino J, Chestnut B, Landek MA. Molecular regulation of DNA damage-induced apoptosis in neurons of cerebral cortex. Cereb Cortex. 2009;19:1273–1293. doi: 10.1093/cercor/bhn167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nijboer CH, Heijnen CJ, van der Kooij MA, Zijlstra J, van Velthoven CT, Culmsee C, van Bel F, Hagberg H, Kavelaars A. Targeting the p53 pathway to protect the neonatal ischemic brain. Ann Neurol. 2011;70:255–264. doi: 10.1002/ana.22413. [DOI] [PubMed] [Google Scholar]
- 36.Gomez-Sanchez JC, Delgado-Esteban M, Rodriguez-Hernandez I, Sobrino T, Perez de la Ossa N, Reverte S, Bolanos JP, Gonzalez-Sarmiento R, Castillo J, Almeida A. The human Tp53 Arg72Pro polymorphism explains different functional prognosis in stroke. J Exp Med. 2011;208:429–437. doi: 10.1084/jem.20101523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong XX, Wang YR, Qin S, Liang ZQ, Liu BH, Qin ZH, Wang Y. p53 mediates autophagy activation and mitochondria dysfunction in kainic acid-induced excitotoxicity in primary striatal neurons. Neuroscience. 2012;207:52–64. doi: 10.1016/j.neuroscience.2012.01.018. [DOI] [PubMed] [Google Scholar]
- 38.Komarov PG, Komarova EA, Kondratov RV, Christov-Tselkov K, Coon JS, Chernov MV, Gudkov AV. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285:1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 39.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474–479. doi: 10.1038/nchembio809. [DOI] [PubMed] [Google Scholar]
- 40.Komarova EA, Neznanov N, Komarov PG, Chernov MV, Wang K, Gudkov AV. p53 inhibitor pifithrin alpha can suppress heat shock and glucocorticoid signaling pathways. The Journal of biological chemistry. 2003;278:15465–15468. doi: 10.1074/jbc.C300011200. [DOI] [PubMed] [Google Scholar]
- 41.Hoagland MS, Hoagland EM, Swanson HI. The p53 inhibitor pifithrin-alpha is a potent agonist of the aryl hydrocarbon receptor. J Pharmacol Exp Ther. 2005;314:603–610. doi: 10.1124/jpet.105.084186. [DOI] [PubMed] [Google Scholar]
- 42.Sohn D, Graupner V, Neise D, Essmann F, Schulze-Osthoff K, Janicke RU. Pifithrin-alpha protects against DNA damage-induced apoptosis downstream of mitochondria independent of p53. Cell death and differentiation. 2009;16:869–878. doi: 10.1038/cdd.2009.17. [DOI] [PubMed] [Google Scholar]
- 43.Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Molecular cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heyne K, Schmitt K, Mueller D, Armbruester V, Mestres P, Roemer K. Resistance of mitochondrial p53 to dominant inhibition. Mol Cancer. 2008;7:54. doi: 10.1186/1476-4598-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakamuro D, Sabbatini P, White E, Prendergast GC. The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene. 1997;15:887–898. doi: 10.1038/sj.onc.1201263. [DOI] [PubMed] [Google Scholar]
- 46.Venot C, Maratrat M, Dureuil C, Conseiller E, Bracco L, Debussche L. The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. EMBO J. 1998;17:4668–4679. doi: 10.1093/emboj/17.16.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J, Jiang J, Zhou W, Zhu K, Chen X. Differential regulation of cellular target genes by p53 devoid of the PXXP motifs with impaired apoptotic activity. Oncogene. 1999;18:2149–2155. doi: 10.1038/sj.onc.1202533. [DOI] [PubMed] [Google Scholar]
- 48.Chipuk JE, Kuwana T, Bouchier-Hayes L, Droin NM, Newmeyer DD, Schuler M, Green DR. Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science. 2004;303:1010–1014. doi: 10.1126/science.1092734. [DOI] [PubMed] [Google Scholar]
- 49.Schuler M, Green DR. Transcription, apoptosis and p53: catch-22. Trends Genet. 2005;21:182–187. doi: 10.1016/j.tig.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 50.Zhao Y, Chaiswing L, Velez JM, Batinic-Haberle I, Colburn NH, Oberley TD, St Clair DK. p53 translocation to mitochondria precedes its nuclear translocation and targets mitochondrial oxidative defense protein-manganese superoxide dismutase. Cancer research. 2005;65:3745–3750. doi: 10.1158/0008-5472.CAN-04-3835. [DOI] [PubMed] [Google Scholar]
- 51.Vaseva AV, Marchenko ND, Ji K, Tsirka SE, Holzmann S, Moll UM. p53 opens the mitochondrial permeability transition pore to trigger necrosis. Cell. 2012;149:1536–1548. doi: 10.1016/j.cell.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crumrine RC, Thomas AL, Morgan PF. Attenuation of p53 expression protects against focal ischemic damage in transgenic mice. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism. 1994;14:887–891. doi: 10.1038/jcbfm.1994.119. [DOI] [PubMed] [Google Scholar]
- 53.Ferecatu I, Bergeaud M, Rodriguez-Enfedaque A, Le Floch N, Oliver L, Rincheval V, Renaud F, Vallette FM, Mignotte B, Vayssiere JL. Mitochondrial localization of the low level p53 protein in proliferative cells. Biochemical and biophysical research communications. 2009;387:772–777. doi: 10.1016/j.bbrc.2009.07.111. [DOI] [PubMed] [Google Scholar]
- 54.Montero J, Dutta C, van Bodegom D, Weinstock D, Letai A. p53 regulates a non-apoptotic death induced by ROS. Cell death and differentiation. 2013;20:1465–1474. doi: 10.1038/cdd.2013.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ. The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer research. 2007;67:3043–3053. doi: 10.1158/0008-5472.CAN-06-4149. [DOI] [PubMed] [Google Scholar]
- 56.Lomonosova E, Chinnadurai G. BH3-only proteins in apoptosis and beyond: an overview. Oncogene. 2008;27(Suppl 1):S2–S19. doi: 10.1038/onc.2009.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maiuri MC, Galluzzi L, Morselli E, Kepp O, Malik SA, Kroemer G. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 58.Okamura S, Arakawa H, Tanaka T, Nakanishi H, Ng CC, Taya Y, Monden M, Nakamura Y. p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Molecular cell. 2001;8:85–94. doi: 10.1016/s1097-2765(01)00284-2. [DOI] [PubMed] [Google Scholar]
- 59.Seillier M, Peuget S, Gayet O, Gauthier C, N'Guessan P, Monte M, Carrier A, Iovanna JL, Dusetti NJ. TP53INP1, a tumor suppressor, interacts with LC3 and ATG8-family proteins through the LC3-interacting region (LIR) and promotes autophagy-dependent cell death. Cell death and differentiation. 2012;19:1525–1535. doi: 10.1038/cdd.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vanlandingham JW, Tassabehji NM, Somers RC, Levenson CW. Expression profiling of p53-target genes in copper-mediated neuronal apoptosis. Neuromolecular Med. 2005;7:311–324. doi: 10.1385/NMM:7:4:311. [DOI] [PubMed] [Google Scholar]
- 61.Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, Gasco M, Garrone O, Crook T, Ryan KM. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 62.Mah LY, O'Prey J, Baudot AD, Hoekstra A, Ryan KM. DRAM-1 encodes multiple isoforms that regulate autophagy. Autophagy. 2012;8:18–28. doi: 10.4161/auto.8.1.18077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang XD, Wang Y, Wang Y, Zhang X, Han R, Wu JC, Liang ZQ, Gu ZL, Han F, Fukunaga K, Qin ZH. p53 mediates mitochondria dysfunction-triggered autophagy activation and cell death in rat striatum. Autophagy. 2009;5:339–350. doi: 10.4161/auto.5.3.8174. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Dong XX, Cao Y, Liang ZQ, Han R, Wu JC, Gu ZL, Qin ZH. p53 induction contributes to excitotoxic neuronal death in rat striatum through apoptotic and autophagic mechanisms. The European journal of neuroscience. 2009;30:2258–2270. doi: 10.1111/j.1460-9568.2009.07025.x. [DOI] [PubMed] [Google Scholar]
- 65.Tasdemir E, Maiuri MC, Galluzzi L, Vitale I, Djavaheri-Mergny M, D'Amelio M, Criollo A, Morselli E, Zhu C, Harper F, Nannmark U, Samara C, Pinton P, Vicencio JM, Carnuccio R, Moll UM, Madeo F, Paterlini-Brechot P, Rizzuto R, Szabadkai G, Pierron G, Blomgren K, Tavernarakis N, Codogno P, Cecconi F, Kroemer G. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Scherz-Shouval R, Weidberg H, Gonen C, Wilder S, Elazar Z, Oren M. p53-dependent regulation of autophagy protein LC3 supports cancer cell survival under prolonged starvation. Proc Natl Acad Sci U S A. 2010;107:18511–18516. doi: 10.1073/pnas.1006124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kreiner G, Bierhoff H, Armentano M, Rodriguez-Parkitna J, Sowodniok K, Naranjo JR, Bonfanti L, Liss B, Schutz G, Grummt I, Parlato R. A neuroprotective phase precedes striatal degeneration upon nucleolar stress. Cell death and differentiation. 2013 doi: 10.1038/cdd.2013.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jaeger PA, Wyss-Coray T. All-you-can-eat: autophagy in neurodegeneration and neuroprotection. Molecular neurodegeneration. 2009;4:16. doi: 10.1186/1750-1326-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ashrafi G, Schwarz TL. The pathways of mitophagy for quality control and clearance of mitochondria. Cell death and differentiation. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Feng D, Liu L, Zhu Y, Chen Q. Molecular signaling toward mitophagy and its physiological significance. Exp Cell Res. 2013;319:1697–1705. doi: 10.1016/j.yexcr.2013.03.034. [DOI] [PubMed] [Google Scholar]
- 71.Gomes LC, Scorrano L. Mitochondrial morphology in mitophagy and macroautophagy. Biochimica et biophysica acta. 2013;1833:205–212. doi: 10.1016/j.bbamcr.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 72.Grenier K, McLelland GL, Fon EA. Parkin- and PINK1-Dependent Mitophagy in Neurons: Will the Real Pathway Please Stand Up? Front Neurol. 2013;4:100. doi: 10.3389/fneur.2013.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang C, Lin M, Wu R, Wang X, Yang B, Levine AJ, Hu W, Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc Natl Acad Sci U S A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang DB, Garden GA, Kinoshita C, Wyles C, Babazadeh N, Sopher B, Kinoshita Y, Morrison RS. Declines in Drp1 and parkin expression underlie DNA damage-induced changes in mitochondrial length and neuronal death. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:1357–1365. doi: 10.1523/JNEUROSCI.3365-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.da Costa CA, Sunyach C, Giaime E, West A, Corti O, Brice A, Safe S, Abou-Sleiman PM, Wood NW, Takahashi H, Goldberg MS, Shen J, Checler F. Transcriptional repression of p53 by parkin and impairment by mutations associated with autosomal recessive juvenile Parkinson's disease. Nat Cell Biol. 2009;11:1370–1375. doi: 10.1038/ncb1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sunico CR, Nakamura T, Rockenstein E, Mante M, Adame A, Chan SF, Newmeyer TF, Masliah E, Nakanishi N, Lipton SA. S-Nitrosylation of parkin as a novel regulator of p53-mediated neuronal cell death in sporadic Parkinson's disease. Molecular neurodegeneration. 2013;8:29. doi: 10.1186/1750-1326-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hoshino A, Mita Y, Okawa Y, Ariyoshi M, Iwai-Kanai E, Ueyama T, Ikeda K, Ogata T, Matoba S. Cytosolic p53 inhibits Parkin-mediated mitophagy and promotes mitochondrial dysfunction in the mouse heart. Nat Commun. 2013;4:2308. doi: 10.1038/ncomms3308. [DOI] [PubMed] [Google Scholar]
- 78.Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009;28:3015–3026. doi: 10.1038/emboj.2009.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim H, Rafiuddin-Shah M, Tu HC, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Hierarchical regulation of mitochondrion-dependent apoptosis by BCL-2 subfamilies. Nat Cell Biol. 2006;8:1348–1358. doi: 10.1038/ncb1499. [DOI] [PubMed] [Google Scholar]
- 80.Zhang J, Ney PA. Mechanisms and biology of B-cell leukemia/lymphoma 2/adenovirus E1B interacting protein 3 and Nip-like protein X. Antioxidants & redox signaling. 2011;14:1959–1969. doi: 10.1089/ars.2010.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Feng X, Liu X, Zhang W, Xiao W. p53 directly suppresses BNIP3 expression to protect against hypoxia-induced cell death. EMBO J. 2011;30:3397–3415. doi: 10.1038/emboj.2011.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fei P, Wang W, Kim SH, Wang S, Burns TF, Sax JK, Buzzai M, Dicker DT, McKenna WG, Bernhard EJ, El-Deiry WS. Bnip3L is induced by p53 under hypoxia, and its knockdown promotes tumor growth. Cancer Cell. 2004;6:597–609. doi: 10.1016/j.ccr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 83.Zhang S, Zhang Z, Sandhu G, Ma X, Yang X, Geiger JD, Kong J. Evidence of oxidative stress-induced BNIP3 expression in amyloid beta neurotoxicity. Brain Res. 2007;1138:221–230. doi: 10.1016/j.brainres.2006.12.086. [DOI] [PubMed] [Google Scholar]
- 84.Prabhakaran K, Li L, Zhang L, Borowitz JL, Isom GE. Upregulation of BNIP3 and translocation to mitochondria mediates cyanide-induced apoptosis in cortical cells. Neuroscience. 2007;150:159–167. doi: 10.1016/j.neuroscience.2007.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao ST, Chen M, Li SJ, Zhang MH, Li BX, Das M, Bean JC, Kong JM, Zhu XH, Gao TM. Mitochondrial BNIP3 upregulation precedes endonuclease G translocation in hippocampal neuronal death following oxygen-glucose deprivation. BMC Neurosci. 2009;10:113. doi: 10.1186/1471-2202-10-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yeh SH, Ou LC, Gean PW, Hung JJ, Chang WC. Selective inhibition of early--but not late--expressed HIF-1alpha is neuroprotective in rats after focal ischemic brain damage. Brain Pathol. 2011;21:249–262. doi: 10.1111/j.1750-3639.2010.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rui Y, Ke K, Li L, Zheng H, Xu W, Tan X, Cao J, Wu X, Cui G, Zhao G, Gao Y, Cao M. Up-regulated expression of Bnip3L after intracerebral hemorrhage in adult rats. J Mol Histol. 2013 doi: 10.1007/s10735-013-9506-7. [DOI] [PubMed] [Google Scholar]
- 88.Miyamoto Y, Kitamura N, Nakamura Y, Futamura M, Miyamoto T, Yoshida M, Ono M, Ichinose S, Arakawa H. Possible existence of lysosome-like organella within mitochondria and its role in mitochondrial quality control. PLoS One. 2011;6:e16054. doi: 10.1371/journal.pone.0016054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kitamura N, Nakamura Y, Miyamoto Y, Miyamoto T, Kabu K, Yoshida M, Futamura M, Ichinose S, Arakawa H. Mieap, a p53-inducible protein, controls mitochondrial quality by repairing or eliminating unhealthy mitochondria. PLoS One. 2011;6:e16060. doi: 10.1371/journal.pone.0016060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakamura Y, Kitamura N, Shinogi D, Yoshida M, Goda O, Murai R, Kamino H, Arakawa H. BNIP3 and NIX mediate Mieap-induced accumulation of lysosomal proteins within mitochondria. PLoS One. 2012;7:e30767. doi: 10.1371/journal.pone.0030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miyamoto T, Kitamura N, Ono M, Nakamura Y, Yoshida M, Kamino H, Murai R, Yamada T, Arakawa H. Identification of 14-3-3gamma as a Mieap-interacting protein and its role in mitochondrial quality control. Sci Rep. 2012;2:379. doi: 10.1038/srep00379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Radhakrishnan VM, Putnam CW, Qi W, Martinez JD. P53 suppresses expression of the 14-3-3 gamma oncogene. BMC Cancer. 2011;11:378. doi: 10.1186/1471-2407-11-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lehar SM, Nacht M, Jacks T, Vater CA, Chittenden T, Guild BC. Identification and cloning of EI24, a gene induced by p53 in etoposide-treated cells. Oncogene. 1996;12:1181–1187. [PubMed] [Google Scholar]
- 94.Zhao YG, Zhao H, Miao L, Wang L, Sun F, Zhang H. The p53-induced gene Ei24 is an essential component of the basal autophagy pathway. The Journal of biological chemistry. 2012;287:42053–42063. doi: 10.1074/jbc.M112.415968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao X, Ayer RE, Davis SL, Ames SJ, Florence B, Torchinsky C, Liou JS, Shen L, Spanjaard RA. Apoptosis factor EI24/PIG8 is a novel endoplasmic reticulum-localized Bcl-2-binding protein which is associated with suppression of breast cancer invasiveness. Cancer research. 2005;65:2125–2129. doi: 10.1158/0008-5472.CAN-04-3377. [DOI] [PubMed] [Google Scholar]
- 96.Bahk YY, Lee J, Cho IH, Lee HW. An analysis of an interactome for apoptosis factor, Ei24/PIG8, using the inducible expression system and shotgun proteomics. J Proteome Res. 2010;9:5270–5283. doi: 10.1021/pr100552y. [DOI] [PubMed] [Google Scholar]
- 97.Merkwirth C, Langer T. Prohibitin function within mitochondria: essential roles for cell proliferation and cristae morphogenesis. Biochimica et biophysica acta. 2009;1793:27–32. doi: 10.1016/j.bbamcr.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 98.Park JY, Wang PY, Matsumoto T, Sung HJ, Ma W, Choi JW, Anderson SA, Leary SC, Balaban RS, Kang JG, Hwang PM. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circulation research. 2009;105:705–712. doi: 10.1161/CIRCRESAHA.109.205310. 711 p following 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wong TS, Rajagopalan S, Freund SM, Rutherford TJ, Andreeva A, Townsley FM, Petrovich M, Fersht AR. Biophysical characterizations of human mitochondrial transcription factor A and its binding to tumor suppressor p53. Nucleic acids research. 2009;37:6765–6783. doi: 10.1093/nar/gkp750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wong TS, Rajagopalan S, Townsley FM, Freund SM, Petrovich M, Loakes D, Fersht AR. Physical and functional interactions between human mitochondrial single-stranded DNA-binding protein and tumour suppressor p53. Nucleic acids research. 2009;37:568–581. doi: 10.1093/nar/gkn974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yoshida Y, Izumi H, Torigoe T, Ishiguchi H, Itoh H, Kang D, Kohno K. P53 physically interacts with mitochondrial transcription factor A and differentially regulates binding to damaged DNA. Cancer research. 2003;63:3729–3734. [PubMed] [Google Scholar]
- 102.Saleem A, Hood DA. Acute exercise induces tumour suppressor protein p53 translocation to the mitochondria and promotes a p53-Tfam-mitochondrial DNA complex in skeletal muscle. J Physiol. 2013;591:3625–3636. doi: 10.1113/jphysiol.2013.252791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Achanta G, Sasaki R, Feng L, Carew JS, Lu W, Pelicano H, Keating MJ, Huang P. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24:3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Souza-Pinto NC, Harris CC, Bohr VA. p53 functions in the incorporation step in DNA base excision repair in mouse liver mitochondria. Oncogene. 2004;23:6559–6568. doi: 10.1038/sj.onc.1207874. [DOI] [PubMed] [Google Scholar]
- 105.Lebedeva MA, Eaton JS, Shadel GS. Loss of p53 causes mitochondrial DNA depletion and altered mitochondrial reactive oxygen species homeostasis. Biochimica et biophysica acta. 2009;1787:328–334. doi: 10.1016/j.bbabio.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kulawiec M, Ayyasamy V, Singh KK. p53 regulates mtDNA copy number and mitocheckpoint pathway. J Carcinog. 2009;8:8. doi: 10.4103/1477-3163.50893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 108.Pontarin G, Ferraro P, Bee L, Reichard P, Bianchi V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc Natl Acad Sci U S A. 2012;109:13302–13307. doi: 10.1073/pnas.1211289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bourdon A, Minai L, Serre V, Jais JP, Sarzi E, Aubert S, Chretien D, de Lonlay P, Paquis-Flucklinger V, Arakawa H, Nakamura Y, Munnich A, Rotig A. Mutation of RRM2B, encoding p53-controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet. 2007;39:776–780. doi: 10.1038/ng2040. [DOI] [PubMed] [Google Scholar]
- 110.Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci U S A. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Almeida A. Regulation of APC/C-Cdh1 and its function in neuronal survival. Mol Neurobiol. 2012;46:547–554. doi: 10.1007/s12035-012-8309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bergeaud M, Mathieu L, Guillaume A, Moll UM, Mignotte B, Le Floch N, Vayssiere JL, Rincheval V. Mitochondrial p53 mediates a transcription-independent regulation of cell respiration and interacts with the mitochondrial F 1F 0-ATP synthase. Cell Cycle. 2013;12 doi: 10.4161/cc.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Austin S, St-Pierre J. PGC1alpha and mitochondrial metabolism--emerging concepts and relevance in ageing and neurodegenerative disorders. Journal of cell science. 2012;125:4963–4971. doi: 10.1242/jcs.113662. [DOI] [PubMed] [Google Scholar]
- 114.Wenz T. Regulation of mitochondrial biogenesis and PGC-1alpha under cellular stress. Mitochondrion. 2013;13:134–142. doi: 10.1016/j.mito.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 115.Tsunemi T, La Spada AR. PGC-1alpha at the intersection of bioenergetics regulation and neuron function: from Huntington's disease to Parkinson's disease and beyond. Prog Neurobiol. 2012;97:142–151. doi: 10.1016/j.pneurobio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Saleem A, Adhihetty PJ, Hood DA. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 117.Sahin E, Colla S, Liesa M, Moslehi J, Muller FL, Guo M, Cooper M, Kotton D, Fabian AJ, Walkey C, Maser RS, Tonon G, Foerster F, Xiong R, Wang YA, Shukla SA, Jaskelioff M, Martin ES, Heffernan TP, Protopopov A, Ivanova E, Mahoney JE, Kost-Alimova M, Perry SR, Bronson R, Liao R, Mulligan R, Shirihai OS, Chin L, DePinho RA. Telomere dysfunction induces metabolic and mitochondrial compromise. Nature. 2011;470:359–365. doi: 10.1038/nature09787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aquilano K, Baldelli S, Pagliei B, Cannata SM, Rotilio G, Ciriolo MR. p53 orchestrates the PGC-1alpha-mediated antioxidant response upon mild redox and metabolic imbalance. Antioxidants & redox signaling. 2013;18:386–399. doi: 10.1089/ars.2012.4615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Villeneuve C, Guilbeau-Frugier C, Sicard P, Lairez O, Ordener C, Duparc T, De Paulis D, Couderc B, Spreux-Varoquaux O, Tortosa F, Garnier A, Knauf C, Valet P, Borchi E, Nediani C, Gharib A, Ovize M, Delisle MB, Parini A, Mialet-Perez J. p53-PGC-1alpha pathway mediates oxidative mitochondrial damage and cardiomyocyte necrosis induced by monoamine oxidase-A upregulation: role in chronic left ventricular dysfunction in mice. Antioxidants & redox signaling. 2013;18:5–18. doi: 10.1089/ars.2011.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sen N, Satija YK, Das S. PGC-1alpha, a key modulator of p53, promotes cell survival upon metabolic stress. Molecular cell. 2011;44:621–634. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 121.Aquilano K, Vigilanza P, Baldelli S, Pagliei B, Rotilio G, Ciriolo MR. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. The Journal of biological chemistry. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 123.Shen L, Sun X, Fu Z, Yang G, Li J, Yao L. The fundamental role of the p53 pathway in tumor metabolism and its implication in tumor therapy. Clin Cancer Res. 2012;18:1561–1567. doi: 10.1158/1078-0432.CCR-11-3040. [DOI] [PubMed] [Google Scholar]
- 124.Contractor T, Harris CR. p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer research. 2012;72:560–567. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- 125.Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci U S A. 2010;107:7455–7460. doi: 10.1073/pnas.1001006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc Natl Acad Sci U S A. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Okamura S, Ng CC, Koyama K, Takei Y, Arakawa H, Monden M, Nakamura Y. Identification of seven genes regulated by wild-type p53 in a colon cancer cell line carrying a well-controlled wild-type p53 expression system. Oncol Res. 1999;11:281–285. [PubMed] [Google Scholar]
- 128.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 129.Zhou S, Kachhap S, Singh KK. Mitochondrial impairment in p53-deficient human cancer cells. Mutagenesis. 2003;18:287–292. doi: 10.1093/mutage/18.3.287. [DOI] [PubMed] [Google Scholar]
- 130.Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, Feraud O, Debili N, Wissing S, Engelhardt S, Madeo F, Piacentini M, Penninger JM, Schagger H, Rustin P, Kroemer G. AIF deficiency compromises oxidative phosphorylation. EMBO J. 2004;23:4679–4689. doi: 10.1038/sj.emboj.7600461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yoon KA, Nakamura Y, Arakawa H. Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J Hum Genet. 2004;49:134–140. doi: 10.1007/s10038-003-0122-3. [DOI] [PubMed] [Google Scholar]
- 132.Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 133.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, Yang X. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13:310–316. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Herrero-Mendez A, Almeida A, Fernandez E, Maestre C, Moncada S, Bolanos JP. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11:747–752. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- 135.Hoshino A, Matoba S, Iwai-Kanai E, Nakamura H, Kimata M, Nakaoka M, Katamura M, Okawa Y, Ariyoshi M, Mita Y, Ikeda K, Ueyama T, Okigaki M, Matsubara H. p53-TIGAR axis attenuates mitophagy to exacerbate cardiac damage after ischemia. J Mol Cell Cardiol. 2012;52:175–184. doi: 10.1016/j.yjmcc.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 136.Xiang H, Hochman DW, Saya H, Fujiwara T, Schwartzkroin PA, Morrison RS. Evidence for p53-mediated modulation of neuronal viability. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1996;16:6753–6765. doi: 10.1523/JNEUROSCI.16-21-06753.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rodriguez-Rodriguez P, Fernandez E, Almeida A, Bolanos JP. Excitotoxic stimulus stabilizes PFKFB3 causing pentose-phosphate pathway to glycolysis switch and neurodegeneration. Cell death and differentiation. 2012;19:1582–1589. doi: 10.1038/cdd.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Katsel P, Tan W, Fam P, Purohit DP, Haroutunian V. Cycle Checkpoint Abnormalities during Dementia: A Plausible Association with the Loss of Protection against Oxidative Stress in Alzheimer's Disease. PLoS One. 2013;8:e68361. doi: 10.1371/journal.pone.0068361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Molecular cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 140.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 141.Hussain SP, Amstad P, He P, Robles A, Lupold S, Kaneko I, Ichimiya M, Sengupta S, Mechanic L, Okamura S, Hofseth LJ, Moake M, Nagashima M, Forrester KS, Harris CC. p53-induced up-regulation of MnSOD and GPx but not catalase increases oxidative stress and apoptosis. Cancer research. 2004;64:2350–2356. doi: 10.1158/0008-5472.can-2287-2. [DOI] [PubMed] [Google Scholar]
- 142.Cano CE, Gommeaux J, Pietri S, Culcasi M, Garcia S, Seux M, Barelier S, Vasseur S, Spoto RP, Pebusque MJ, Dusetti NJ, Iovanna JL, Carrier A. Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer research. 2009;69:219–226. doi: 10.1158/0008-5472.CAN-08-2320. [DOI] [PubMed] [Google Scholar]
- 143.Esworthy RS, Ho YS, Chu FF. The Gpx1 gene encodes mitochondrial glutathione peroxidase in the mouse liver. Archives of biochemistry and biophysics. 1997;340:59–63. doi: 10.1006/abbi.1997.9901. [DOI] [PubMed] [Google Scholar]
- 144.Mari M, Morales A, Colell A, Garcia-Ruiz C, Kaplowitz N, Fernandez-Checa JC. Mitochondrial glutathione: features, regulation and role in disease. Biochimica et biophysica acta. 2013;1830:3317–3328. doi: 10.1016/j.bbagen.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Holley AK, Dhar SK, St Clair DK. Manganese superoxide dismutase vs. p53: regulation of mitochondrial ROS. Mitochondrion. 2010;10:649–661. doi: 10.1016/j.mito.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 146.Pani G, Galeotti T. Role of MnSOD and p66shc in mitochondrial response to p53. Antioxidants & redox signaling. 2011;15:1715–1727. doi: 10.1089/ars.2010.3499. [DOI] [PubMed] [Google Scholar]
- 147.Dhar SK, Xu Y, St Clair DK. Nuclear factor kappaB- and specificity protein 1-dependent p53-mediated bi-directional regulation of the human manganese superoxide dismutase gene. The Journal of biological chemistry. 2010;285:9835–9846. doi: 10.1074/jbc.M109.060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bakthavatchalu V, Dey S, Xu Y, Noel T, Jungsuwadee P, Holley AK, Dhar SK, Batinic-Haberle I, St Clair DK. Manganese superoxide dismutase is a mitochondrial fidelity protein that protects Polgamma against UV-induced inactivation. Oncogene. 2012;31:2129–2139. doi: 10.1038/onc.2011.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Polyak K, Xia Y, Zweier JL, Kinzler KW, Vogelstein B. A model for p53-induced apoptosis. Nature. 1997;389:300–305. doi: 10.1038/38525. [DOI] [PubMed] [Google Scholar]
- 150.Liu G, Chen X. The ferredoxin reductase gene is regulated by the p53 family and sensitizes cells to oxidative stress-induced apoptosis. Oncogene. 2002;21:7195–7204. doi: 10.1038/sj.onc.1205862. [DOI] [PubMed] [Google Scholar]
- 151.Maxwell SA, Rivera A. Proline oxidase induces apoptosis in tumor cells, and its expression is frequently absent or reduced in renal carcinomas. The Journal of biological chemistry. 2003;278:9784–9789. doi: 10.1074/jbc.M210012200. [DOI] [PubMed] [Google Scholar]
- 152.Liu Y, Borchert GL, Donald SP, Surazynski A, Hu CA, Weydert CJ, Oberley LW, Phang JM. MnSOD inhibits proline oxidase-induced apoptosis in colorectal cancer cells. Carcinogenesis. 2005;26:1335–1342. doi: 10.1093/carcin/bgi083. [DOI] [PubMed] [Google Scholar]
- 153.Gertz M, Steegborn C. The Lifespan-regulator p66Shc in mitochondria: redox enzyme or redox sensor? Antioxidants & redox signaling. 2010;13:1417–1428. doi: 10.1089/ars.2010.3147. [DOI] [PubMed] [Google Scholar]
- 154.Su K, Bourdette D, Forte M. Mitochondrial dysfunction and neurodegeneration in multiple sclerosis. Frontiers in physiology. 2013;4:169. doi: 10.3389/fphys.2013.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi PP, Lanfrancone L, Pelicci PG. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 156.Trinei M, Giorgio M, Cicalese A, Barozzi S, Ventura A, Migliaccio E, Milia E, Padura IM, Raker VA, Maccarana M, Petronilli V, Minucci S, Bernardi P, Lanfrancone L, Pelicci PG. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–3878. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 157.Kim CS, Jung SB, Naqvi A, Hoffman TA, DeRicco J, Yamamori T, Cole MP, Jeon BH, Irani K. p53 impairs endothelium-dependent vasomotor function through transcriptional upregulation of p66shc. Circulation research. 2008;103:1441–1450. doi: 10.1161/CIRCRESAHA.108.181644. [DOI] [PubMed] [Google Scholar]
- 158.Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F, Bernier G. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nature medicine. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Forsberg K, Wuttke A, Quadrato G, Chumakov PM, Wizenmann A, Di Giovanni S. The tumor suppressor p53 fine-tunes reactive oxygen species levels and neurogenesis via PI3 kinase signaling. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2013;33:14318–14330. doi: 10.1523/JNEUROSCI.1056-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Barone E, Cenini G, Sultana R, Di Domenico F, Fiorini A, Perluigi M, Noel T, Wang C, Mancuso C, St Clair DK, Butterfield DA. Lack of p53 decreases basal oxidative stress levels in the brain through upregulation of thioredoxin-1, biliverdin reductase-A, manganese superoxide dismutase, and nuclear factor kappa-B. Antioxidants & redox signaling. 2012;16:1407–1420. doi: 10.1089/ars.2011.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chatoo W, Abdouh M, Bernier G. p53 pro-oxidant activity in the central nervous system: implication in aging and neurodegenerative diseases. Antioxidants & redox signaling. 2011;15:1729–1737. doi: 10.1089/ars.2010.3610. [DOI] [PubMed] [Google Scholar]
- 163.Sompol P, Ittarat W, Tangpong J, Chen Y, Doubinskaia I, Batinic-Haberle I, Abdul HM, Butterfield DA, St Clair DK. A neuronal model of Alzheimer's disease: an insight into the mechanisms of oxidative stress-mediated mitochondrial injury. Neuroscience. 2008;153:120–130. doi: 10.1016/j.neuroscience.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou D, Zhan C, Zhong Q, Li S. Upregulation of sestrin-2 expression via P53 protects against 1-methyl-4-phenylpyridinium (MPP+) neurotoxicity. Journal of molecular neuroscience : MN. 2013;51:967–975. doi: 10.1007/s12031-013-0081-x. [DOI] [PubMed] [Google Scholar]
- 165.Su KG, Savino C, Marracci G, Chaudhary P, Yu X, Morris B, Galipeau D, Giorgio M, Forte M, Bourdette D. Genetic inactivation of the p66 isoform of ShcA is neuroprotective in a murine model of multiple sclerosis. The European journal of neuroscience. 2012;35:562–571. doi: 10.1111/j.1460-9568.2011.07972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Su K, Bourdette D, Forte M. Genetic inactivation of mitochondria-targeted redox enzyme p66ShcA preserves neuronal viability and mitochondrial integrity in response to oxidative challenges. Frontiers in physiology. 2012;3:285. doi: 10.3389/fphys.2012.00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Smith WW, Norton DD, Gorospe M, Jiang H, Nemoto S, Holbrook NJ, Finkel T, Kusiak JW. Phosphorylation of p66Shc and forkhead proteins mediates Abeta toxicity. The Journal of cell biology. 2005;169:331–339. doi: 10.1083/jcb.200410041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brown JE, Zeiger SL, Hettinger JC, Brooks JD, Holt B, Morrow JD, Musiek ES, Milne G, McLaughlin B. Essential role of the redox-sensitive kinase p66shc in determining energetic and oxidative status and cell fate in neuronal preconditioning. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:5242–5252. doi: 10.1523/JNEUROSCI.6366-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.DuBoff B, Feany M, Gotz J. Why size matters - balancing mitochondrial dynamics in Alzheimer's disease. Trends Neurosci. 2013;36:325–335. doi: 10.1016/j.tins.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 170.Itoh K, Nakamura K, Iijima M, Sesaki H. Mitochondrial dynamics in neurodegeneration. Trends Cell Biol. 2013;23:64–71. doi: 10.1016/j.tcb.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.McInnes J. Insights on altered mitochondrial function and dynamics in the pathogenesis of neurodegeneration. Transl Neurodegener. 2013;2:12. doi: 10.1186/2047-9158-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS One. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Uo T, Dworzak J, Kinoshita C, Inman DM, Kinoshita Y, Horner PJ, Morrison RS. Drp1 levels constitutively regulate mitochondrial dynamics and cell survival in cortical neurons. Exp Neurol. 2009;218:274–285. doi: 10.1016/j.expneurol.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. The Journal of cell biology. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.DuBoff B, Gotz J, Feany MB. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75:618–632. doi: 10.1016/j.neuron.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. EMBO J. 2006;25:3900–3911. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, Wang Y, Casadesus G, Zhu X. Amyloid-β overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proceedings of the National Academy of Sciences. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Knott AB, Perkins G, Schwarzenbacher R, Bossy-Wetzel E. Mitochondrial fragmentation in neurodegeneration. Nat Rev Neurosci. 2008;9:505–518. doi: 10.1038/nrn2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Liot G, Bossy B, Lubitz S, Kushnareva Y, Sejbuk N, Bossy-Wetzel E. Complex II inhibition by 3-NP causes mitochondrial fragmentation and neuronal cell death via an NMDA- and ROS-dependent pathway. Cell death and differentiation. 2009;16:899–909. doi: 10.1038/cdd.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Chen H, Vermulst M, Wang YE, Chomyn A, Prolla TA, McCaffery JM, Chan DC. Mitochondrial fusion is required for mtDNA stability in skeletal muscle and tolerance of mtDNA mutations. Cell. 2010;141:280–289. doi: 10.1016/j.cell.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen H, Chan DC. Emerging functions of mammalian mitochondrial fusion and fission. Hum Mol Genet. 2005;14(Spec No. 2):R283–R289. doi: 10.1093/hmg/ddi270. [DOI] [PubMed] [Google Scholar]
- 183.Wang W, Cheng X, Lu J, Wei J, Fu G, Zhu F, Jia C, Zhou L, Xie H, Zheng S. Mitofusin-2 is a novel direct target of p53. Biochemical and biophysical research communications. 2010;400:587–592. doi: 10.1016/j.bbrc.2010.08.108. [DOI] [PubMed] [Google Scholar]
- 184.Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet. 2010;6:e1000795. doi: 10.1371/journal.pgen.1000795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wang JX, Jiao JQ, Li Q, Long B, Wang K, Liu JP, Li YR, Li PF. miR-499 regulates mitochondrial dynamics by targeting calcineurin and dynamin-related protein-1. Nature medicine. 2011;17:71–78. doi: 10.1038/nm.2282. [DOI] [PubMed] [Google Scholar]
- 186.Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guo X, Disatnik MH, Monbureau M, Shamloo M, Mochly-Rosen D, Qi X. Inhibition of mitochondrial fragmentation diminishes Huntington's disease-associated neurodegeneration. The Journal of clinical investigation. 2013;123:5371–5388. doi: 10.1172/JCI70911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Smith AC, Blackshaw JA, Robinson AJ. MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic acids research. 2012;40:D1160–D1167. doi: 10.1093/nar/gkr1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]