Abstract
Background
Appendage regeneration in amphibians is regulated by the combinatorial actions of signaling molecules. The requirement of molecules secreted from specific tissues is reflected by the observation that the whole process of regeneration can be inhibited if a certain tissue is removed from the amputated stump. Interestingly, urodeles and anurans show different tissue dependencies during tail regeneration. The spinal cord is essential for tail regeneration in urodele but not in anuran larva, whereas the notochord but not the spinal cord is essential for tail regeneration in anuran tadpoles. Sonic hedgehog is one of the signaling molecules responsible for such phenomenon in axolotl, as hedgehog signaling is essential for overall tail regeneration and sonic hedgehog is exclusively expressed in the spinal cord. In order to know whether hedgehog signaling is involved in the molecular mechanism underlying the inconsistent tissue dependency for tail regeneration between anurans and urodeles, we investigated expression of hedgehog signal-related genes in the regenerating tail of Xenopus tadpole and examined the effect of the hedgehog signal inhibitor, cyclopamine, on the tail regeneration.
Results
In Xenopus, sonic hedgehog is expressed exclusively in the notochord but not in the spinal cord of the regenerate. Overall regeneration was severely impaired in cyclopamine-treated tadpoles. Notochord maturation in the regenerate, including cell alignment and vacuolation, and myofiber formation were inhibited. Proliferation of spinal cord cells in the neural ampulla and of mesenchymal cells was also impaired.
Conclusion
As in the axolotl, hedgehog signaling is required for multiple steps in tail regeneration in the Xenopus tadpole, although the location of the Shh source is quite different between the two species. This difference in Shh localization is the likely basis for the differing tissue requirement for tail regeneration between urodeles and anurans.
Keywords: Xenopus, Tail regeneration, Hedgehog, Cyclopamine, Notochord, Spinal cord
Background
Regeneration of amphibian appendages, including the tail, represents a valuable model system to analyze molecular mechanisms underlying cell growth, cell differentiation, morphogenesis and pattern formation in epimorphic regeneration (reviewed in [1-5]). Multiple signaling molecules have critical roles both in anuran and urodele tail regeneration. For example, larval tail regeneration in Xenopus is regulated by bone morphogenetic protein (BMP), Notch, fibroblast growth factors (FGFs), transforming growth factor (TGF)–β, canonical Wnt and non-canonical Wnt [6-12]. Hedgehog signaling is required for axolotl tail regeneration [13]. A significant role of FGFs and Wnts has been suggested by their expression patterns during tail regeneration in adult or larval salamander [14-17]. Therefore, a considerable number of signaling mechanisms are thought to be shared between anurans and urodeles during tail regeneration.
The requirement for signaling molecules in regeneration should be related, at least in part, to the phenomenon of the tissue dependency in appendage regeneration, which has been shown by ablating or destroying a specific tissue in the amputated stump [17-20]. The tissue dependency in tail regeneration is quite different between anurans and urodeles, although both species expressed overlapping set of genes for signaling molecules during tail regeneration. Urodele tail regeneration is completely dependent on the presence of the spinal cord [21], which is now known to be the source of FGFs and sonic hedgehog (Shh), while an ablation of the spinal cord results in a normal or partially defective regenerating tail in anuran larvae [12,18,19]. As hedgehog signalling is essential for overall tail regeneration in urodele [13], the exclusive expression of shh in the spinal cord is likely to be one of major reasons for the requirement of the spinal cord in urodele tail regeneration. It is interesting to know whether hedgehog signal is essential for the tail regeneration in anuran larvae, especially since the presence of the spinal cord is not required for anuran tail regeneration.
In this work we showed that tail regeneration in Xenopus larva is dependent on hedgehog signaling as in axolotl and that shh is expressed in the notochord but not in the spinal cord in the regenerating tadpole tail. Our results suggest that the different tissue specificity of shh expression is the major cause leading to the differences in the tissue dependency between anurans and urodeles.
Results
Expression of genes related to hedgehog signal during tail regeneration
Xenopus laevis have four hedgehog genes, sonic hedgehog (shh), banded hedgehog (bhh), and the closely related cephalic and desert hedgehogs[22]. RT-PCR showed that shh and bhh were expressed in the regenerating tail but cephalic/desert hedgehogs were not (Figure 1A). In axolotl shh is exclusively expressed in the regenerating spinal cord during tail regeneration [13], whereas a previous study suggested that it is expressed in the regenerating notochord but not in the regenerating spinal cord in Xenopus larva [23]. In situ hybridization of whole mount samples and sagittal sections confirmed that shh signal was detected exclusively in the notochord in the entire regenerating region (Figure 1B, C). Expression signal for bhh was not detected in any tissue by in situ hybridization probably due to a low expression level (data not shown). These results show that the hedgehog ligands are produced exclusively or dominantly by the notochord in the regenerating tail in Xenopus in contrast to its exclusive expression in the spinal cord in axolotl tail.
Figure 1.
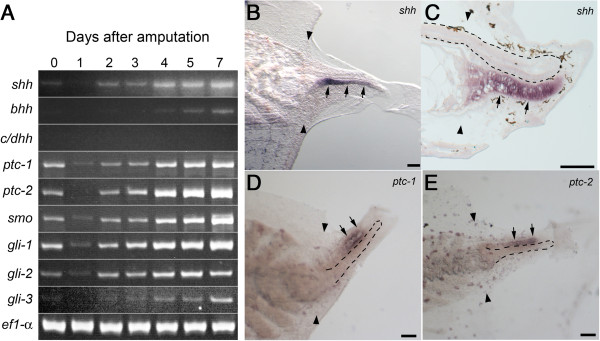
Expression of hedgehog signal-related genes in regenerating tail. (A) Semi-quantitative gene expression analysis by reverse transcription–polymerase chain reaction (RT–PCR) was carried out. RNA was isolated from amputated tails at the indicated day. Specific primer pairs and PCR conditions are indicated in Additional file 2: Table S1. (B-E) Whole mount (B, D, E) and section (C) in situ hybridization of regenerated tail. The spatial expression of shh(B, C)ptc-1(D) and ptc-2(E) was analyzed on the regenerating tail at day 4 (C-E) or day 5 (B). Arrows indicate hybridization signal in the regenerating notochord (B, C) or spinal cord (D, E). A pair of arrowheads marks the amputation plane. Black dashed lines indicate the shapes of the regenerating spinal cord (C) or notochord (D, E). Bars, 100 μm.
RT-PCR showed that two patched genes (ptc-1 and ptc-2), smoothened (smo) and three gli genes (gli-1, gli-2 and gli-3) were also expressed in the regenerating tail (Figure 1A). Whole mount in situ hybridization revealed that ptc-1 and ptc-2 were expressed in the regenerating spinal cord (Figure 1D, E). Hybridization signal in other tissues, for example, notochord and muscle, was indistinguishable from the background staining. Expression of gli genes was not detected in any tissues by in situ hybridization (data not shown). These results support a possibility that hedgehog signal regulates growth and/or differentiation of regenerating tissues, including the spinal cord in Xenopus.
Cyclopamine treatment suppresses tail regeneration in Xenopus
The DV patterning, blastema cell proliferation and cartilage differentiation in the regenerating axolotl tail are severely affected by cyclopamine, a widely used specific inhibitor for hedgehog signaling [13]. To test roles of hedgehog signal in Xenopus tail regeneration, the tail-amputated tadpoles were treated with cyclopamine. Growth of the regenerating tail was significantly retarded by cyclopamine supplied in the breeding water but was not affected by the same dosage of tomatidine, a control compound for cyclopamine (Figure 2A-C, I, Table 1). Similarly the length of the regenerating notochord was reduced by the cyclopamine treatment (Figure 2E-G, J, Table 2). Administration of pumorphamine, an agonist for hedgehog pathway, in the breeding water suppressed the effect of cyclopamine resulting in the nearly normal tail regeneration (Figure 2D, H-J), showing that the defects observed in the cyclopamine-treated tail were caused by the specific inhibition of the hedgehog signal.
Figure 2.
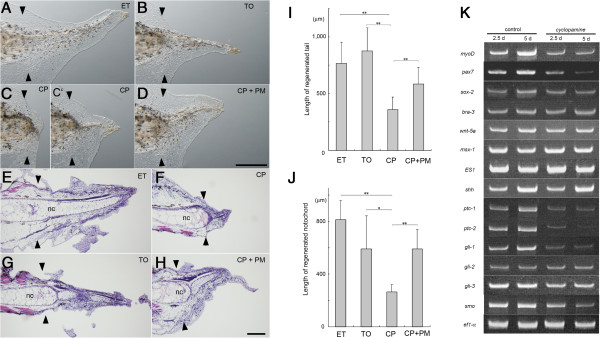
Effect of cyclopamine on the tail regeneration. The tail-amputated tadpole was maintained in the presence of the indicated compound. ET, 0.1% ethanol. TO, 2.5 μM tomatidine. CP, 2.5 μM cyclopamine. CP + PM, 2.5 μM cyclopamine and 0.25 μM purmorphamine. (A-D) Regenerated tails were observed under a stereoscopic microscope at day 7. Bar, 500 μm. (E-H) Histological analysis of the regenerated tail at day 7. Sagittal sections were stained with hematoxylin and eosin. nc, notochord. Bar, 100 μm. A pair of arrowheads marks the amputation plane. (I, J) The length of the regenerated tail and notochord at day 5. Mean length of the regenerated tail (I) or the notochord (J) was indicated with the standard deviation. Detail is shown in Tables 1 and 2. *, p-value < 0.05. **, p-value <0.01. (K) Gene expression analysis with RT-PCR. RNA from the control or cyclopamine-treated tails was analyzed at day 2.5 or day 5.
Table 1.
Effect of cyclopamine on length of regenerating tail
| |
Day 3 |
Day 5 |
||||||
|---|---|---|---|---|---|---|---|---|
| Chemicals | ET | TO | CP | CP + PM | ET | TO | CP | CP + PM |
| Number |
11 |
11 |
12 |
11 |
9 |
9 |
12 |
10 |
| Mean (μm) |
725.3 |
785.2 |
401.2 |
487 |
1528.8 |
1751.6 |
718.4 |
1168.2 |
| SD |
209.3 |
225.7 |
171.9 |
134.1 |
375.2 |
408.1 |
219 |
293.1 |
| P-value (ET) |
- |
0.26 |
0.2 E-4 |
2.4 E-3 |
- |
0.12 |
2.7 E-6 |
1.6 E-2 |
| P-value (TO) |
- |
- |
7.5 E-5 |
6.1 E-4 |
- |
- |
2.2 E-7 |
1.1 E-2 |
| P-value (CP) | - | - | - | 0.1 | - | - | - | 2.7 E-4 |
Tail-amputated st. 48 tadpoles were maintained in water containing ethanol (ET, 0.1%), tomatidine (TO, 2.5 μM) or cyclopamine (CP, 2.5 μM) or cyclopamine plus purmorphamine (PM, 0.25 μM). Mean length of the regenerated tails was determined at day 3 and day 5. Student’s t-test was used to identify differences between indicated groups.
Table 2.
Effect of cyclopamine on length of regenerating notochord
| |
Day 3 |
Day 5 |
||||||
|---|---|---|---|---|---|---|---|---|
| Chemicals | ET | TO | CP | CP + PM | ET | TO | CP | CP + PM |
| Number |
8 |
8 |
8 |
8 |
8 |
8 |
7 |
8 |
| Mean (μm) |
301.5 |
274.5 |
198.5 |
254.9 |
811.3 |
590.7 |
263.3 |
591.9 |
| SD |
81.8 |
78.4 |
28.6 |
59.9 |
147.5 |
251.4 |
56.4 |
146.4 |
| P-value (ET) |
- |
0.26 |
2.3 E-3 |
0.12 |
- |
2.5 E-2 |
2.3 E-7 |
4.9 E-3 |
| P-value (TO) |
- |
- |
1.1 E-2 |
0.3 |
- |
- |
2.6 E-3 |
0.5 |
| P-value (CP) | - | - | - | 0.98 | - | - | - | 4.6 E-5 |
Tail-amputated st. 48 tadpoles were maintained in water containing ethanol (ET, 0.1%), tomatidine (TO, 2.5 μM), cyclopamine (CP, 2.5 μM), or cyclopamine plus purmorphamine (PM, 0.25 μM). Mean length of the regenerated notochord was determined at day 3 and day 5. Student’s t-test was used to identify differences between indicated groups.
TUNEL staining showed apoptotic cells in the distal region of the amputated tail at day 1 to day 3 in both cyclopamine-treated and control tadpoles (Additional file 1: Figure S1). The signals were found mainly in epidermis but rarely in notochord, spinal cord and mesenchymal region. It appeared that cyclopamine do not cause a significant change of the apoptotic cell number in this condition.
Gene expression in the regenerating tail was analyzed by RT-PCR (Figure 2K). Cyclopamine treatment resulted in down-regulation of several component genes for hedgehog signaling cascade, including ptc-1, ptc-2, gli-1 and smo, indicating that these genes are targets of hedgehog signaling, although the expression level of shh was not affected. The down-regulation of ptc-1 is consistent with the reduced expression of ptc-1 in cyclopamine-treated axolotl tails [13]. The cyclopamine treatment down-regulated expression of myoD representing muscle tissue and slightly affected sox-2 representing nerve tissues, although it did not affect bra-3 representing the notochord or genes expressed in a broad range of regenerating tissues, such as msx-1 and wnt-5a. Formation of normal wound epidermis was observed morphologically and confirmed by the unaffected expression of ES-1, a marker for the wound epidermis [11] (Figure 2K).
Cyclopamine inhibits notochord differentiation
In the normal tail regeneration process, undifferentiated notochord cells accumulate at the distal edge of the amputated notochord to make a mass of immature notochord cells at day 2 and proliferate extensively. The cells then align with a perpendicular orientation to the anterior to posterior axis and start to finally differentiate into large vacuole-containing cells after day 3 [5,23]. The cell mass of the notochord was formed in the cyclopamine-treated tail (Figures 3C, 4C, F), but the perpendicular alignment and vacuolation of the notochord cells was poorly accomplished in the cyclopamine-treated tails (Figure 2F, Figure 3C). The proliferation rate of the undifferentiated notochord cells was not affected by cyclopamine (Figure 4C, G, Table 3). These results together with the unaffected bra3 expression (Figure 2K) show that cyclopamine does not affect the early stage of the notochord regeneration, but we concluded that hedgehog signaling is required for a later step of notochord differentiation, including cell alignment and vacuolation.
Figure 3.
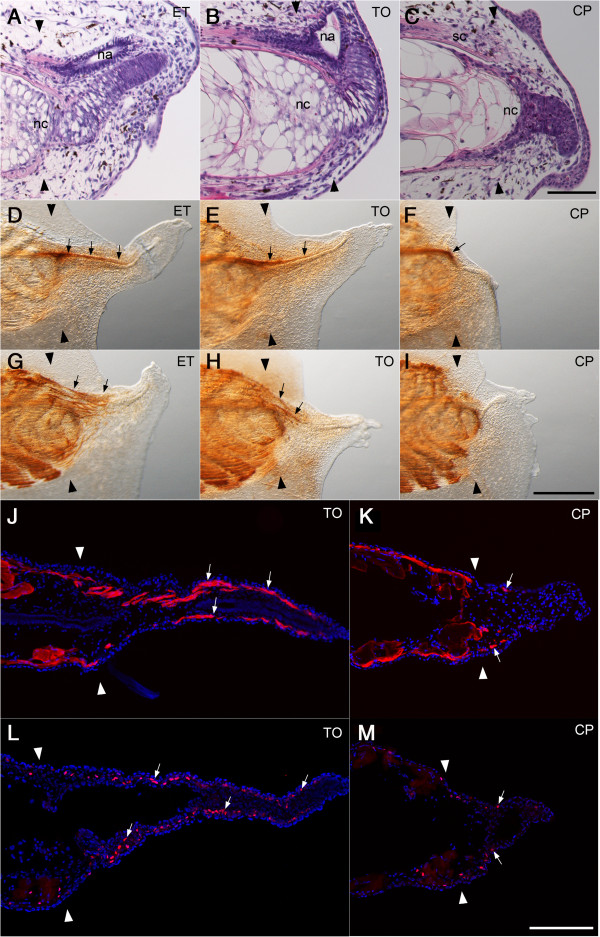
Cyclopamine affects cell differentiation in the regenerating tail. (A-C) Histological analysis of the regenerating tail at day 3. Sagittal sections were stained with hematoxylin and eosin. Cyclopamine suppressed the elongation and the vacuolation of the regenerating notochord cells. It also suppressed the formation of the neural ampulla (C). nc, notochord. na, neural ampulla. sc, spinal cord. Bar, 200 μm. (D-I) Immunohistochemical detection of nerve cells and myofibers in the regenerating tail at day 5. Whole mount sample was immunostained with an anti-NCAM monoclonal antibody (4d) and a monoclonal antibody 12/101 to detect the nerve cells (D-F) and the myofibers (G-I), respectively. Arrows indicate the regenerated spinal cord (D-F) and the regenerated myofibers (G, H). Bar, 500 μm. (J-K) Immunofluorescent detection of muscle cells. The frontal cryosection was immunostained with a monoclonal antibody 12/101 (J, K) or an anti-PAX7 monoclonal antibody (L, M) to detect myofibers (white arrows in J, K) or myoblasts (white arrows in L, M) in the regenerated tail at day 5. Bar, 200 μm. A pair of black and white arrowheads marks the amputation plane.
Figure 4.
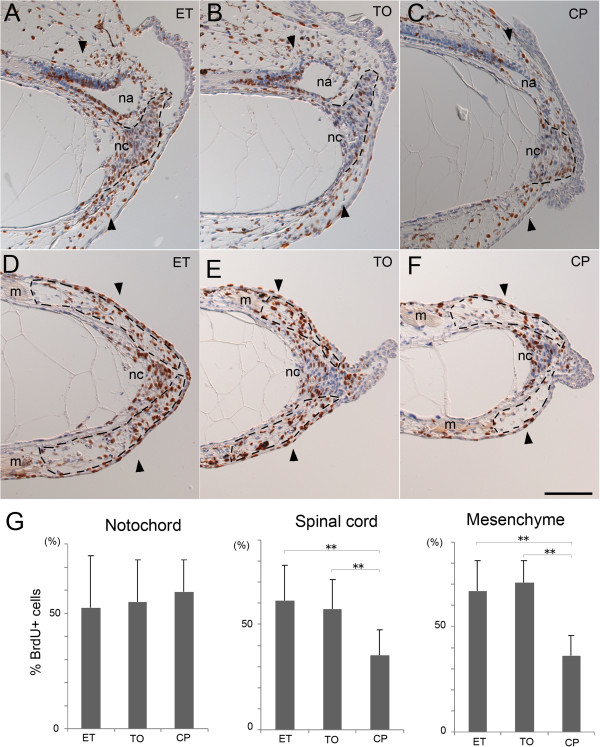
Cyclopamine affects cell proliferation in the regenerating tail. Tail-amputated tadpoles maintained in the presence of the indicated compound were incubated with BrdU for 12 h before fixation. (A-F) Immunohistochemical detection of proliferation cells at day 2.5. BrdU-incorporated cells (brown) were detected on sagittal (A-C) or frontal (D-F) section. Un-labeled nuclei were counter-stained with hematoxylin (blue). Dashed lines indicate the shapes of the regenerating notochords (A-C) and the mesenchymal regions (D-F) containing the myoblasts. nc, notochord. na, neural ampulla of the regenerating spinal cord. m, muscle. A pair of arrowheads marks the amputation plane. Bar, 100 μm. (G) Proliferation rate of cells in the regenerating tail. Mean rate of the BrdU-labeled cells was determined for the indicated tissue and shown with standard deviation. Detail is shown in Table 3. **, p-value <0.01.
Table 3.
Effect of Cyclopamine on proliferation of regenerating cells
|
Tissues |
Notochord |
Spinal cord |
Mesenchyme |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Chemicals | ET | TO | CP | ET | TO | CP | ET | TO | CP |
| % BrdU+ cells |
52.3 |
54.9 |
59.3 |
61.2 |
57.5 |
35.6 |
66.9 |
70.7 |
36.2 |
| SD |
22.5 |
18.2 |
13.8 |
16.7 |
13.6 |
11.6 |
14.5 |
10.5 |
9.6 |
| number of sections |
18 |
20 |
21 |
18 |
16 |
15 |
14 |
27 |
36 |
| p-value (ET) |
- |
0.65 |
0.19 |
- |
0.24 |
1.0 E-5 |
- |
0.83 |
8.4 E-12 |
| p-value (TO) | - | - | 0.81 | - | - | 2.1 E-5 | - | - | 2.3 E-20 |
Tail-amputated st. 48 tadpoles were maintained in water containing ethanol (ET, 0.1%), tomatidine (TO, 2.5 μM) or cyclopamine (CP, 2.5 μM). Four tadpoles for each condition were fixed at day 2.5 after 12 h BrdU labeling. Mean rate of proliferating cells were determined on sagittal (notochord and spinal cord) or frontal (mesenchymal region) sections. Student’s t-test was used to identify differences between indicated groups.
Cyclopamine affects spinal cord regeneration
The spinal cord grows posteriorly in close proximity to the regenerating notochord during tail regeneration. Immunostaining with an anti-NCAM antibody showed an impaired growth of the spinal cord in the cyclopamine-treated regenerated tail (Figure 3F). Expression of sox2, which is abundant in the regenerating spinal cord, is slightly reduced by the cyclopamine treatment (Figure 2K). A sagittal section showed that the neural ampulla of the regenerated spinal cord was small or not formed in the cyclopamine-treated tail (Figures 3C, 4C). Cyclopamine treatment significantly reduced the proliferation rate of cells in the neural ampulla region (Figure 4C, G, Table 3) but not in the proximal region of the spinal cord (data not shown).
Cyclopamine inhibits muscle regeneration
Cyclopamine treatment greatly impaired the myofiber formation in the regenerated tail (Figure 3G-K). It suppressed up-regulation of myoD during tail regeneration (Figure 2K). It is known that Pax7-expressing myogenic cells migrate and proliferate to form new muscle tissue after tail amputation [5,24]. A few myogenic cells labeled with anti-Pax7 antibody were found in the regenerating tail treated with cyclopamine (Figure 3M), while many more Pax7 positive cells are in the control tadpole (Figure 3L). This is consistent with the result of RT-PCR showing that expression of pax7 was down-regulated by cyclopamine treatment (Figure 2K). The proliferation rate of mesenchymal cells was reduced by cyclopamine in the region distal to the damaged muscle where Pax7-positive myogenic cells reside (Figure 4D-G, Table 3). We conclude that the hedgehog signal is required for muscle regeneration in the larval tail.
Discussions
Cyclopamine inhibits tail regeneration of Xenopus tadpole
In this study we showed that cyclopamine treatment resulted in a wide range of cellular defects including proliferation and differentiation during Xenopus tail regeneration. Muscle regeneration was especially inhibited. The fact that the number of Pax-7 positive myogenic cells and myofibers were significantly reduced is consistent with a previous report in which cyclopamine-treatment during axolotl tail regeneration results in a reduced number of Pax7-positive cells in regenerating blastema [13]. Our results suggest that the proliferation of the myogenic cells requires hedgehog signaling. The Pax-7 positive cells in the regenerating tail are derived from the satellite cells residing in the remaining muscle of the Xenopus tadpole tail [5,24,25]. As the promotive effect of Shh on proliferation and differentiation of the satellite cells has been reported in mouse and chicken [26,27], our results suggest that vertebrate muscle regeneration is regulated by a common mechanism involving hedgehog signaling.
We found a different effect of cyclopamine on the spinal cord regeneration between axolotl and Xenopus. Cyclopamine treatment does not affect proliferation of the regenerating spinal cord cells in axolotl tail regeneration [13]. On the other hand, it reduced the proliferation rate of cells in the neural ampulla region of the Xenopus spinal cord. Expression of ptc-1 and ptc-2 in the regenerating spinal cord (see Figure 1D, E) suggests a direct influence of hedgehog signal on spinal cord growth.
Cyclopamine treatment affected late steps of the notochord regeneration including the alignment of the immature cells and their terminal differentiation, although it did not affect the early step including the proliferation of the immature notochord cells. This result may be consistent with a previous report that shh is required for maintenance but not formation of the embryonic mouse notochord [28]. As the regenerating notochord cells express the hedgehog ligand, Shh, the hedgehog signal may regulate differentiation of notochord cells in an autocrine manner. Alternatively, Shh secreted by the notochord may regulate notochord regeneration indirectly by affecting other tissues, for example, the spinal cord that has been shown to be required for correct regeneration of the notochord [12]. The precise role of hedgehog signaling in the notochord cells will be determined using defined experimental systems, for example, culturing isolated notochord cells.
Localized expression of shh causes the different tissue dependency in tail regeneration
Despite the differences in the affected tissues by cyclopamine treatment, this study together with previous reports show that hedgehog signaling is essential for the overall tail regeneration both in urodele and anuran [13]. The critical difference between Xenopus and axolotl is in the tissue-specific expression of shh in regenerating tail: shh is expressed exclusively in spinal cord in axolotl while it is expressed exclusively or predominantly in notochord in Xenopus. Genes for other signaling molecules, including FGFs, Wnts and BMPs, are also required for tail regeneration in Xenopus and are expressed in a broader range of tissues in contrast to shh[7,12,23]. The different location of shh expressing cells is likely to be a main reason underlying the different tissue dependency for tail regeneration between urodele and anuran. shh is expressed in both the ventral spinal cord, namely the floor plate, and the notochord in vertebrate embryos. The original spinal cord in the Xenopus tadpole tail has a floor plate that expresses shh, while the regenerated one has no identifiable floor plate [23]. Therefore, the absence of shh expression in the regenerated spinal cord reflects the incomplete regeneration of the Xenopus spinal cord. Notochord, the main source of Shh in the regenerating Xenopus tadpole tail, is not formed in the regenerated tail of axolotl larva [13]. Regenerating tail in urodele larva forms cartilaginous tissue in place of the notochord, reflecting the normal developmental process in which notochord is replaced by cartilaginous vertebrae. On the other hand cartilage is never formed in the anuran tadpole tail. The difference in tissue-specific expression of shh between axolotl and Xenopus is, therefore, caused by the incomplete spinal cord regeneration in Xenopus and the absence of regenerated notochord in axolotl.
Conclusion
In this study we clearly showed that overall regeneration of the Xenopus tadpole tail is inhibited by cyclopamine, a specific inhibitor for hedgehog signaling, consistent with a previous report using axolotl larva. We also showed that shh is exclusively or predominantly expressed in regenerating notochord but not in spinal cord in Xenopus, while it is expressed exclusively in spinal cord in axolotl. Our findings together with the previous report explain why the tissue dependency for tail regeneration is different between anuran and urodele larvae.
Methods
Surgery and animals
Xenopus laevis larvae were maintained at 22°C in 0.1% NaCl. The developmental stages were determined according to Nieuwkoop and Faber [29]. The stage 49 tadpoles were used for the tail amputation described previously [12]. The animals were handled in with a protocol approved by the animal care and use committee in University of Hyogo.
Treatment with chemicals
Stock solutions (5 mM) of Cyclopamine (Enzo Life Science) and Tomatidine hydrochloride (Enzo Life Science) were prepared with ethanol and added into the feeding water at final concentration of 2.5 μM. Pumorphamine (Calbiochem) was dissolved in dimethyl sulfoxide at 10 mM and used at 0.25 μM. The concentrations used were determined empirically. Tadpoles were transferred into the water containing the chemicals immediately after amputation and the feeding water was replaced every day. Length of regenerated tail was measured from the center of the amputation plane to the distal epidermal tip. Length of regenerated notochord was measured using the different batch of tadpoles.
Histological and immunological analyses
Histological analysis for staining with hematoxylin and eosin was performed as described previously [12]. Procedure for whole mount immunostaining was also described previously [23]. For frozen sections a tadpole were fixed in 7.4% formaldehyde, infiltrated in 18% sucrose, embedded in OCT compound and frozen at -80°C. Cryosection was made with cryostat (Leica CM3050S) and treated with a primary antibody and followed with Alexa Fluor 594-labeled secondary antibody (Molecular probes). Monoclonal antibodies 4d (anti-N-CAM) [30], 12/101 [31] and anti-PAX7 [32] were obtained from the Developmental Studies Hybridoma Bank at the University of Iowa (Iowa City, IA, USA).
Labeling of proliferating cells using bromodeoxyuridine (BrdU) was performed as described previously [12]. BrdU-labeled cells were detected with horseradish peroxidase-labeled secondary antibody and diaminobenzidine (DAB) coloring.
Apoptotic cells were detected by whole mount TUNEL staining using the In Situ Cell Death Detection Kit, POD (Roche Applied Science) according to manufacture’s protocol.
Reverse transcription-polymerase chain reaction (RT-PCR)
The procedure for RNA extraction, cDNA synthesis and polymerase chain reaction was described previously [12]. The reaction conditions were empirically determined in each case within the linear range of amplification. Primers, annealing temperature and cycle number were as described in Additional file 2: Table S1.
In situ hybridization
Procedures for whole mount and section in situ hybridizations were described previously [11,23]. Bluescript plasmids encoding Xenopus laevis shh (xl096o20), patched-1 (xl008p21), patched-2 (xl045b21), gli-1 (xl088k18), gli-2 (xl063k18) and gli-3 (xl074e23) were characterized using the NIBB/NIG/NBRP Xenopus laevis EST database [33]. Complementary DNAs for Xenopus bhh was amplified from cDNA of the taildud embryo using primers (US: 5′-TGGAAGCTGGATTTGACTGGGTCTAC-3′; DS: 5′-GCTATACCAGTGAATGCCCATCTGCT-3′) and cloned into the PCR II-TOPO VECTOR (Life technologies) according to the manufacture’s protocol.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
YT and MM performed the experiments. The paper was written by YT, KW and MM. The content is agreed by all the authors.
Supplementary Material
Primer pairs and cycling conditions of the reverse transcription–polymerase chain reaction.
Apoptotic cells in the regenerating tail. Tail-amputated tadpoles maintained in the presence of 2.5 μM tomatidine (TO, A-F) or 2.5 μM cyclopamine (CP, D-F) were fixed at indicated days. Apoptotic cells were detected by the TUNEL staining (brown dots). Bar, 100 μm.
Contributor Information
Yuka Taniguchi, Email: yuka.taniguchi@crt-dresden.de.
Kenji Watanabe, Email: kirowato@sci.u-hyogo.ac.jp.
Makoto Mochii, Email: mmochii@sci.u-hyogo.ac.jp.
Acknowledgements
We thank Hidekazu Yasudi and Yuriko Nakahata for providing preliminary data for this study. We also thank Drs. Akira Tazaki, Josh Currie and Elly Tanaka for critical reading of the manuscript. This work is supported by JSPS KAKENHI Grant numbers 21570232, 24570239 to MM.
References
- Carlson BM. Principles Of Regenerative Biology. London: Academic; 2007. An Introduction To Regeneration; pp. 1–30. [Google Scholar]
- Beck CW, Izpisúa Belmonte JC, Christen B. Beyond early development: xenopus as an emerging model for the study of regenerative mechanisms. Dev Dyn. 2009;238:1226–1248. doi: 10.1002/dvdy.21890. [DOI] [PubMed] [Google Scholar]
- Slack JMW, Lin G, Chen Y. The Xenopus tadpole: a new model for regeneration research. Cell Mol Life Sci. 2008;65:54–63. doi: 10.1007/s00018-007-7431-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka EM, Reddien PW. The cellular basis for animal regeneration. Dev Cell. 2011;21:172–185. doi: 10.1016/j.devcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochii M, Taniguchi Y, Shikata I. Tail regeneration in the Xenopus tadpole. Dev Growth Differ. 2007;49:155–161. doi: 10.1111/j.1440-169X.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- Beck CW, Christen B, Slack JMW. Molecular pathways needed for regeneration of spinal cord and muscle in a vertebrate. Dev Cell. 2003;5:429–439. doi: 10.1016/s1534-5807(03)00233-8. [DOI] [PubMed] [Google Scholar]
- Beck CW, Christen B, Barker D, Slack JMW. Temporal requirement for bone morphogenetic proteins in regeneration of the tail and limb of Xenopus tadpoles. Mech Dev. 2006;123:674–688. doi: 10.1016/j.mod.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Ho DM, Whitman M. TGF-beta signaling is required for multiple processes during Xenopus tail regeneration. Dev Biol. 2008;315:203–216. doi: 10.1016/j.ydbio.2007.12.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Slack JMW. Requirement for Wnt and FGF signaling in Xenopus tadpole tail regeneration. Dev Biol. 2008;316:323–335. doi: 10.1016/j.ydbio.2008.01.032. [DOI] [PubMed] [Google Scholar]
- Lin G, Chen Y, Slack JMW. Transgenic analysis of signaling pathways required for Xenopus tadpole spinal cord and muscle regeneration. Anat Rec. 2012;295:1532–1540. doi: 10.1002/ar.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura T, Tazaki A, Ueno N, Watanabe K, Mochii M. Xenopus Wnt-5a induces an ectopic larval tail at injured site, suggesting a crucial role for noncanonical Wnt signal in tail regeneration. Mech Dev. 2009;126:56–67. doi: 10.1016/j.mod.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Taniguchi Y, Sugiura T, Tazaki A, Watanabe K, Mochii M. Spinal cord is required for proper regeneration of the tail in Xenopus tadpoles. Dev Growth Differ. 2008;50:109–120. doi: 10.1111/j.1440-169X.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- Schnapp E, Kragl M, Rubin L, Tanaka EM. Hedgehog signaling controls dorsoventral patterning, blastema cell proliferation and cartilage induction during axolotl tail regeneration. Development. 2005;132:3243–3253. doi: 10.1242/dev.01906. [DOI] [PubMed] [Google Scholar]
- Caubit X, Nicolas S, Shi DL, Le Parco Y. Reactivation and graded axial expression pattern of Wnt-10a gene during early regeneration stages of adult tail in amphibian urodele Pleurodeles waltl. Dev Dyn. 1997;208:139–148. doi: 10.1002/(SICI)1097-0177(199702)208:2<139::AID-AJA1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Caubit X, Nicolas S, Le Parco Y. Possible roles for Wnt genes in growth and axial patterning during regeneration of the tail in urodele amphibians. Dev Dyn. 1997;210:1–10. doi: 10.1002/(SICI)1097-0177(199709)210:1<1::AID-AJA1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Zhang F, Clarke JD, Ferretti P. FGF-2 Up-regulation and proliferation of neural progenitors in the regenerating amphibian spinal cord in vivo. Dev Biol. 2000;225:381–391. doi: 10.1006/dbio.2000.9843. [DOI] [PubMed] [Google Scholar]
- Zhang F, Clarke JD, Santos-Ruiz L, Ferretti P. Differential regulation of fibroblast growth factor receptors in the regenerating amphibian spinal cord in vivo. Neuroscience. 2002;114:837–848. doi: 10.1016/s0306-4522(02)00321-4. [DOI] [PubMed] [Google Scholar]
- Goldfarb AJ. The influence of the nervous system in regeneration. J Exp Zool. 1909;7:643–722. [Google Scholar]
- Goss RJ. Principles of regeneration. New York: Academic; 1969. [Google Scholar]
- Géraudie J, Ferretti P. Gene expression during amphibian limb regeneration. Int Rev Cytol. 1998;180:1–50. doi: 10.1016/s0074-7696(08)61769-6. [DOI] [PubMed] [Google Scholar]
- Holtzer H, Holtzer S, Avery G. An experimental analysis of the development of the spinal column. IV. Morphogenesis of tail vertebrae during regeneration. J Morph. 1955;96:145–168. [Google Scholar]
- Takabatake T, Ogawa M, Takahashi TC, Mizuno M, Okamoto M, Takeshima K. Hedgehog and patched gene expression in adult ocular tissues. FEBS Lett. 1997;410:485–489. doi: 10.1016/s0014-5793(97)00645-5. [DOI] [PubMed] [Google Scholar]
- Sugiura T, Taniguchi Y, Tazaki A, Ueno N, Watanabe K, Mochii M. Differential gene expression between the embryonic tail bud and regenerating larval tail in Xenopus laevis. Dev Growth Differ. 2004;46:97–105. doi: 10.1111/j.1440-169x.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lin G, Slack JMW. Control of muscle regeneration in the Xenopus tadpole tail by Pax7. Development. 2006;133:2303–2313. doi: 10.1242/dev.02397. [DOI] [PubMed] [Google Scholar]
- Gargioli C, Slack JMW. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–2679. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- Koleva M, Kappler R, Vogler M, Herwig A, Fulda S, Hahn H. Pleiotropic effects of sonic hedgehog on muscle satellite cells. Cell Mol Life Sci. 2005;62:1863–1870. doi: 10.1007/s00018-005-5072-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia D, Madhala D, Ardon E, Reshef R, Halevy O. Sonic hedgehog promotes proliferation and differentiation of adult muscle cells: Involvement of MAPK/ERK and PI3K/Akt pathways. Biochim Biophys Acta. 2007;1773:1438–1446. doi: 10.1016/j.bbamcr.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal table of Xenopus laevis (daudin): a systematical and chronological survey of the development from the fertilized Egg till the End of metamorphosis. New York: Garland; 1956. [Google Scholar]
- Watanabe M, Frelinger AL 3rd, Rutishauser U. Topography of N-CAM structural and functional determinants. I. Classification of monoclonal antibody epitopes. J Cell Biol. 1986;103:1721–1727. doi: 10.1083/jcb.103.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies identify blastemal cells derived from dedifferentiating limb regeneration. Nature. 1984;308:67–69. doi: 10.1038/308067a0. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Kimura-Kawakami M, Nomura T, Fujisawa H. Distributions of PAX6 and PAX7 proteins suggest their involvement in both early and late phases of chick brain development. Mech Dev. 1997;66:119–130. doi: 10.1016/s0925-4773(97)00097-x. [DOI] [PubMed] [Google Scholar]
- NIBB/NIG/NBRP Xenopus laevis EST database (XDB3) [ http://xenopus.nibb.ac.jp/]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primer pairs and cycling conditions of the reverse transcription–polymerase chain reaction.
Apoptotic cells in the regenerating tail. Tail-amputated tadpoles maintained in the presence of 2.5 μM tomatidine (TO, A-F) or 2.5 μM cyclopamine (CP, D-F) were fixed at indicated days. Apoptotic cells were detected by the TUNEL staining (brown dots). Bar, 100 μm.


