Abstract
The recent synthesis of pyrimidine ribonucleoside-2′,3′-cyclic phosphates under prebiotically plausible conditions has strengthened the case for the involvement of RNA at an early stage in the origin of life. However, a prebiotic conversion of these weakly activated monomers, and their purine counterparts, to the 3′,5′-linked RNA polymers of extant biochemistry has been lacking – previous attempts leading only to short oligomers with mixed linkages. Here we show that the 2′-hydroxyl group of oligoribonucleotide-3′-phosphates can be chemoselectively acetylated in water under prebiotically credible conditions, allowing rapid and efficient template-directed ligation. The 2′-O-acetyl group at the ligation junction of the product RNA strand can be removed under conditions that leave the internucleotide bonds intact. Remarkably, acetylation of mixed oligomers possessing either 2′- or 3′-terminal phosphates is selective for the 2′-hydroxyl group of the latter. This newly discovered chemistry thus suggests a prebiotic route from ribonucleoside-2′,3′-cyclic phosphates to predominantly 3′,5′-linked RNA via partially 2′-O-acetylated-RNA.
RNA is postulated to have had a catalytic role during the origin of life in addition to its role as an information carrier1-4. Numerous studies have demonstrated the catalytic capacity of RNA, but the question as to how RNA might have first arisen has remained largely unanswered. Our recent demonstration of an efficient, predisposed synthesis of pyrimidine ribonucleoside-2′,3′-cyclic phosphates (N>P’s; see Supplementary Fig. S1 for a key to abbreviations)5,6 is contributory experimental evidence that these RNA building blocks might have existed on early Earth solely as a consequence of prebiotic chemistry. However, before RNA itself can be shown to be a potentially prebiotic product, a route to the purine ribonucleotides, and a means of converting the full set of N>P’s to the polymer must also be demonstrated.
The direct polymerisation of N>P’s is thermodynamically unfavourable and kinetically sluggish because the 2′,3′-cyclic phosphate ring is only slightly activated towards nucleophilic ring-opening by a hydroxyl group7. These fundamental limitations pertain whatever the reaction phase, but oligomerisation in water suffers additionally because pseudo-first-order monomer hydrolysis – to 2′- and 3′-monophosphates (N2′P’s and N3′P’s)8 – is inherently faster than second-order oligomerisation. Thus, in earlier work it was shown that oligomerisation of A>P only occurred productively in the dry-state with prebiotically plausible general acid/base catalysts9. Reasonable yields of oligomers up to the hexamer were obtained with traces of longer products - maximally fourteen nucleotides long10. The dry-state mixtures were prepared by drying down solutions, and during this process some 2′,3′>P hydrolysis took place such that the final oligomers products terminated in 2′- and 3′-monophosphates (2′/3′P). Lacking inherent regiocontrol, these non-templated reactions resulted in both 2′,5′- and 3′,5′-phosphodiester linkages in a ratio of approximately 1:29.
One potential route to longer RNA strands is through the templated ligation of these short oligomers. However, activation of the mixed 2′/3′P termini with an electrophile would rapidly generate a 2′,3′>P terminus – owing to the high effective molarity of the adjacent 3′/2′-hydroxyl group (3′/2′-OH) – before ligation with the 5′-OH group of a neighbouring oligomer could occur. The template-induced proximity effect should accelerate strand ligation over hydrolysis, and chelation by the template should additionally improve the thermodynamic favourability of the reaction. However, templated ligation of oligomers with 2′,3′>P termini, though accelerated, is still very slow and low yielding over a period of days to weeks11,12, and the newly formed junctions are almost exclusively 2′,5′-phosphodiester linkages11,12. Taken with the lack of regiocontrol of non-templated N>P oligomerisation, this has suggested that prebiotic ribonucleic acids would have had mixed 2′,5′- and 3′,5′-linkages. However, it is not clear what degree of linkage isomer heterogeneity will allow evolution to take place – a certain amount might be tolerated for any particular genetic sequence if those isomers that adopt the structure required for a selectable phenotype constitute a big enough fraction of the overall population13. Whatever the case, the evolutionary transition to exclusively 3′,5′-linked extant RNA is likely to be easier if the starting point is RNA significantly enriched in 3′,5′-linkages, and mechanisms for this have thus been sought.
One factor that might have contributed to such enrichment is the increased stability of 3′,5′-linked duplexes relative to 2′,5′-linked duplexes14, which should favour the templated ligation of 3′,5′-linked oligomers. Another factor is the preferential hydrolysis of 2′,5′-over 3′,5′-phosphodiester bonds in the context of a right-handed double-helix 15,16, wherein 2′,5′-linkages hydrolyse between two and three orders of magnitude faster than 3′,5′-linkages16,17. However, in the absence of a nick repair mechanism, selective hydrolysis can only bring about enrichment for 3′,5′-linkages at the expense of chain cleavage. Thus, although there are tantalising hints to the contrary, some degree of linkage isomer heterogeneity has appeared inevitable in the self-assembly of ribonucleic acid of useful length from N>P’s.
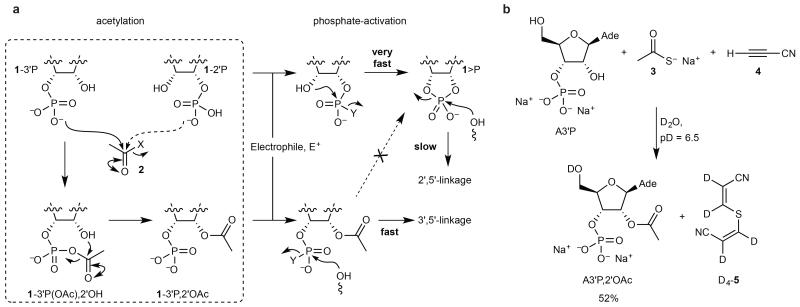
In conventional synthetic RNA chemistry, 2′-OH protecting groups ensure efficient chain extension and regiocontrol18,19, and we were intrigued by the possibility of achieving similar protection under prebiotically plausible conditions. This might then enable efficient templated ligation because electrophilic activation of a terminal phosphate would permit direct reaction with a neighbouring oligomer by preventing cyclisation to a 2′,3′>P. Although subsequent deprotection would then be required to liberate unmodified RNA, such chemistry could overcome the kinetic and thermodynamic barriers inherent to the ligation of 2′,3′>P’s. Furthermore, any selectivity for the protection of 2′-OH over 3′-OH groups (as shown for a(n) (oligo)ribonucleotide with mixed terminal 2′- and 3′-phosphates, 1-3′/2′P, in Fig. 1a) would enrich the ligation junctions in 3′,5′-linkages. We have now found a way to effect such protection and ligation, with an attendant selectivity for 3′,5′-linkages. The protection reaction was discovered through a systems chemistry approach and simply involves the reaction of 1-3′/2′P with an acetylating agent 2 in water at near-neutral pH.
Figure 1. Chemoselective acetylation of RNA.
a, Protection of the 2′-OH group of 1-3′P facilitates rapid template-directed 3′,5′-ligation following electrophilic phosphate activation. The 3′-OH group of 1-2′P is protected to a lesser extent such that 1>P is the major product of phosphate activation and slow template-directed 2′,5′-ligation follows. b, Treatment of adenosine-3′-phosphate (A3′P, 100 mM) with sodium thioacetate 3 (100 mM) and cyanoacetylene 4 (200mM) in D2O at neutral pD for 24 h results in selective acetylation of the 2′-OH group. Curly arrows indicate electrophilic activation/acetylation steps. X, leaving group; Y, leaving group generated by electrophilic activation of phosphate oxygen, with or without a subsequent nucleophilic displacement; Ade, N9-linked adenine; yield judged by 1H-NMR integration.
Results and Discussion
Chemoselective acetylation of nucleotides
We decided to use adenosine-3′-phosphate (A3′P, Fig. 1b & Supplementary Fig. S1) as a surrogate for a 2′/3′P terminated oligomer mixture in our screen for a prebiotic protection reaction, for reasons of economy and ease of screening by 1H-NMR spectroscopy. We were drawn to acetyl as a protecting group: it functions well in conventional synthesis18,19; thioacetate 3 (Fig. 1b) is considered to be prebiotically available via NiS-catalysed reaction of CO and H2S20,21; and subsequent electrophilic22,23 or oxidative24 activation of 3 should generate an acetylating agent. Accordingly, we explored the systems chemistry of mixtures of A3′P, thioacetate 3 and prebiotically plausible electrophiles. We discovered the reaction we sought with the first electrophile we tried, cyanoacetylene 4 – selected because of its role as a nucleobase precursor in our pyrimidine N>P synthesis5. Thus, treatment of a solution of the sodium salt of A3′P and sodium thioacetate 3 with cyanoacetylene 4 in D2O (to facilitate 1H-NMR spectroscopic analysis of reaction products) at pD = 6.5 resulted in selective acetylation of the 2′-OH group, giving 2′-O-acetyladenosine-3′-phosphate (A3′P,2′OAc) in 52% yield within 24 h (Fig. 1b and Supplementary Table S1). A precipitate which formed rapidly at the outset of the reaction proved to be tetradeuterio-β,β-dicyanovinyl-thioether D4-5.
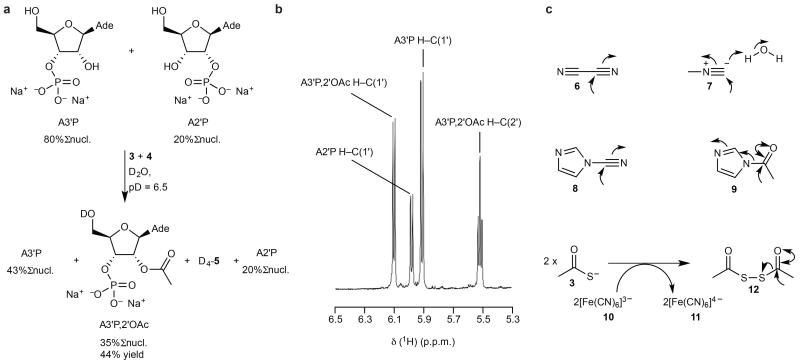
Extension of this chemistry to the other nucleoside-3′-phosphates (N3′P′s: C3′P, U3′P, G3′P, and I3′P; Supplementary Table S1) revealed selective 2′-OH group acetylation in all cases, and in no case was any nucleobase amino group acetylation observed. The reaction was most efficient with the purine N3′P′s, and direct competition experiments between different N3′P′s revealed the following rough reactivity trend: A3′P ≃ I3′P > G3′P > C3′P > U3′P. We then explored the chemistry with N2′P′s (Supplementary Table S1) and found that 3′-OH group acetylation occurred, but with significantly reduced efficiency. Furthermore, relative to the previous experiments with N3′P′s, an increased amount of cyclisation to N>P’s was observed. The reduced efficiency of acetylation of the 3′-OH group of N2′P’s results in the selective acetylation of the 2′-OH group of N3′P’s in mixtures of N2′P’s and N3′P’s (Supplementary Table S1). Thus, for example, in a 4:1 mixture of A3′P and A2′P, 44% of the A3′P is converted to the 2′-acetate but acetylation of the A2′P is undetectable by 1H-NMR spectroscopic analysis (Fig. 2a and b).
Figure 2. Chemoselective acetylation: mixtures and alternative electrophiles.
a, Treatment of A3′P (80 mM) and A2′P (20 mM) as above (Fig. 1b) results in the exclusive 2′-acetylation of the former nucleotide. b, Partial 1H-NMR spectrum of the reaction products described in a. c, Additional electrophiles (6-8) shown to drive the acetylation of ribonucleotides with thioacetate 3. Direct acetylation with 9 is also possible, as is oxidative activation of 3 with ferricyanide 10 to afford ferrocyanide 11 and a dimeric acetylating agent 12. Curly arrows indicate electrophilic activation/acetylation steps. Ade, N9-linked adenine; %Σnucl., yields expressed as percentage of total nucleotide, otherwise yield of product expressed as a percentage of the specific starting material from which it derives (as judged by 1H-NMR integration).
Electrophiles other than cyanoacetylene 4 also functioned efficiently and selectively in the reaction (Fig. 2c). Treatment of A3′P with thioacetate 3 and cyanogen 6 gave A3′P,2′OAc in 51% yield as the only nucleotide product. Methyl isocyanide 7 brought about the efficient acetylation of the 2′-OH group of N3′P’s by 3, although with increased yields of accompanying N>P (Supplementary Table S23). N-cyanoimidazole (NCI) 8 – which we later used as a phosphate activation agent in ligation experiments – also brought about efficient nucleotide acetylation using 3 (Supplementary Table S24), likely through the intermediacy of N-acetylimidazole (NAI) 9. Oxidative activation was also effective; a combination of thioacetate 3 and ferricyanide 10 afforded high yields of A3′P,2′OAc, but low yields of A2′P,3′OAc (Supplementary Table S26). Direct acetylation without using 3 was possible with N-acetylimidazole (NAI) 9 (Supplementary Table S27), and we later used this reagent extensively as a generic prebiotic acetylating agent. We note also that the chemoselective acetylation described might extend to other acylating reagents, as has already been reported for aminoacylation with N-carboxyanhydrides25.
The chemistry (as shown for a(n) (oligo)ribonucleotide-3′-phosphate, 1-3′P in Fig. 1a and Supplementary Fig. S12) is thought to proceed by an acyl-transfer cascade via a mixed carboxy-phosphate anhydride. Thus, an acetylating agent reacts with the phosphate dianion of 1-3′P to give 1-3′P(OAc),2′OH which then rearranges to give 1-3′P,2′OAc. In support of the proposed intermediacy of mixed carboxy-phosphate anhydrides, we found that when methyl phosphate was treated with thioacetate 3 and cyanoacetylene 4, acetyl methyl phosphate was formed and persisted for several days - there being no proximal intramolecular hydroxyl group to allow rearrangement to an acetate ester (Supplementary Tables S28 & S29). This mechanism contrasts with the direct acylation of the 2′-OH groups of flexible nucleotides that has been extensively studied by Weeks and colleagues and applied to the ‘SHAPE’ (i.e. ‘selective 2′-hydroxyl acylation and primer extension’) analysis of nucleotide structure and dynamics26,27.
A clear-cut explanation for the noteworthy selective acetylation of the 2′-OH group of N3′P’s in mixtures of N2′P’s and N3′P’s is lacking, but several potential contributory factors can be identified. Firstly, the phosphate dianion, rather than the monoanion, is the presumed nucleophile, and the reaction pH (or pD) is close to the pKa of a generic monoalkylphosphate monoanion. The pKa of a 3′-phosphate is ca. 0.2-0.5 units lower than that of a 2′-phosphate, and these small differences might influence selectivity28,29. Secondly, the acetylation of N2′P’s gives more N>P’s than does the acetylation of N3′P’s, suggesting that the intermediate carboxy-phosphate anhydrides behave differently. These anhydrides are ambident electrophiles, and this is more clearly manifest for N2′P’s, wherein the 3′-OH attacks either carbon or phosphorus, than for N3′P’s, wherein the 2′-OH almost exclusively attacks carbon.
Oligomer acetylation and regioselective ligation
We next sought to apply this aqueous acetylation chemistry to oligomers of the kind generated by dry-state reactions of N>P’s. For simplicity we investigated reactions of all 3′,5′-linked oligomers. The capacity of this chemistry for selective acetylation of oligomers was first assessed by 1H-NMR spectroscopy using a trimer, AGA3′P; acetylation with thioacetate 3 and NCI 8 gave AGA3′P,2′OAc in a remarkable 64% yield along with 25% AGA>P (Supplementary Table S24 and Fig. S13). No acetylation of the two internal 2′-OH groups or nucleobase exocyclic amino functionality was observed. In addition, the terminal 2′-OH group of AGA3′P was more efficiently acetylated than the terminal 3′-OH group of AGA2′P in a competition experiment (Supplementary Table S24). To further probe the 3′/2′-selectivity, a 1.6:1 mixture of CC3′P and CC2′P was treated with one equivalent of thioacetate 3 and cyanoacetylene 4, which gave CC3′P,2′OAc in 20% yield along with CC>P in 4% yield, but importantly with no detectable CC2′P,3′OAc (Supplementary Table S1). This result also suggested that the nature of the 3′-terminal nucleobase has an effect on the efficiency of the reaction, and that the purine > pyrimidine reactivity trend observed for monoribonucleotides also extends to oligoribonucleotides.
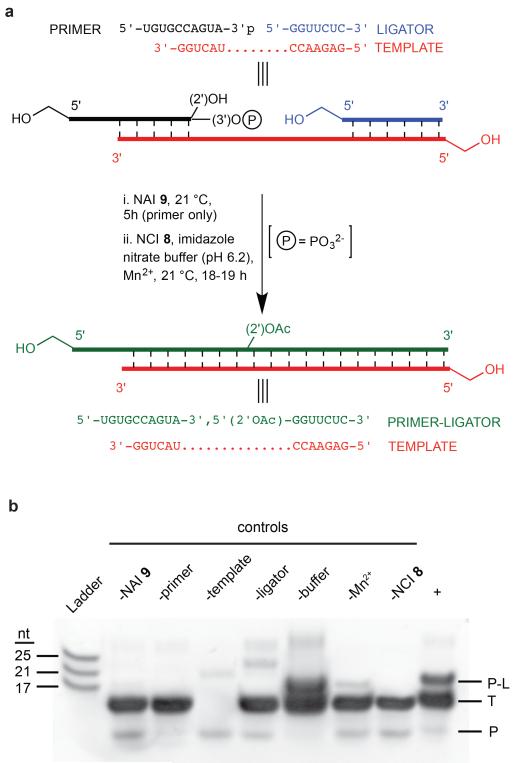
To determine whether or not the selective acetylation of 3′P-terminated RNA strands would expedite their templated ligation, oligomer sequences were chosen which would later allow direct comparison with previous work by Szostak and co-workers, who described the extremely slow but highly 3′,5′-selective templated ligation between a 5′-overhanging primer bearing a 2′,3′-diol and a 5′-triphosphate activated ligator17,30. Thus, an upstream 10 ribonucleotide (nt) ‘primer’ RNA strand terminating in adenosine-3′-phosphate was treated with NAI 9, then mixed with a downstream 7 nt ‘ligator’ strand and 13 nt ‘template’ RNA, and the resultant gapped duplex activated for ligation with NCI 8 (Fig. 3a). Mass spectrometry of the acetylated primer before ligation indicated that > 70% of the oligomer was 2′-O-acetylated (estimated from peak integrals; see Supplementary Fig. S14 & Table S31). Denaturing gel electrophoresis of the ligation reaction showed the successful joining of primer and ligator to yield the 17 nt product (Fig. 3b), with control reactions demonstrating that acetylation and subsequent phosphate activation are essential for efficient ligation.
Figure 3. Chemoselective acetylation of 3′P oligoribonucleotides expedites templated ligation.
a, Sequences and reaction conditions employed for acetylation (i) and subsequent templated ligation (ii). The acetylation mixture contained 80 μM primer and 50 mM NAI 9; the ligation mixture contained 4 μM primer from the acetylation reaction, 25 μM template, 30 μM ligator, 200 mM imidazole nitrate buffer (pH 6.2), 10 mM MnCl2, and 100 mM NCI 8. Ligation conditions were based on those reported previously for the conversion of A3′P to A>P35, and for the ligation of oligomers with 5′P and 2′,3′-diol termini36,37. b, Denaturing PAGE analysis of a ligation reaction (+), and controls without the reaction component indicated. The gel was imaged by UV transillumination after treatment with SYBR® Gold nucleic acid gel stain, which does not reveal the ligator strand. MALDI-TOF MS further evidenced the formation of the monoacetylated 17 nt ligation product isolated from an excised gel band: (m/z) [M+H]+ calcd average mass for C162H202O122N59P16 found 5422.42; requires 5423.24. NAI, N-acetylimidazole; NCI, N-cyanoimidazole; P, 10 nt primer; T, 13 nt template; P-L, 17 nt primer-ligator ligation product.
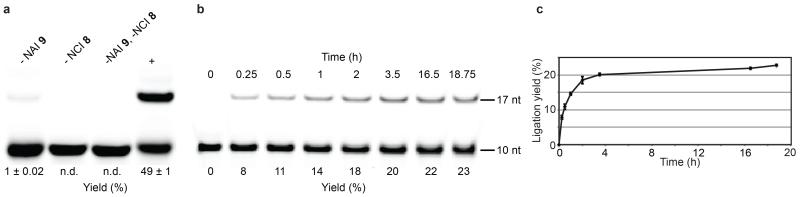
In order to quantify the ligation product, further reactions were performed with a primer fluorescently labelled at the 5′-terminus. A 49% yield of the 17 nt ligation product was apparent after 19 h (Fig. 4a). This contrasted with a control reaction without acetylation; in this case, an average yield of 1% ligated material was evident, likely possessing a 2′,5′-linked ligation junction derived from the slow reaction of the 2′,3′>P terminated primer. Ligation was also promoted when the primer was acetylated as part of a preformed gapped duplex; in this case yields were lower (23 %, Fig. 4b and c) than had been observed when the primer was acetylated in isolation, but still an order of magnitude higher than for the ligation of 2′,3′>P RNA. While we do not attempt a detailed analysis of the kinetics for the templated ligation of 2′,3′>P as compared to 3′P,2′OAc oligoribonucleotides, it is clear that, upon electrophilic activation, the acetylated material ligates at a far greater rate.
Figure 4. Quantification of ligation products.
a, Deanturing PAGE analysis of the templated ligation (+) of the fluorescently 5′-(6FAM)-labelled 10 nt primer and 7 nt ligator (concentrations as per Fig. 3a), with controls lacking either NAI 9, NCI 8, or both. Only labelled primer and products deriving therefrom are visible. Yields are presented as mean ± s.d.; n = 3 (controls) or 6 (+). b, Time-course gel analysis for the ligation of the fluorescently-labelled primer that has been acetylated in the presence of the template and ligator strands. Concentrations for the acetylation were 6 μM primer, 38 μM template, 46 μM ligator and 50 mM NAI 9. Ligation reaction concentrations were as above (Fig. 3a). Yields are averages of three reactions. c, Graph derived from the time-course analysis (b) depicting the development of ligation product with time. Error bars are ± s.d.; n = 3. Yields were obtained by fluorescence scanning; n.d. not detectable. NAI, N-acetylimidazole; NCI, N-cyanoimidazole.
To assess the relative selectivity for ligation of 2′P and 3′P terminated oligoribonucleotides, ligation reactions were performed using the original 10 nt 3′P primer and a 7 nt 2′P primer, separately and in competition (Fig. 5a and b). The shorter (one nt) 5′-overhang of the 7 nt primer served to permit resolution of the ligation products by gel electrophoresis and mass spectrometry, and a fluorescent label at the 3′-terminus of the 7 nt ligator allowed quantitation of reaction products. To control for the effect of different 5′-overhangs on ligation efficiency, the reactions were also performed with a 10 nt 2′P primer, and a 7 nt 3′P primer. Mass spectrometric analysis revealed the selective acetylation of those primers bearing a 3′P (Supplementary Fig. S15); estimates from integrated peak areas consistently showed a 2- to 3-fold greater yield for acetylation of the 3′P primer (Supplementary Fig. S16). Gel analysis and quantitation revealed selective ligation of the acetylated 3′P primers; yields were up to 700-fold higher for 3′P primers than 2′P primers, including when the primers were in direct competition (Fig. 5c). Ligation yields were higher for 7 nt primers than for 10 nt primers, and we attribute this to the formation of primer dimers for the latter (see Supplementary Fig. S17), though we have not investigated why longer 10 nt primers might dimerise more efficiently than their shorter 7 nt counterparts.
Figure 5. Chemoselective acetylation favours ligation of 3′P oligomers over 2′P oligomers.
a, Denaturing PAGE analysis of reactions to assess ligation selectivity. The gel was imaged by fluorescence scanning (top), before it was stained with SYBR® Gold and imaged by UV transillumination (bottom) to reveal the 13 nt template (primers could not be detected). Unreacted 7 nt (dye-labelled) ligator is also present in the right-hand lane. The acetylation mixtures contained 40 μM of (each of) the indicated primer(s) and 50 mM NAI 9; the ligation mixture contained 4 μM (of each) primer from the acetylation reaction, 4 μM template, 4 μM 3′-(6FAM)-labelled ligator, 200 mM imidazole nitrate buffer (pH 6.2), 10 mM MnCl2, and 100 mM NCI 8. b, Sequences of oligomers used to assess linkage selectivity are shown (top), and an illustration of the acetylation-ligation reactions conducted with each primer pair is given (bottom), depicting the preferential ligation of 3′P oligomers. c, Mean yields (± s.d.; n = 3) of ligation products determined by fluorescence scanning. n.d. not detectable; the ligation yield in this case was below the detection limit of the fluorescence scanner when gel loading was reduced to prevent detector saturation and allow accurate quantification.
RNA molecules produced by the ligation chemistry described require a deacetylation step to furnish native RNA. Per-2′-O-acetylated RNA has been described in the patent literature31, and ammonolysis or hydrolysis without phosphodiester backbone cleavage was reported. These observations suggested that partially 2′-O-acetylated RNA, as produced by acetylation-ligation chemistry, would also be convertible to RNA. A 13 nt RNA strand possessing a 2′-O-acetyl group at an internal junction (5′-GCAGUA(2′OAc)GGUUCUC-3′, prepared using a solid-phase synthesis protocol and phosphoramidites newly developed in our laboratory, manuscript in preparation) was therefore subjected to ammonolysis and the products were examined by MALDI-TOF MS. Near complete removal of the acetyl group without conspicuous backbone hydrolysis was effected within 1 h by aqueous (~ 5 M) ammonia at pH 9.2 and 40 °C (Supplementary Fig. S18a-d). These conditions were chosen for expediency, but more prebiotically plausible lower concentrations and lower temperatures were also conducive to the (slower) removal of residual acetates (Supplementary Fig. S18e,f). Importantly, this is clear demonstration that acetyl group removal is substantially faster than the hydrolysis of extant 3′,5′-phosphodiester bonds.
Although it seemed likely that acetylation-ligation of 3′P and 2′P oligomers would generate 3′,5′- and 2′,5′-linked ligation junctions respectively, we sought experimental proof for this. Thus, acetylation-ligation products for 10 nt 2′P and 3′P (non-labelled) primers were desalted and ammonolysed (MALDI-TOF MS of the desalted 10 nt 3′P reaction mixture was again consistent with the presence of a 17 nt monoacetylated ligation product, and ammonolysis resulted in loss of 42 Da as expected for deacetylation; see Supplementary Fig. S19). HPLC analysis, with and without co-injections of synthetic standards, revealed formation of the expected linkage isomers (Fig. 6 & Supplementary Fig. S20). Further HPLC of 17 nt synthetic standards acetylated at a position corresponding to the ligation junction (both 2′,5′- and 3′,5′-linkage isomers prepared by solid-phase synthesis, manuscript in preparation), pre- and post-ammonolysis in the presence of the complementary 13 nt template, demonstrated that 3′,5′-phosphodiester bonds were stable to the deacetylation conditions, whereas the single 2′,5′-junction was susceptible to hydrolysis, but at a rate lower than that of the deacetylation (Supplementary Fig. S21). It has previously been proposed that prebiotic RNA synthesis gave molecules possessing a mixture of 2′,5′- and 3′,5′-linkges, which gradually ‘evolved’ to enrich for 3′,5′-bonds that are more resistant to hydrolysis within a duplex. Our findings suggest that geologic conditions that could enable this evolution would also result in deacetylation of partially 2′-O-acetylated RNA as produced by the acetylation-ligation chemistry we have described. Native 3′,5′-linked RNA would thus persist beyond deacetylation and 2′,5′-linkage hydrolysis, providing a time window for any catalytic properties of such RNA to manifest.
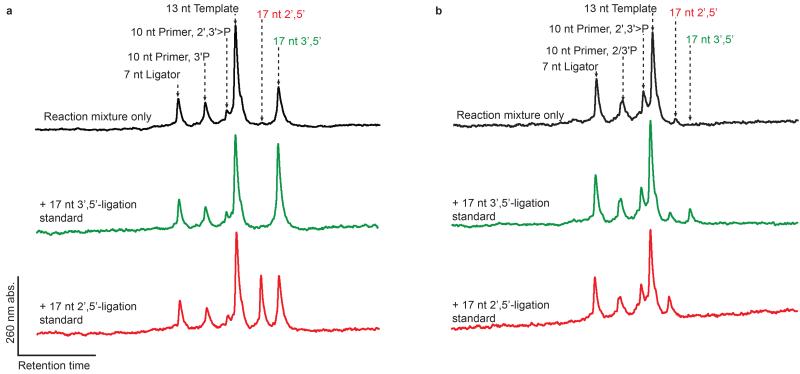
Figure 6. Templated ligation of acetylated 3′P and 2′P oligomers affords 3′,5′- and 2′,5′-linkages respectively.
a, b, HPLC traces of ammonolysed (deacetylated) ligation reactions, using 3′P (a) or 2′P (b) primer, template and ligator strands (sequences as per Fig. 3a). Following ligation, ammonolysis was performed as described in the text and the mixtures analysed by HPLC (top, black). Co-injection with 17 nt product standards possessing either a 3′,5′-(middle, green) or 2′,5′-(bottom, red) linkage at the ligation position revealed the selective formation of all-3′,5′-linked RNA from the 3′P primer, and RNA 2′,5′-linked (at the ligation junction only) from the 2′P primer. Smaller amounts of 17 nt standards were used for co-injections in b to allow easier comparison with the smaller ligation product peak. The split 10 nt primer peak indicated in b consists of a mixture of 2′P- and 3′P-terminated oligomers, owing to ammonolysis of the 2′,5′-linked product and the primer 2′,3′>P. See Supplementary Fig. S20 and S21 for peak assignments; yields of 44 % and 5 % were estimated from peak integrals for 3′P and 2′P ligation respectively (adjusting to account for partial hydrolysis of the 2′,5′-linkage under ammonolysis conditions).
Conclusions and outlook
In summary, we have discovered that chemoselective acetylation of RNA oligomers is possible in water under plausible prebiotic conditions. This chemistry permits the rapid and efficient template-directed ligation of short RNA oligomers, such as those generated by the dry-state reaction of N>P’s, to longer RNA molecules. Natural 3′,5′-linkages at the ligation junctions are favoured by the selective acetylation of 3′P-terminated oligomers. RNA strands thus generated are partially acetylated at internal 2′-positions, and these acetyl groups are readily removed by ammonolysis to liberate native RNA. The deacetylation conditions cause partial hydrolysis of non-natural 2′,5′-junctions but leave 3′,5′-phosphodiester bonds intact thus providing further, albeit slight enrichment for 3′,5′-linkages. Prior to hydrolysis, the partial 2′-O-acetylation is expected to favour duplex structure over variable tertiary structures due to reduced A-minor interactions 32 and increased North-type sugar puckering33,34. Reduced secondary (and thus tertiary) structural variability should facilitate replication of partially 2′-O-acetylated RNA relative to RNA. Thus the prebiotic synthesis of RNA might have proceeded through a partially protected intermediate with superior genotypic properties.
Finally, we speculate that the chemistry described herein might reconcile RNA-based models for the origin of life with those that emphasize metabolism. Thus, realistic experimental support for the ‘iron-sulfur world’ only goes so far as the synthesis of activated acetyl groups on the surface of (Fe,Ni)S20, but catalytic cycles fuelled by these acetyl groups have not been demonstrated despite many years of effort. Without the benefits that accrue to the system from this subsequent metabolism, the iron-sulfur world has had no raison d’être – the reliance of RNA synthesis and replication on a supply of activated acetyl groups would provide one.
Supplementary Material
Acknowledgements
This work has been funded by the Engineering and Physical Sciences Research Council through the provision of postdoctoral fellowships (C.D.D. & B.G.) and PhD studentships (M.W.P., S.I. & C.K.W.C.), the Medical Research Council through the provision of career development fellowships (F.R.B. & J.X., project no. MC_UP_A024_1009), and the Origin of Life Challenge – we thank Harry Lonsdale for the latter. We thank Christine Hilcenko, Mark J. Churcher and Vitor B. Pinheiro for advice on PAGE and fluorescence scanning, and Donna Williams for advice on solid-phase oligonucleotide synthesis.
Footnotes
Additional information. The authors have no competing interests.
Supplementary information and chemical compound information are available in the online version of this paper. Reprints and permission information is available online at http://www.nature.com/reprints.
References
- 1.Joyce GF. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- 2.Woese C. The genetic code. Harper & Row; New York: 1967. pp. 179–195. [Google Scholar]
- 3.Crick FHC. The origin of the genetic code. J. Mol. Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 4.Orgel LE. Evolution of the genetic apparatus. J. Mol. Biol. 1968;38:381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- 5.Powner MW, Gerland B, Sutherland JD. Synthesis of activated pyrimidine ribonucleotides in prebiotically plausible conditions. Nature. 2009;459:239–242. doi: 10.1038/nature08013. [DOI] [PubMed] [Google Scholar]
- 6.Szostak JW. Systems chemistry on early Earth. Nature. 2009;459:171–172. doi: 10.1038/459171a. [DOI] [PubMed] [Google Scholar]
- 7.Renz M, Lohrmann R, Orgel LE. Catalysts for the polymerisation of adenosine cyclic 2′,3′-phosphate on a poly (U) template. Biochim. Biophys. Acta. 1971;240:463–471. doi: 10.1016/0005-2787(71)90703-9. [DOI] [PubMed] [Google Scholar]
- 8.Eftink MR, Biltonen RL. Energetics of ribonuclease A catalysis. 2. Nonenzymatic hydrolysis of cytidine cyclic 2′,3′-phosphate. Biochemistry. 1983;22:5134–5140. doi: 10.1021/bi00291a012. [DOI] [PubMed] [Google Scholar]
- 9.Verlander MS, Lohrmann R, Orgel LE. Catalysts for the self-polymerization of adenosine cyclic 2′,3′-phosphate. J. Mol. Evol. 1973;2:303–316. doi: 10.1007/BF01654098. [DOI] [PubMed] [Google Scholar]
- 10.Verlander MS, Orgel LE. Analysis of high molecular weight material from the polymerization of adenosine cyclic 2′, 3′-phosphate. J. Mol. Evol. 1974;3:115–120. doi: 10.1007/BF01796557. [DOI] [PubMed] [Google Scholar]
- 11.Usher DA, McHale AH. Nonenzymic joining of oligoadenylates on a polyuridylic acid template. Science. 1976;192:53–54. doi: 10.1126/science.1257755. [DOI] [PubMed] [Google Scholar]
- 12.Bolli M, Micura R, Pitsch S, Eschenmoser A. Pyranosyl-RNA: Further Observations on Replication. Helv. Chim. Acta. 1997;80:1901–1951. [Google Scholar]
- 13.Trevino SG, Zhang N, Elenko MP, Lupták A, Szostak JW. Evolution of functional nucleic acids in the presence of nonheritable backbone heterogeneity. Proc. Natl. Acad. Sci. USA. 2011;108:13492–13497. doi: 10.1073/pnas.1107113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kierzek R, He L, Turner DH. Association of 2′-5′ oligoribonucleotides. Nucleic Acids Res. 1992;20:1685–1690. doi: 10.1093/nar/20.7.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Usher DA. Early chemical evolution of nucleic acids: a theoretical model. Science. 1977;196:311–313. doi: 10.1126/science.583625. [DOI] [PubMed] [Google Scholar]
- 16.Usher DA, McHale AH. Hydrolytic stability of helical RNA: a selective advantage for the natural 3′,5′-bond. Proc. Natl. Acad. Sci. USA. 1976;73:1149–1153. doi: 10.1073/pnas.73.4.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohatgi R, Bartel DP, Szostak JW. Nonenzymatic, template-directed ligation of oligoribonucleotides is highly regioselective for the formation of 3′-5′ phosphodiester bonds. J. Am. Chem. Soc. 1996;118:3340–3344. doi: 10.1021/ja9537134. [DOI] [PubMed] [Google Scholar]
- 18.Rammler DH, Lapidot Y, Khorana HG. Studies on polynucleotides. XIX. The specific synthesis of C3′-C5′ inter-ribonucleotidic linkage. A new approach and its use in the synthesis of C3′-C5′-linked uridine oligonucleotides. J. Am. Chem. Soc. 1963;85:1989–1997. [Google Scholar]
- 19.Coutsogeorgopoulos C, Khorana HG. Studies on polynucleotides. XXXI. The specific synthesis of C3′-C5′-linked ribopolynucleotides (6). A further study of the synthesis of uridine polynucleotides. J. Am. Chem. Soc. 1964;86:2926–2932. [Google Scholar]
- 20.Huber C, Wächtershäuser G. Activated acetic acid by carbon fixation on (Fe,Ni)S under primordial conditions. Science. 1997;276:245–247. doi: 10.1126/science.276.5310.245. [DOI] [PubMed] [Google Scholar]
- 21.Loison A, Dubant S, Adam P, Albrecht P. Elucidation of an iterative process of carbon-carbon bond formation of prebiotic significance. Astrobiology. 2010;10:973–988. doi: 10.1089/ast.2009.0441. [DOI] [PubMed] [Google Scholar]
- 22.de Duve C. Blueprint for a cell: the nature and origin of life. Neil Patterson Publishers, Carolina Biological Supply Company; Burlington, NC: 1991. [Google Scholar]
- 23.Hagan WJ., Jr. Uracil-catalyzed synthesis of acetyl phosphate: a photochemical driver for protometabolism. ChemBioChem. 2010;11:383–387. doi: 10.1002/cbic.200900433. [DOI] [PubMed] [Google Scholar]
- 24.Liu R, Orgel LE. Oxidative acylation using thioacids. Nature. 1997;389:52–54. doi: 10.1038/37944. [DOI] [PubMed] [Google Scholar]
- 25.Biron J-P, Parkes AL, Pascal R, Sutherland JD. Expeditious, potentially primordial, aminoacylation of nucleotides. Angew. Chem. Int. Ed. 2005;44:6731–6734. doi: 10.1002/anie.200501591. [DOI] [PubMed] [Google Scholar]
- 26.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 27.McGinnis JL, Dunkle JA, Cate JHD, Weeks KM. The mechanisms of RNA SHAPE chemistry. J. Am. Chem. Soc. 2012;134:6617–6624. doi: 10.1021/ja2104075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta SC, Islam NB, Whalen DL, Yagi H, Jerina DM. Bifunctional catalysis in the nucleotide-catalyzed hydrolysis of (±)-7β,8α-dihydroxy-9 α,10 α - epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene. J. Org. Chem. 1987;52:3812–3815. [Google Scholar]
- 29.Cavalieri LF. Studies on the structure of nucleic acids. VII. On the identification of the isomeric cytidylic and adenylic acids. J. Am. Chem. Soc. 1953;75:5268–5270. [Google Scholar]
- 30.Rohatgi R, Bartel DP, Szostak JW. Kinetic and mechanistic analysis of nonenzymatic, template-directed oligoribonucleotide ligation. J. Am. Chem. Soc. 1996;118:3332–3339. doi: 10.1021/ja953712b. [DOI] [PubMed] [Google Scholar]
- 31.Goldsborough AS. Modified polynucleotides and uses thereof. US 6,867,290. 2005
- 32.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- 34.Guschlbauer W, Jankowski K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Res. 1980;8:1421–1433. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferris JP, Huang C-H, Hagan WJ., Jr. N-cyanoimidazole and diimidazole imine: water-soluble condensing agents for the formation of the phosphodiester bond. Nucleosides and Nucleotides. 1989;8:407–414. doi: 10.1080/07328318908054184. [DOI] [PubMed] [Google Scholar]
- 36.Kanaya E, Yanagawa H. Template-directed polymerization of oligoadenylates using cyanogen bromide. Biochemistry. 1986;25:7423–7430. doi: 10.1021/bi00371a026. [DOI] [PubMed] [Google Scholar]
- 37.Horowitz ED, et al. Intercalation as a means to suppress cyclization and promote polymerization of base-pairing oligonucleotides in a prebiotic world. Proc. Natl. Acad. Sci. USA. 2010;107:5288–5293. doi: 10.1073/pnas.0914172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.