Summary
Recurrent amplification of 8p12 is observed in Squamous Cell Lung Cancer (SQLC) and FGFR1 is thought to be the main oncogenic driver in this region. In this issue of Cancer Discovery, Malchers et al., perform a detailed characterization of 8p12 in SQLC and find remarkable genomic heterogeneity in this region raising the possibility that other genes in addition to FGFR1 may play a role in SQLC. Mechanistic studies of the FGFR1 amplified subset of SQLC revealed potential roles for FGF ligands and MYC expression levels in modulating the response of these tumors to FGFR inhibition.
Lung cancer is the leading cause of cancer-related mortality in the world. A lack of effective chemotherapeutic strategies available to treat advanced tumors combined with the fact that many lung cancers are diagnosed at advanced stages both contribute to the poor prognosis of this disease. However, lung cancer is not a single disease but instead a collection of phenotypically and genotypically diverse malignancies, associated with unique mechanisms of pathogenesis and likely, cells of origin. Based on clinical and histological criteria, lung cancer is separated into two major types: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC). This original distinction was important in the clinical management of the disease as SCLC was found to display acute sensitivity to initial treatment with conventional cytotoxic agents. However, NSCLC is an antiquated classification as it consists of multiple, diverse histological types and subtypes, with adenocarcinoma (AC) and squamous cell lung cancer (SQLC) representing the vast majority of all cases. Despite this heterogeneity, NSCLC has traditionally been treated as a single entity, with tumor stage serving as the primary factor in determining which treatment regimen to apply. Yet it has recently become clear that treating AC and SQLC as a single disease ignores important biological factors underlying their differential development, which may lead to suboptimal response rates to therapy.
Recent studies that comprehensively characterized the genomic landscape of AC and SQLC have led to an important understanding of the genetic basis of these diseases and identified genes associated with the pathogenesis of the specific lung cancer subtypes (1–3). For example, mutations or rearrangements in genes encoding receptor tyrosine kinases (RTKs) such as EGFR and ALK as well as downstream signaling components (e.g. KRAS) are frequent in AC whereas SQLC contains frequent disruption of phosphatidylinositol 3-kinase (PI3K) pathway components as well as TP53. Amplification of the lineage-specific oncogenes NKX2-1 and SOX2 in AC and SQLC, respectively, are also frequent events that distinguish these subtypes of lung cancer (4, 5). In AC, these genetic changes - mainly the recurrent kinase alterations - have successfully been translated into the clinical management of the disease; EGFR and ALK tyrosine kinase inhibitors (TKIs) are routinely used to treat patients with alterations in these genes. In comparison, the identification of clinically targetable alterations in significant fractions of SQLCs has lagged significantly. For example, mutation of the DDR2 kinase gene in SQLC is associated with sensitivity to the multitargeted kinase inhibitor dasatinib in preclinical studies but occurs in less than four percent of tumors (6). Thus, the recent finding that amplification of the proximal portion of chromosome arm 8p encompassing the gene encoding the RTK FGFR1 in >20% of SQLC cases, and that amplification of FGFR1 was associated with response to FGFR1 TKIs in experimental models, was of great interest from a clinical standpoint, as it suggested that SQLC patients with this alteration could be candidates for targeted therapy (7, 8). Subsequently, several clinical trials have been initiated in lung and other cancer types with FGFR1 amplification in order to test this hypothesis. Preliminary information from these studies has revealed activity in a subset of FGFR amplified cancers; however, complete data from these studies have yet to be reported (9, 10). Even with these promising initial experimental and clinical findings numerous questions remain. For example, although multiple lung cancer cell lines contain amplification of FGFR1, only half are sensitive to FGFR1 TKIs suggesting that additional factors influence drug response (7). Further, of the cell lines with FGFR1 amplification that responded to TKIs, none were SQLCs confounding the association between histology, FGFR1 amplification and drug response. Together, these issues could have significant implications in identifying the patients most likely to benefit from FGFR targeted therapy.
In this issue of Cancer Discovery, Malchers et al. present an innovative study that aims to identify the factors that influence sensitivity to FGFR1 inhibition in lung cancer (11). They started out by performing an in depth study of SNP array data from 306 SQLCs which had revealed broad genomic alterations (12). Using an approach called Focal Amplification Peak Purification or FAPP, they were able to smooth out the confounding effects of broad alterations to refine the boundaries and target genes of recurrent regions of focal amplification. As one would expect, most known amplicons became more focused on their suspected target genes (ie. EGFR) when using this method; however, the 8p12 amplicon yielded an extremely heterogenous pattern of amplification unlike any other region, with chromosomal breakpoints spanning an unusually large region surrounding the peak. Surprisingly, only 28% of samples with 8p12 amplification had peaks that centered on FGFR1, with some tumors having amplifications of this region that excluded FGFR1 all together. These genomic findings have major implications as they suggest that FGFR1, although increased in copy number, may not be the only driver or even be a bystander, in the majority of cases with 8p12 amplification and not the primary target of the alteration. Thus, screening for increased FGFR1 gene dosage alone using methods like fluorescence in situ hybridization (FISH) would have poor predictive value in identifying patients with tumors driven by activated FGFR1, and accordingly, candidates to respond to therapies targeting this receptor. Interestingly, through this analysis the authors found amplification of FRS2, an adapter protein that connects FGFRs to intracellular signaling pathways, in 2% of SQLCs revealing a parallel strategy used by the cells to engage FGFR signaling.
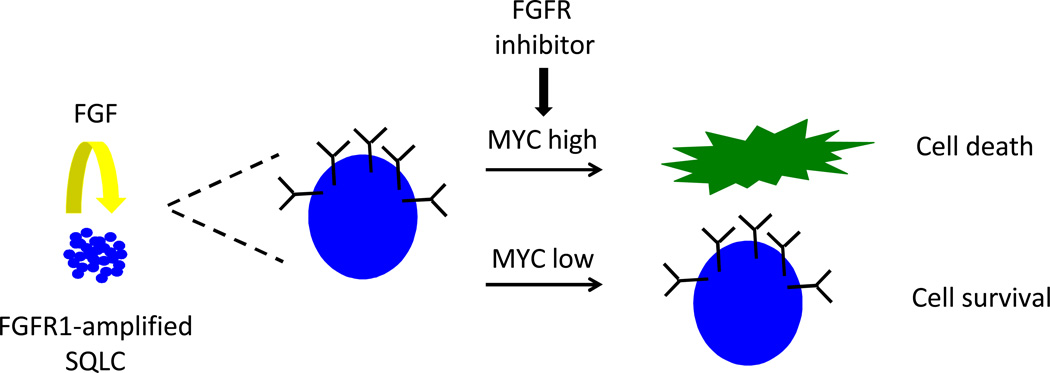
These complicated genomics associated with FGFR1 amplification highlight the need for in depth mechanistic studies into the biology of FGFR1-amplified lung cancer to understand if and how this oncogene transforms cells and to identify predictive biomarkers of response to FGFR1 inhibition (Figure 1). In their manuscript, Malchers and colleagues, tackle both of these topics. First, they set out to explore the importance of FGF ligands for the growth of FGFR1-amplified cell lines. Interestingly, they observed that all of the FGFR1-amplified cell lines examined, irrespective of their sensitivity to FGFR inhibitors, were responsive to exogenously added ligand, in particular FGF1, FGF2 and FGF4 and/or produced their own ligands. When FGFR1 amplified cells were injected into mice, tumor growth was prevented by adenoviral expression of the extracellular domain of FGFR1 in “FGF trap” competition experiments, further supporting the ligand dependence of cells with FGFR1 amplification. Predictably, increased levels of ligand (e.g. FGF2) decreased the sensitivity of FGFR1 amplified cells to FGFR1 inhibition, raising the possibility that ligand levels or the ability of tumor cells to produce ligands may be a factor that affects the sensitivity of FGFR1 amplified tumors to this class of drugs. The role of growth factors in mediating resistance to RTK-directed therapies was recently explored and although FGF was shown to rescue many different cancer cell lines treated with a wide variety of kinase inhibitors, it did not show much effect in FGFR2 or FGFR3-amplified cell lines examined (13). Additional experiments in human tumors and FGFR1-amplified cell lines will be required to assess whether the levels of FGF ligands in these tumors influence the response to FGFR inhibitors.
Figure 1. Mechanisms of transformation of FGFR1-amplified SQLCs.
FGF ligands produced by tumor cells support the growth of FGF-amplified cancer cells and can potentially modulate sensitivity to FGFR inhibitors in a dose-dependent manner. High expression levels of MYC in these tumor cells confer greater sensitivity to FGFR inhibitors than low levels leading to cell death and tumor regression.
Second, Malchers et al explore the role of cooperating oncogenes in transformation by FGFR1. SQLC-associated FGFR1 splicing isoforms are only weakly oncogenic when transfected into cells. The authors co-transfected FGFR1 with other SQLC-associated oncogenes and found a synergistic effect of FGFR1 and MYC on cell transformation. Most surprisingly, when these cells were used to form tumors in mice, FGFR1 and MYC expressing tumors exhibited sensitivity to FGFR inhibitors with consequent tumor regression. In contrast, tumors that only expressed FGFR1 grew more slowly but they did not shrink in size. To further study the relationship between MYC levels and FGFR inhibitor sensitivity, the authors examined the levels of MYC expression in FGFR1-amplified sensitive and resistant cell lines and found that high levels of MYC were observed in the sensitive lines. Knockdown of MYC in one of these cell lines altered the sensitivity of the cell line and caused it to become resistant to FGFR inhibition. These studies would predict that high MYC levels are required for FGFR1-amplified tumors to respond to FGFR1 inhibitors. The authors, indeed, present two cases of patients with FGFR1-amplified lung cancer and high levels of MYC (using immunohistochemistry) that responded to these drugs. These data highlight the need for further studies to test the hypothesis that MYC expression is a predictive biomarker for responsiveness of FGFR1-amplified SQLC to FGFR inhibitors and to understand the mechanism that underlies this response. One possibility is that the pro-apotoptic functions of MYC facilitate FGFR inhibitor mediated cell death.
In contrast to what is observed in EGFR mutant and ALK-rearranged lung adenocarcinomas, where the genetic alterations are highly predictive of a response to appropriate inhibitors, the case of FGFR1-amplified SQLC is more complex. Given the complexity of the genomic alterations observed in the 8p12 region, genes other than FGFR1 may represent the drivers in a fraction of SQLCs, with potential candidates including WHSC1L1 and BRF2 previously described (14, 15). Data from clinical trials in which FGFR1 amplification is measured by FISH will be important to determine how predictive this assay is in determining response to FGFR1 inhibition, and whether quantitative thresholds can be elucidated. It is crucial that a thorough genomic analysis of specimens from these trials be performed to establish the specific regions of amplification and correlate these with response to the FGFR targeted therapies. In the cases where FGFR1 is a clear oncogenic driver, this study from Malchers et al., identifies two potential modulators of sensitivity to FGFR inhibition: cells-ligand levels and MYC expression (Figure 1). Even with the compelling data presented here, studies in large patient cohorts will be required to establish either of these as a biomarker of response or resistance to these drugs. Moreover, neither of these factors are straightforward to quantify in patients samples further complicating these studies.
In conclusion, as genomic studies reveal new targetable cancer drivers, it is likely that in many cases, the relationship between the genetic alteration and response to therapy is not clear cut, as exemplified by FGFR1-amplified lung cancer. Functional studies, such as those presented in the study by Malchers et al, paired with in depth genomic analyses, will be necessary to understand the biological context in which the genomic alteration confers sensitivity to specific drugs.
Acknowledgements
The authors acknowledge support from the NIH/National Cancer Institute (KP), Uniting Against Lung Cancer (KP), the Labrecque Foundation (KP), the Canary Foundation (KP), the Lung Cancer Research Foundation (KP) and the Yale Cancer Center (KP).
References
- 1.Hammerman PS, Hayes DN, Wilkerson MD, Schultz N, Bose R, Chu A, et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass AJ, Watanabe H, Mermel CH, Yu S, Perner S, Verhaak RG, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007 doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hammerman PS, Sos ML, Ramos AH, Xu C, Dutt A, Zhou W, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer discovery. 2011;1:78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2:62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dutt A, Ramos AH, Hammerman PS, Mermel C, Cho J, Sharifnia T, et al. Inhibitor-sensitive FGFR1 amplification in human non-small cell lung cancer. PLoS ONE. 2011;6:e20351. doi: 10.1371/journal.pone.0020351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolf J, LoRusso PM, Camidge RD, Perez JM, Tabernero J, Hidalgo M, et al. A phase I dose escalation study of NVP-BGJ398, a selective pan FGFR inhibitor in genetically preselected advanced solid tumors. Proceedings of the 103rd Annual Meeting of the American Association for Cancer Research. 2012;72(8 Suppl) Abstract nr LB-122. [Google Scholar]
- 10.Andre F, Ranson M, Dean E, Varga A, Noll Rvd, Stockman PK, et al. Results of a phase I study of AZD4547, an inhibitor of fibroblast growth factor receptor (FGFR), in patients with advanced solid tumors . Proceedings of the 104th Annual Meeting of the American Association for Cancer Research. 2013;73(8 Suppl):LB-145. [Google Scholar]
- 11.Malchers F, Dietlein F, Schottle J, Lu X, Nogova L, Albus K, et al. Cell-autonomous and non-cell-autonomous mechanisms of transformation by amplified FGFR1 in lung cancer. Cancer discovery. 2013 doi: 10.1158/2159-8290.CD-13-0323. [DOI] [PubMed] [Google Scholar]
- 12.Clinical Lung Cancer Genome P, Network Genomic M. A genomics-based classification of human lung tumors. Sci Transl Med. 2013;5:209ra153. doi: 10.1126/scitranslmed.3006802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lockwood WW, Chari R, Coe BP, Thu KL, Garnis C, Malloff CA, et al. Integrative genomic analyses identify BRF2 as a novel lineage-specific oncogene in lung squamous cell carcinoma. PLoS medicine. 2010;7:e1000315. doi: 10.1371/journal.pmed.1000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tonon G, Wong KK, Maulik G, Brennan C, Feng B, Zhang Y, et al. High-resolution genomic profiles of human lung cancer. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9625–9630. doi: 10.1073/pnas.0504126102. [DOI] [PMC free article] [PubMed] [Google Scholar]