SUMMARY
Patients with EGFR-mutant lung adenocarcinomas (LUADs) who initially respond to first-generation TKIs develop resistance to these drugs. A combination of the irreversible TKI afatinib and the EGFR antibody cetuximab can be used to overcome resistance to first-generation TKIs; however, resistance to this drug combination eventually emerges. We identified activation of the mTORC1 signaling pathway as a mechanism of resistance to dual inhibition of EGFR in mouse models. Addition of rapamycin reversed resistance in vivo. Analysis of afatinib+cetuximab-resistant biopsy specimens revealed the presence of genomic alterations in genes that modulate mTORC1 signaling including NF2 and TSC1. These findings pinpoint enhanced mTORC1 activation as a mechanism of resistance to afatinib+cetuximab and identify genomic mechanisms that lead to activation of this pathway, revealing a potential therapeutic strategy for treating patients with resistance to these drugs.
INTRODUCTION
Targeted therapies effectively treat subsets of solid cancers. However, the inevitable development of acquired resistance (AR) has hampered their success. A paradigm for this concept is the case of Epidermal Growth Factor Receptor (EGFR)-mutant lung cancer. EGFR mutations (Exon 19 deletions or the L858R point mutation) are associated with sensitivity to the first-generation tyrosine kinase inhibitors (TKIs) gefitinib and erlotinib (Pao and Chmielecki, 2010), but drug resistance emerges on average 1 year after TKI treatment. In ~50% of resistant tumors, the mutant EGFR allele has acquired a secondary mutation in exon 20 (T790M) (Pao and Chmielecki, 2010). Additional mechanisms of resistance include amplification of other receptor tyrosine kinases (RTKs) like MET and HER2 (ERBB2), mutations in genes encoding downstream signaling components or phenotypic transformations such as epithelial-to-mesenchymal transition (EMT) and neuroendocrine differentiation (Ohashi et al., 2013).
In a previous study using transgenic mice with EGFRL858R+T790M-induced LUADs, we showed that resistance due to EGFR T790M could be overcome using a combination of afatinib+cetuximab (A+C) (Regales et al., 2009). Afatinib is a second-generation TKI that covalently binds EGFR at cysteine 797, while cetuximab is an anti-EGFR antibody. This preclinical study prompted a Phase IB/II clinical trial testing this drug combination in patients with progressive disease after TKI treatment. The trial showed an overall 32% response rate with a median duration of response of eight months (Janjigian et al., 2012). Unfortunately, patients responding to the drug combination still develop progressive disease.
We used xenografts and transgenic mice to model AR to the combination of A+C. Molecular analysis of resistant tumors revealed activation of the mTOR signaling pathway. Consistent with these findings, two separate patients with A+C-resistant tumors exhibited alterations in genes (NF2 and TSC1), that when silenced in EGFR-mutant cells led to activation of the mTOR pathway. In vitro and in vivo, A+C resistance can be overcome by addition of an mTOR pathway inhibitor. These studies are the first to demonstrate mechanisms of AR to dual inhibition of EGFR in EGFR-mutant lung cancer and provide new insight into the biology of this subset of lung cancers, with immediate therapeutic implications for patients.
RESULTS
Acquired resistance to A+C combination therapy in xenografts
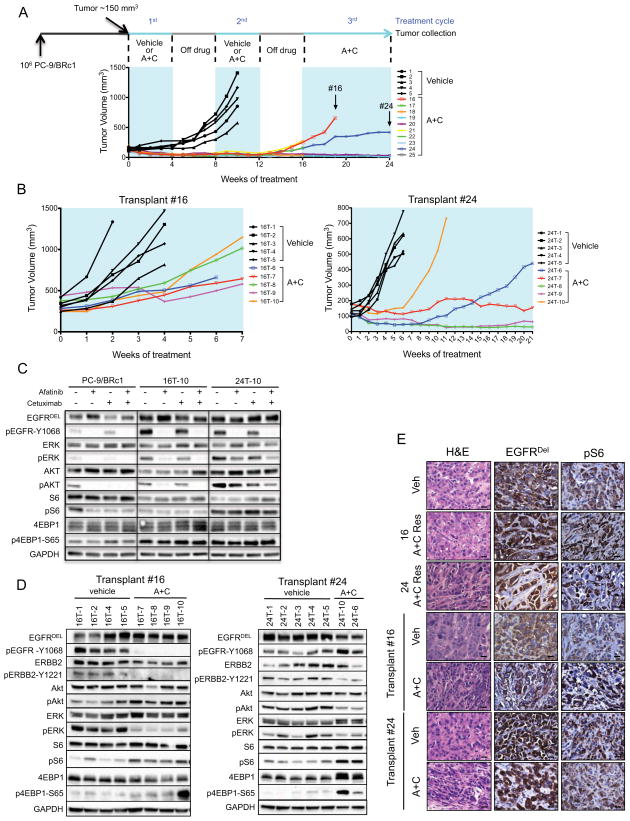
We previously modeled AR to erlotinib in tetracycline-inducible mouse models of EGFR-dependent lung cancer by intermittently treating mice with the TKI (Politi et al., 2010). We observed clinically relevant mechanisms of AR, such as the EGFR T790M mutation and Met amplification, validating this experimental approach. We adopted the same strategy to establish models of resistance to A+C, first in xenograft models using the PC-9/BRc1 human LUAD cell line that harbors an EGFRΔE746-A750+T790M mutation (Chmielecki et al., 2011). Immunocompromised mice with PC-9/BRc1-induced tumors were randomized to receive either vehicle (n=5) or A+C (n=10). After 1 month of treatment, drug administration was interrupted for 1 month, and this on/off drug treatment regimen was repeated 3 times (Figure 1A). All tumors in control mice grew continuously. In the A+C treated cohort, tumors initially regressed. During the third cycle of treatment, 2 tumors (#16 and #24) became resistant (Figure 1A). These were re-implanted into mice and treated with A+C or vehicle alone for 4 weeks (Figure 1B). Eventually, we collected 4 A+C resistant transplants from tumor #16 (labeled 16T-7, 16T-8, 16T-9 and 16T-10) and two from tumor #24 (24T-6, 24T-10) (Table S1). Cell lines were established from tumors 16T-10 and 24T-10. Resistance to A+C in these cell lines compared to parental PC-9 and PC-9/BRc1 cells was confirmed in a 3D colony assay (Figure S1A).
Figure 1. Activation of the mTOR pathway in afatinib+cetuximab-resistant xenografts.
A. Representation of the intermittent dosing protocol used to generate acquired resistance to afatinib and cetuximab in xenografts. 106 PC-9/BRc1 cells were injected s.c. into the flanks of immunocompromised mice. When tumors reached a volume of ~150 mm3, mice were treated with vehicle (n=5, in black) or A+C (n=10, in color). After one month of treatment, drug administration was stopped for one month. The intermittent drug cycle was repeated three times. Tumor volume measurements are shown. Tumors indicated by the arrows (#16 and #24) acquired resistance to A+C.
B. Tumor growth of the transplants derived from A+C resistant tumors #16 (left panel) and #24 (right panel). The resistant tumors were further transplanted into 10 nude mice and treated continuously with vehicle (in black, n=5) or with A+C (in color, n=5). Transplants are labeled with the number of the original tumor they were derived from (#16 or 24), the letter “T” and a number.
C. Immunoblotting analysis of extracts from PC-9/BRc1, 16T-10 and 24T-10 cells treated with afatinib (100 nM), cetuximab (10 μg/ml) or the A+C combination. Lysates were probed with the indicated antibodies; p, phospho.
D. Immunoblotting analyses of tumor lysates from vehicle- and A+C-treated transplants derived from A+C-resistant tumors 16 and 24. Lysates were probed with the indicated antibodies; p, phospho.
E. Hematoxylin and Eosin staining (H&E) and IHC performed on paraffin sections of tumors derived from vehicle- and A+C-treated mice as indicated. Sections were stained with antibodies to EGFR exon 19 deletion mutant (EGFRDEL) and phospho-S6 (pS6) as indicated. 40X magnification is shown. Bars, 20 μm.
Evidence for mTOR pathway activation in A+C resistant xenografts
To identify mechanisms of resistance to A+C, we performed molecular analyses of the tumors collected. We first asked whether resistance to A+C could be explained by the acquisition of new mutations in EGFR or ERBB2, both of which are targets of A+C. Sequencing of control and A+C resistant tumors did not detect any mutations in EGFR and ERBB2 (data not shown). Analysis of the tumors revealed increased EGFR copy number in both vehicle-treated and A+C-resistant tumors compared to the parental PC-9/BRc1 cell line with tumor #16, but not #24, exhibiting high level EGFR amplification (Figure S1B). Minor fluctuations in ERBB2, MET and IGF1R copy number were also observed, the significance of which is likely limited given the magnitude of these changes. Together, the copy number data suggested that RTK amplification alone could not explain the resistance phenotype observed in our samples.
These results prompted us to further investigate RTK levels and pathway activation in A+C-resistant samples. The xenograft-derived cell lines exhibited higher levels of phospho (p)-EGFR, pERK and pAKT compared to parental PC-9/BRc1 cells. However, the levels of activation of these proteins decreased in the presence of A+C, suggesting that the drugs retained the ability to block these pathways in A+C-resistant cells (Figure 1C). Interestingly, drug treatment did not affect the levels of pS6 or p4EBP1, markers of mTOR pathway activation, in the A+C-resistant lines in contrast to parental PC-9/BRc1 cells. This evidence suggests that pathway re-wiring in resistant tumor cells leads to sustained activation of the mTORC1 pathway. Similarly, in vivo, the mTOR pathway was consistently engaged in all A+C-resistant xenografts, as measured by pS6 and p4EBP1 (Figure 1D and 1E). The levels of pAKT and pERK in the xenografts did not reveal a consistent pattern that would support either playing a major role in resistance to A+C in this model (Figure 1D). Together these results suggest that whilst pAKT and pERK can be inhibited in A+C-resistant tumors, the tumors retain sustained activation of mTOR signaling that may play a role in resistance to A+C combination therapy. Whole exome sequencing (WES) of the A+C-resistant #16 and #24 tumors did not detect mutations in 23 mTOR-pathway related genes, strongly suggesting that non-mutational processes account for sustained activation of this pathway in these tumors.
Highly penetrant resistance to A+C in genetically engineered mouse models of EGFR-mutant lung cancer
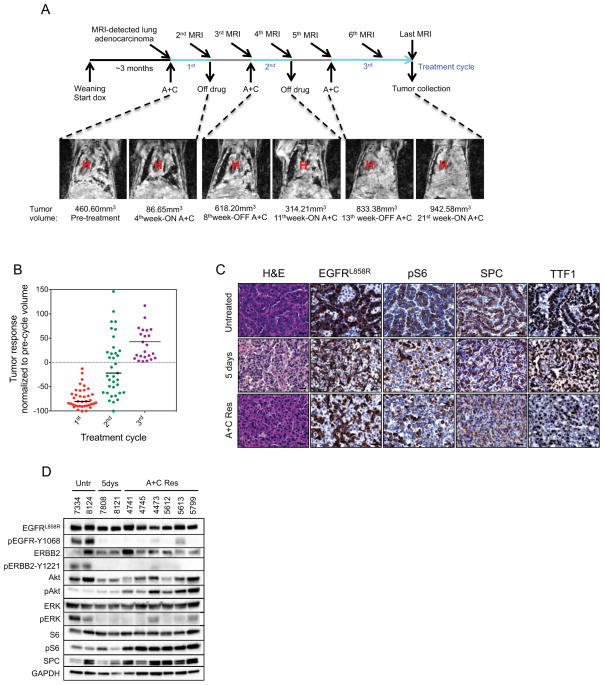
In parallel, we developed models of resistance to A+C using transgenic mice with EGFRL858R+T790M-induced LUADs. Tumors in these mice are resistant to erlotinib but sensitive to A+C (Regales et al., 2009). Thirty-eight CCSP-rtTA; TetO-EGFRL858R+T790M tumor-bearing mice were cycled on and off A+C using the protocol used for the xenograft experiments (Figure 2A). Tumor burden before and during treatment was tracked using magnetic resonance imaging (MRI) at the beginning and end of each drug cycle. This on/off drug treatment schedule was repeated until lung tumors no longer responded to treatment and increased in size on MR images (Figure 2A). All 38 mice that underwent the intermittent dosing treatment protocol developed resistance to the A+C combination. The majority of mice developed resistance after three cycles of A+C (21 out of 38); 15 mice developed resistance after 2 cycles and 2 mice after 4 cycles of drug treatment (Table S2A). The median tumor shrinkage during the first cycle of A+C was 80%, but this was attenuated during the second and third cycles of treatment (Figure 2B). Six mice that were treated without interruption with A+C also developed resistance to the drug combination (Figure S2A and Table S2B). Consistent with the emergence of resistance, tumors from the mice with AR displayed higher levels of proliferation and lower levels of apoptosis, compared to tumors from mice that had undergone short-term A+C treatment (Figure S2B).
Figure 2. Activation of the mTOR pathway in A+C-resistant mouse LUADs.
A. Intermittent dosing of A+C was performed on a 1 month on drug and 1 month off drug cycle. Doxycycline administration was initiated at weaning and subsequently kept constant throughout the life of the animal. Tumor response was evaluated by MRI at the beginning and at the end of every drug treatment cycle. Coronal MR images of a CCSP-rtTA; TetO-EGFRL858R+T790M mouse subjected to intermittent A+C treatment are shown. Tumor volume measurements are found below each image. H, heart.
B. Tumor response in CCSP-rtTA; TetO-EGFRL858R+T790M mice subjected to the A+C intermittent treatment protocol. Tumor volume is plotted as the percentage of the tumor volume detected on the pre-cycle MRI. Median tumor volume change is −80% in the first cycle, −22% in the second cycle and +29% at the third cycle of A+C.
C. Hematoxylin and Eosin staining (H&E) and IHC performed on paraffin sections of LUADs derived from CCSP-rtTA; TetO-EGFRL858R+T790M untreated mice and mice treated with A+C for 5 days or at resistance to the drug combination (A+C Res). Sections were stained with antibodies to EGFRL858R, pS6, surfactant protein C (SPC) and thyroid transcription factor (TTF1) as indicated. 40X magnification is shown. Bars, 20 μm.
D. Immunoblotting analyses of tumor lysates from LUADs derived from untreated (Untr), A+C-treated (5dys) or resistant (A+C res) CCSP-rtTA; TetO-EGFRL858R+T790M mice. Lysates were probed with the indicated antibodies; p, phospho.
We explored whether A+C resistant tumors showed any phenotypic differences compared to untreated tumors. CCSP-rtTA; TetO-EGFRL858R+T790M mice developed solid and papillary LUADs, positive for the type II pneumocyte marker surfactant protein-C (SP-C) and for thyroid transcription factor-1 (TTF1) (Figure 2C). While most untreated adenocarcinomas were papillary, A+C treated and resistant tumors almost invariably were solid and more poorly differentiated (Figure 2C).
Evidence for mTOR pathway activation in the A+C-resistant mouse LUADs
To elucidate further the pathways that may account for resistance to A+C, we sequenced the EGFR transgene and Erbb2 from 23 resistant tumors. Similar to our observations in xenografts, we did not find mutations in these genes or in Pik3ca, Pik3cb and Kras (n=15, data not shown). A+C-resistant mouse LUADs did not show copy number alterations in the EGFR transgene or in endogenous Egfr, Erbb2, Met and Igf1r (Figure S2C).
We then examined which signaling events might promote AR to A+C. As expected, we found that upon short-term (5 days) A+C treatment, phosphorylation of EGFR and Erbb2 were decreased. As a consequence reduced levels of phosphorylated Erk were observed, however Akt phosphorylation did not change (Figure 2D). Phosphorylation of EGFR was greatly reduced or completely abrogated in all of the resistant tumors, and phosphorylation of Akt was consistently higher than in untreated tumors, suggesting the presence of compensatory mechanisms of activation of the PI3K pathway in these tumors. Similarly, phosphorylation of Erbb2 was not restored to untreated levels in the A+C-resistant tumors. Similar to the xenografts discussed previously, A+C-resistant tumors consistently showed increased pS6, suggesting that increased activation of mTORC1 may play a role in AR to A+C (Figure 2C and 2D).
Mutations in mTOR signaling pathway genes are associated with resistance to A+C in human tumors
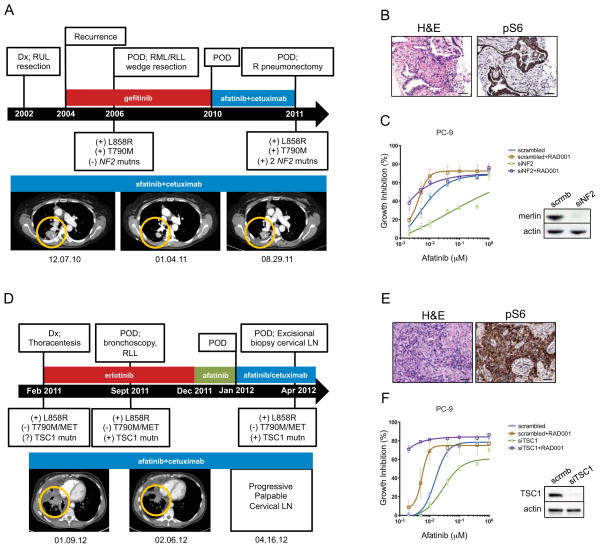
Consistent with the preclinical modeling, we found genetic evidence for potential activation of the mTOR signaling pathway in tumor samples from 2 (of 4 analyzed) separate patients with AR to A+C (Figure 3, Table 1 and Supplemental Information for patient details). Strikingly, the mutated tumor genes were not shared between the two patients, but they both converged on the mTOR pathway. In the first patient, targeted resequencing of 182 genes using the FoundationOne platform (Table 1 and Table S3) as well as WES revealed that the resected A+C-resistant tumor (Figure 3A and S3A) still harbored both the L858R and T790M mutations (frequencies of 0.38 of 1162 reads and 0.24 of 1279 reads, respectively in the FoundationOne Assay) (Jeselsohn et al., 2014). Unexpectedly, 2 additional mutations were found in NF2 (c.592C>T_p.R198* at frequency 0.15 of 631 reads and c.811-2A>T: splice at 0.13 frequency of 1168 reads). These two mutations were not detected in the 2006 tumor specimen, as assessed by deep amplicon-based resequencing (Table 1).
Figure 3. Genetic alterations associated with activation of mTORC1 in human lung tumors resistant to A+C.
A. Top, disease milestones for Patient 1. Time from diagnosis to A+C resistance is indicated by the black arrow. Clinical findings, procedures and drug-treatments are indicated above the arrow. Molecular findings are shown below the black arrow. Dx, diagnosis; POD, progression of disease; RUL, right upper lobe; RML, right middle lobe; RLL, right lower lobe; R, right; Mutns, mutations. Bottom, computed tomography scans of the lungs are shown prior (12.07.10), during (01.04.11) and at resistance (08.29.11) to A+C. Tumors areas are circled.
B. Hematoxylin and eosin (H&E, left panel) and IHC for pS6 (right panel) of the A+C resistant tumor harboring the NF2 mutations. 20X magnification is shown. Bars, 50 μm.
C. Growth inhibition of PC-9 cells after knockdown of NF2 in response to afatinib (left). Viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. Data are presented as the mean ± SE. Immunoblotting of PC-9 cells showing efficient knock-down of Merlin expression is shown on the right. Lysates were probed with the indicated antibodies.
D. Top, disease milestones for Patient 2. Time from diagnosis to A+C resistance is indicated by the black arrow. Clinical findings, procedures and drug treatments are indicated above the arrow. Molecular findings are shown below the black arrow. Dx, diagnosis; POD, progression of disease; RLL, right lower lobe; Mutn, mutation; LN, lymph node. Bottom, computed tomography scans of the lungs are shown prior (01.09.12) and during treatment with A+C (02.06.12). Tumor areas are circled.
E. Hematoxylin and eosin (H&E) staining of the excised cervical lymph node from Patient 2 (left) and IHC showing phosphorylation of S6 (pS6, right). Bars, 50 μm.
F. Growth inhibition of PC-9 cells after knockdown of TSC1 in response to afatinib (left). Viable cells were measured after 72 hours of treatment and plotted relative to untreated controls. Data are presented as the mean ± SE. Immunoblotting of PC-9 cells showing efficient knock-down of TSC1 expression is shown on the right. Lysates were probed with the indicated antibodies.
Table 1.
List of mutations detected by targeted sequencing in Patient 1 and Patient 2
| Patient 1 | ||||
|---|---|---|---|---|
| Gene | Nucleotide | Protein | Pre A+C Frequency, Total reads | Post A+C Frequency, Total reads |
| EGFR | c.2753>G | p.L858R | Positive* | 38%, 1162 |
| EGFR | c.2369C>G | p.T790M | Positive* | 24%, 1279 |
| NF2 | c.592>T | p.R198* | 0%, 2301 | 15%, 631 |
| NF2 | c.811-2A>T | splice | 0%, 2569 | 13%, 1168 |
| Patient 2 | ||||
|---|---|---|---|---|
| Gene | Nucleotide | Protein | Pre A+C Frequency, Total reads | Post A+C Frequency, Total reads |
| EGFR | c.2753>G | p.L858R | 3%, 827 | 19%, 715 |
| TSC1 | c.345_345delT | p.L116fs* | 1%, 613 | 15%, 399 |
as per 2009 retrospective clinical report (Genzyme) of 2006 biopsy sample
The NF2 gene encodes Merlin, a protein with putative tumor-suppressive function. Both mutations are predicted to cause loss of protein function. The R198* mutation is a truncating mutation that causes loss of two-thirds of Merlin and has previously been described in cancers including ependymoma (Lamszus et al., 2001). The c.811-2A>T alteration is a splice-site mutation that at a minimum affects the FERM domain, important for Merlin’s localization and activation. In support of a functional role of this mutation, a 69bp deletion encompassing this exon 9 splice site and causing NF2 exon 9 skipping has been associated with familial autosomal dominant intramedullary ependymoma (Zemmoura et al., 2014).
In different cellular contexts, NF2 has been shown in independent studies to negatively regulate EGFR signaling and mTORC1 (Curto et al., 2007; James et al., 2009; Lopez-Lago et al., 2009). To determine whether the mTORC1 pathway was activated in this sample, we used immunohistochemistry (IHC) to stain the biopsy collected at the time of resistance to A+C with a pS6 antibody and observed a strong signal (Figure 3B). In support of a role for NF2 on TKI-sensitivity, knockdown of NF2 led to a decrease in the sensitivity of PC-9 cells to afatinib (Figure 3C). Importantly, the addition of an mTOR inhibitor, everolimus (RAD001), re-sensitized PC-9 cells with NF2 knockdown to afatinib in vitro (Figure 3C). Notably, everolimus alone was not able to inhibit cell proliferation in cells treated with either control (scrambled) or NF2 siRNAs (Figure S3C). The same effect was observed in HCC827 cells upon cetuximab treatment (Figure S3B). Moreover, A+C treatment in LUAD HCC827 cells did not decrease the levels of pS6 upon NF2 knock down (Figure S3B). Taken together, this patient and in vitro data suggest that the NF2 mutations were acquired during treatment on A+C and that NF2 loss leads to activation of the mTORC1 signaling pathway to mediate drug resistance.
In the second patient, initial molecular analysis of the A+C-resistant tumor (Figure 3D) did not detect the T790M mutation, MET amplification or ERBB2 amplification. Further analysis using a more recent FoundationOne panel (Frampton et al., 2013) revealed the presence of the L858R mutation (c.2573T>G; frequency of 0.19 of 715 reads) plus a mutation in the Tuberous Sclerosis 1 (TSC1) gene (c.345_345delT_p.L116fs*; frequency 0.15 of 399 reads; this sample contained approximately 70% tumor cells) (Table 1, Table S3). The observed L116fs* frame-shift mutation leads to the creation of a stop codon immediately downstream of codon 116, truncating the protein. The somatic status and zygosity of the TSC1 L116fs*2 alteration (see Supplemental Experimental Procedures) were consistent with a somatic alteration clonally present on a single TSC1 copy in the tumor, indicating that LOH occurred. The TSC1 mutation status of the pleural fluid collected at diagnosis was not assessed due to insufficient tumor material. FoundationOne analysis of the erlotinib-resistant lung specimen (before A+C) identified the presence of the L858R mutation (c.2573T>G; frequency of 0.03 of 827 reads) and of the TSC1 mutation (c.345_345delT_p.L116fs*; frequency 0.01 of 613 reads), indicating that it did pre-exist treatment with A+C (Table 1, Table S3). The low allele frequency of both of the mutations is due to low tumor purity of this sample (10% purity). These data suggest that selection of the deleterious TSC1 mutant may have occurred during A+C treatment.
The TSC1 gene encodes for Hamartin, which together with Tuberin (TSC2) forms a complex that suppresses mTORC1 signaling (Laplante and Sabatini, 2012). To determine whether this pathway was active in the sample collected after A+C treatment, we performed IHC for pS6 and observed strong staining (Figure 3E). The functional role of disruption of TSC1 on drug response was tested using siRNAs. Knockdown of TSC1 in PC-9 cells led to a decrease in sensitivity of the cells to afatinib, and sensitivity to afatinib was restored by the addition of everolimus upon TSC1 loss (Figure 3F). Moreover, cells treated with either scrambled or TSC1 siRNAs were not sensitive to everolimus treatment (Figure S3C). These results indicate that the absence of TSC1 mediates resistance to EGFR-directed therapies by activating the mTORC1 signaling pathway.
Xenografts and LUADs resistant to A+C are sensitive to concurrent EGFR and mTOR inhibition
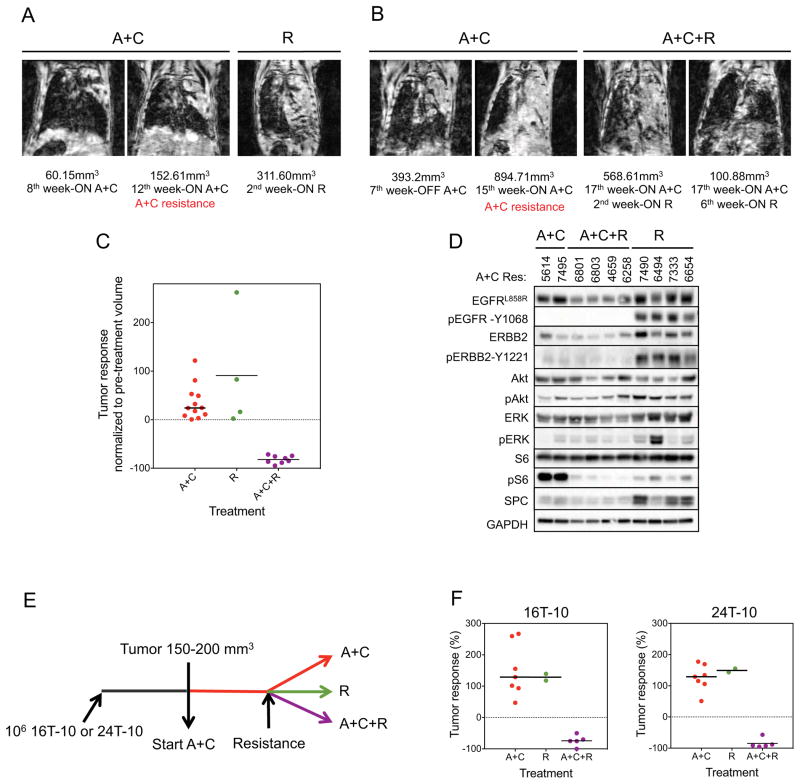
Activation of the mTOR pathway in mouse models and patient samples led us to explore whether A+C resistant tumors responded to inhibition of this pathway. To test this, we treated 4 CCSP-rtTA; TetO-EGFRL858R+T790M mice with A+C resistant tumors with rapamycin as a single agent. Rapamycin treatment alone was ineffective in all 4 cases (Figures 4A and 4C). This result is in line with previous findings showing that inhibition of mTOR alone is not sufficient to abolish Akt signaling and that the combination of an mTOR inhibitor with an RTK inhibitor is more likely to have anti-tumor activity (Li et al., 2008; Rodrik-Outmezguine et al., 2011). To test whether combined inhibition of EGFR and mTOR could overcome resistance to A+C, we added rapamycin to the treatment regimen of CCSP-rtTA; TetO-EGFRL858R+T790M mice at the time of emergence of resistance to A+C (A+C+R). All 8 mice with LUADs resistant to A+C responded dramatically to the addition of rapamycin (Figures 4B and 4C). In the mice with tumor burden lower than 600 mm3 (6/8), tumor shrinkage was greater than 72% after one month of treatment with A+C+R (Figure 4B and Table S4). We also stained paraffin-embedded sections of A+C-resistant LUADs treated with rapamycin alone or in combination with A+C with antibodies against phospho-histone H3 and cleaved caspase 3 (Figure S4). Tumors treated with rapamycin alone were not growth inhibited whilst tumors treated with A+C+R exhibited both proliferation arrest and cell death. We further found that addition of rapamycin to A+C decreased pS6 in the LUADs (Figure 4D). All 4 of the A+C-resistant tumors treated with rapamycin alone showed activation of EGFR and Erbb2, as expected by the absence of EGFR-directed therapies (Figure 4D).
Figure 4. Tumors resistant to A+C are sensitive to concurrent EGFR and mTOR inhibition.
A. Coronal MR images of CCSP-rtTA; TetO-EGFRL858R+T790M mouse lungs prior to and upon treatment with rapamycin (R) following the development of resistance to A+C as indicated in red. Tumor volume measurements are shown.
B. Coronal MR images of CCSP-rtTA; TetO-EGFRL858R+T790M mouse lungs prior to and upon treatment with rapamycin in combination with afatinib and cetuximab (A+C+R) following the development of resistance to A+C as indicated in red. Tumor volume measurements are shown.
C. Response of A+C-resistant LUADs from CCSP-rtTA; TetO-EGFRL858R+T790M mice to 4 weeks of treatment with Rapamycin alone (R) or in combination with afatinib and cetuximab (A+C+R). The increase in tumor volume in the presence of A+C before the randomization to R or A+C+R is shown on the left. Tumor volumes are plotted as the percentage of the tumor volume detected in the pre-cycle MRI. Median tumor volume change to R is +52%; to A+C+R is −79%.
D. Immunoblotting analysis of LUADs resistant to A+C, following 4 weeks of treatment with rapamycin alone or in combination with afatinib and cetuximab. Lysates were probed with the indicated antibodies; p, phospho.
E. Strategy used to test the response of A+C-resistant xenograft tumors to concurrent EGFR and mTOR inhibition. 106 16T-10 or 24T-10 cells were injected s.c. into immunocompromised mice. When tumors reached a volume between 150–200 mm3, mice were treated with A+C (n=14). When resistance emerged, 2 mice were switched to rapamycin treatment (R, 2mg/Kg/day) and 5 mice received A+C+R. The rest of the mice were maintained on A+C. Mice were treated for 4 weeks from the randomization point.
F. Tumor response to A+C, rapamycin alone (R) or in combination (A+C+R) in xenografts. Data are plotted as percentage of tumor volume change from the randomization point. The median response to A+C+R was −75% in xenografts derived from 16T-10 cells (left) and −91% in xenografts derived from 24T-10 cells (right).
To evaluate the effect of concurrent EGFR and mTOR inhibition in xenografts, we injected 106 cells derived from the A+C-resistant xenografts 16T-10 and 24T-10 into immunodeficient mice. Upon the growth of A+C-resistant tumors, mice were divided into 3 groups (Figure 4E). One group was maintained on A+C for 4 weeks (n=7). The second group was treated with rapamycin alone (R, n=2) and the third group was treated with rapamycin in addition to A+C (A+C+R, n=5). Tumors in the A+C combination and rapamycin arms grew throughout the 4 weeks. In contrast, all of the tumors in mice that received A+C+R shrank (Figure 4F). Together these data indicate that inhibition of mTORC1 can re-sensitize cells to A+C treatment.
DISCUSSION
We show that resistance to dual inhibition of EGFRL858R+T790M with A+C is due to activation of mTORC1 signaling in mouse models. Addition of drugs targeting mTOR re-sensitizes tumors to A+C treatment. Consistent with these findings, we have identified mutations in genes that affect the mTOR signaling cascade in A+C-resistant biopsy samples from two separate patients with EGFR-mutant lung cancer.
Previous studies have shown that the presence of active mTORC1 in untreated EGFR mutant tumors is a direct consequence of mutant EGFR signaling. Effective therapies that target mutant EGFR lead to a decrease in mTORC1 signaling and consequent tumor regression. Indeed, in cell lines harboring EGFR TKI-sensitizing mutations (e.g. EGFRL858R), EGFR blockade using TKIs leads to a decrease in pS6 equivalent to that observed with rapamycin, accompanied by a decrease in cell viability (Li et al., 2007). Further supporting the critical role of mTORC1 signaling in the maintenance of EGFR-mutant lung tumors, the combination of either afatinib or HKI-272 with rapamycin together was required to elicit regression of EGFRL858R+T790M-induced tumors (Li et al., 2008; Li et al., 2007). Our study shows that in addition to playing a role in the maintenance of EGFR-mutant lung tumors, the mTORC1 pathway also plays a role in resistance to EGFR-directed therapies, specifically following A+C treatment. First, pS6 is observed in cell lines, xenografts and GEM models of A+C-resistant EGFR-mutant lung cancer. Second, these tumors regress following the addition of rapamycin to A+C. These data highlight the importance of mTORC1 for the survival of lung cancer cells with EGFR mutations and suggest that as resistance emerges, tumors increasingly rely on mTORC1 activation to survive.
The presence of a TSC1 frameshift mutation coupled with LOH at the same locus in a sample from a patient biopsied upon progression with A+C provides further evidence for dysregulation of the mTOR pathway as a mechanism of resistance to A+C. Indeed, strong pS6 staining was observed in tumor cells in this sample and disruption of TSC1 in human EGFR-mutant lung cancer cell lines increased their viability in the presence of EGFR TKIs. Unexpectedly, acquired NF2 inactivating mutations were observed in A+C-resistant specimens from a separate patient on the same trial. Recent work has found that a downstream biochemical consequence of NF2 loss is activation mTORC1 (Lopez-Lago et al., 2009). We show that the effects of both TSC1 and NF2 loss can be reversed in cells by treatment with a rapalog, suggesting that the presence of genomic changes in these genes indicates sensitivity to mTOR inhibition. Further studies to determine the prevalence of NF2 and TSC1 mutations in EGFR-mutant lung cancer are ongoing.
mTORC1 represents the output of several signaling pathways and external stimuli. In addition to genetic mechanisms like those described above that lead to its activation, it can be engaged through non-genetic mechanisms. Increased growth factor receptor signaling, through, for example, IGF1R, activates mTORC1 through the PI3K pathway. In this setting, one would expect to observe higher levels of phosphorylation of mTORC1 and AKT. Consistent with the possibility of similar mechanisms occurring in some of our models, we observed increased phosphorylation of AKT in the #16 xenograft-derived tumors and in the A+C-resistant GEMM tumors (Figures 1D and 2D). An increase in the levels of phosphorylation of IGF1R was indeed observed in the #16 derived tumors (data not shown) and may explain the increased mTORC1 signaling found in these tumors. These results also highlight how activation of mTORC1 can occur both via signals upstream and downstream of AKT. Moreover, our data from cell lines, xenografts and patient samples suggest that mTORC1 activation acts cell autonomously in the tumor cells to confer resistance. Whether this pathway is also activated in other cells in the tumor microenvironment cannot be excluded and is under further investigation.
Our findings suggest that patients with AR to A+C may benefit from drug combinations that include EGFR-directed therapies and mTOR inhibitors. In this regard, a phase IB trial of afatinib with the rapalog sirolimus in patients with EGFR-mutant lung cancer is currently ongoing. Phase III trials of A+C in patients with TKI-naïve and refractory EGFR-mutant lung cancer are planned. Inhibition of mTOR in this context may delay resistance. Due to concerns about the toxicity of this multi-drug combination, it will be important to use preclinical models to determine whether continuous or intermittent dosing of the mTOR inhibitor are equally effective at countering drug resistance. Moreover, rapalogs only partially block downstream functions of mTOR in contrast to mTORC1/2 kinase inhibitors. Investigation of these latter novel agents will be informative to determine their efficacy in the context of EGFR-mutant lung cancer. Finally, the recent development of mutant specific EGFR inhibitors that induce reduced toxicity due to less inhibition of wild-type EGFR may open the door to use of drug combinations including those of EGFR inhibitors with mTOR inhibitors (Walter et al., 2013).
In summary, resistance to targeted therapies remains the major hurdle to the long-term success of EGFR-directed therapies. Our data in multiple preclinical models and human tumor samples show increased mTORC1 signaling after long-term treatment with A+C, identifying this node as a critical vulnerability of drug-resistant cells.
EXPERIMENTAL PROCEDURES
Transgenic mice and xenografts
All animals were kept in pathogen-free housing under guidelines approved by the MSKCC and Yale Institutional Animal Care & Use Committees (IACUC). TetO-EGFRL858R+T790M mice (Regales et al., 2007) and CCSP-rtTA mice were previously described (Tichelaar et al., 2000). For xenografts, 8-week-old nu/nu athymic nude mice (Harlan Labs) were injected subcutaneously with 10×106 PC-9/BRc1 cells together with Matrigel (BD Biosciences). Mice were randomized to receive either drug diluent alone (“vehicle”) or A+C. Tumor size was measured twice a week using calipers. To further propagate A+C-resistant tumors, these were minced and immediately injected subcutaneously with Matrigel (tumor #16) or cultured for two weeks then re-injected subcutaneously into immunodeficient mice (tumor #24). Afatinib (produced by the Organic Synthesis Core Facility at MSKCC) was suspended in 0.5% (w/v) methylcellulose and administered orally (p.o., 25mg/kg/d 5 days a week). Cetuximab (Erbitux; Bristol-Myers Squibb and Eli Lilly Pharmaceuticals) was purchased and administered intraperitoneally (i.p., 1mg twice a week). Rapamycin (LC Laboratories) was suspended in 0.5% carboxy-methylcellulose and given p.o. at 2mg/kg/d 5 days a week.
Cell culture
Human LUAD cell lines, PC-9, PC-9/BRc1 and HCC827 were used (Chmielecki et al., 2011). A+C-resistant cells, obtained from xenograft tumors, were kept in culture in presence of afatinib (250 nM) and cetuximab (10 μg/mL).
Immunoblotting
Cells and crushed tumors were lysed in ice-cold RIPA lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 5 mM MgCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, protease and phosphatase inhibitor cocktail (Thermo Scientific)). For cell treatment studies, cells were starved overnight, treated with drugs for 8h and washed twice with cold PBS before lysate preparation. Equal amounts of total protein were separated by SDS-PAGE and probed as indicated. Signals were detected using either SuperSignal West Pico or Femto chemiluminescent substrates (Pierce Biotechnology). For a list of antibodies, see Supplemental experimental procedures.
Supplementary Material
HIGHLIGHTS.
Activation of mTORC1 is found in EGFR-mutant tumors resistant to afatinib+cetuximab
Afatinib+cetuximab-resistant tumors have genomic alterations inNF2 and TSC1
NF2 loss and TSC1 loss lead to mTORC1 activation
mTOR inhibition re-sensitizes resistant tumors to afatinib+cetuximab therapy
Acknowledgments
The authors acknowledge support from the NIH/National Cancer Institute grants R00-CA131488 (K.P.), R01-CA120247 (K.P.), R01-CA121210 (W.P., K.P.), P01-CA129243 (W.P., M. Kris PI), U54-CA143798 (W.P., F. Michor PI), F30-CA180353 (C.N.), DP2OD004362 (E.M.S) and P30 CA008748 (EdS). This work was also supported by the American Italian Cancer Foundation (V.P.), Uniting Against Lung Cancer (K.P., X.S.), the VICC Cancer Center Core grant P30-CA68485 (W.P., J. Pietenpol PI), VICC Melly Family Scholarship (C.N.) and the Ingram Professorship (Z.Z.).
Footnotes
AUTHOR CONTRIBUTIONS
VP and CN designed the study, performed experiments, analyzed the data and wrote the manuscript. XS, EdS, ZZ, PJ, MG, ES and RY performed experiments and analyzed data. AW, ZW, GC, LH, DC, PS, VM and SG contributed to the analysis of patient samples. WP and KP supervised and designed the study and wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chmielecki J, Foo J, Oxnard GR, Hutchinson K, Ohashi K, Somwar R, Wang L, Amato KR, Arcila M, Sos ML, et al. Optimization of Dosing for EGFR-Mutant Non-Small Cell Lung Cancer with Evolutionary Cancer Modeling. Science translational medicine. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curto M, Cole BK, Lallemand D, Liu CH, McClatchey AI. Contact-dependent inhibition of EGFR signaling by Nf2/Merlin. J Cell Biol. 2007;177:893–903. doi: 10.1083/jcb.200703010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James MF, Han S, Polizzano C, Plotkin SR, Manning BD, Stemmer-Rachamimov AO, Gusella JF, Ramesh V. NF2/merlin is a novel negative regulator of mTOR complex 1, and activation of mTORC1 is associated with meningioma and schwannoma growth. Molecular and cellular biology. 2009;29:4250–4261. doi: 10.1128/MCB.01581-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janjigian YY, Smit EF, Horn L, Groen HJM, Camidge DR, Gettinger S, Fu Y, Denis LJ, Miller V, Pao W. Activity of afatinib/cetuximab in patients with EGFR mutant non-small cell lung cancer and acquired resistance to EGFR inhibitors. Annals of Oncology. 2012:23. [Google Scholar]
- Jeselsohn R, Yelensky R, Buchwalter G, Frampton GM, Meric-Bernstam F, Gonzalez-Angulo AM, Ferrer-Lozano J, Perez-Fidalgo A, Cristofanilli M, Gomez H, et al. Emergence of constitutively active estrogen receptor-alpha mutations in pretreated advanced estrogen receptor positive breast cancer. Clin Cancer Res. 2014 doi: 10.1158/1078-0432.CCR-13-2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamszus K, Lachenmayer L, Heinemann U, Kluwe L, Finckh U, Hoppner W, Stavrou D, Fillbrandt R, Westphal M. Molecular genetic alterations on chromosomes 11 and 22 in ependymomas. International journal of cancer. 2001;91:803–808. doi: 10.1002/1097-0215(200002)9999:9999<::aid-ijc1134>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR Signaling. Cold Spring Harbor perspectives in biology. 2012:4. doi: 10.1101/cshperspect.a011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Ambrogio L, Shimamura T, Kubo S, Takahashi M, Chirieac LR, Padera RF, Shapiro GI, Baum A, Himmelsbach F, et al. BIBW2992, an irreversible EGFR/HER2 inhibitor highly effective in preclinical lung cancer models. Oncogene. 2008;27:4702–4711. doi: 10.1038/onc.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Shimamura T, Ji H, Chen L, Haringsma HJ, McNamara K, Liang MC, Perera SA, Zaghlul S, Borgman CL, et al. Bronchial and peripheral murine lung carcinomas induced by T790M-L858R mutant EGFR respond to HKI-272 and rapamycin combination therapy. Cancer cell. 2007;12:81–93. doi: 10.1016/j.ccr.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Lopez-Lago MA, Okada T, Murillo MM, Socci N, Giancotti FG. Loss of the tumor suppressor gene NF2, encoding merlin, constitutively activates integrin-dependent mTORC1 signaling. Molecular and cellular biology. 2009;29:4235–4249. doi: 10.1128/MCB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–1080. doi: 10.1200/JCO.2012.43.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nature reviews Cancer. 2010;10:760–774. doi: 10.1038/nrc2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politi K, Fan PD, Shen R, Zakowski M, Varmus H. Erlotinib resistance in mouse models of epidermal growth factor receptor-induced lung adenocarcinoma. Dis Model Mech. 2010;3:111–119. doi: 10.1242/dmm.003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regales L, Balak MN, Gong Y, Politi K, Sawai A, Le C, Koutcher JA, Solit DB, Rosen N, Zakowski MF, Pao W. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS ONE. 2007;2:e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regales L, Gong Y, Shen R, de Stanchina E, Vivanco I, Goel A, Koutcher JA, Spassova M, Ouerfelli O, Mellinghoff IK, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. The Journal of clinical investigation. 2009;119:3000–3010. doi: 10.1172/JCI38746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrik-Outmezguine VS, Chandarlapaty S, Pagano NC, Poulikakos PI, Scaltriti M, Moskatel E, Baselga J, Guichard S, Rosen N. mTOR kinase inhibition causes feedback-dependent biphasic regulation of AKT signaling. Cancer discovery. 2011;1:248–259. doi: 10.1158/2159-8290.CD-11-0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichelaar JW, Lu W, Whitsett JA. Conditional expression of fibroblast growth factor-7 in the developing and mature lung. The Journal of biological chemistry. 2000;275:11858–11864. doi: 10.1074/jbc.275.16.11858. [DOI] [PubMed] [Google Scholar]
- Walter AO, Sjin RT, Haringsma HJ, Ohashi K, Sun J, Lee K, Dubrovskiy A, Labenski M, Zhu Z, Wang Z, et al. Discovery of a Mutant-Selective Covalent Inhibitor of EGFR that Overcomes T790M-Mediated Resistance in NSCLC. Cancer discovery. 2013;3:1404–1415. doi: 10.1158/2159-8290.CD-13-0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmoura I, Vourc’h P, Paubel A, Parfait B, Cohen J, Bilan F, Kitzis A, Rousselot C, Parker F, Francois P, Andres CR. A deletion causing NF2 exon 9 skipping is associated with familial autosomal dominant intramedullary ependymoma. Neuro-oncology. 2014;16:250–255. doi: 10.1093/neuonc/not165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.