Abstract
Eukaryotic cells are divided into distinct membrane-bound organelles with unique identities and specialized metabolic functions. Communication between organelles must take place to regulate the size, shape, and composition of individual organelles, as well as to coordinate transport between organelles. The endoplasmic reticulum (ER) forms an expansive membrane network that contacts and participates in crosstalk with several other organelles in the cell, most notably the plasma membrane (PM). ER–PM junctions have well-established functions in the movement of small molecules, such as lipids and ions, between the ER and PM. Recent discoveries have revealed additional exciting roles for ER–PM junctions in the regulation of cell signaling, ER shape and architecture, and PM domain organization.
Introduction
The transmission of signals between cells and tissues is fundamental for normal cell growth and development. Only recently have we begun to appreciate the significance of information transfer between intracellular organelles at membrane contact sites. The endoplasmic reticulum (ER) is the origin of the secretory pathway and it has essential roles in protein modification and quality control, lipid biosynthesis, and calcium signaling. Numerous proteins and lipids synthesized in the ER are ultimately destined for transport to the plasma membrane (PM). The PM undergoes remodeling, in response to both intrinsic and extrinsic cues, by the delivery of new material via the secretory pathway and removal of proteins and lipids by endocytosis. Consequently, biosynthesis in the ER must be modulated, corresponding to changes in PM composition. However, secretory vesicles derived from the ER do not directly fuse with the PM, and endocytic vesicles derived from the PM do not fuse with the ER, implying that rapid transfer of information between the ER and PM occurs independent of vesicle trafficking (Figure 1a). To balance ER metabolism with changes in PM composition, the ER and PM engage in crosstalk at membrane junctions — where these two organelles become closely apposed without undergoing membrane fusion (Figure 1a). The ER consists of a continuous system of membrane sheets and tubules that form contacts with several organelles in the cell (e.g. the PM, Golgi compartments, endosomes, lysosomes, and mitochondria) [1-3]. In this way, the ER coordinates with multiple membrane compartments along the secretory and endocytic systems. Of particular interest, ER–PM contacts have fascinated cell biologists for decades and recent findings have highlighted the importance of these conserved structures in membrane transport and cell signaling pathways.
Figure 1.
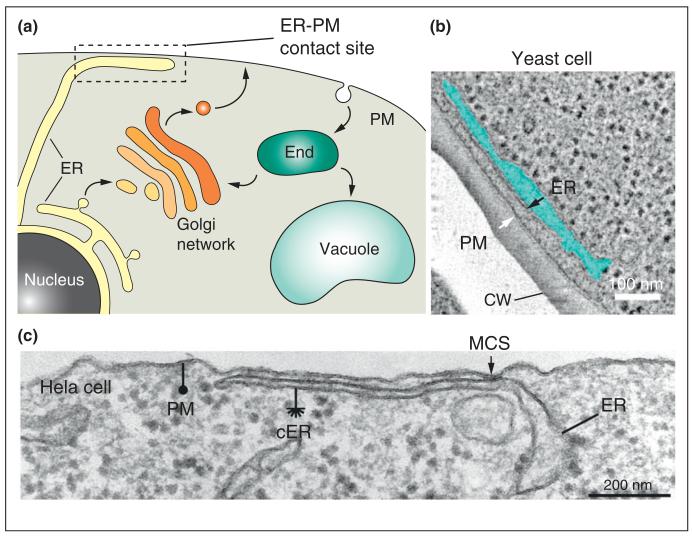
Membrane contact sites between the endoplasmic reticulum (ER) and plasma membrane (PM) are conserved cellular structures. (a) The ER consists of a continuous membrane meshwork throughout the cell. Peripheral ER membranes form close associations with the PM without undergoing membrane fusion. These membrane contact sites allow for direct ER–PM crosstalk independently of membrane trafficking through the secretory and endocytic pathways. (b) In yeast cells, the PM is extensively associated with cortical ER (cER) membranes, as revealed by electron microscopy [12•], © West et al., 2011. Originally published in J Cell Biol, 193:333-346. The cortical ER (cER) membrane (black arrow) and PM (white arrow) are in close apposition, approximately 30 nm apart. The lumen of the cER compartment is shaded in blue. Notably, electron-dense ribosomes in the cytoplasm are excluded from the ER–PM contact zone. The cell wall (CW) and scale bar (100 nm) are indicated. ER–PM contact sites share conserved features and also display unique properties in different cell types. (c) Electron microscopy of a mammalian Hela cell expressing the STIM proteins [14], © Orci et al. and the National Academy of Sciences, 2009. Originally published in Proc Natl Acad Sci USA 2009, 106:19358-19362. Similar to yeast cells, the cortical ER (cER, asterisk) is depleted of ribosomes as compared to internal ER membranes. Notably, the PM-associated cER and internal ER membranes display additional distinctions. The cER structures associated with the PM appear more flattened as compared to internal ER. Remarkably, the distance between the opposing membranes at ER–PM contacts can narrow to within 10 nm. An example of a tight membrane contact site (MCS) enriched in electron-dense material is shown (arrow). The scale bar indicates 200 nm.
Discovery and features of ER–PM contacts
ER–PM contact sites are ubiquitous structures that take on unique architectures in different cell types [4]. Junctions between peripheral ER structures and the PM were first discovered by Porter and Palade in electron microscopy studies on muscle cells [5]. These structures, termed dyads and triads, were later shown to control calcium fluxes involved in excitation–contraction coupling [6]. Subsequently, ER–PM junctions have been described in several cell types including neurons, immune cells, insect photoreceptor cells, and in plants [7-11]. While ER–PM contacts are not abundant in some cell types, large regions of the PM in yeast cells (approximately 20–45% of the PM) have an underlying network of peripheral ER, with an average distance of 30 nm between the organelles (Figure 1b) [12•,13•]. Owing to the close association of the PM and ER, ribosomes are excluded from the face of the ER adjacent to the PM [12•]. In mammalian cells, this distance can even narrow to within 10 nm (Figure 1c) [14], suggesting that the spacing of ER–PM contacts is precisely regulated.
While ER–PM contacts are ubiquitous structures, they adopt distinct shapes and architectures. Even in the same cell type, such as neurons, ER–PM junctions take on different forms. ER–PM contacts appear as discrete punctate regions at the synapse [7]. In other regions (the cell body, at bases of axons and dendrites, and the PM of cells neighboring synaptic boutons) ER–PM associations are more extensive (>1 μm) [8]. In yeast cells, the cortical ER is a meshwork of sheets and tubules shaped by the membrane-inserting reticulon proteins [12•], suggesting that ER curvature may be particularly important for some processes at ER–PM contacts. The many features (shapes and sizes) displayed by distinct ER–PM contacts also suggest multiple functions for these conserved structures. Consistent with this, ER–PM contacts have well-established roles in both calcium and lipid transport. Within the last few years, ER–PM contacts have become even more prominent in cell biology and are no longer limited to calcium and lipid homeostasis. As such, recent studies have uncovered additional roles in the control of organelle shape and morphology, inter-organelle communication, cell stress responses and signaling networks, and membrane trafficking pathways.
Calcium transport
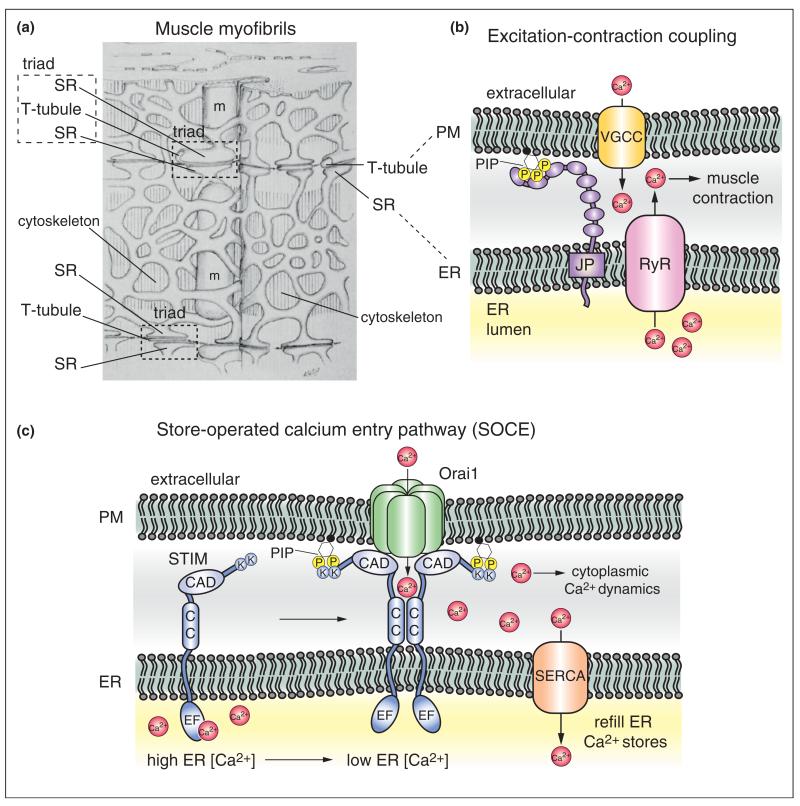
A well-known function of ER–PM contacts is control of calcium (Ca2+) dynamics. In metazoans, the ER is a major storage site for Ca2+ and release of this intracellular pool is triggered by various stimuli at the PM to generate cytoplasmic Ca2+ signals. At ER–PM junctions in muscle (termed triads, Figure 2a), voltage-gated Ca2+ channels (VGCCs) in PM invaginations (named T-tubules,Figure 2a) activate closely apposed ryanodine receptors (RyR) in the ER to elicit Ca2+ release during excitation–contraction coupling (Figure 2b) [6]. An accessory protein named junctophilin serves as a bridge to maintain the close association of the ER and PM in muscle (Figure 2b) [15]. Junctophilin is an ER-localized integral membrane protein with several cytoplasmic MORN (membrane occupation and recognition nexus) repeats suggested to bind phosphoinositide (PIP) lipids at the PM. Interestingly, loss of the PIP 3-kinase isoforms, PI3KC1A and PI3KC1B, in mice results in mislocalization of both junctophilin and VGCCs, as well as a reduction in T-tubules [16•].
Figure 2.
ER–PM membrane contact sites regulate calcium (Ca2+) dynamics. Associations between the ER and other organelles, such as the PM and mitochondria (m), were first observed by Keith Porter and George Palade in electron microscopy experiments on muscle cells [5]. (a) An illustration of cardiac muscle from this study is shown, © Rockefeller University Press, 1957. Originally published in J Biophys Biochem Cytol, 3:269-300. Large invaginations of the sarcolemma (the PM), termed transverse tubules (T-tubules), were found to be closely associated with membranes of the sarcoplasmic reticulum (SR, the ER in muscle cells). These structures were termed triads (a T-tubule closely apposed by two terminal SR compartments) and were later shown to be involved in Ca2+-mediated and cytoskeletal-mediated muscle contraction, known as excitation–coupled contraction. (b) Molecular components responsible for excitation-coupled contraction have been identified. At ER–PM contacts, voltage-gated Ca2+ channels (VGCC) localized in T-tubules interact with and activate ryanodine receptors (RyR) in the ER [6]. Upon activation, the RyR releases Ca2+ stores from the ER lumen to generate Ca2+ signals in the cytoplasm necessary for cytoskeletal-mediated muscle cell contraction. The protein junctophilin (JP) is involved in ER–PM contact formation in muscle cells [15]. Junctophilin is an integral ER membrane protein with several cytoplasmic MORN repeats proposed to bind PIP lipids in the PM. (c) Following release, luminal ER Ca2+ stores must be replenished for normal ER function (e.g. lipid synthesis, protein folding and secretion). In many cell types, the STIM proteins function in the store-operated Ca2+ entry (SOCE) pathway to maintain Ca2+ homeostasis in the ER lumen [4]. The ER-localized STIM protein senses Ca2+ levels in the ER lumen via a Ca2+-binding EF hand domain. Upon depletion of Ca2+ in the ER lumen, STIM proteins oligomerize via cytoplasmic coiled-coil domains (CC) and translocate to ER–PM contact sites. A polybasic region rich in lysine residues (K) within the cytoplasmic tail of STIM binds to PIP lipids in the PM. The STIM proteins subsequently interact with and recruit PM-localized Orai1 Ca2+ channels (also known as Ca2+ release-activated channels, CRAC) to ER–PM junctions. The CRAC-activation domain (CAD) within the STIM proteins directly binds and activates Orai1 channels, resulting in Ca2+ influx necessary for cytoplasmic Ca2+ signals and for reloading Ca2+ stores into the ER lumen through SERCA (smooth ER Ca2+ ATPase) transporters.
ER–PM contacts came into the spotlight in cell biology upon the discovery that ER-localized STIM (stromal interaction molecule) proteins couple with the PM Ca2+ channel Orai1, also named the Ca2+ release-activated Ca2+ channel (CRAC), in the store-operated Ca2+ entry (SOCE) pathway [17,18]. ER Ca2+ pools must be replenished to sustain excitation–coupling contraction, as well as for Ca2+ fluxes mediated by the ER-localized inositol trisphosphate (IP3) receptor in response to activation of signaling receptors and phospholipase C isoforms at the cell surface [4]. The STIM proteins sense Ca2+ concentrations in the ER lumen and oligomerize at ER–PM contacts when Ca2+ levels are depleted (Figure 2c) [11]. Cytoplasmic regions in the STIM proteins include a polybasic domain that binds PIP lipids at the PM and the CRAC activation domain (CAD) that directly interacts with the PM-localized Ca2+ channel Orai1 (Figure 2c). STIM–Orai1 interaction both activates and bolsters the Ca2+ selectivity of the Orai1 ion channel [19]. Interestingly, the crystal structure of the Orai1 channel has been recently solved, revealing further insights into Orai1 channel activity and STIM interactions [20••].
In addition to Orai1, the STIM proteins interact with several other factors. A protein termed junctate serves as an accessory Ca2+ sensor and structural component of STIM–Orai1 assemblies at ER–PM contacts [21]. The ER resident protein SARAF associates with STIM to attenuate ER Ca2+ reloading, providing another regulatory layer to SOCE [22]. Interestingly, STIM proteins are known to inhibit L-type voltage gated Ca2+ channel (VGCC) activity [23•,24•]. Reciprocal regulation of VGCC (inhibition) and Orai1 (activation) by STIM1 ensures that SOCE predominates in cells expressing both Ca2+ channel types, such as immune cells. STIM proteins are linked to Ca2+ transporter activities as well. POST (partner of STIM) binds STIM upon depletion of Ca2+ ER stores, but does not regulate Orai1 channel activity [25•]. Instead, POST inhibits PM Ca2+ ATPase (PMCA) transporter activity at the PM and binds a SERCA (smooth ER Ca2+ ATPase) isoform. The effect on SERCA transporter activity is unknown, but one possibility is that POST specifically couples Orai1 activation to SERCA-mediated Ca2+ reloading in the ER.
Lipid transport
ER–PM contacts are also implicated in nonvesicular sterol lipid transport [1,26]. A conserved family of oxysterol-binding (OSBP)-related proteins (ORPs) is thought to carry out sterol lipid transfer. ORP family members from both yeast and mammalian cells localize to ER–PM junctions [27,28], and in addition to the conserved sterol-binding domain, specific domains or motifs target ORPs to ER–PM contacts (Figure 3a). In a subset of ORPs, a pleckstrin homology (PH) domain binds PIP lipids in the PM [29], and a motif containing two phenylalanine residues in an acidic tract (FFAT) binds to integral ER membrane proteins named VAPs (VAMP-associated proteins) [30].
Figure 3.
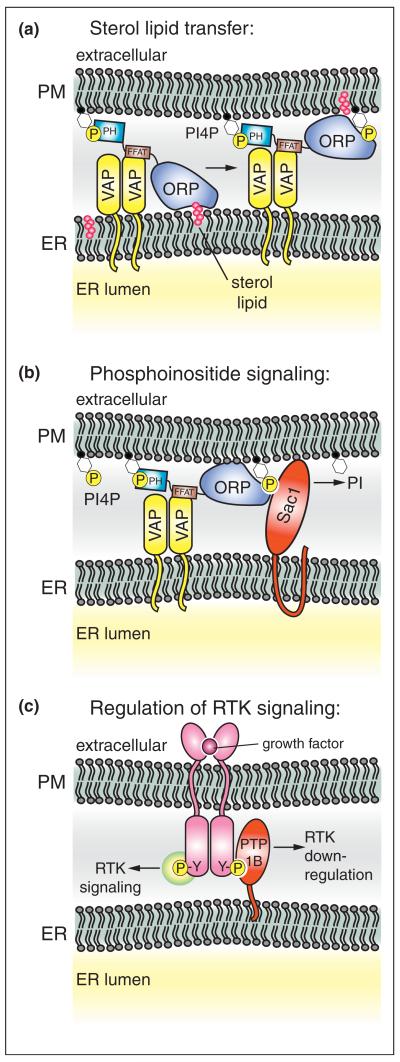
Essential cellular processes occur at ER–PM contact sites. (a) Non-vesicular sterol lipid transfer is facilitated at ER–PM junctions. Oxysterol-binding protein related proteins (ORPs) are recruited to ER–PM junctions by protein–protein and protein–lipid interactions [28]. ORP family members contain a FFAT motif that binds integral ER membrane proteins termed VAPs and a PH domain that binds the PIP isoform PI4P in the PM [29,30]. The conserved ORP sterol-binding domain extracts newly synthesized sterol lipids (magenta) in the ER. Upon subsequent delivery of sterol to the PM, interactions between the mouth of the ORP sterol-binding domain and PI4P may inhibit extraction of sterol from the PM [32•]. (b) ER–PM contacts are also sites for regulation of PI4P levels in the PM [39•,50••]. The ER-localized PIP phosphatase Sac1 is activated by ORPs family members at ER–PM contacts, resulting in PI4P turnover at the PM. This process is facilitated by VAPs in the ER. (c) In addition to lipid transfer and metabolism, growth factor receptor signaling is regulated at ER–PM junctions. The ER-anchored protein tyrosine phosphatase isoform PTP1B interacts with and dephosphorylates ligand-bound receptor tyrosine kinases (RTKs) at ER–PM contacts. Dephosphorylation of active RTKs terminates signaling events at the PM and promotes RTK endocytosis and down-regulation [42,43,44•,58].
The structure of an ORP family member from yeast (named Osh4/Kes1) bound to sterol has been determined [31]. The core of the sterol-binding domain forms a barrel containing hydrophobic residues critical for function. The sterol-binding domain also binds negatively charged phospholipids on the surface of the protein [28], and a crystal structure of Osh4/Kes1 bound to the PIP lipid isoform phosphatidylinositol 4-phosphate (PI4P) has been recently solved [32•]. Interestingly, interactions between basic residues at the top of the barrel and PI4P prevent sterol loading in vitro, suggesting a model whereby PI4P drives the directional transport of sterol from the ER to the PM (Figure 3a). At ER–PM contacts, ORPs may readily extract newly synthesized sterol lipid in the ER, where PI4P levels are relatively low. Following delivery, PI4P may prevent re-extraction of sterol from the PM. Consistent with this, loss of a PI 4-kinase (PI4KIIIα) that generates PI4P at the PM results in slightly reduced PM cholesterol levels [33•]. While this is an intriguing model, overexpression of ORPs and VAPs promotes the transport of sterol lipids from the PM to the ER [34]. Thus, additional regulatory factors (e.g. involved in PI4P metabolism) may control the directionality of sterol lipid transfer at ER–PM contacts. In addition, direct evidence for ORPs as sterol transfer proteins in vivo remains controversial [35], and ORPs are also suggested to function as sterol-sensing signal transduction proteins [36].
Regulation of cell signaling networks
It is becoming increasingly evident that ER–PM contacts are sites for the transmission of signals, in addition to Ca2+ and sterol lipids. In particular, the PIP isoform PI4P serves as an essential signaling molecule in the control of cell growth and polarity, hormone and calcium signaling, and regulated secretion and endocytosis [37•,38]. Notably, an ER-localized PIP phosphatase, named Sac1, regulates PI4P levels at ER–PM contact sites [39•]. This process is mediated by the ER-localized VAP proteins and by ORP family members localized to ER–PM junctions (Figure 3b). ORPs serve as sensors of PI4P in the PM, and in turn control PI4P levels by activating the Sac1 PIP phosphatase. While sterol binding is not required for ORP-mediated Sac1 activation, Sac1 may promote ORP sterol-binding and signaling functions. PI4P turnover by Sac1 may relieve PI4P inhibition of sterol extraction by ORP family members, and thus drive ORP-stimulated PM to ER sterol lipid transfer [34]. In addition, the founding member of the ORP family, OSBP, attenuates MAPK signaling upon binding cholesterol and downstream protein phosphatases [40]. Thus, turnover of PI4P by Sac1 at ER–PM contacts might facilitate ORP cholesterol binding and subsequent downstream signaling activities. Regulation of PI4P by Sac1 may be important for other processes at ER–PM contacts as well. A PI4K isoform (PI4KIIIα) that generates PI4P at the PM is involved in the formation of STIM-dependent ER–PM junctions and activation of the Orai1 Ca2+ channel [33•,41]. Consequently, Sac1 function at ER–PM contacts may be important in control of the SOCE pathway.
The ER-anchored protein tyrosine phosphatase PTP1B functions at ER–PM junctions as well. Several potential PM-localized substrates for PTP1B have been identified, including receptor tyrosine kinases (RTKs) such as the insulin receptor, epidermal growth factor receptor (EGFR), and ephrin (Eph) receptors (Figure 3c) [42]. Activated, internalized RTKs are dephosphorylated by PTP1B at ER-endosome membrane contact sites, and this process is necessary for RTK down-regulation and degradation in lysosomes [43]. However, new evidence suggests that PTP1B directly dephosphorylates PM-localized RTKs at ER–PM contacts, at least in the case of Eph receptors [44•]. In addition to integral membrane RTKs, PTP1B interacts with and regulates the nonreceptor tyrosine kinase Src at ER–PM contact sites [45]. Remarkably, PTP1B-Src interactions recruit Src to cell adhesion complexes [45], and PTP1B dephosphorylates Eph receptors at ER–PM junctions associated with cell–cell contacts [44•]. These findings imply that inter-organelle signaling at ER–PM contacts may control cell–cell communication during normal cell development and disease states, such as tumor cell progression. ER–PM crosstalk at cell adhesion sites may even mediate communication between ER membrane networks between neighboring cells. PTP1B activity is regulated by reactive oxygen species (ROS) [46], and the ER is a major site for ROS production upon protein misfolding [47]. Thus, ER-mediated ROS signaling may allow a cell to convey its ER status (i.e. oxidative stress, protein misfolding, Ca2+ depletion, lipid misregulation) to neighboring cells by modulating PTP1B and RTK activity at ER–PM contacts. While these ideas are still speculative, inter-organelle communication between plant cells is known to occur through specialized ER–PM junctions termed plasmodesmata [48].
ER–PM tether proteins control ER organization and stress responses
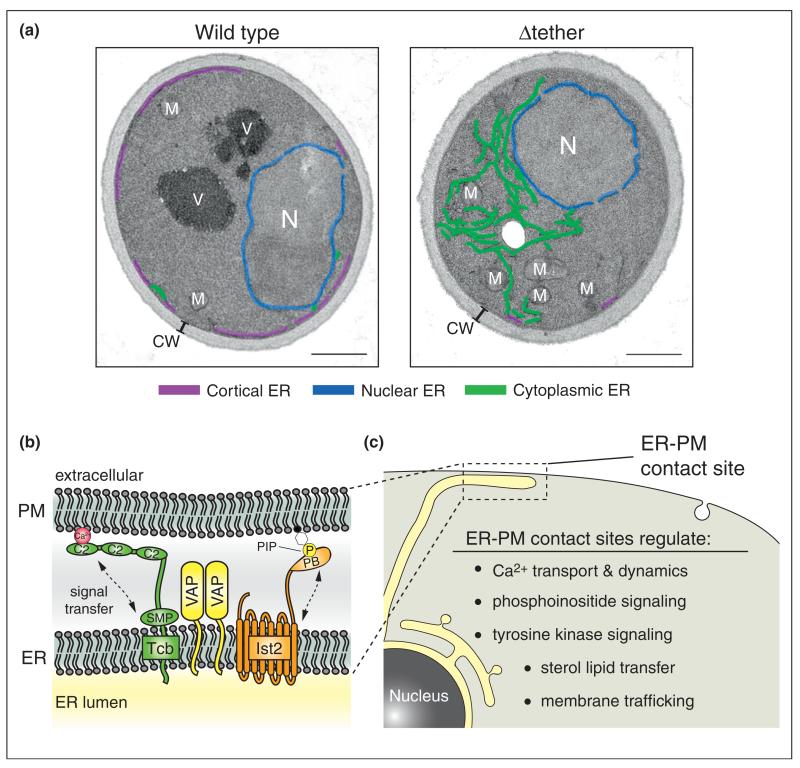
The proteins primarily responsible for facilitating ER–PM contacts have remained unclear until recently. In yeast, where there are substantial ER–PM contacts (Figure 4a), three conserved protein families serve as ER–PM tethers [49•,50••,51••,52•]: the yeast VAP proteins Scs2/22, Ist2 (related to the TMEM16 channel family [53,54]), and the tricalbin proteins Tcb1/2/3 (orthologs of the extended synaptotagmin-like proteins E-Syt1/2/3 [55]). The ER–PM tethering proteins are anchored in the ER and interact with the PM via cytoplasmic lipid-binding and protein-binding domains (Figure 4b) [50••,51••]. While each of the tethering protein families is sufficient for ER–PM contacts, loss of all six ER–PM tethers results in a massive reduction in ER–PM contacts, decreasing the percentage of the PM associated with ER from ~40% to ~5% (Figure 4a) [50••]. Sequential loss of ER–PM tethers also drastically changes the morphology of the peripheral ER network and results in accumulation of internal ER structures (Figure 4a) [50••,56]. The ER is arranged into functional domains, and changes in ER architecture may cause mislocalization (i.e. mixing or dilution) of resident ER proteins resulting in ER dysfunction. Cells lacking ER–PM tethering proteins accumulate high levels of PI4P on the PM, consistent with impaired Sac1-mediated PI4P turnover at ER–PM contacts [50••]. Moreover, yeast cells lacking ER–PM contacts constitutively induce the unfolded protein response (UPR) pathway and require an intact UPR system for survival [50••]. The UPR is critical for maintaining the homeostasis of the ER [57], suggesting a role for ER–PM contacts in ER function and stress signaling pathways.
Figure 4.
Three conserved protein families are involved in ER–PM contact site formation and function. The tricalbin proteins (Tcb, orthologs of the extended synaptotagmin-like proteins), VAP family members, and Ist2 (related to the TMEM16 channel family) tether the ER to the PM in yeast cells [49•,50••,51••,52•]. (a) Electron microscopy of wild type and Δtether yeast cells [50••]. Originally published in Dev Cell, 2012, 23: 1129–1140. In wild type cells, large regions of the PM are associated with cortical ER (purple). In cells lacking the ER–PM tether proteins (Δtether), cortical ER structures are depleted, and a meshwork of cytoplasmic ER accumulates (green). The nuclear ER (blue), mitochondria (M), nucleus (N), cell wall (CW) and vacuole (V) are also labeled. Scale bars, 500 nm. (b) All of the ER–PM tether proteins are integral ER membrane proteins that contain conserved cytoplasmic lipid-binding and protein-binding domains. The tricalbin proteins possess C2 domains that display Ca2+-stimulated lipid binding activity and a conserved SMP domain that is sufficient for lipid binding and ER targeting. The Ist2 protein contains a carboxyl terminal polybasic domain (PB) proposed to bind PIP lipids at the PM. The lipid binding activities in the tether proteins may facilitate the transmission of signals between the PM and ER. The VAP proteins recruit lipid-binding ORP family members to ER–PM contact sites; additional interactions with PM proteins and lipids may be involved in VAP tethering function. (c) ER–PM contact sites serve key roles in Ca2+ dynamics, sterol and phosphoinositide lipid signaling, growth factor receptor signaling, and membrane trafficking pathways. In addition to specific signaling roles, the ER–PM tether proteins may have general housekeeping functions in several of these essential cellular processes.
Vesicle trafficking and plasma membrane domain organization
Vesicle-mediated trafficking controls cell signaling through the regulated secretion of hormones and neuro-transmitters, and by endocytosis of signaling receptors from the cell surface. Roles for ER–PM contacts in membrane trafficking, especially PM internalization, are now being recognized. PTP1B activity at ER–PM contacts is necessary for efficient internalization of active ligand-bound Eph receptors (Figure 3c) [58]. Interestingly, PTP1B regulates interferon receptor (IFNAR1) internalization by dephosphorylating a tyrosine residue in an endocytic motif within IFNAR1 recognized by the clathrin adaptor protein-2 (AP-2) complex [59], providing some insight into how PTP1B activity at ER–PM contacts modulates rates of PM protein internalization. In addition to regulatory roles in endocytosis, the ER-localized STIM proteins are recruited to sites of phagocytosis where they regulate Ca2+ and actin cytoskeletal dynamics necessary for phagosome formation [60•]. Interestingly, certain pathogens that escape the innate immune response co-opt phagosomal-ER membrane contacts for survival of the pathogen within phagosomes [61,62].
The role for ER–PM contacts in supplying local Ca2+ signals during phagocytosis suggests additional roles for ER–PM junctions in Ca2+-regulated membrane trafficking events. The SOCE pathway provides capacitive Ca2+ stores necessary for protein cargo sorting in the ER and Golgi compartments [4], but it is unknown whether Orai1 regulates endocytosis or fusion of secretory vesicles at the PM. In neurons, L-type VGCCs are responsible for Ca2+ influx necessary for synaptic vesicle fusion and neurotransmitter release. Notably, STIM proteins interact with L-type VGCCs and promote their clearance from the PM, in addition to acutely inhibiting channel activity [23•]. Because STIM proteins inhibit VGCC activity and promote VGCC down-regulation [23•], ER–PM contacts may have important regulatory roles in neurotransmission.
The very close association of the ER and PM may preclude clathrin-coated pit (CCP) formation as well as secretory vesicle delivery at ER–PM contacts. However, the peripheral ER membrane network is a highly dynamic meshwork [3]. In yeast, constant re-organization of the peripheral ER appears to play a role in determining the distribution of CCPs at the PM [63]. Thus, the PM may be organized into ER-free zones where vesicle trafficking takes place and ER-bound regions that form productive ER–PM connections for the regulation of PM lipid composition, as well as control of PM channels, transporters, and signaling receptors (Figure 4c).
Conclusions and future directions
Organelle identity and specialization are established and maintained by membrane trafficking pathways. However, direct ER–PM crosstalk occurs at sites of close apposition between these two organelles independent of vesicle transport. The ER is a major site for protein quality control and secretion, lipid synthesis, and calcium homeostasis. Signaling between the ER and PM at membrane contact sites may regulate these important ER functions in response to changes in the cell’s environment or metabolic requirements. In addition, ER–PM contacts may allow the ER to sense the state of the PM (damage, composition) and launch responses (Ca2+ dynamics, protein and lipid synthesis) to ensure the integrity and function of the PM even under stress conditions. As such, future studies may reveal PM proteins that bind to the recently described ER tether proteins and aid in this process.
Interestingly, the ER forms connections with several compartments in the cell (the PM, Golgi compartments, endosomes, lysosomes, mitochondria, and peroxisomes). ER-mediated inter-organelle signaling may allow cross-talk between distinct organelles to strengthen cellular responses to various stimuli. Thus, the ER may serve as a conduit for information transfer to synergize the functions of specialized organelles. For instance, mitochondria associate with the peripheral ER network at ER–PM contacts and this aids in coupling Ca2+ influx to mitochondrial Ca2+ import [64] (see the accompanying article by B. Kornmann). The signals (e.g. Ca2+, lipids, ROS, phosphorylation events) and effector proteins that transmit information along the ER network will be an important area for future studies in cell biology.
Acknowledgements
We thank members of the Emr laboratory, Dave Holowka, Tamas Balla, and Will Prinz for helpful discussions and comments on the manuscript.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
•of special interest
•• of outstanding interest
- 1.Toulmay A, Prinz WA. Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr Opin Cell Biol. 2011;23:458–463. doi: 10.1016/j.ceb.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–623. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Friedman JR, Voeltz GK. The ER in 3D: a multifunctional dynamic membrane network. Trends Cell Biol. 2011;21:709–717. doi: 10.1016/j.tcb.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrasco S, Meyer T. STIM proteins and the endoplasmic reticulum-plasma membrane junctions. Annu Rev Biochem. 2011;80:973–1000. doi: 10.1146/annurev-biochem-061609-165311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porter KR, Palade GE. Studies on the endoplasmic reticulum. III. Its form and distribution in striated muscle cells. J Biophys Biochem Cytol. 1957;3:269–300. doi: 10.1083/jcb.3.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Endo M. Calcium-induced calcium release in skeletal muscle. Physiol Rev. 2009;89:1153–1176. doi: 10.1152/physrev.00040.2008. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi M, Raimondi A, O’Toole E, Paradise S, Collesi C, Cremona O, Ferguson SM, De Camilli P. Cell- and stimulus-dependent heterogeneity of synaptic vesicle endocytic recycling mechanisms revealed by studies of dynamin 1-null neurons. Proc Natl Acad Sci U S A. 2008;105:2175–2180. doi: 10.1073/pnas.0712171105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenbluth J. Subsurface cisterns and their relationship to the neuronal plasma membrane. J Cell Biol. 1962;13:405–421. doi: 10.1083/jcb.13.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sparkes IA, Frigerio L, Tolley N, Hawes C. The plant endoplasmic reticulum: a cell-wide web. Biochem J. 2009;423:145–155. doi: 10.1042/BJ20091113. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki E, Hirosawa K. Immunolocalization of a Drosophila phosphatidylinositol transfer protein (rdgB) in normal and rdgA mutant photoreceptor cells with special reference to the subrhabdomeric cisternae. J Electron Microsc (Tokyo) 1994;43:183–189. [PubMed] [Google Scholar]
- 11.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.West M, Zurek N, Hoenger A, Voeltz GK. A 3D analysis of yeast ER structure reveals how ER domains are organized by membrane curvature. J Cell Biol. 2011;193:333–346. doi: 10.1083/jcb.201011039. See anotation to Ref. [13•].
- 13•.Wei D, Jacobs S, Modla S, Zhang S, Young CL, Cirino R, Caplan J, Czymmek K. High-resolution three-dimensional reconstruction of a whole yeast cell using focused-ion beam scanning electron microscopy. Biotechniques. 2012;53:41–48. doi: 10.2144/000113850. This study along with Ref. [12•] reveal, in great detail, the architecture of the ER membrane network in yeast cells using advanced three dimensional electron microscopy techniques.
- 14.Orci L, Ravazzola M, Le Coadic M, Shen WW, Demaurex N, Cosson P. From the Cover: STIM1-induced precortical and cortical subdomains of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2009;106:19358–19362. doi: 10.1073/pnas.0911280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeshima H, Komazaki S, Nishi M, Iino M, Kangawa K. Junctophilins: a novel family of junctional membrane complex proteins. Mol Cell. 2000;6:11–22. doi: 10.1016/s1097-2765(00)00003-4. [DOI] [PubMed] [Google Scholar]
- 16•.Wu CY, Jia Z, Wang W, Ballou LM, Jiang YP, Chen B, Mathias RT, Cohen IS, Song LS, Entcheva E, Lin RZ. PI3Ks maintain the structural integrity of T-tubules in cardiac myocytes. PLoS One. 2011;6:e24404. doi: 10.1371/journal.pone.0024404. This study implicates PI3K activity and the phosphoinositide isoform PI(3,4,5)P3 in the regulation of junctophilin, providing additional mechanistic insight into how this protein links the ER and PM in muscle cells.
- 17.Liou J, Kim ML, Heo WD, Jones JT, Myers JW, Ferrell JE, Jr, Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park CY, Hoover PJ, Mullins FM, Bachhawat P, Covington ED, Raunser S, Walz T, Garcia KC, Dolmetsch RE, Lewis RS. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNally BA, Somasundaram A, Yamashita M, Prakriya M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 2012;482:241–245. doi: 10.1038/nature10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20••.Hou X, Pedi L, Diver MM, Long SB. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. This study provides the first molecular model for Orai1 Ca2+ channel activity and selectivity. The crystal structure shows that Orai1 assembles into a hexamer forming the channel pore and additional details for Orai1–STIM protein interactions are proposed.
- 21.Srikanth S, Jew M, Kim KD, Yee MK, Abramson J, Gwack Y. Junctate is a Ca2+-sensing structural component of Orai1 and stromal interaction molecule 1 (STIM1) Proc Natl Acad Sci U S A. 2012;109:8682–8687. doi: 10.1073/pnas.1200667109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palty R, Raveh A, Kaminsky I, Meller R, Reuveny E. SARAF inactivates the store operated calcium entry machinery to prevent excess calcium refilling. Cell. 2012;149:425–438. doi: 10.1016/j.cell.2012.01.055. [DOI] [PubMed] [Google Scholar]
- 23•.Park CY, Shcheglovitov A, Dolmetsch R. The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science. 2010;330:101–105. doi: 10.1126/science.1191027. See anotation to Ref. [25•].
- 24•.Wang Y, Deng X, Mancarella S, Hendron E, Eguchi S, Soboloff J, Tang XD, Gill DL. The calcium store sensor, STIM1, reciprocally controls Orai and CaV1. 2 channels. Science. 2010;330:105–109. doi: 10.1126/science.1191086. See anotation to Ref. [25•].
- 25•.Krapivinsky G, Krapivinsky L, Stotz SC, Manasian Y, Clapham DE. POST, partner of stromal interaction molecule 1 (STIM1), targets STIM1 to multiple transporters. Proc Natl Acad Sci U S A. 2011;108:19234–19239. doi: 10.1073/pnas.1117231108. This study along with Refs. [23•,24•] have uncovered additional roles for the STIM proteins in the inhibition of Ca2+ channel and transporter activities at the PM, thereby allowing Orai1 to predominate as the primary source for Ca2+ influx, as needed, in cells expressing multiple Ca2+ channels and transporters. The study by Park et al. [23•] also found that STIM proteins promote the down-regulation of voltage-gated Ca2+ channels by the endocytic pathway.
- 26.Lev S. Non-vesicular lipid transport by lipid-transfer proteins and beyond. Nat Rev Mol Cell Biol. 2010;11:739–750. doi: 10.1038/nrm2971. [DOI] [PubMed] [Google Scholar]
- 27.Lehto M, Hynynen R, Karjalainen K, Kuismanen E, Hyvarinen K, Olkkonen VM. Targeting of OSBP-related protein 3 (ORP3) to endoplasmic reticulum and plasma membrane is controlled by multiple determinants. Exp Cell Res. 2005;310:445–462. doi: 10.1016/j.yexcr.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 28.Schulz TA, Choi MG, Raychaudhuri S, Mears JA, Ghirlando R, Hinshaw JE, Prinz WA. Lipid-regulated sterol transfer between closely apposed membranes by oxysterol-binding protein homologues. J Cell Biol. 2009;187:889–903. doi: 10.1083/jcb.200905007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy A, Levine TP. Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J Biol Chem. 2004;279:44683–44689. doi: 10.1074/jbc.M401583200. [DOI] [PubMed] [Google Scholar]
- 30.Loewen CJ, Levine TP. A highly conserved binding site in vesicle-associated membrane protein-associated protein (VAP) for the FFAT motif of lipid-binding proteins. J Biol Chem. 2005;280:14097–14104. doi: 10.1074/jbc.M500147200. [DOI] [PubMed] [Google Scholar]
- 31.Im YJ, Raychaudhuri S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32•.de Saint-Jean M, Delfosse V, Douguet D, Chicanne G, Payrastre B, Bourguet W, Antonny B, Drin G. Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J Cell Biol. 2011;195:965–978. doi: 10.1083/jcb.201104062. This study determined the molecular structure of the ORP family member Osh4 bound to the phosphoinositide isoform PI4P. The structure indicated that the headgroup of PI4P interacts with basic residues at the mouth of the Osh4 sterol-binding barrel, suggesting that PI4P and sterol binding are mutually exclusive. Biochemical experiments supported this model, demonstrating that PI4P inhibited the ability of Osh4 to extract sterol lipids from the membrane bilayer. These findings suggest a model for the directional transport of sterol lipids from the ER to PIP-enriched membranes, such as the PM.
- 33•.Nakatsu F, Baskin JM, Chung J, Tanner LB, Shui G, Lee SY, Pirruccello M, Hao M, Ingolia NT, Wenk MR, De Camilli P. PtdIns4P synthesis by PI4KIIIalpha at the plasma membrane and its impact on plasma membrane identity. J Cell Biol. 2012;199:1003–1016. doi: 10.1083/jcb.201206095. This study shows that the PI4K isoform type IIIα, previously found on ER membranes, in fact localizes to the PM and generates pools of PI4P involved in STIM protein translocation to ER–PM contacts, as well as regulation of PI(4,5)P2 and cholesterol levels in the PM.
- 34.Jansen M, Ohsaki Y, Rita Rega L, Bittman R, Olkkonen VM, Ikonen E. Role of ORPs in sterol transport from plasma membrane to ER and lipid droplets in mammalian cells. Traffic. 2011;12:218–231. doi: 10.1111/j.1600-0854.2010.01142.x. [DOI] [PubMed] [Google Scholar]
- 35.Georgiev AG, Sullivan DP, Kersting MC, Dittman JS, Beh CT, Menon AK. Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic. 2011;12:1341–1355. doi: 10.1111/j.1600-0854.2011.01234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beh CT, McMaster CR, Kozminski KG, Menon AK. A detour for yeast oxysterol binding proteins. J Biol Chem. 2012;287:11481–11488. doi: 10.1074/jbc.R111.338400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Hammond GR, Fischer MJ, Anderson KE, Holdich J, Koteci A, Balla T, Irvine RF. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–730. doi: 10.1126/science.1222483. This study implicates the PIP isoform PI4P in key cellular processes at the PM previously assigned to the more extensively characterized PIP isoform PI(4,5)P2, including regulation of PM channels and cell signaling
- 38.Minogue S, Waugh MG. The phosphatidylinositol 4-kinases: Don’t Call it a Comeback. Subcell Biochem. 2012;58:1–24. doi: 10.1007/978-94-007-3012-0_1. [DOI] [PubMed] [Google Scholar]
- 39•.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. This study uncovered a pathway for regulation of the PIP isoform PI4P at ER–PM contact sites by the ER-localized PIP phosphatase Sac1, ORP family members, and VAP orthologs. PI4P regulation may control additional processes that occur at ER–PM contacts, such as sterol lipid transport and Orai1 channel activation.
- 40.Wang PY, Weng J, Anderson RG. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 41.Korzeniowski MK, Popovic MA, Szentpetery Z, Varnai P, Stojilkovic SS, Balla T. Dependence of STIM1/Orai1-mediated calcium entry on plasma membrane phosphoinositides. J Biol Chem. 2009;284:21027–21035. doi: 10.1074/jbc.M109.012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderie I, Schulz I, Schmid A. Direct interaction between ER membrane-bound PTP1B and its plasma membrane-anchored targets. Cell Signal. 2007;19:582–592. doi: 10.1016/j.cellsig.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Eden ER, Burgoyne T, Edgar JR, Sorkin A, Futter CE. The relationship between ER-multivesicular body membrane contacts and the ESCRT machinery. Biochem Soc Trans. 2012;40:464–468. doi: 10.1042/BST20110774. [DOI] [PubMed] [Google Scholar]
- 44•.Haj FG, Sabet O, Kinkhabwala A, Wimmer-Kleikamp S, Roukos V, Han HM, Grabenbauer M, Bierbaum M, Antony C, Neel BG, Bastiaens PI. Regulation of signaling at regions of cell–cell contact by endoplasmic reticulum-bound protein–tyrosine phosphatase 1B. PLoS One. 2012;7:e36633. doi: 10.1371/journal.pone.0036633. This study found that the protein tyrosine phosphatase PTP1B dephosphoryates active receptor tyrosine kinases (RTK) at ER–PM contacts. PTP1B attenuates RTK signaling at the PM and stimulates clearance of cell-surface RTKs by endocytosis [see [58]]. Remarkably, PTP1B-RTK interactions at ER–PM contacts were also associated with cell–cell adhesion sites, implying that ER–PM crosstalk further regulates cell–cell signaling pathways.
- 45.Monteleone MC, Gonzalez Wusener AE, Burdisso JE, Conde C, Caceres A, Arregui CO. ER-bound protein tyrosine phosphatase PTP1B interacts with Src at the plasma membrane/substrate interface. PLoS One. 2012;7:e38948. doi: 10.1371/journal.pone.0038948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haque A, Andersen JN, Salmeen A, Barford D, Tonks NK. Conformation-sensing antibodies stabilize the oxidized form of PTP1B and inhibit its phosphatase activity. Cell. 2011;147:185–198. doi: 10.1016/j.cell.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Appenzeller-Herzog C. Glutathione- and non-glutathione-based oxidant control in the endoplasmic reticulum. J Cell Sci. 2011;124:847–855. doi: 10.1242/jcs.080895. [DOI] [PubMed] [Google Scholar]
- 48.Lucas WJ, Ham BK, Kim JY. Plasmodesmata — bridging the gap between neighboring plant cells. Trends Cell Biol. 2009;19:495–503. doi: 10.1016/j.tcb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 49•.Loewen CJ, Young BP, Tavassoli S, Levine TP. Inheritance of cortical ER in yeast is required for normal septin organization. J Cell Biol. 2007;179:467–483. doi: 10.1083/jcb.200708205. See anotation to Ref. [51••].
- 50••.Manford AG, Stefan CJ, Yuan HL, Macgurn JA, Emr SD. ER-to-plasma membrane tethering proteins regulate cell signaling and ER morphology. Dev Cell. 2012;23:1129–1140. doi: 10.1016/j.devcel.2012.11.004. See anotation to Ref. [51••].
- 51••.Toulmay A, Prinz WA. A conserved membrane-binding domain targets proteins to organelle contact sites. J Cell Sci. 2012;125:49–58. doi: 10.1242/jcs.085118. Proteins that tether the ER and PM have been long sought. two These studies [50••,51], along with studies by the Levine and Seedorf laboratories [49•,52•], implicate three conserved protein families as critical ER–PM tethering proteins in yeast: the VAP isoforms Scs2/22, the tricalbins (orthologs of the mammalian extended-synaptotagmin proteins E-Syt1/2/3), and Ist2 (related to the TMEM16 channel family). Interestingly, the study by Toulmay et al. [51••] found that the mammalian E-Syt1 protein localizes to membrane contact sites in yeast, suggesting conserved functions for this protein family. The study by Manford et al. found [50••] that loss of all ER–PM tether proteins results in a drastic reduction in ER–PM contact sites. This study also found that ER–PM contacts are essential for PI4P turnover at the PM as well as for normal ER homeostasis and function.
- 52•.Wolf W, Kilic A, Schrul B, Lorenz H, Schwappach B, Seedorf M. Yeast Ist2 recruits the endoplasmic reticulum to the plasma membrane and creates a ribosome-free membrane microcompartment. PLoS One. 2012;7:e39703. doi: 10.1371/journal.pone.0039703. See anotation to Ref. [51••].
- 53.Tian Y, Schreiber R, Kunzelmann K. Anoctamins are a family of Ca2+-activated Cl-channels. J Cell Sci. 2012;125:4991–4998. doi: 10.1242/jcs.109553. [DOI] [PubMed] [Google Scholar]
- 54.Duran C, Qu Z, Osunkoya AO, Cui Y, Hartzell HC. ANOs 3-7 in the anoctamin/Tmem16 Cl-channel family are intracellular proteins. Am J Physiol Cell Physiol. 2012;302:C482–C493. doi: 10.1152/ajpcell.00140.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Min SW, Chang WP, Sudhof TC. E-Syts, a family of membranous Ca2+-sensor proteins with multiple C2 domains. Proc Natl Acad Sci U S A. 2007;104:3823–3828. doi: 10.1073/pnas.0611725104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D, Vjestica A, Oliferenko S. Plasma membrane tethering of the cortical ER necessitates its finely reticulated architecture. Curr Biol. 2012;22:2048–2052. doi: 10.1016/j.cub.2012.08.047. [DOI] [PubMed] [Google Scholar]
- 57.Walter P, Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- 58.Nievergall E, Janes PW, Stegmayer C, Vail ME, Haj FG, Teng SW, Neel BG, Bastiaens PI, Lackmann M. PTP1B regulates Eph receptor function and trafficking. J Cell Biol. 2010;191:1189–1203. doi: 10.1083/jcb.201005035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carbone CJ, Zheng H, Bhattacharya S, Lewis JR, Reiter AM, Henthorn P, Zhang ZY, Baker DP, Ukkiramapandian R, Bence KK, Fuchs SY. Protein tyrosine phosphatase 1B is a key regulator of IFNAR1 endocytosis and a target for antiviral therapies. Proc Natl Acad Sci U S A. 2012;109:19226–19231. doi: 10.1073/pnas.1211491109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60•.Nunes P, Cornut D, Bochet V, Hasler U, Oh-Hora M, Waldburger JM, Demaurex N. STIM1 juxtaposes ER to phagosomes, generating Ca(2)(+) hotspots that boost phagocytosis. Curr Biol. 2012;22:1990–1997. doi: 10.1016/j.cub.2012.08.049. It has been known for some time that ER membranes associate with sites of phagocytosis. However, roles for ER–PM contacts during phagocytosis are not well-understood. This study indicates that the ER-localized STIM proteins generate localized Ca2+ signals that drive actin dynamics necessary for phagocytic cup formation.
- 61.Campbell-Valois FX, Trost M, Chemali M, Dill BD, Laplante A, Duclos S, Sadeghi S, Rondeau C, Morrow IC, Bell C, Desjardins M. Quantitative proteomics reveals that only a subset of the endoplasmic reticulum contributes to the phagosome. Mol Cell Proteomics. 2012;11 doi: 10.1074/mcp.M111.016378. M111 016378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Derre I, Swiss R, Agaisse H. The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites. PLoS Pathog. 2011;7:e1002092. doi: 10.1371/journal.ppat.1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stradalova V, Blazikova M, Grossmann G, Opekarova M, Tanner W, Malinsky J. Distribution of cortical endoplasmic reticulum determines positioning of endocytic events in yeast plasma membrane. PLoS One. 2012;7:e35132. doi: 10.1371/journal.pone.0035132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Korzeniowski MK, Szanda G, Balla T, Spat A. Store-operated Ca2+ influx and subplasmalemmal mitochondria. Cell Calcium. 2009;46:49–55. doi: 10.1016/j.ceca.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]