Abstract
We use broadband coherent anti-Stokes Raman scattering (BCARS) microscopy to characterize lineage commitment of individual human mesenchymal stem cells cultured in adipogenic, osteogenic, and basal culture media. We treat hyperspectral images obtained by BCARS in two independent ways, obtaining robust metrics for differentiation. In one approach, pixel counts corresponding to functional markers, lipids, and minerals, are used to classify individual cells as belonging to one of the three lineage groups: adipocytes, osteoblasts, and undifferentiated stem cells. In the second approach, we use multivariate analysis of Raman spectra averaged exclusively over cytosol regions of individual cells to classify the cells into the same three groups, with consistent results. The exceptionally high speed of spectral imaging with BCARS allows us to chemically map a large number of cells with high spatial resolution, revealing not only the phenotype of individual cells, but also population heterogeneity in the degree of phenotype commitment.
Introduction
There is a significant need for reliable, quantitative, and noninvasive methods for cell-by-cell functional characterization in many areas of medicine, biology, and biotechnology. In the emerging field of single-cell analysis, medical researchers seek to identify rare cell types and functional changes in specific cells that may have significant impact on tissue organization and disease state. Scientists in the field of tissue engineering and regenerative medicine (TERM) and stem cell sourcing have similar needs. In TERM, researchers seek to guide cellular responses such as differentiation and matrix production using engineered tissue scaffolds and other extrinsic cues to produce specifically tailored tissues.1 The presentation of these cues is necessarily spatially heterogeneous. Thus, characterization of cell responses is best done at the level of individual cells, and in the context of their unique environment. Analogously, those involved in cell sourcing desire stable maintenance of stem cell pluripotency in embryoid bodies,2,3 where changes in phenotype of individual cells can be important seeding events, and should thus be tracked.
Characterization of stem cell pluripotency and phenotype commitment is vitally important in many of these research areas. Although it is currently possible to characterize cell pluripotency and phenotype commitment, the methods in use are typically intrusive or provide information only averaged over entire populations. Use of invasive, endpoint methods necessitate assumptions about correlations between materials sampled and those not sampled, and these assumptions can lead to incorrect findings when working with heterogeneous tissues and cell cultures.
Conventional techniques for characterizing stem cell lineage commitment generally rely on cell surface markers, genetic activity profiles, or production of specific proteins. These techniques include immunolabeling proteins of interest, gene microarrays, and insertion of functional DNA markers, and each of these has drawbacks. Reliable surface markers are sometimes difficult to establish, cell characterization by immunostaining for phenotype-specific proteins significantly perturbs the cells, requiring membrane permeabilization, and gene microarray analyses can assess only average lineage commitment for a large number of cells. Although single-cell genetic analysis is becoming feasible,4 these techniques are destructive.
In addition to the markers typically measured, many other cell properties change with specific lineage commitment. Some of these can be monitored noninvasively for phenotype and functional characterization. These include changes in nucleic acid levels,5–7 spatial distribution, and metabolic activity of mitochondria,8,9 and spatial distribution and expression of key cytoskeletal proteins.10 Accordingly, chemical11 and morphological10 imaging have been effective in classifying differentiation state using these metrics. Recently, Treiser et al. demonstrated that morphological analyses of cell shape and cytoskeletal organization can be used as early as 24 h to forecast the long-term (2 week) lineage commitment of individual human mesenchymal stem cells (hMSCs).10 Their approach was rapid, but was only realized in fluorescently labeled cells.
Spontaneous Raman spectroscopy has been used as a label-free approach for characterizing stem cell differentiation. Raman spectra of biological macromolecules such as nucleic acids, proteins, lipids, and carbohydrates, exhibit unique spectral features that contain information indicative of cell phenotype and function. For example, Notingher et al. analyzed Raman spectral changes of differentiating murine embryonic stem cells using multivariate analysis and found that major spectral differences were consistent with variations in mRNA concentration.12 Hung and colleagues used Raman scattering to determine the degree of osteogenic (OS) differentiation of hMSCs.13 More recently, Chan et al. used multivariate analysis of Raman spectra to identify spectral differences between human embryonic stem cells and differentiated cardiomyocytes.6
Although spontaneous Raman studies have demonstrated that Raman spectra from cells can be used to distinguish different phenotypes, these studies required spectral averages over entire cells or multiple cells to obtain sufficient signals. On the other hand, characteristic functional markers are generally localized to their relevant subcellular organelles,8,12 so the ability to perform spectroscopic imaging with a higher spatial resolution would allow for a more complete characterization of phenotype commitment through the collection of Raman spectra on an organelle-by-organelle basis. Confocal Raman microscopy provides the spatial resolution needed, and has been used to acquire spatially resolved Raman spectra from biological samples. However, the weak signal levels and concomitant long image acquisition time makes it impractical to use for sampling large numbers of cells required for characterization of heterogeneous biological systems.14 Coherent Raman imaging methods offer a potential solution to the need for rapid spectroscopic imaging with a high spatial resolution.
Coherent anti-Stokes Raman scattering (CARS) is a coherent Raman process based on four-wave mixing, where a blue-shifted anti-Stokes signal is enhanced when the frequency difference between two input light fields matches a molecular vibrational resonance. Under the right conditions, coherent Raman scattering provides a much stronger signal generation than spontaneous Raman processes. Single-frequency CARS microscopy has been extensively developed for video rate imaging in biological, physical, and material research.15–18 However, single-frequency and multifrequency configurations commonly used are not suitable for detecting subtle changes in many chemical components of complex systems, such as biological cells and tissues. In 2004, Kee and Cicerone19 and Kano and Hamaguchi20 demonstrated that a simple laser configuration with a narrowband probe field and a broadband continuum field could be used to acquire broadband CARS spectra ranging from 500 to 3500 cm−1 with a diffraction limit spatial resolution. Since then, broadband CARS (BCARS) microscopy has been used for quantitative spectroscopic imaging of polymeric materials and biological systems.21–25 BCARS provides significantly more chemical specificity than narrowband CARS through the broad spectral response, and is much faster than spontaneous Raman scattering. In this study, we show that BCARS imaging provides sufficient spectral coverage, sensitivity, and reproducibility to accurately track stem cell differentiation. Additionally, we show that our technique can be performed at sufficient speeds to facilitate high-resolution imaging of hundreds of cells, as required for characterization of heterogeneous biological systems.
Dynamic population heterogeneity is intrinsic to biological systems,26 so it is important that a sufficient number of cells be analyzed to sample the range of cell states. In this study, we acquired high-resolution BCARS images comprising a total of 1.3 million spectra over a total of 9 h of imaging time. A similar study using confocal spontaneous Raman imaging would have been possible, but perhaps not practical, as it would require >150 h of imaging time using integration times required for sufficient signal quality that individual spectra can be analyzed and used for image contrast,27,28 as is done here. Shorter dwell times have been reported for confocal spontaneous Raman imaging,29 but only when subsequent image contrast is based on multivariate analysis of spectral clusters. We point out that the spectral quality obtained in the present study is probably not sufficient to detect very subtle spectral changes that accompany some phenotype changes,30 but this is not due to fundamental limitations of BCARS.
Materials and Methods
BCARS microscopy
The experimental setup of the broadband CARS microscopy has been described previously.21,22 Briefly, the output (70 fs, centered at 830 nm, 80 MHz) of a Ti:S laser oscillator (MaiTai-DeepSee; Spectra-Physics) (certain commercial equipment, instruments, or materials are identified in this article to specify the experimental procedure adequately. Such identification is not intended to imply recommendation or endorsement by the National Institute of Standards and Technology, nor is it intended to imply that the materials or equipment identified are necessarily the best available for the purpose) was split into two parts. One part was introduced into a photonic crystal fiber (Femtowhite; Crystal Fiber) to generate a continuum pulse. The other part was spectrally narrowed by a 4-f dispersionless filter to 10 cm−1 full-width-half-maximum with the center wavelength at 830 nm. The two beams were introduced collinearly and with parallel polarization into a 60×1.35 NA oil immersion objective lens (Olympus) and focused on the sample. The CARS signal generated from the sample was collected in the forward direction and passed through an 830 nm notch filter and an 810 nm short-pass filter and was analyzed using a spectrometer (SP-2300; Acton) with a charge-coupled device (DU920-BR-DD; Andor). The average laser power at the sample was kept below 20 mW for each pulse, to avoid visible damage at the sample. The sample was scanned with a spacing of 1 μm for 100×100 image pixels, and the charge coupled device exposure time was typically set to 30 ms per pixel. The acquired CARS spectrum was processed using time-domain Kramers-Kronig phase retrieval31 with separately measured nonresonant CARS spectra in the glass coverslip.
Cell culture
hMSCs were obtained from commercial sources (Lonza). Cells were cultured in a humidity-controlled environment under 5% CO2 and 37°C and fed every 3–4 days with growth media (basal [BA] media) supplemented with commercial SingleQuot's (PT-3001; Lonza). Cells were received at passage 1 and used up to passage 5. Cells were subcultured upon reaching ∼80% confluence. OS and adipogenic (AD) induction media were reconstituted as per manufacturer's instructions (Lonza). For differentiation assays, cells were cultured for 2 weeks and then stained for alkaline phosphatase (ALP) and lipid production with Fast Blue RR/napthol (Kit number 85; Sigma-Aldrich) and 5 μL AdipoRed (Lonza) per mL of PBS, respectively. For total cell counts, cell nuclei were stained with 2 μg/mL Hoechst (Invitrogen). For BCARS imaging, cells were cultured for 2 weeks then fixed according to protocols described elsewhere, without staining.22
Spectral analysis
BCARS image data of 130 hMSCs were acquired as a 3D (x, y, w) matrix, where the x- and y-axes are parallel to the substrate and w is the Raman shift frequency dimension. Raw hyperspectral data were first processed to generate masks for specific biochemical species. To do so, the following workflow was implemented: (1) singular value decomposition was performed for noise reduction, (2) a time-domain Kramers-Kronig transform was applied to retrieve the Raman susceptibility spectra,31 (3) second derivatives of the individual spectra were calculated, and (4) mask images were constructed using intensity thresholds at Raman shifts that are specific to the chemical species of interest. Principal component analysis (PCA) was performed using Solo+MIA (Eigenvector) after mean centering of the Raman spectra.
Results and Discussion
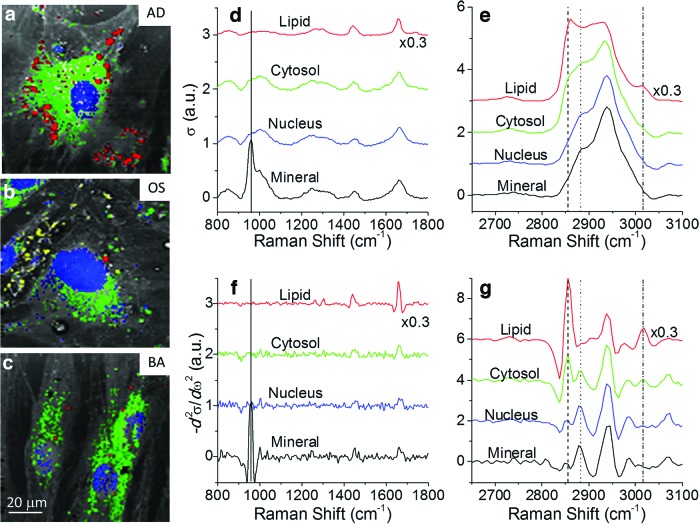
Figure 1a–c shows BCARS images of representative hMSCs cultured in three different types of differentiation-inducing media: AD, OS, and BA growth media. Cells cultured in each of the three media types were confluent at 14 days. We obtained ∼35 images of cells from each of the three culture conditions, each image containing one or two cells. All images were collected over a 3-day period, with images for each culture group acquired over a 2-day period to check for potential day-to-day variation in instrumentation. No indication of day-to-day variation was detected, as results of image analysis were entirely independent of the day on which the images were acquired. Each hyperspectral image consists of 100×100 spatial pixels at a 1 μm lateral resolution, and each spatial pixel contains a Raman-equivalent BCARS spectrum with 560 spectral elements covering the spectral range (500–3400 cm−1).
FIG. 1.
Broadband coherent anti-Stokes Raman scattering (BCARS) images of human mesenchymal stem cells (hMSCs) cultured in (a) adipogenic (AD), (b) osteogenic (OS), and (c) basal (BA) growth media for 2 weeks. Spectral images were acquired at 30 ms/pixel over 100×100 pixels. BCARS spectral maps are pseudo-colored for nuclei (blue), cytosol (green), lipids (red), and mineralization (yellow). The unstained gray-scale images are constructed with the square root of total CARS intensity. (d, e) Raman spectra, σ (ω), and (f, g) their second derivatives of the stained regions are plotted. The vertical lines in the spectra indicate Raman frequencies where threshold values are applied to second derivative spectra, as in (f) and (g), to discriminate nuclei (dot), cytosol (dash), lipids (dash-dot), and minerals (solid). The way the threshold values were determined is displayed in Figure 2a. The ordinate scale is arbitrary units (a.u.). Color images available online at www.liebertpub.com/tec
Raman spectra from different regions of Figure 1a–c are shown in Figure 1d–g. The image of the AD sample exhibits regions inside cells where the CH2 Raman peak at 2860 cm−1 is strong, corresponding to the presence of lipid droplets (pseudo-colored red). The image from the OS sample contains small aggregates outside cells, characterized by a distinctive peak at 955 cm−1, corresponding to minerals containing calcium phosphate (pseudo-colored yellow). Similarly, nuclear (pseudo-colored blue) and cytosolic regions (pseudo-colored green) exhibit differences in Raman spectra, largely due to differences in the relative abundance of aromatic and aliphatic carbon–hydrogen bonds.
The second derivative of the Raman spectrum can help to reveal subtle spectral differences. For example, although the aromatic C-H bonds from nucleotides in the nuclear regions exhibit Raman peaks for C-H at higher frequencies than cytosolic regions, it is difficult to discriminate this difference by inspecting the Raman spectra in Figure 1e. On the other hand, these differences show up clearly in the second derivative spectra of Figure 1f. Whereas this spectral processing step can accentuate spectral changes of interest and can substantially improve chemometric analysis of spectra with slowly varying baselines, it also accentuates high-frequency noise. Thus, to use this processing step, it is crucial that individual spectra have sufficiently high signal-to-noise ratios. Figure 1f and g show second derivatives of the Raman spectra from Figure 1d and e. The broad peaks in the fingerprint region appear less noticeable, while the relative amplitude of the high-frequency noise is increased. On the other hand, the second derivative emphasizes the narrow mineral peak at 955 cm−1, while the broad band near the mineral peak is suppressed. The second derivative has a much more dramatic effect on the C-H region between 2800 and 3100 cm−1. The broad peaks in this region are clearly decomposed into distinct components, with unique spectral features for nuclear and cytosolic regions as well as lipids and minerals.
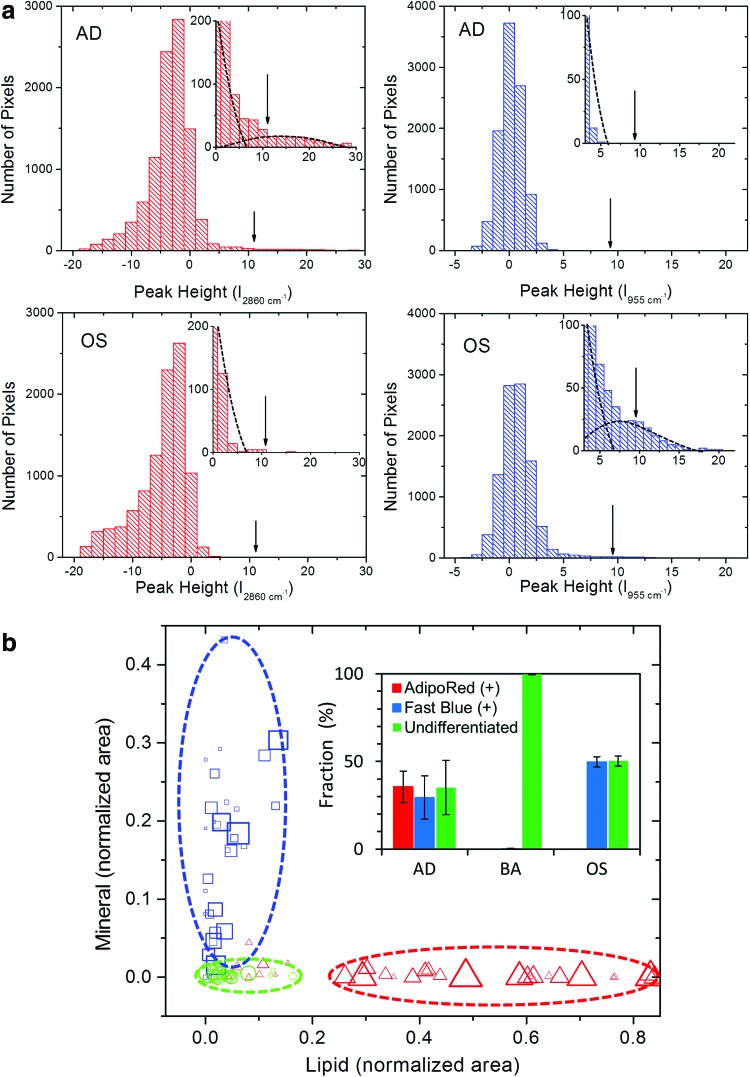
In this study, we utilized two complementary approaches to analyze the spectral images, obtaining objective and unsupervised discrimination between multipotent cells and those committed to osteoblastic or AD lineages. In one approach, we quantify the image area associated with functional markers and use these values, normalized by the total number of image pixels associated with the cell, to determine which lineage commitment a particular cell has made. The presence of functional markers, lipid droplets for AD lineage, and minerals for osteoblastic lineage, is determined in a binary manner by simple criteria based on spectral amplitudes at a small number of discrete frequencies. A given pixel is assigned a mineral value of 1 if the associated second derivative Raman spectral amplitude at 955 cm−1 is above a threshold determined by histogram analysis. The criterion for the presence of lipid is analogous, with a threshold value for spectral amplitude at 2860 cm−1. The thresholds for mineral and lipid are determined based on the distribution of the peak amplitudes at 955 and 2860 cm−1, respectively. Figure 2a shows an example of how the threshold values of minerals and lipids are determined in the intensity histograms. The threshold values indicated with vertical arrows are located not to be affected by contribution from the presumed nonspecific mode edge. For consistency, a single threshold value was used for each functional marker over the whole image sets. Assignment of nucleus and cytosol pixels is based on previous work.22,23 The threshold for the nucleus is set for the difference between amplitudes of 2950 and 2850 cm−1 peaks, which correspond to aromatic and aliphatic C-H frequencies, respectively. Cytosol status is assigned to pixels with average amplitude over the entire C-H region (2800–3000 cm−1) above the level of background, which was not assigned as mineral, lipid, or nucleus. These assignments result in spatial maps such as those shown in Figure 1a–c, where each component of interest has been pseudo-colored according to the scheme indicated above.
FIG. 2.
(a) Histograms of peak amplitudes at 955 and 2860 cm−1 are plotted from the images in Figure 1a and b, where the vertical arrows indicate locations of the thresholds used for minerals and lipids. In the insets, with expanded y-axes, the dashed lines are drawn for guidance of bimodal distributions of minerals and lipids. (b) BCARS demarcates phenotypic markers. The scatter plot of the number of lipids and mineral pixels observed in BCARS images of the AD, OS, and BA samples. The amount of lipids and minerals per cell is represented as the number of phenotypic image pixels divided by the total number of pixels assigned to the cell (the sum of lipids, minerals, cytosols, and nuclear pixels). The size of the symbols represents the total number of pixels. (b, inset) hMSCs were cultured for 2 weeks in AD, BA, or OS media and stained with Fast Blue (alkaline phosphatase), AdipoRed (intracellular triglycerides), and Hoechst (nuclear marker). To determine the percentage of osteogenic and AD differentiation, Fast Blue- and AdipoRed-positive cells were divided by the total number of cells. Color images available online at www.liebertpub.com/tec
Figure 2b shows lineage commitment sorting results from analysis of the pixel assignment protocol described above. On the abscissa and ordinate, we plot the number of pixels associated with lipids and minerals, respectively, for each cell image analyzed. These values are normalized by the total number of pixels associated with the nucleus, cytosol, mineral, and lipid in each image. The normalization is performed to account for variability in properties such as cell density in an imaged region and thin z-section relative to cell thickness. The symbol size is proportional to the total number of spectral pixels used to analyze the cell, and thus represents the statistical significance.
A wide range of mineral signals (5%–30% of pixels assigned a classification) was found in images of cells from OS culture. Two cells in OS culture showed no indication of matrix generation. hMSCs cultured under AD conditions showed a similarly wide range of lipid content. Most of the cells in AD culture had between (30% and 85%) of the total pixel count classified as lipids. However, 7 of 35 cells cultured under AD conditions had a lipid pixel count of <18%, in the same range as cells cultured under BA conditions, and significantly less than other cells cultured under AD conditions. We classified these cells as undifferentiated. Based on this pixel counting analysis, we determined that 20% and 6% of the hMSCs cultured under AD and OS conditions, respectively, did not differentiate even after 2 weeks. These results are quantitatively consistent with the findings of Pittenger et al., who reported that a smaller percentage of hMSCs differentiated to the intended lineage when using AD induction media as compared with OS induction media.32
In contrast to the BCARS results, conventional image-based phenotype determination methods may yield unreliable results. The inset to Figure 2b shows results from such a method that is in common use. In this study, Fast Blue was used to stain for ALP and AdipoRed was used to stain for lipids, and lineage commitment was assigned based on positive staining for either of the two functional markers. Although the Fast Blue stain is intended to target only ALP, evidence of Fast Blue staining was detected on all samples. We therefore set a detection threshold such that trace amount of Fast Blue staining found on cells in the BA condition was not considered Fast Blue positive. In spite of this, there was significant Fast Blue staining above threshold for cells in AD culture conditions. AdipoRed staining was only seen in AD cultures. Cell images found positive for Fast Blue were counted as committed to an OS lineage, and those found positive for AdipoRed were counted as committed to an AD lineage. This conventional analysis resulted in 50% of cells cultured under OS conditions being classified as uncommitted, and 30% of cells cultured under AD conditions being classified as osteoblastic. These results are in clear disagreement with our BCARS results, and the results of previous work that used cell surface antigens for phenotype assignments.32 Culture characterization using stains such as Fast Blue and AdipoRed is quite common; however, the poor specificity of these stains relegates these methods to the realm of qualitative assays. Furthermore, even when analyzed quantitatively, results from their use can be misleading.
The Raman signal is very specific for the functional markers of interest, and its amplitude is linearly proportional to the molecular concentration throughout the focal volume.14 In the foregoing, BCARS analysis takes full advantage of the specificity, but probably not the signal linearity; the binary pixel counting approach is limited in its ability to properly reflect a large dynamic range in analyte concentrations. Below we use an objective spectral analysis approach that takes advantage of both linearity and specificity. This approach yields results consistent with the functional marker analysis above, although in the following, we exclude all spectral pixels associated with functional markers, on which we based the previous analysis.
In this study, we use PCA, an objective but agnostic approach to cell classification. We have shown previously that BCARS can be used to objectively quantify individual chemical species in a complex system using classical least square (CLS), provided the spectra of the individual species are available.21 CLS analysis of the data presented here is possible in principle, but would require individual spectra from all the major cells and extracellular components, and is thus beyond the scope of the present work. PCA does not require knowledge of individual chemical components. Rather, this method reconstructs each input spectrum, φj, as a linear combination of fixed set of basis functions called “principal components” (PCi), according to φj=ΣNi=1 ai,j PCi. The principal components are constructed so as to maximize differences in their amplitudes (ai,j) among the groups of spectra being analyzed. The principal components are ranked according to their amplitude, averaged over all the input spectra, and labeled accordingly as PCn, where n runs from 1 to N, with N being the number of spectral elements in the φj. Input spectra are then arranged in an M-dimensional space (M≤N) by the magnitudes of the amplitude scores associated with the PCs, so that groupings can be explored.
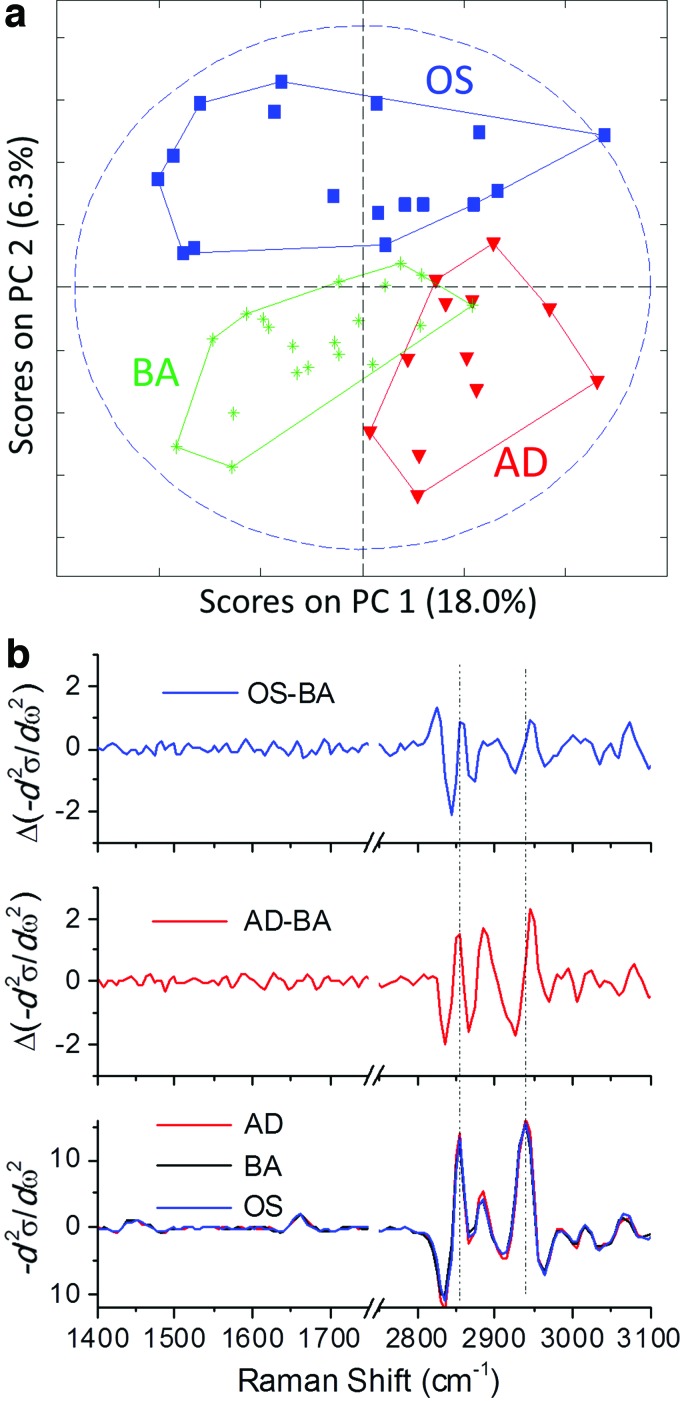
Figure 3a shows results of a PC analysis using spectra from selected regions of cells imaged in this study. In this study, we explore whether spectral differences among the cells can be used to classify them in the absence of the obvious functional markers used in the previous analysis. The input spectra for this analysis were obtained by averaging spectra from cytosol regions in individual cells after removing spectra that showed signs of influence from the functional markers used above—lipid droplets or mineral content. Additionally, when the cytosol area was smaller than 2% of the total imaged area, the cytosol spectra were not considered in this analysis. Because it uses a complimentary set of spectra compared with the previous analysis, the PC analysis used here is an essentially orthogonal characterization. We ensured that the grouping observed in Figure 3a is not a trivial result of differences in the spectral signature of the various media types by noting that BCARS images of hMSCs cultured in the three distinct media for 7 days or less showed no discernable spectral differences.
FIG. 3.
(a) Principal component analysis of Raman spectra averaged in the cytosol region of individual images. Each spectrum was preprocessed by second derivative followed by mean centering. The symbols represent cells in specific culture conditions as follows: blue squares—OS, green cross-stars—BA, and red triangles—AD. The dashed ellipse indicates 95% confidence of the grouping boundaries. (b) Bottom Panel: Second derivative spectra of cytosol regions from AD, OS, and BA samples. Top Panels: Differences in the second derivative spectra between BA and the other two cultures, as indicated. The ordinate scale is a.u. Color images available online at www.liebertpub.com/tec
The result of the PC analysis is consistent with that of the functional marker analysis, but appears to provide some additional information. In Figure 3a, as in Figure 2b, we observe that cells from the three culture conditions are grouped separately, with some overlap between the BA and AD groups. On the other hand, we see no grouping in Figure 3a of cells cultured under OS and BA conditions, and significantly less cogrouping of the cells cultured under AD and BA conditions than in the previous analysis. The fact that the PC analysis better separates these groups suggests that there are differences in the cells cultured in AD and BA media that do not directly or immediately translate to differences in the content of the commonly targeted functional markers, lipid droplets, and mineral. As we note above, it is possible (and seems likely) that some of the stem cells falling in the BA grouping in Figure 2b have committed to the AD or OS line, but not yet started to produce mineral or significant amounts of lipid droplets. The early changes in these differentiating cells seem to be picked up by the PC analysis before they are detected by the functional marker analysis.
The PC analysis can provide direct information as to the origin of these early spectral differences through the PC vectors. However, the fact that we obtain good separation using PCs with significant amplitude suggests that the differences among the various cell spectra are reasonably large, and should be resolvable at the level of overall average spectra. Accordingly, we show average second derivative cytosol spectra for all cells from the AD, BA, and OS culture conditions in the bottom panel of Figure 3b. The top two panels in Figure 3b show differences between the BA and OS or AD spectra. The differences among these spectra are small but significant. The uncertainty in the spectral averages is determined by the random deviations in the fingerprint region of the difference spectra in the top two panels of Figure 3b to be ∼0.2 arbitrary units (a.u.), whereas the differences between the spectra are 10 times larger, on the order of 2 a.u.
The difference spectra, plotted in the top panels of Figure 3b, correspond to changes within the cytosol upon differentiation to AD and OS phenotype. Whereas we do not account for these changes here, we do mention that differentiation into a specific phenotype involves a number of biochemical changes inside cells. Previous spontaneous Raman studies demonstrated that overall biochemical changes during differentiation could be detected in the Raman spectra, and these differences could be attributed to significant changes in the concentration of major species, such as RNA, chymotrypsin, phosphatidyl choline, and cholesterol.33 The spontaneous Raman studies use spectra averaged over entire cells or tissues, while BCARS microscopy provides similar Raman spectra, but makes feasible the imaging of subcellular organelles with the diffraction-limit spatial resolution. Thus, BCARS is sensitive not only to changes in cell chemistry, but also changes in the distribution of cellular components. We suggest that here, relocation of mitochondria could be important since they redistribute from being highly paranuclear for multipotent cells to somewhat spread throughout the cytosol in lineage-committed cells.8
We mention that Raman spectra of the nuclear regions were averaged for each cell and analyzed in the same way as the cytosol spectra in Figure 3. Unlike the cytosol spectra, the PC analysis results of the nucleus spectra do not generate recognizable separation among groups with any possible combinations of the eight largest principal components either with or without second-order differentiation. The lack of spectral differences of nucleus regions among different lineages indicates that the overall chemical composition in the nucleus does not change sufficiently to be detected by BCARS. On the other hand, differentiation does lead to extensive changes in the structural reorganization within the nucleus, including chromatin condensation and nucleoprotein immobilization.10 These changes do not involve major production or consumption of new chemical species in the nucleus, but BCARS may be able to pick up the structural changes for label-free analysis of the shapes associated with a nuclear substructure.
In summary, we have demonstrated that BCARS microscopy can be used as an imaging tool to characterize differentiation lineages of hMSCs cultured in three different (AD, OS, and BA) growth media for 2 weeks. The statistical analysis of more than 100 images reveals large population heterogeneity in the cells through the wide distribution of expression of the lineage-specific functional markers. We have analyzed the spectral image data in two independent ways, by counting image pixels corresponding to lineage-specific functional markers, and by performing a multivariate analysis of Raman spectra of the cytosol. Both approaches provide similar outcomes, with some complimentary information.
These results could not have been obtained by either narrowband (or several-frequency) CARS, or practically by spontaneous Raman imaging; the former does not provide sufficient spectral information, and the latter is too slow to practically image the required number of cells. Our results show that BCARS is sufficiently sensitive to detect subtle changes in cells upon differentiation, and is sufficiently reproducible from day to day that spectral data from individual cells acquired at distinct times can be reliably compared. This work opens the way for application of BCARS to label-free characterization of cell responses to controlled culture conditions, such as in TERM applications, and characterization of cell populations for cell-sourcing applications at the single-cell level. It further opens the possibilities of using spatio-chemical contrast from BCARS imaging in conjunction with morphometric analysis tools that independently have been shown to provide very powerful indicators of cell phenotype.10 Development of reliable spectral markers for mitochondria, actin networks, and various vesicles would improve not only the classification capability, but provide a unique tool for investigating biological mechanisms of cell function and response.
Acknowledgments
The project described was supported by Award Number P41EB001046 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIBIB or the National Institutes of Health. This work is an official contribution of the National Institute of Standards and Technology, and is not subject to copyright in the United States.
Disclosure Statement
No competing financial interests exist.
References
- 1.Gentleman E., Swain R.J., Evans N.D., Boonrungsiman S., Jell G., Ball M.D., et al. . Comparative materials differences revealed in engineered bone as a function of cell-specific differentiation. Nat Mater 8, 763, 2009 [DOI] [PubMed] [Google Scholar]
- 2.James D., Nam H., Seandel M., Nolan D., Janovitz T., Tomishima M., et al. . Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFβ inhibition is Id1 dependent. Nat Biotechnol 28, 161, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shafa M., Sjonnesen K., Yamashita A., Liu S., Michalak M., Kallos M.S., et al. . Expansion and long-term maintenance of induced pluripotent stem cells in stirred suspension bioreactors. J Tissue Eng Regen Med 6, 462, 2012 [DOI] [PubMed] [Google Scholar]
- 4.Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., et al. . Tumour evolution inferred by single-cell sequencing. Nature 472, 90, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notingher I., Jell G., Lohbauer U., Salih V., and Hench L.L.In situ non-invasive spectral discrimination between bone cell phenotypes used in tissue engineering. J Cell Biochem 92, 1180, 2004 [DOI] [PubMed] [Google Scholar]
- 6.Chan J.W., Lieu D.K., Huser T., and Li R.A.Label-free separation of human embryonic stem cells and their cardiac derivatives using Raman spectroscopy. Anal Chem 81, 1324, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ami D., Neri T., Natalello A., Mereghetti P., Doglia S.M., Zanoni M., et al. . Embryonic stem cell differentiation studied by FT-IR spectroscopy. Biochim Biophys Acta 1783, 98, 2008 [DOI] [PubMed] [Google Scholar]
- 8.Lonergan T., Brenner C., and Bavister B.Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J Cell Physiol 208, 149, 2006 [DOI] [PubMed] [Google Scholar]
- 9.Varum S., Rodrigues A.S., Moura M.B., Momcilovic O., Ramalho-Santos J., Van Houten B., et al. . Energy metabolism in human pluripotent stem cells and their differentiated counterparts. Plos One 6, e20914, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Treiser M.D., Yang E.H., Gordonov S., Cohen D.M., Androulakis I.P., Kohn J., et al. . Cytoskeleton-based forecasting of stem cell lineage fates. Proc Natl Acad Sci U S A 107, 610, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan J.W., and Lieu D.K.Label-free biochemical characterization of stem cells using vibrational spectroscopy. J Biophotonics 2, 656, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Notingher I., Bisson I., Bishop A.E., Randle W.L., Polak J.M.P., and Hench L.L.In situ spectral monitoring of mRNA translation in embryonic stem cells during differentiation in vitro. Anal Chem 76, 3185, 2004 [DOI] [PubMed] [Google Scholar]
- 13.Chiang H.K., Peng F.-Y., Hung S.-C., and Feng Y.-C.In situ Raman spectroscopic monitoring of hydroxyapatite as human mesenchymal stem cells differentiate into osteoblasts. J Raman Spectrosc 40, 546, 2009 [Google Scholar]
- 14.Kunstar A., Leijten J., van Leuveren S., Hilderink J., Otto C., van Blitterswijk C.A., et al. . Recognizing different tissues in human fetal femur cartilage by label-free Raman microspectroscopy. J Biomed Opt 17, 116012, 2012 [DOI] [PubMed] [Google Scholar]
- 15.Zumbusch A., Holtom G.R., and Xie X.S.Three-dimensional vibrational imaging by coherent anti-stokes Raman scattering. Phys Rev Lett 82, 4142, 1999 [Google Scholar]
- 16.Evans C.L., and Xie X.S.Coherent anti-stokes Raman scattering microscopy: chemical imaging for biology and medicine. Annu Rev Anal Chem 1, 883, 2008 [DOI] [PubMed] [Google Scholar]
- 17.Konorov S.O., Glover C.H., Piret J.M., Bryan J., Schulze H.G., Blades M.W., et al. . In situ analysis of living embryonic stem cells by coherent anti-stokes Raman microscopy. Anal Chem 79, 7221, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Downes A., Mouras R., Bagnaninchi P., and Elfick A.Raman spectroscopy and CARS microscopy of stem cells and their derivatives. J Raman Spectrosc 42, 1864, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kee T.W., and Cicerone M.T.Simple approach to one-laser, broadband coherent anti-stokes Raman scattering microscopy. Opt Lett 29, 2701, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kano H., and Hamaguchi H.Femtosecond coherent anti-Stokes Raman scattering spectroscopy using supercontinuum generated from a photonic crystal fiber. Appl Phys Lett 85, 4298, 2004 [Google Scholar]
- 21.Lee Y.J., Moon D., Migler K.B., and Cicerone M.T.Quantitative image analysis of broadband CARS hyperspectral images of polymer blends. Anal Chem 83, 2733, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Parekh S.H., Lee Y.J., Aamer K.A., and Cicerone M.T.Label-free cellular imaging by broadband coherent anti-stokes Raman scattering microscopy. Biophys J 99, 2695, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Afonso P.V., Janka-Junttila M., Lee Y.J., McCann C.P., Oliver C.M., Aamer K.A., et al. . LTB4 is a signal-relay molecule during neutrophil chemotaxis. Dev Cell 22, 1079, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kano H., and Hamaguchi H.Supercontinuum dynamically visualizes a dividing single cell. Anal Chem 79, 8967, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Bonn M., Müller M., Rinia H.A., and Burger K.N.J.Imaging of chemical and physical state of individual cellular lipid droplets using multiplex CARS microscopy. J Raman Spectrosc 40, 763, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sisan D.R., Halter M., Hubbard J.B., and Plant A.L.Predicting rates of cell state change caused by stochastic fluctuations using a data-driven landscape model. Proc Natl Acad Sci U S A 109, 19262, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pully V.V., Lenferink A., and Otto C.Raman-fluorescence hybrid microspectroscopy of cell nuclei. Vib Spectrosc 53, 12, 2010 [Google Scholar]
- 28.Bräutigam K., Bocklitz T., Schmitt M., Rösch P., and Popp J.Raman spectroscopic imaging for the real-time detection of chemical changes associated with docetaxel exposure. Chem Phys Chem 14, 550, 2013 [DOI] [PubMed] [Google Scholar]
- 29.Zuser E., Chernenko T., Newmark J., Miljković M., and Diem M.Confocal Raman microspectral imaging (CRMI) of murine stem cell colonies. Analyst 135, 3030, 2010 [DOI] [PubMed] [Google Scholar]
- 30.Pascut F.C., Goh H.T., George V., Denning C., and Notingher I.Toward label-free Raman-activated cell sorting of cardiomyocytes derived from human embryonic stem cells. J Biomed Opt 16, 045002, 2011 [DOI] [PubMed] [Google Scholar]
- 31.Liu Y., Lee Y.J., and Cicerone M.T.Broadband CARS spectral phase retrieval using a time-domain Kramers-Kronig transform. Opt Lett 34, 1363, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pittenger M.F., Mackay A.M., Beck S.C., Jaiswal R.K., Douglas R., Mosca J.D., et al. . Multilineage potential of adult human mesenchymal stem cells. Science 284, 143, 1999 [DOI] [PubMed] [Google Scholar]
- 33.Notingher L., Jell G., Notingher P.L., Bisson I., Tsigkou O., Polak J.M., et al. . Multivariate analysis of Raman spectra for in vitro non-invasive studies of living cells. J Mol Struct 744, 179, 2005 [Google Scholar]