Abstract
T cell receptor (TCR) signal strength determines the differentiation outcome of naïve CD4+ T cells: low signal strength favors Foxp3pos regulatory T cells (Treg) whereas high TCR signals are required to induce IL-2-producing helper T cells (Th). To better understand the signaling requirements for this cell-fate decision, we constructed a logic circuit model of the TCR signaling pathways. A major feature of this model is an incoherent feed-forward loop involving activation of Foxp3 and its inhibition by mTOR, which leads to transient appearance of Foxp3pos cells under simulation conditions that drive IL-2 producing Th cells. This behavior along with the predicted ability of TGF-β to induce Treg despite continued activation of the Akt/mTOR pathway were confirmed experimentally. The latter provides a possible reason for the observed instability of TGF-β-induced Treg. The model also predicted and experiments confirmed that transient high dose Ag stimulation results in three stable T cell fates (Th, Treg, and non-activated) with relative proportions depending on the duration of stimulation. Experimental analysis of the cell population at the time of Ag removal identified three distinct populations based on CD25 abundance and Akt/mTOR activation that correlated with these T cell fates. Further analysis of corresponding simulation trajectories implicated a negative feedback loop involving Foxp3, PTEN, and Akt/mTOR. Taken together, these results suggest that there is a critical period following TCR stimulation during which heterogeneity in the differentiating population leads to increased plasticity of cell fate.
Introduction
CD4+ T cells can be grouped into two main sub-types: those which exert an activating effect on the immune response (helper T cells, Th), and those which suppress immune responses (regulatory T cells, Treg). Treg cells play an important role in suppressing T cell mediated immunity, limiting damage caused by the immune response and preventing autoimmune diseases (1). However, Treg cells may play a detrimental role in cancer by acting as an effector of immune suppression by tumors. In several recent studies of human Treg cells in patients with cancer, the number of Treg cells within tumors and their suppressive activity were found to be elevated and to predict poor survival (2). Understanding the factors that control the induction of Treg cells has the potential to advance therapies involving either elimination of antigen-specific Treg cells in the context of cancer (3), or enhancement of Treg-mediated suppression in the context of autoimmune responses (4).
Treg cells are characterized by the transcription factor forkhead box P3 (Foxp3), a specific pattern of cytokine production, and immunosuppressive function (5–7). Treg cells suppress other effector T cells through several mechanisms (8), including deletion of effector cells via granzyme B (9), secretion of immunosuppressive cytokines such as transforming growth factor β (TGF-β), interleukin (IL-)10 and IL-35 (10), metabolic disruption through the production of adenosine (11) or competition for IL-2 (12, 13) and finally, inhibition of dendritic cell maturation(8, 14).Two main groups of Treg cells are currently known: natural and adaptive (induced) Treg cells. Natural regulatory T (nTreg) cells become committed to a regulatory fate while still in the thymus (14), whereas induced Treg (iTreg) cells, arise from naïve T cells in the periphery under stimulation by specific factors including IL-10 (15), TGF-β (16), low antigen (Ag) dose (17, 18), and certain dendritic cell (DC) subsets (19–21). iTreg and Th both develop in the periphery from common, “naïve”, uncommitted T cell precursors, which differentiate upon encountering cognate peptide:major histocompatibility complex (pMHC) on the surface of antigen-presenting cells (APC). Previous studies have indicated that both Ag dose and the duration of Ag stimulation strongly influence the choice between regulatory and helper cells in the T cell response specific for a particular Ag, such that high dose favors Th and low dose favors Treg (17, 18, 22). Understanding the molecular determinants of this process has major implications for the development of targeted immunomodulation therapies (23). For example, it is important in a cancer vaccination strategy to deliver a high enough dose to induce tumor-specific Th1 cells. A more delicate balance must be achieved in the context of autoimmunity, because many patients have increased numbers of auto-reactive T cells, which any therapy must avoid activating. Thus, a strategy designed to induce Ag-specific Treg must deliver a dose that will favor the induction of self-Ag-specific Treg without activating or inducing autoreactive Th1/Th17 cells. Few vaccination strategies in cancer or autoimmunity consider the dose of the Ag and this may be one reason for the limited success of these efforts to date (24, 25).
Treg differentiation is influenced by multiple signaling pathways including those stimulated by engagement of T cell receptor (TCR), the co-stimulatory molecule CD28, IL-2 receptor (IL-2R) and TGF-β receptor (TGF-βR). We recently reported that the culture of CD4+ T cells with DC presenting low dose Ag resulted in both the expansion of preexisting nTreg and the conversion of naïve T cells into iTreg (18), and the induction of Treg was inversely correlated with the strength of the TCR signal as measured by phosphorylation of the S6 ribosomal protein (pS6). pS6 is downstream of the phosphoinositide 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR) signaling pathway and several other studies have found that activation of the PI3K/Akt/mTOR pathway inhibits the induction of Treg (26, 27). Activation of Akt/mTOR was shown to inhibit Treg development (26), whereas inhibition of Akt/mTOR enhances Treg development (27). Specific components of the Akt/mTOR pathway have also been recently implicated in the differentiation of Th populations, with mTOR Complex 1 (mTORC1)-deficient animals failing to generate Th1 and Th17 cells and mTOR Complex 2 (mTORC2)-deficient animals failing to generate Th2 cells (28, 29). In vitro studies demonstrated that the mTORC1 inhibitor rapamycin enables expansion of functional Treg of murine (30) or human (31, 32) origin.
Mathematical modeling of T cell activation, and signaling pathways in general, provides a means to identify important regulatory elements, including feedback and feedforward loops, to test hypotheses, and to provide testable predictions (33, 34). Modeling TCR activation has so far mainly involved the use of reaction network models focusing on early steps in T cell activation (35–38). Because parameterization of reaction network models requires detailed quantitative data that are not currently available for all of the signaling processes involved in T cell differentiation, we have taken an alternative approach based on logical modeling (39–41). This approach uses discrete variables to represent network elements and logical rules to describe element interactions. Logical models do not require quantitative parameters, but rather allow the development of complex qualitative networks. Other groups have developed logical models to investigate T cell activation (42, 43) and differentiation (44–46), although none of these have explicitly modeled the effects of Ag dose.
We have developed a logical model of T cell differentiation that focuses on both early steps in T cell activation and events leading to the generation or suppression of the Treg phenotype. Using the logical modeling approach allowed us to capture multiple layers of the cellular response to signals, including extracellular stimulation, intracellular signaling leading to activation of transcription factors, gene transcription and autocrine or paracrine responses to secreted factors. Model simulations reproduced known experimental results and made novel predictions that were confirmed experimentally. Importantly, our results suggest that differential activation of signaling pathways downstream of TCR by high and low dose Ag are better modeled as changes in the duration of contact time rather than changes in the strength of activation of pathways downstream of TCR. Restricting the time of TCR activation resulted in a mixed population of Treg and Th cells, as well as inactive cells, an observation we confirmed experimentally. Analysis of the dynamics of this scenario revealed that the Th phenotype stabilized more rapidly than the Treg phenotype, and demonstrated an important role for a feedback loop involving Foxp3, phosphatase and tensin homolog (PTEN), and Akt/mTOR. The logical model presented here demonstrates heterogeneity on the cell population level that recapitulates experimental results and suggests that differential timing of key TCR signaling events plays a major role in determination of T cell fate.
Results
Naïve T cell differentiation control network
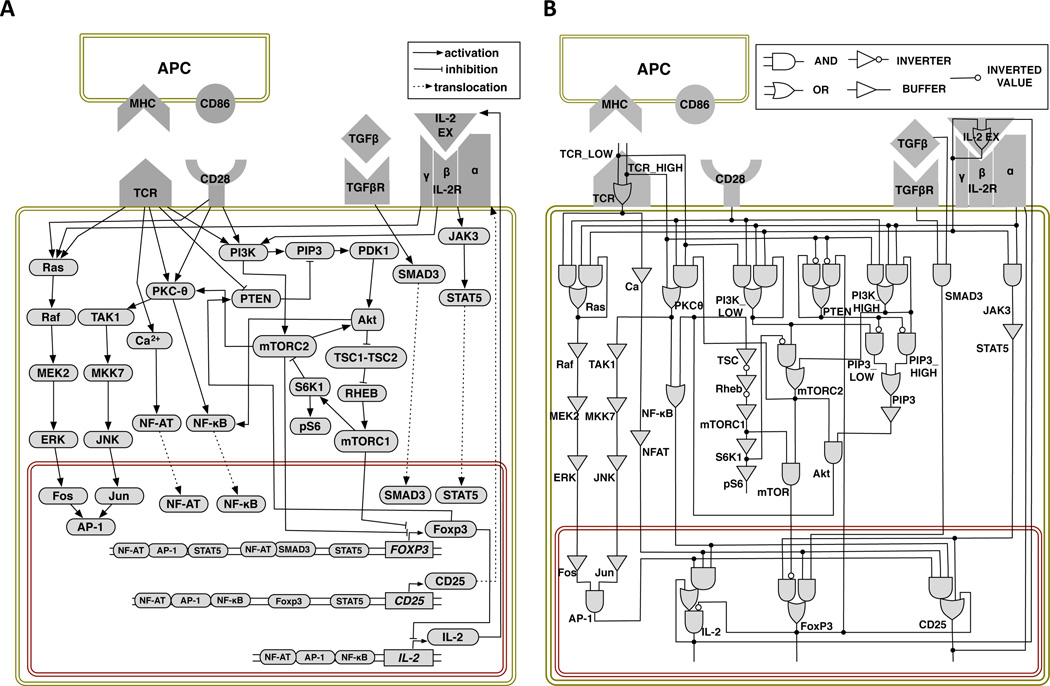
We conducted an extensive literature survey to identify the key signaling events and molecules involved in determining T cell fate upon exposure to antigen. The model presented in Fig. 1A captures critical signaling events, from stimulatory signals via cell surface receptors, through activation of transcription factors, to production of proteins critical to specific phenotypes. The two primary phenotypes that arise in the model are (1) cells that express the transcription factor FOXP3 but not the cytokine IL2 and (2) cells that do not express FOXP3 but have the ability to produce and secrete IL-2. We identify these cells as Treg and Th, respectively.
Figure 1.
Computational model of T cell differentiation: (A) Signaling network connections. Pointed arrowheads represent activation, blunt arrowheads represent inhibition, and dashed edges represent translocation. (B) Corresponding logic circuit model. Elements are represented by single logic gates (e.g., Raf) or combinations of multiple gates (e.g., Ras). Element values are computed at each time step based on the values of their inputs with instantaneous updating of gates within elements.
The model includes four receptors: the TCR, the co-stimulatory receptor CD28, and the receptors for IL-2, and TGF-β. The two signals necessary for naïve T cell activation (47) are the binding of pMHC complexes on the surface of the APC to the TCR, and the binding of co-stimulatory ligands CD80 or CD86 on the APC to CD28 receptors on the T cell. The combined signals from TCR and CD28 activate the transcription factors: nuclear factor of activated T-cells (NF-AT), activator protein 1 (AP-1) and nuclear factor -κ-light-chain-enhancer of activated B cells (NF-κB). These transcription factors promote expression of the IL2 gene and the α subunit of IL-2 receptor (IL2RA, also called CD25) (48, 49). The β and γ subunits of IL2-R are constitutively present and are represented as a single variable in the model. Binding of IL-2 to IL-2R results in activation of another transcription factor, signal transducer and activator of transcription 5 (STAT5) (50, 51), which in turn activates Foxp3, the key transcription factor that drives differentiation into Treg (52). STAT5 can also modulate the expression of IL2 through the induction of Blimp-1 (53), but we have not included this interaction in the current version of the model because it complicates analysis of the trajectories by making IL2 expression transient. We confirmed by additional simulations that including this interaction does not otherwise affect our results.
FOXP3 is expressed after one of the following combinations of transcription factors binds to its promoter: NF-AT, AP-1, and STAT5; NF-AT and mothers against decapentaplegic homolog 3 (Smad3); or STAT5 alone (54, 55). Foxp3 itself inhibits transcription of the IL-2 gene while maintaining IL2RA (CD25) expression (7). In parallel with activation of NF-AT, AP-1, NF-κB, and STAT5, the PI3K/Akt/mTOR pathway can also be activated. The latter depends on the activity of PTEN, a phosphatase constitutively present in naïve T cells, which inhibits the function of PI3K, and which is reduced following TCR activation (56). Reduced PTEN protein abundance or activity results in increased signaling via the PI3K/Akt/mTOR pathway (further in the text referred to as the mTOR pathway), and, importantly, it has been shown that PTEN is more abundant in Tregs (57). Based on the reported effects of mTOR inhibition following T cell activation (58), we assume that active mTORC1 and mTORC2 can override other signals driving Foxp3 expression. However, since it is well-established that TGF-β, in combination with high Ag doses, induces Treg (16, 59), we also assume that the combined signals from Smad3, downstream of the TGF-β receptor, and NF-AT drive expression of FOXP3 even when mTOR is active.
Logical Model of Naïve T Cell Differentiation
In Boolean network modeling each node in the graph is represented by a single Boolean variable that takes a value of OFF if the element is inactive and ON if the element is active. For some nodes, however, this two-state representation may not be sufficient. For example, since we are studying the impact of varying the TCR signal strength, we need to model at least three possible stimulation values, corresponding to OFF, LOW, and HIGH, representing the effective antigen dose. In such cases, depending on the modeling formalism used, node values may be represented by multiple Boolean variables (the approach taken here) or a single discrete variable. We also model three values of PI3K to avoid oscillations in mTOR pathway (see Methods for additional details).
The interaction map shown in Fig. 1A is not a complete dynamical model because it does not specify how multiple input signals to a node are to be combined. For example, the diagram shows that mTORC2 has two regulators — activation by PI3K and inhibition by ribosomal protein S6 kinase beta-1 (S6K1) — but does not specify how the regulations work together. In the logical modeling approach (39–41) a dynamical model is constructed using logical functions that determine the next state of each element in the system as a function of its regulators. These functions are constructed from the primitives of Boolean logic—AND, OR, and NOT. For example, mTORC2 activation is described by the rule
MTORC2' = PI3K_HIGH or (PI3K_LOW and not S6K1),
meaning that mTORC2 is activated by either a HIGH PI3K value that overcomes any inhibition by S6K1, or by a LOW PI3K value in the absence of S6K1.
We defined the full set of elements (fig. S1) and their corresponding Boolean variables and logic update rules (fig. S2) using information from our own experiments and the literature. We tuned the logical functions used in several rules in order to match experimental observations. References and observations used to determine the regulatory relationships in the model are given in fig. S2. In addition, we set varying thresholds for elements PTEN, PI3K, mTORC2 and protein kinase C theta (PKCθ), in order to match observed differences in the effects of high and low dose Ag. For example, high dose Ag, which produces a HIGH TCR activation, is required to inactivate PTEN, which is assumed to be constitutively active in naïve T cells. Because full activation of phosphatidylinositol (3,4,5)-trisphosphate (PIP3) and the Akt pathway can only occur if PTEN is inactive, the LOW TCR value that represents low Ag dose cannot fully activate either of the mTOR complexes, mTORC1 or mTORC2. As shown above, a HIGH PI3K value is required to overcome inhibition of mTORC2 activation by S6K1.
Fig. 1B presents a complete description of the logical model using logic gates that represent the AND and OR rules. This representation is similar to diagrams used in digital circuits and helps to clarify the overall input-output structure of the network.1 Besides implementing logic functions, logic gates may also have associated delays that we use to model differences in signal propagation between network elements. For example, by including delay gates on the AP-1 pathway the time of transcription factor activation can be fine-tuned.
We simulate the dynamics of the system using the method of random order asynchronous Boolean updates (60). Each simulation represents a sample from the set of possible trajectories that a cell can undergo. Trajectories are started with each variable in a defined initial state. At each round in the simulation, the next state value of each variable is assigned using the logic rules described above. Within a round, variables are updated in random order, mimicking the randomness in event timing that may occur because of cell-to-cell variation in the abundance of many proteins and the intrinsic stochasticity of chemical kinetics involving a small number of molecules (61). Time is reported in number of update rounds, and based on comparison with our experimental results each update corresponds to roughly 3 hours of real time (see Methods for additional details).
Table 1 summarizes the behaviors of the model and their correspondence with experimental observations. The model reproduces most of the experimentally known behaviors of the system and makes novel predictions, several of which we have tested in this work. In addition, an extensive sensitivity analysis (see Methods for more detail) shows that qualitative results do not depend sensitively on the choice of initial conditions or timing parameters (see Event Timing and Delays below). In the following sections, we discuss these results in more detail.
Table 1.
Summary of observations from model simulations and data analysis.
| Scenario | Model observation | Supporting evidence | |
|---|---|---|---|
| High antigen dose | 1 | Differentiation into Th cells | (18) |
| 2 | Transient FOXP3 expression | (69), This paper) | |
| 3 | pS6 high | (18, 27) | |
| 4 | IL-2 increases early after stimulation and remains high | (64) | |
| Low antigen dose | 5 | Differentiation into Treg cells | (17, 18, 22) |
| 6 | Transient IL2 expression | Prediction, | |
| 7 | pS6 low | (18, 27) | |
| 8 | Delayed FOXP3 expression (compared with HD) | Prediction | |
| High antigen dose + TGFβ | 9 | Differentiation into Treg cells | (15, 20, 27) |
| 10 | pS6 high, Foxp3+ | (This paper) | |
| 11 | Foxp3 increases very early, before pS6 | Prediction | |
| 12 | Unstable Treg phenotype – TGFβ removal switches cells back to Th phenotype | (70) | |
| High antigen dose + inhibitor addition | 13 | NFAT inhibition still results in Th phenotype | (27) |
| 14 | NFAT inhibition leads to several different phenotypes (Th and inactive) | Prediction | |
| 15 | mTORC1/Akt inhibition induces Treg phenotype | (27, 30) | |
| High antigen dose + antigen removal | 16 | Mixed population of Treg, Th and inactivated cells | (27) |
| 17 | 11 different phenotypes observed | Prediction | |
| 18 | Treg cell population size changes with removal time (never 100%) | Prediction, (27) | |
| 19 | Th cell population size increases slowly to 100% with increasing removal time | Prediction | |
| 20 | Feedback between PTEN and Foxp3 critical for stable Treg phenotype | Prediction | |
| 21 | Race between CD25/STAT5 and mTOR pathway activation critical for early FOXP3 expression | Prediction | |
| 22 | Slower response to activation allows for differentiation intoTreg phenotype after Ag removal | Prediction | |
| 23 | Variability in timing of events combined with effective duration of receptor-antigen binding leads to mixed population | Prediction |
Architecture of the Differentiation Control Network
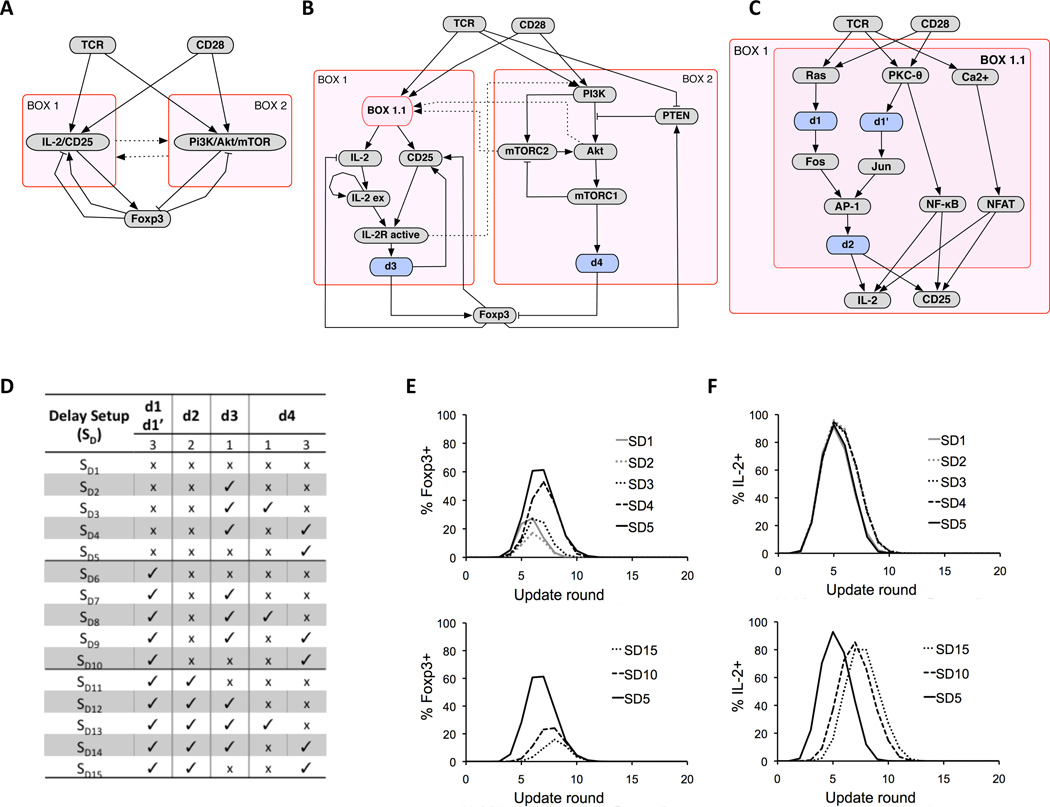
The behavior of our model is governed by control loops involving Foxp3. These include both positive and negative feedback and coherent and incoherent feed-forward loops (FFL) (Fig. 2, A to C), which affect both the transient and steady state behavior of the model.
Figure 2.
Network architecture and implementation of delay circuits. (A) Overview of the model control structure. Signaling through TCR and CD28 leads to induction of IL-2 and CD25 (Box 1) and activation of PI3K/Akt/mTOR (Box 2), which have opposing effects on the induction of Foxp3, which also exhibits multiple feedbacks. Pointed arrowheads represent activation, blunt arrowheads represent inhibition, and dashed edges represent interactions between boxes. (B) More detailed view of the control network highlighting several places where delay circuits have been introduced. Pathways leading to activation of IL-2 and CD25 are represented as sub-box, Box 1.1, and delays in Foxp3 activation by STAT5 and Foxp3 inhibition by mTOR, are represented by d3 and d4, respectively. (C) Details of Box 1.1. Delay d1 replaces network connections Ras->Raf, Raf->MEK2, MEK2->ERK, and ERK->Fos. Delay d1’ replaces connections PKC-θ->TAK1, TAK1->MKK7, MKK7->JNK, and JNK->Jun. The two delays, d1 and d1’ are always assumed to be equal, since activation of AP-1 in the model will always depend on the longer delay on the two pathways. (D) Delay setups (SD1-SD15) are defined as different combinations of delay lengths for d1-d4. Absence of delay is marked with ‘x’, and the presence of delay is marked with ‘✓’. For each delay d1(d1’)-d4, we show the maximum number of rounds that the delay can take, according to the random asynchronous update scheme (see Methods). (E) Effect of delay setups on transient induction of Foxp3 along high Ag dose trajectories. (F) Effect of delay setups on transient induction of IL-2 along high Ag dose trajectories. (see Methods for additional details).
The main FFL begins with signals from TCR/CD28 propagating to either the IL-2/CD25 pathway (Box 1 in Fig. 2A) or the mTOR pathway (Box 2), and then converging on Foxp3. Because the link from Box 2 to Foxp3 is inhibitory, the signals propagating on parallel branches have opposite sign, leading to designation of this motif as an incoherent FFL (iFFL) (62). A general feature of iFFL's is that they may exhibit transient activation of the output, in this case Foxp3.
In addition to the iFFL, there exist feedback loops between Foxp3 and each of the FFL branches. (Fig. 2, A and B). The mutual inhibition between Foxp3 and mTOR (Fig. 2A) produces a toggle switch (62), which forms the central circuit governing the T cell fate decision. By inhibiting Foxp3 (Fig. 2B), sustained activation of the mTOR pathway leads to a stable Th phenotype. On the other hand, activation of Foxp3 activates PTEN, which then turns off the mTOR pathway, producing a Treg phenotype. Positive feedback between Foxp3 and CD25 (Fig. 2A) also stabilizes FOXP3 expression. Negative regulation of mTOR by Foxp3 in our model is supported by the experimental observation that TCR-mediated mTOR activation is attenuated in Foxp3pos Tregs (18).
As discussed in detail below, the simulation results suggest that relative timing of several pathways is critical for determining the final state of the toggle switch and hence the T cell fate decision. These relationships include time of activation of the mTOR pathway relative to activation of IL-2/CD25 pathway, and time of activation of Foxp3/PTEN feedback relative to signaling from the TCR.
Event Timing and Delays
The model describes regulatory interactions that can propagate through the network at different speeds. In order to capture these differences in the logical model, we introduced buffers within key regulatory elements that control the delay between receipt of an input signal and a change in the output state (blue boxes in Fig. 2, B and C). The model setups used to investigate the effects of introducing delays, labeled SD1-SD15, are presented in Fig. 2D, and the resulting time courses following high and low dose Ag stimulation are shown in Fig. 2, E and F, respectively. Delaying mTOR inhibition of Foxp3 (SD3-SD5, SD8-SD10, and SD13-SD15) increases the magnitude of the Foxp3 transient (Fig. 2E, top). Fast signal propagation through the AP-1 activation pathway (SD1-SD5) and, consequently, fast transcription of IL2, leads to fast activation of STAT5 and transcription of FOXP3 before the inhibitory effect of mTOR activation reaches Foxp3, resulting in a large percentage of cells transiently becoming Foxp3pos (Fig. 2E). Conversely, when the delay in AP-1 is long with respect to the delay along the mTOR pathway (SD11-SD15, Fig. 2E, bottom, and fig. S3), the percentage of Foxp3pos cells decreases, becoming closer to the experimentally observed value of 10% (see below).
The model also predicts that cells destined to become Foxp3pos Treg transiently become IL-2pos (Fig. 2F). The magnitude of the transient increase in IL-2pos cells is similar for all delay scenarios (Fig. 2F, top), but can shift to earlier times depending on the timing of AP-1 activation and its effect on Foxp3 (Fig. 2F, bottom). For the simulations described in the rest of the paper, we used the delay setup defined as SD14 in Fig. 2D, which best reproduces data for transient appearance of Foxp3pos cells, as discussed below (see fig. S3 for additional results). Sensitivity analysis (see Methods for more detail) shows that qualitative results do not depend on the choice of delay setup.
Main Differentiation Scenarios
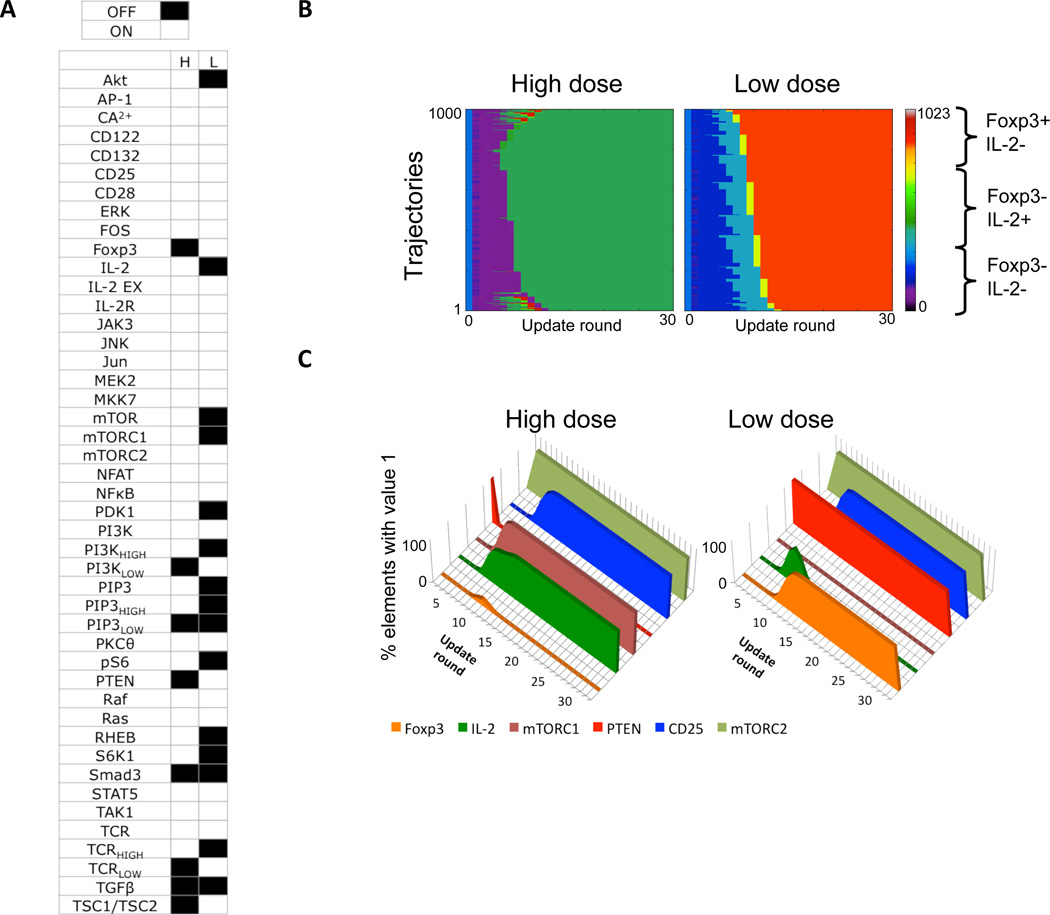
We validated our model based on its ability to reproduce the main results of several key experiments namely high Ag dose alone, high Ag dose + inhibitors, high Ag dose + TGF-β, and low Ag dose alone. For both low and high Ag dose stimulation, all trajectories start from a single initial state (fig. S4) and eventually reach a state from which no further transitions are possible. This state is known as a fixed-point attractor, because it represents a point in the state space to which many trajectories converge. High and low Ag dose trajectories reach distinct fixed points, characterized by the two sets of element values shown in Fig. 3A. In addition to the phenotype marker molecules, Foxp3 and IL-2, elements that have different values for the two scenarios include PTEN, Akt, and mTORC1, emphasizing their role in differentiating between the two phenotypes. A total of 16 variables had different values and thus distinguished between the two scenarios (Fig. 3A). We selected a subset of ten key variables – Foxp3, IL-2, PTEN, TCR, Ras, CD25, PI3K, Akt, mTORC1, and mTORC2 – that included all of the major pathways differing between the scenarios, and removed those that were redundant.
Figure 3.
Results for high and low Ag dose scenarios. (A) Values of individual elements in the steady state attractors that are reached following high (H) and low (L) Ag dose (black=OFF, white=ON). (B) Heat map representation of 1000 simulation trajectories for each scenario. (C) Average values of six key elements along the trajectories in (B).
The dynamics of the system are shown using trajectory heat maps (Fig. 3B), for which we used a Gray coding scheme (63) to map the ten-element binary vector representing the system state at a given time into a color on the heat map (see fig. S5 for further details). This approach allows for compact representation of trajectories, in which green and yellow shades represent states with active IL-2, orange and red shades represent states with active Foxp3, and purple and dark blue shades represent states with most elements being inactive. Values for six of the key elements, averaged from the 1000 trajectories computed for each dose, are shown in Fig. 3C.
High Ag dose
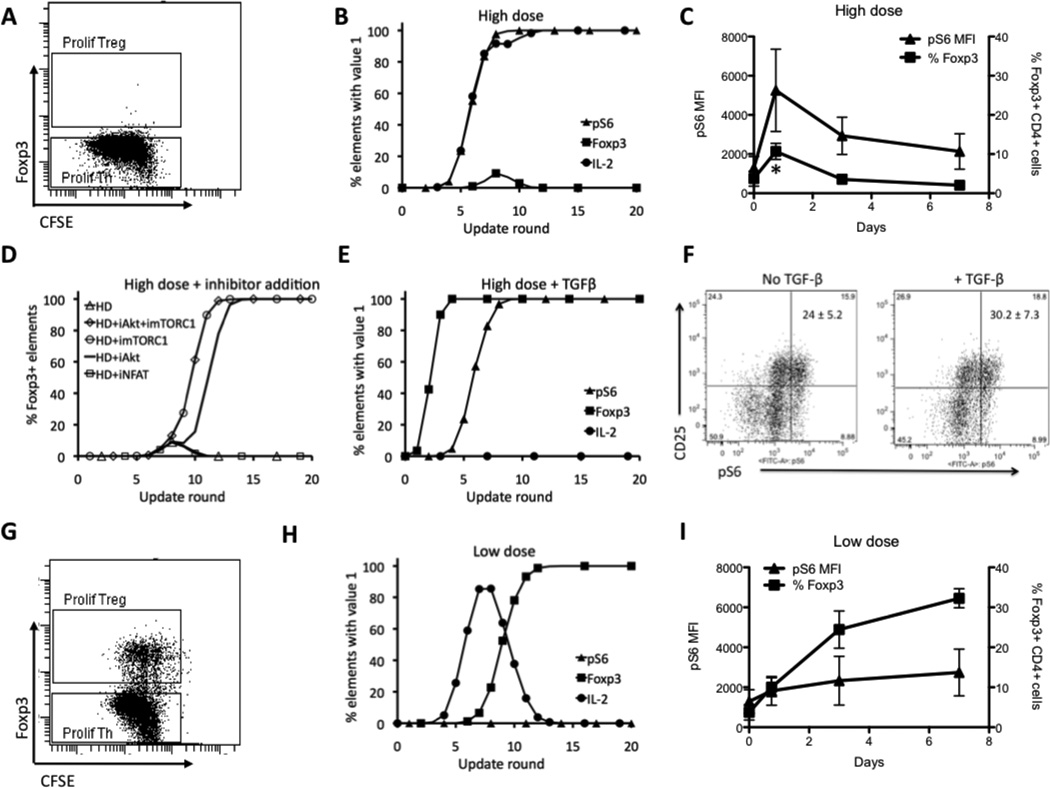
Our previous experimental results (18) showed that high Ag dose produces mostly Th cells, as demonstrated by the presence of proliferating (CFSE low) Foxp3pos cells (Fig. 4A). Our simulations of high Ag dose stimulation also showed predominance of the Th phenotype, and revealed in addition that a substantial number of trajectories pass through a Foxp3pos state (Fig. 4B). We subsequently confirmed in experiments that about 10–20% of cells become Foxp3pos cells 18 hours following stimulation with high Ag dose (Fig. 4C. p<0.05 18 hours compared to all other time points; one way ANOVA with Tukey post test). This information was used to identify the best-fitting delay scenario as SD14 (see Fig. 2D, left). Our previous experiments (18) also showed that pS6, a surrogate measure of mTOR activity, peaks at 18 hours following high Ag dose stimulation and correlates inversely with the number of Foxp3pos cells remaining after the 7-day culture (Fig. 4C). Simulation results also recapitulate the negative correlation between pS6 and the percentage of Foxp3pos cells (Fig. 4B)
Figure 4.
Logical model recapitulates behavior of key elements in the system. (A) Experimental results for a population of cells stimulated with high Ag dose show that almost all cells in the population are Foxp3neg. (B) Simulations results for the same conditions as in B, showing behavior of Foxp3, pS6, and IL-2. (C) Experimental data showing transient increase in the % of Foxp3pos T cells (squares) following high Ag dose stimulation. Mean fluorescence intensity (MFI) values of pS6 (triangles) are maximal at early time points and are maintained throughout the stimulation * p<0.05 18 hours compared to all other time points; one way ANOVA with Tukey post test). (D) Simulation results for Foxp3 for stimulation with high Ag dose only (HD) and with addition of inhibitors at round 6, Akt inhibitor (HD+iAkt), mTOR inhibitor (HD+imTOR), both Akt and mTOR inhibitors (HD+iAkt+imTOR), and NFAT inhibitor (HD+iNFAT). (E) Simulation results for Foxp3, pS6 and IL-2 for stimulation with high Ag dose in the presence of TGF-β. (F) Experimental results showing CD25 and pS6 measured at 18 hours in the population of cells for high Ag dose stimulation without (left) and with (right) TGF-β. Results shown are representative of three independent experiments and the numbers in the upper right quadrants represent the mean ± SEM of CD25pos pS6pos cells of the 3 experiments. (G) Experimental results for a population of cells stimulated with low Ag dose show that there is a fraction of cells that have high value of Foxp3. (H) Simulations results for the same conditions as in B, showing behavior of Foxp3, pS6, and IL-2. (I) Experimental results showing the % of Foxp3+ Treg (squares) and pS6 MFI (triangles) at the indicated time points following T cell stimulation with low Ag dose. Data presented in panels A and F are representative of 3 independent experiments and those shown in panels C and I depict the mean ± SEM of 3 independent experiments.
The trajectory heat map for high Ag dose (Fig. 3B, left, and fig. S6A) reveals three distinct clusters of behavior. Although all trajectories reach the same IL-2pos attractor, approximately 10% of the trajectories fall into the top cluster, characterized by the appearance of red pixels, which indicate cells that are transiently Foxp3pos. The 80% of trajectories in the middle cluster never become Foxp3pos. About 10% of the trajectories fall into the bottom cluster, in which cells are transiently Foxp3pos followed by a loss of both Foxp3 and IL-2 (purple pixels) before becoming IL-2pos. The transient appearance of Foxp3pos cells arises from the governing incoherent FFL involving IL-2/CD25 and PI3K/Akt/mTOR, as discussed above (Fig. 2A).
High Ag dose + inhibitors
Experimental results have shown that the addition of Akt and mTOR inhibitors after 18 hours of strong TCR stimulation enhances the generation of Foxp3pos cells, while the addition of NF-AT inhibitors has no effect (27). As in the experiments, when we modeled inhibition by setting either Akt or mTORC1 to OFF after six simulation rounds, all trajectories produced the Treg phenotype (Fig. 4D). Conversely, when the NF-AT inhibitor was added after six rounds, there was no increase in Foxp3pos cells (Fig. 4D).
High Ag dose + TGF-β
TGF-β activates Smad3, which, along with TCR-induced NF-AT, contributes to the induction of Foxp3 by promoting acetylation at the Foxp3 enhancer (64). Smad3 activation induced by TGF-β apparently counteracts the negative influence exerted on Foxp3 by active mTORC1, leading to strong Treg induction even at high antigen dose (58). Simulations (Fig. 4E) recapitulate these experimental observations, and also predict that Foxp3 rises with faster kinetics upon co-stimulation with TGF-β than with high Ag dose alone (Fig. 4C), because activation of Foxp3 is more direct (see Fig. 1). In addition, the model predicts that pS6 remains activated in this scenario (in contrast to the low Ag dose case discussed below), because sustained high Ag dose stimulation of the TCR overrides negative regulation of mTOR signaling through the Foxp3->PTEN feedback loop (Fig. 2, A and B).
We conducted further experiments to test the prediction that induction of Treg by TGF-β occurs despite sustained mTOR activity as indicated by pS6. T cells stimulated with high dose anti-CD3 in the presence or absence of TGF-β exhibited equivalent abundance of pS6 following 18 hours of stimulation (Fig. 4F). As expected, the addition of TGF-β induced a significant increase in the number of Foxp3+ CD4+ cells following five days of culture compared to cultures without TGF-β (31.7% ± 10.1 with TGF-β vs. 8.8% ± 4.2 without TGF-β; p <0.01 one way ANOVA with Tukey’s Multiple Comparison post test). Thus, as predicted by the model, the induction of Treg by TGF-β does not involve or require inhibition of the mTOR pathway.
Low Ag dose
Experimental results show that low Ag dose generates a high fraction of Foxp3pos Treg cells (Fig. 4G). Simulation trajectories following low Ag dose stimulation uniformly result in Foxp3pos cells without production of pS6 (Fig. 4H). This agrees with experimental measurements showing a reduced induction of pS6 at 18 hours compared with high Ag dose stimulation, which is followed by an increased number of Foxp3pos cells at seven days (Fig. 4I). On the other hand, whereas 100% of the model trajectories reach the Treg steady state (phenotype) at low Ag dose, in experiments only about 35–50% of cells are Foxp3pos (Fig. 4G). Thus, the model fails to capture the observed heterogeneity in differentiation outcome. A possible source for this discrepancy is the known variability in the abundance of TCR or other signaling molecules (65) within the naïve T cell population, which is not directly accounted for in the model. Alternatively, variability in the activation timing could also account for this, an issue we address below.
Varying the Duration of TCR Stimulation
The failure of the model to reproduce the observed heterogeneity of differentiation outcomes in the low Ag dose scenario prompted us to explore potential mechanisms, beyond changing activation thresholds between low and high Ag dose stimulation. Sauer et al. (27) had shown that Ag removal at 18 hours after stimulation results in a significant expansion of Treg in the culture, which they attributed to a reduction in the signaling via the mTOR pathway. Other recent reports have shown that the duration of TCR signaling depends on the dose of the stimulating Ag (66, 67).
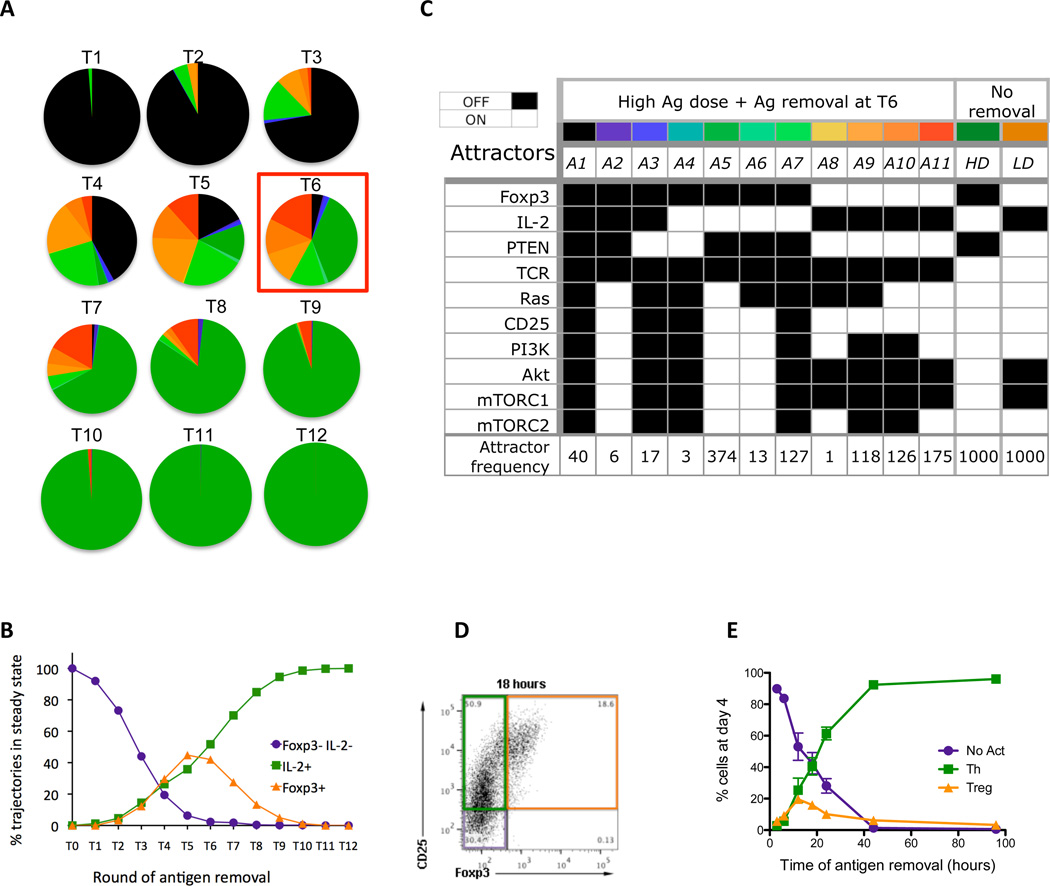
We examined the effect of varying the duration of TCR stimulation by removing the Ag at various times post stimulation but keeping the same update rules as for high Ag dose. Under these conditions, simulations reach eleven different fixed-point attractors, and their relative frequency changes with the time of Ag removal (Fig. 5, A and B). These attractors fall into three major cell phenotypes; Treg, Th and non-activated (Fig. 5B). If Ag is removed too early, e.g., before time point 2 (T2), only non-activated (IL-2neg Foxp3neg) cells result (Fig. 5A). If Ag is removed after T8, only Th (IL-2pos Foxp3neg) cells are observed. Ag removal between T3 and T8 results in increasing numbers of Treg (IL-2neg Foxp3pos) cells, with a peak at T5 and T6 (Fig. 5B, see fig. S7 for complete results).
Figure 5.
Analysis of transient stimulation with high antigen dose. (A) Distribution of attractors when antigen is removed at different times, starting with removal at simulation round 1 (Tl), and incrementing the time of removal by one round, up to round 12 (T12). (B) Changes in sizes of lumped attractors (Foxp3- IL-2- population, IL-2+ population and Foxp3+ population (C) 11 attractors that are observed in simulations of antigen removal scenario T6 (colors correspond to those in part (A)), presented in terms of steady state values of ten elements (Foxp3, IL-2, PTEN, TCR, Ras, CD25, PI3K, Akt, mTORCl, mTORC2) and the number of simulation trajectories ending in each given attractor for 1000 simulation runs. (D) Flow cytometry plot identifying the three major populations induced. Purified CD4+ T cells were stimulated on plates coated with anti-CD3 (lug/ml) in the presence of soluble anti-CD28 mAb. After 18 hours of culture the cells were removed from the coated plates and placed in new wells and cultured for a total of 96 hours. Cells were analyzed by flow cytometry and results show the CD25 and Foxp3 staining on gated CD3+ CD4+ cells. CD25- Foxp3- cells (purple box) are non-activated T cells, CD25+ Foxp3- cells (green box) are Th cells and CD25+ Foxp3+ (orange box) are Treg cells. (E) Purified CD4+ T cells were stimulated as before and the cells were removed from the stimulation at the indicated times (3, 6, 12, 18, 24, 44 and 96 hours). All cells were cultured for a total of 96 hours and then were analyzed by flow cytometry to identify non-activated (No Act, purple circles) Th (Th, green squares) and Treg (Treg, orange triangles) cells, as defined in Panel D. The results are the mean ± SEM of 4–8 replicates from 3 independent experiments.
We selected the scenario T6 for further analysis, because it generates all eleven attractors (A1–11), which can be distinguished by variations in the state of key element values (Fig. 5C). Attractors A1-A3 correspond to non-activated cells (Fig. 5C). Attractors A4-A7 and A8-A11 represent Th and Treg phenotypes respectively. Of the Th attractors, A5 is the most common outcome and is nearly identical to the high dose attractor (Fig. 5C). The Treg attractors A9-A11 occur with equal frequency, and A11 is most closely related to the low dose attractor (Fig. 5C). A9 and A10 differ from A11 and low dose attractor in the elements Ras (ON in A10 and A11, OFF in A9), PI3K and mTORC2 (ON in A11 and OFF in A9 and A10). These results suggest that the state values of Ras, PI3K and mTORC2 are not critical to the outcome, whereas Akt and mTORC1 are more important for the Treg phenotype.
The presence of 11 different cell fates following the removal of antigen prompted us to determine whether these could be identified experimentally. Thus, we activated purified CD4pos T cells with a high dose of immobilized anti-CD3 mAb and removed the cells from the stimulation at various time points following the activation. When cells were removed from antigen after 18 hours and analyzed after an additional 78 hours without stimulation, we observed three distinct populations of cells (Fig. 5D). As in the simulations CD25neg Foxp3neg, CD25pos Foxp3neg, and CD25pos Foxp3pos cells representing non-activated, Th and Treg cells respectively were observed. Varying the duration of antigen stimulation from 3 to 96 hours revealed a distribution of phenotypes (Fig. 5E) in excellent agreement with modeling results (Fig. 5B).
Dynamics of the Cell Fate Decision
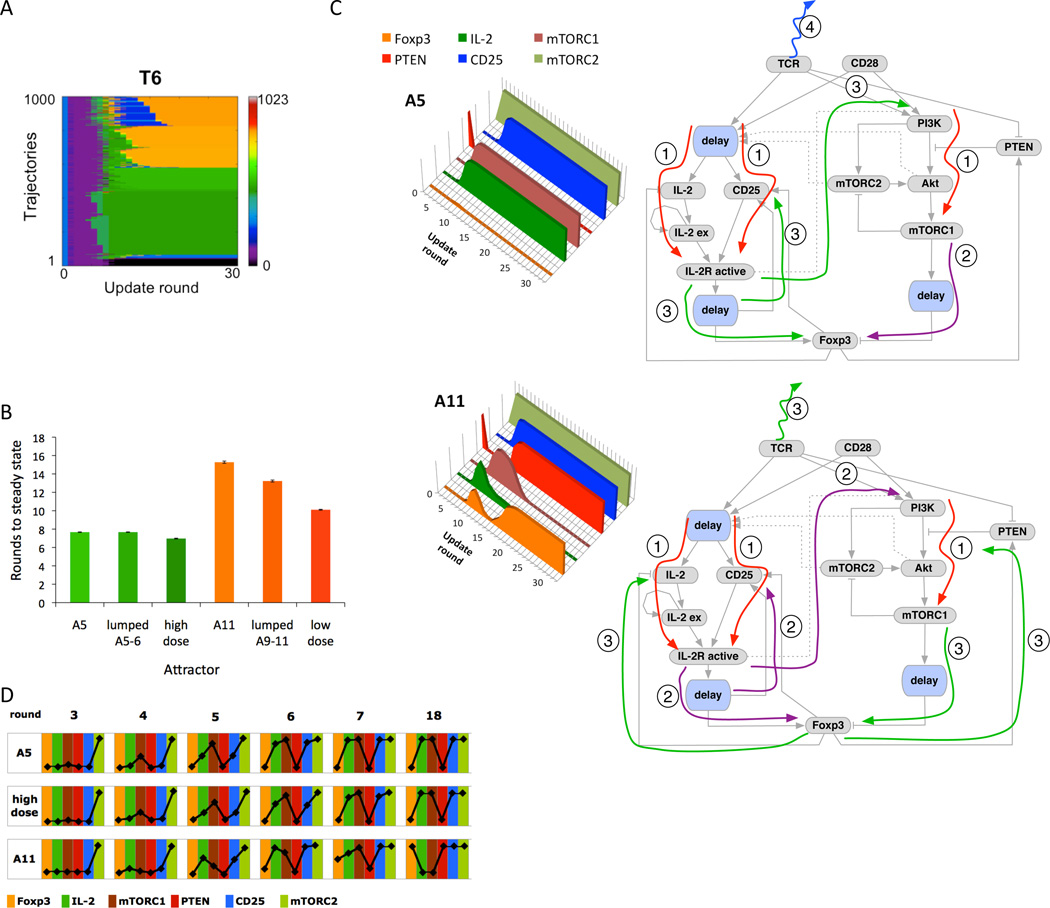
The observation that removal of the antigen at T6 resulted in eleven different possible outcomes gave us the opportunity to analyze in detail the factors that affect cell fate selection. Fig. 6A presents a heat map of 1000 simulation trajectories for this scenario, clustered by steady state attractor sorted from A1 (bottom rows) to A11 (top rows). Additional heat maps showing system trajectories in scenario T6, for different delay setups, are presented in fig. S8B. Differentiation into Th phenotype appears to occur faster than differentiation into Treg, so we calculated the mean times required to reach steady state. We found that on average it took eight rounds longer to reach the Treg (Foxp3pos) phenotype than the Th (IL-2pos) phenotype (Fig. 6B). This held true for all attractors that led to IL-2pos vs. Foxp3pos- fate (Fig. 6B).
Figure 6.
Analysis of timing effects for Ag removal scenario. (A) Trajectories of 1000 model simulations for scenario T6 (Ag removal at round 6) showing different transient behavior and different steady-states. (B) Average number of simulation rounds to reach steady state for dominant attractors for scenario T6 and high- and low-Ag dose, shown with standard error marks. (C) Left: Average values for six key elements leading to attractor A5 (top) and attractor A11 (bottom) computed from 374 and 175 trajectories respectively. Right: Diagrams summarizing the order of events along trajectories leading to attractor A5 (top) and A11 (bottom). Corresponding plots and diagrams for the remaining attractors are shown in fig. S6. (D) Values of six elements from part (C), averaged across simulations leading to each of the attractors (374 for A5, 1000 for high dose, and 175 for A11) for scenario T6, in rounds 3,4,5 (before antigen removal), 6 (when antigen is removed), 7 (transient round), and 18 (when all trajectories reach steady state).
The differences in the speed of reaching the Foxp3pos and IL-2pos attractors arise from differences in the order in which feedback and feed-forward loops are activated (Fig. 6C, right), which in turn is a result of the stochasticity inherent in our simulations as mentioned above. IL-2pos phenotypes result from situations where antigen removal effects propagate slowly enough to permanently shut off Foxp3 (Fig. 6C top). Foxp3pos phenotypes have two main requirements: (1) early (transient) activation of Foxp3 occurs via the CD25/STAT5 pathway; (2) early activation of Foxp3 leads to activation of PTEN and inhibition of mTOR. Foxp3neg/IL-2neg phenotypes (fig. S9 and fig. S10) arise from either (1) slow propagation of the TCR ON signal or (2) rapid shutoff of activation following Ag removal.
Our analysis shows that the critical factor distinguishing trajectories leading to A5 (the dominant IL2pos attractor) and A11 (the dominant Foxp3pos attractor) is transient induction of Foxp3 in the rounds just following Ag removal (Fig. 6, C and D). Although the fraction of Foxp3pos trajectories is less than 100% for any specific round, in fact all trajectories leading to A11 are transiently Foxp3pos at some point following Ag removal. Foxp3 begins to activate PTEN at round 8, and this activation is complete by round 11. After this, PTEN remains active because of the absence of further TCR stimulation, and mTORC1 activation falls rapidly, which paves the way for Foxp3 to be turned on again and for stabilization of the Treg phenotype (Fig. 6C).
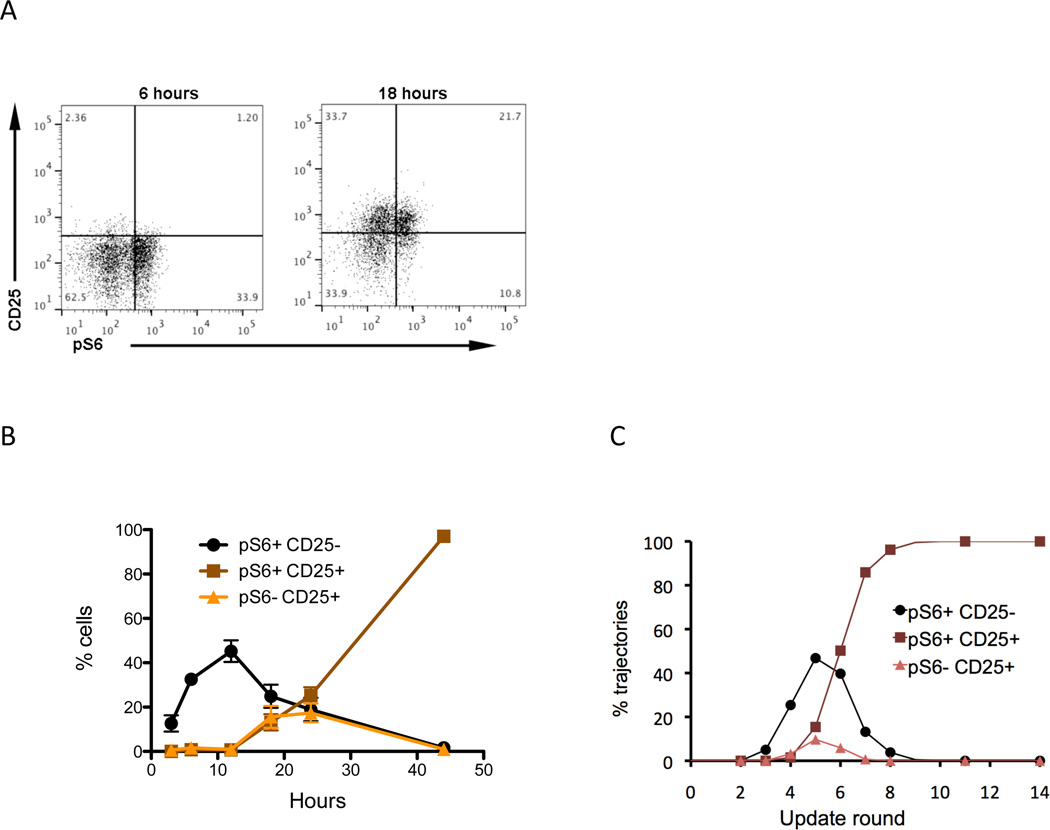
We questioned whether we could predict the subsequent behavior of the A5 and A11 attractors from the state of the cell at the time of antigen removal (Fig. 6D). The increase in mTOCR1 activation appears to occur a little slower in the A11 trajectory such that by round 6 the average mTORC1 activity is lower in A11 than in A5 or in the high dose trajectories (Fig. 6D). We measured the level of mTORC1 activity experimentally by assessing the presence of pS6, which is downstream of mTORC1 activation. Following 6 hours of activation we observed a significant proportion of pS6pos cells but no CD25pos cells (Fig. 7A, left panel). After 18 hours activation CD25pos cells were visible and some were also pS6pos, whereas others were pS6neg (Fig. 7A, right panel). We followed the relative proportions of pS6pos, CD25pos, single and double positive cells over time and observed an early peak of mTORC1 activation (pS6pos) in the absence of CD25 (Fig. 7B). There was a peak of CD25pos pS6neg cells between 12 and 18 hours and the double positive cells increased progressively from 12 hours on (Fig. 7B). We analyzed the same populations in the model and observed very similar kinetics for these three populations in the simulations (Fig. 7C).
Figure 7.
State of cells at the time of removal. (A) Flow cytometry demonstrating the activation state of T cells stimulated for 6 and 18 hours. Purified CD4+ T cells were stimulated on plates coated with anti-CD3 (1ug/ml) in the presence of soluble anti-CD28 mAb. After 6 or 18 hours of culture the cells were analyzed by flow cytomtery for the presence of pS6 and CD25 on gated CD3+ CD4+ cells. (B) Time course of activation state of T cells activated with a high dose (1–2 µg/ml) anti-CD3 and anti-CD28. Cells were analyzed at the indicated times for the presence of pS6 (a measure of mTORC1 activation) and CD25. Three populations are followed pS6+ CD25- (green circle), pS6+ CD25+ (red square) and pS6- CD25+ (blue triangle) over time. The results are the mean ± SEM of 4–8 replicates from 3 independent experiments. (C) Distribution of pS6+ CD25-, pS6+ CD25+, and pS6- CD25 negative trajectories for high Ag dose stimulation, at different rounds before antigen is removed.
Discussion
We have developed a logical network model of TCR signaling (Fig. 1) aimed at investigating the role of Ag dose in determining T cell fate following activation of critical signaling pathways downstream of TCR, CD28, IL-2, and TGF-β receptors. The model reproduced important experimental results (Table 1), including the induction of Foxp3neg Th cells by high dose stimulation and the induction of Foxp3pos Treg by low dose stimulation (Fig. 3). In addition, it captured the experimentally observed inverse correlation between induction of Treg and activation of the mTOR pathway, as measured by S6 phosphorylation (Fig. 4, B, C, H, and I). It also made several novel predictions that we have verified experimentally (Table 1), including the transient appearance of Foxp3pos cells in a proportion of T cells activated with high Ag dose (Fig. 4, B and C) and sustained mTOR activation in Treg induced by a combination of TGF-β and high Ag dose (Fig. 4, E and F). The model also predicted and experiments confirmed that transient high dose Ag stimulation results in three stable T cell fates (Th, Treg, and inactive) with relative proportions depending on the duration of stimulation (Fig. 5). Experimental analysis of the cell population at the time of Ag removal identified three distinct populations based on CD25 abundance and mTOR activation that correlated with these T cell fates (Fig. 7). Overall this model has provided new insights into how TCR signal strength influences T cell differentiation and has demonstrated an important role for the differential timing of key signaling elements in determining cell fate.
We observed in simulations that a proportion of T cells exposed to high Ag dose transiently became Foxp3pos (Fig. 4B). The early increase in the number of Foxp3pos cells following stimulation with high Ag dose, had been shown previously in human (68), and more recently, in mouse T cells (69). This can be explained by an incoherent feedforward loop starting with TCR activation and passing through the activation of IL-2/CD25/STAT5 and PI3K/Akt/mTOR pathways (Fig. 2A). Upon stimulation by high Ag dose, Foxp3 can be transiently induced by STAT5, but is ultimately blocked by activation of the mTOR pathway. We tested this prediction of the model in our experimental system using mouse T cells and observed that a proportion of T cells became transiently Foxp3pos upon stimulation with high antigen dose (Fig. 4C). A more detailed analysis of these activation-induced Foxp3pos T cells has revealed that these cells do not have suppressor function and exhibit increased methylation of the Foxp3 locus (69). As discussed in more detail below, however, our simulations also revealed that the transient increase in Foxp3pos cells plays a critical role for Treg differentiation in situations of low or unsustained TCR signaling.
The model also suggests a possible explanation for the apparent instability of Treg induced by the TGF-β cytokine. It is well established that the TGF-β can induce Treg when combined with high dose Ag stimulation (16, 20). Tregs induced in this fashion are Foxp3pos and are suppressive, but recent data suggest that this phenotype is not stable following TGF-β removal, and is associated with incomplete demethylation of the Foxp3 locus (70). Simulations of the model in which TGF-β was present along with high Ag dose show rapid induction of Foxp3pos cells accompanied by sustained activation of the mTOR pathway (Fig. 4E). This was in contrast to our experiments and model simulations with low Ag dose in the absence of TGF-β showing that the induction of Foxp3+ cells was associated with reduced mTORC1 activation (Fig. 4H). We tested this prediction experimentally and made the novel observation that the proportion of cells with active pS6, a marker of mTORC1 activity, was maintained in the presence of TGF-β, despite the induction of significant numbers of Treg cells in these cultures (Fig. 4F). These results suggest that the addition of TGF-β overrides the negative effects of mTOR signaling on Foxp3 induction, and provides a mechanism by which depletion of TGF-β can lead to suppression of Foxp3 by activated mTORC1. The continued activation of the mTOR pathway in this setting may thus provide an explanation for the instability of Treg induced by TGF-β under conditions of high TCR signal.
The logical modeling approach allowed us to model complex behaviors using a simple approach with two values for each variable. A few logical models of T cell differentiation and activation have been developed previously (43–46, 71). Two of these models (43, 71) considered TCR signaling in detail but did not examine the ultimate fate of the activated T cell. Other models describing Th differentiation (44–46) have focused on the interplay between cytokines that drive specific Th cell differentiation, but have not treated molecular pathways in detail or the effect TCR signal strength. Thus, our model bridges a gap in the existing literature by connecting TCR signaling cascades to the fate of the differentiating T cell. A limitation of the current model is that it considers only two major outcomes – Treg and Th – without tracking further differentiation of Th cells into Th1, Th2 or Th17. It is known that Ag dose has a profound influence on the differentiation of Th1 and Th2 cells (66), and we plan to further develop the existing model to include these cell fates in the future.
Although all elements of the model were initially given only two values (ON or OFF), we observed that this could give rise to oscillatory behavior in the low dose scenario, where a steady state was never reached. This was overcome by allowing two of the elements (PI3K and PIP3) to have three values (OFF, LOW and HIGH), and by setting different activation thresholds for low and high Ag doses. These were centered on the ability of the TCR signal to inhibit the critical negative regulator PTEN and to activate PKCθ and the PI3K/Akt/mTOR pathway. These initial attempts to model Treg induction following low dose simulation resulted in a model that was too rigid – producing 100% Foxp3pos cells (Fig. 4H), instead of mixed population with 35–50% Tregs observed experimentally (Fig. 4G).
We subsequently used an alternative interpretation of low dose stimulation in which the duration of TCR activation was varied while keeping the same activation threshold. Sauer et al. (27) have shown that the removal of Ag after a short period of stimulation results in a significant expansion of Treg in the culture, which was attributed to a reduction in the signaling via the mTOR pathway. Recapitulating this scenario by using the high Ag dose activation thresholds but varying the length of time that the TCR signal was on, we found an optimal duration of TCR signaling that generated both Th and Treg phenotypes in roughly the same proportions that we observe experimentally (Fig. 5, A and B). We confirmed this prediction experimentally by showing that removing T cells from the activating stimulus at various times also generating the same pattern of T cell differentiation outcomes (Figure 5, D and E). This result suggests that differential activation of signaling pathways downstream of TCR by high and low dose Ag can arise from changes in the duration of contact time, which is consistent with several recent experimental findings. These reports showed that Treg induction in vivo by low dose Ag requires a high affinity pMHC/TCR interaction (22, 66). This difference was related to the induction of IL-2 responsiveness, such that, in contrast to low doses of high affinity peptides, high doses of low affinity peptides failed to induce CD25 robustly and thus cells activated in this way exhibited reduced STAT5 signaling in response to IL-2 (66). Imaging studies have also revealed that only a small percentage of T cells sensing low affinity pMHC had long-lasting stable contacts with DC, compared to T cells sensing low doses of high affinity pMHC (66). A recent paper (72) has shown that a single Ag-specific T cell in vivo can differentiate into different Th cell types and that this is dependent on the dose of the Ag delivered.
Analysis of individual simulation trajectories allowed us to identify critical elements that contribute to determination of cell fate (Fig. 6 and Fig. 7). Two main findings emerged from this analysis. The first was the importance of the feedback loop (Fig. 6C) involving IL-2R, PTEN, Foxp3 and Akt/mTOR. Trajectories leading to Th phenotype have high mTORC1 activation before antigen removal takes place, thus allowing for inhibition of Foxp3 (Fig. 6D, A5 trajectories). Cells that become Treg cells have lower mTORC1 activity, which allows Foxp3 to be induced and to increase activity of PTEN. At later times, PTEN activation leads to inactivation of mTORC1 paving the way for sustained Foxp3pos cells. Experimentally we showed that the mTORC1 activation occurs very early prior to the induction of CD25, and at later time points the culture became dominated by CD25pos cells with high mTORC1 activation. At intermediate time points an additional cell type was identified: CD25pos without mTORC1 activation (Figure 7B). We hypothesize that the heterogeneity in T cell activation status observed at these intermediate time points contribute to plasticity in cell fate observed when activation is interrupted at these times.
The second major finding from trajectory analysis was that differentiation into Treg took much longer than differentiation into Th (Fig. 6B and Fig. 7). This suggests that there is fundamentally more instability in the pathway towards Treg differentiation. Th cells switch more-or-less directly from the naïve state to the final Th phenotype, whereas cells destined for Treg pass through an additional intermediate state where they produce IL-2 before turning on PTEN, which enables permanent activation of Foxp3.
PTEN, which emerged as a critical regulator in this analysis, is known to play a critical role in modulating the activity of the mTOR pathway, but the regulation of its abundance and activity is not well understood. Two potential mechanisms for the regulation of PTEN are included in the current version of the model: high Ag dose decreases activity of PTEN, whereas Foxp3 activation increases it. This is consistent with experimental observations that Tregs have higher amounts of PTEN than other T cells and that they signal poorly via AKT/mTOR (73). Although we are not aware of direct evidence that Foxp3 causes increase in abundance of PTEN, the work in (73) shows that PTEN is responsible for the reduced activation of mTOR in Tregs, and that activation of Treg causes substantial increase in PTEN abundance. The combination of transient FOXP3 induction coupled with the ability of Foxp3 to increase PTEN was responsible for the generation of Treg cells in the antigen removal scenarios, suggesting one possible mechanism for stabilization of the Foxp3pos phenotype. Further experimental work is necessary to define the mechanisms by which Foxp3 leads to increases in PTEN protein and function.
In conclusion these studies suggest that logical modeling can provide important new insights into the role of TCR strength in determining T cell differentiation outcome. The model made several predictions that were confirmed experimentally, and further refinements to the model will increase its power and usefulness to immunologists. In the future this model can be refined to aid in the design and development of targeted immunotherapy for diseases, such as autoimmunity, cancer and transplant rejection, in which the Th subsets that are induced play a critical role in disease outcome.
Materials and Methods
Discrete Logical Model Development
In order to study the control of T cell differentiation, we created a logic circuit model following an iterative procedure. First, we defined key elements in the system to include in the model. The next step was defining element regulatory sets, and the number of values that are necessary to describe each element. As we already discussed in previous sections, we aimed to restrict the number of values to the minimum that still allows us to reproduce experimental results. Therefore, we initially chose all discrete variables that represent individual elements to have two values (OFF and ON), except TCR variable, which needs to have three values (OFF, LOW, and HIGH) to implement low and high dose scenarios. Since the negative loop between mTORC1 and mTORC2 includes elements with only two values, it always oscillates. To avoid oscillations that do not exist in the real loop, we decided to model PI3K (and consequently, PIP3), which is upstream of the mTORC1/mTORC2 loop and, in our model, is a direct regulator of mTORC2 and an indirect regulator of mTORC1, with three values, similar to TCR. Another solution would be to model all elements in the loop with three values, and we plan to include such analysis in the future.
For those elements that are implemented with multi-valued variables, translation of a discrete variable into a vector of Boolean variables is necessary. In our case, this is simple, since it includes translating from three-valued to two-valued variables, that is, defining LOW and HIGH variables for TCR, PI3K and PIP3. When developing logic rules, we decided how to include LOW vs. HIGH variables in the rules according to experimental results and literature review.
Model Simulation Method
Having built the model, we ran simulations in order to determine whether this model could accurately capture the experimental results that we obtained previously. Simulations were carried out using BooleanNet tool (60). Logical rules were updated in each simulation step using a random asynchronous approach, where in each step, only one variable is chosen and the rule associated with that variable is evaluated to obtain new value for the variable. The choice of rule to be evaluated next can follow different algorithms. The procedure that is applied in the BooleanNet tool is a random choice of the order of rules in one update round. In other words, in each update round all rules are accessed and evaluated once, but the order in which this is done is random. We simulated and analyzed several scenarios, and for each scenario, 1000 independent simulations were conducted. Each run consisted of up to 30 update rounds. Stable presence of Foxp3 is used as a marker for the Treg phenotype, and stable presence of IL-2 is used as a marker for the Th phenotype.
In order to run simulations, it is first necessary to decide the initial conditions, that is, initial variable values when the simulation starts. Since we are interested in studying the differentiation of naïve T cells and understanding the rationale behind the differentiation, we start with the variable values that best represent the naïve T cell state. Most of the variables are assumed to initially have a logical ‘0’ value, which means that they are inactive or absent in the beginning. The only elements and their corresponding variables that are assumed to be active or present initially are:
-
-
TCR (either low or high variable, depending on the scenario we are analyzing)
-
-
CD28 (stimulation without co-stimulation is suggested to lead to anergy)
-
-
CD122, CD132 (assumed to be constitutively active)
-
-
PTEN (suggested to be have a non-zero value in naïve T cells)
-
-
TSC1-TSC2 (assumed to be ON, such that it inhibits Rheb until Akt is activated by TCR signaling).
Analysis of Simulation Trajectories
To emphasize stochastic behavior of the system leading to different trajectories for the same initial conditions and same stimulation scenario, we visualize the computed system trajectories using heat maps (Fig. 3B and Fig. 6A). Each color in a heat map represents a different value of a vector of ten Boolean variables implementing key elements of the model (Foxp3, IL-2, PTEN, TCR, Ras, CD25, PI3K, Akt, mTORC1, mTORC2). We selected these elements based on the fact that they can be used to characterize the system at any time point with minimal loss of information. A heat map shows 1000 simulation trajectories of the same scenario (same stimulation and initial state of the ten-element vector), with each row representing a single trajectory (changes in ten-element vector from initial state, to simulation round 30). In order to group similar trajectories in the map, we assume that values of the ten-element binary vector encode decimal values using Gray code, a binary number system where two successive values differ in only one bit (see fig. S4). Different from binary code, Gray code is a non-weighted code. For example, 4-digit binary numbers 0111 and 1000 in binary code encode decimal numbers 7 and 8, while the same decimal numbers are encoded using 0100 and 1100 in Gray code. Computation of decimal values according to the Gray code allows for grouping simulation trajectories that are closest in terms of individual element behaviors, since two consecutive decimal numbers will always represent vectors that differ in a single element. The grouping of trajectories is performed by sorting trajectories according to decimal vector values starting from end of the trajectory (last simulated state or steady state) towards beginning (initial state). This sorting approach allows for grouping all trajectories leading to same steady state together (Fig. 6A), and then, within these groups, cluster closer the trajectories that have similar behavior.
To emphasize differences across studied scenarios, we studied individual element behavior by averaging element values in each round across 1000 trajectories, from initial state to round 30. We then compared average trajectories of different elements within same scenario and trajectories of the same element across different scenarios.
Sensitivity Analysis
We conducted an extensive analysis of the sensitivity of our model in the scenarios described above to changes in internal model delays, to turning individual model elements OFF or ON, and to different initial element values. Changes in model delays can be interpreted as variations of model parameters. As expected, such changes lead only to quantitative differences in average values of individual model elements, but not in the qualitative difference in overall system behavior and observed phenotypes (Fig. 2, E and F; fig. S3, fig. S6 and fig. S8). On the other hand, similar to our study of inhibitor addition at simulation round 6, switching elements of the model permanently OFF affects logical rules and can lead to qualitative differences in observed phenotypes (fig. S11). We expand our study of high Ag dose + inhibitor addition scenario, and show that timing does not play an important role in the case of turning model elements OFF (fig. S11B). Interestingly, assuming random initial values for model elements does not affect the steady-state values in high Ag dose and high Ag dose + TGF-β scenarios (fig. S12, first and second column), but it results in a mixed population for low Ag dose stimulation (fig. S12, third column), and can lead to a small percentage of activated cells in the absence of stimulation (fig. S12, fourth column).
DC and CD4+ T cell Purification and Activation
DC were generated as previously described (18), by growing bone marrow precursors for five days in RPMI containing 5ng/ml GM-CSF and 2ng/ml IL-4 (G4DC). G4DC were purified using a 13.5% Histodenz gradient (Sigma), as previously described, to enrich for the mature population. CD4+ T cells were purified from dissociated spleens and LN of BDC2.5 mice by negative selection, using a CD4+ T cell isolation kit (Miltenyi Biotech). DC and T cells were co-cultured in 96-well U-bottom plates (BD Falcon). DC were seeded at 2 × 104 cells/well and peptides added at least two hours prior to the addition of CFSE-labeled CD4+ T cells (1 × 105/well). Low dose and high dose stimulation consisted of 0.01ng/ml and 100ng/ml of the AV10 peptide. Cultures were maintained in complete DMEM, containing 10% FBS, Penicillin & Streptomycin, L-Glutamine, Sodium Pyruvate, Non-Essential Amino Acids, Hepes Buffer, and β-Mercaptoethanol. Cells were fed on days 3, 4, 5, and 6 by replacing half of the medium. Cells were harvested at the indicated times for flow cytometric analysis.
In experiments with TGF-β or for antigen removal purified CD4+ T cells were stimulated on tissue culture wells precoated with a high dose (1µg/ml) anti-CD3 antibody with soluble anti-CD28 in the presence or absence of TGF-β (5ng/ml). In the case of antigen removal cells were removed from the coated plate at various time points and placed in a fresh uncoated plates for the remainder of the culture period. Cells were analyzed by flow cytometry for the presence of pS6 at various time points and for Foxp3 4–5 days following stimulation.
Cell Staining and Flow Cytometry
Cell surface staining was performed in PBS containing 2% FBS, with the following antibodies: anti-CD11c–PE, anti-CD25-APC (eBioscience) and anti-CD4-PerCP (BD Bioscience). Intracellular Foxp3 was stained with anti-Foxp3-PacificBlue antibody and staining buffers from eBioscience, according to the manufacturer’s instructions. Staining with antibodies against phospho-S6 (Cell Signaling Technologies) was performed in conjunction with the Foxp3 antibody and staining buffers. Stained cells were analyzed on an LSR II (BD Bioscience) using FACSDiva software (BD Bioscience).
Supplementary Material
One sentence summary.
Computational modeling and experiments on the factors affecting T cell differentiation in response to antigen dose suggest that differential timing in the activation of central regulators, which is strongly modulated by the duration of T cell receptor signaling, is an important mechanism driving T cell fate decisions.
Acknowledgments
This work was supported by NIH grants P01CA73743 (PM) and UL1-RR024153 (NMZ and JRF), American Diabetes Association Grant 1-06-RA-94 (PM), and NSF Expeditions in Computing Grant 0926181 (NMZ and JRF). MT was supported by a NIH training grant 5T32 CA82084-10. We also acknowledge helpful conversations with John Sekar in the construction of the model
Footnotes
The primary difference between the two representations is that we do not include memory elements in Fig. 1B for clarity of representation. Memory elements are necessary to keep the state of model elements, as all element states are part of the overall system state.
Author contributions
The author(s) have made the following declarations about their contributions: Conceived and designed the experiments: NMZ, PAM, JRF. Performed the experiments: MST, PAM, Developed and tested the logical model: NMZ, JRF. Analyzed the data: NMZ, PAM, JRF. Contributed reagents/materials/analysis tools: LK. Wrote the paper: NMZ, PAM, JRF.
References
- 1.Shevach EM. Regulatory T Cells in Autoimmmunity*. Annual Review of Immunology. 2000;18:423–449. doi: 10.1146/annurev.immunol.18.1.423. [DOI] [PubMed] [Google Scholar]
- 2.Curiel TJ, Coukos G, Zou LH, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, Zhu Y, Wei S, Kryczek I, Daniel B, Gordon A, Myers L, Lackner A, Disis ML, Knutson KL, Chen LP, Zou WP. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10:942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 3.Facciabene A, Motz GT, Coukos G. T-Regulatory Cells: Key Players in Tumor Immune Escape and Angiogenesis. Cancer Research. 2012;72:2162–2171. doi: 10.1158/0008-5472.CAN-11-3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Long SA, Buckner JH. CD4+FOXP3+ T Regulatory Cells in Human Autoimmunity: More Than a Numbers Game. The Journal of Immunology. 2011;187:2061–2066. doi: 10.4049/jimmunol.1003224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sakaguchi S, Wing K, Miyara M. Regulatory T cells - a brief history and perspective. Eur J Immunol. 2007;37(Suppl 1):S116–S123. doi: 10.1002/eji.200737593. [DOI] [PubMed] [Google Scholar]
- 6.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: An Effector CD4 T Cell Lineage with Regulatory T Cell Ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 7.Ziegler SF. FOXP3: of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt A, Oberle N, Krammer PH. Molecular mechanisms of treg-mediated T cell suppression. Frontiers in immunology. 2012;3:51. doi: 10.3389/fimmu.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gondek DC, Lu LF, Quezada SA, Sakaguchi S, Noelle RJ. Cutting edge: contact-mediated suppression by CD4+CD25+ regulatory cells involves a granzyme B-dependent, perforin-independent mechanism. J Immunol. 2005;174:1783–1786. doi: 10.4049/jimmunol.174.4.1783. [DOI] [PubMed] [Google Scholar]
- 10.Banchereau J, Pascual V, O’Garra A. From IL-2 to IL-37: the expanding spectrum of anti-inflammatory cytokines. Nat Immunol. 2012;13:925–931. doi: 10.1038/ni.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bodor J, Bopp T, Vaeth M, Klein M, Serfling E, Hünig T, Becker C, Schild H, Schmitt E. Cyclic AMP underpins suppression by regulatory T cells. European Journal of Immunology. 2012;42:1375–1384. doi: 10.1002/eji.201141578. [DOI] [PubMed] [Google Scholar]
- 12.Hofer T, Krichevsky O, Altan-Bonnet G. Competition for IL-2 between Regulatory and Effector T Cells to Chisel Immune Responses. Frontiers in immunology. 2012;3:268. doi: 10.3389/fimmu.2012.00268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandiyan P, Zheng L, Ishihara S, Reed J, Lenardo MJ. CD4+CD25+Foxp3+ regulatory T cells induce cytokine deprivation-mediated apoptosis of effector CD4+ T cells. Nat Immunol. 2007;8:1353–1362. doi: 10.1038/ni1536. [DOI] [PubMed] [Google Scholar]
- 14.Raimondi G, Turner MS, Thomson AW, Morel PA. Naturally occurring regulatory T cells: recent insights in health and disease. Crit Rev Immunol. 2007;27:61–95. doi: 10.1615/critrevimmunol.v27.i1.50. [DOI] [PubMed] [Google Scholar]
- 15.Bergmann C, Strauss L, Zeidler R, Lang S, Whiteside T. Expansion and characteristics of human T regulatory type 1 cells in co-cultures simulating tumor microenvironment. Cancer Immunology, Immunotherapy. 2007;56:1429–1442. doi: 10.1007/s00262-007-0280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF. Cutting Edge: TGF-Œ≤ Induces a Regulatory Phenotype in CD4+CD25ñ T Cells through Foxp3 Induction and Down-Regulation of Smad7. The Journal of Immunology. 2004;172:5149–5153. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 17.Kretschmer K, Apostolou I, Hawiger D, Khazaie K, Nussenzweig MC, Boehmer Hvon. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 18.Turner MS, Kane LP, Morel PA. Dominant role of antigen dose in CD4+Foxp3+ regulatory T cell induction and expansion. J Immunol. 2009;183:4895–4903. doi: 10.4049/jimmunol.0901459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tarbell KV, Yamazaki S, Olson K, Toy P, Steinman RM. CD25+ CD4+ T Cells, Expanded with Dendritic Cells Presenting a Single Autoantigenic Peptide, Suppress Autoimmune Diabetes. The Journal of Experimental Medicine. 2004;199:1467–1477. doi: 10.1084/jem.20040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamazaki S, Bonito AJ, Spisek R, Dhodapkar M, Inaba K, Steinman RM. Dendritic cells are specialized accessory cells along with TGF- for the differentiation of Foxp3+ CD4+ regulatory T cells from peripheral Foxp3 precursors. Blood. 2007;110:4293–4302. doi: 10.1182/blood-2007-05-088831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki S, Dudziak D, Heidkamp GF, Fiorese C, Bonito AJ, Inaba K, Nussenzweig MC, Steinman RM. CD8+CD205+ Splenic Dendritic Cells Are Specialized to Induce Foxp3+ Regulatory T Cells. The Journal of Immunology. 2008;181:6923–6933. doi: 10.4049/jimmunol.181.10.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gottschalk RA, Corse E, Allison JP. TCR ligand density and affinity determine peripheral induction of Foxp3 in vivo. J Exp Med. 2010;207:1701–1711. doi: 10.1084/jem.20091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*) Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palucka K, Banchereau J. Cancer immunotherapy via dendritic cells. Nature reviews. Cancer. 2012;12:265–277. doi: 10.1038/nrc3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michels AW, Herrath Mvon. 2011 Update: antigen-specific therapy in type 1 diabetes. Current opinion in endocrinology, diabetes, and obesity. 2011;18:235–240. doi: 10.1097/MED.0b013e32834803ae. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haxhinasto S, Mathis D, Benoist C. The AKT-mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med. 2008;205:565–574. doi: 10.1084/jem.20071477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer S, Bruno L, Hertweck A, Finlay D, Leleu M, Spivakov M, Knight ZA, Cobb BS, Cantrell D, O’Connor E, Shokat KM, Fisher AG, Merkenschlager M. T cell receptor signaling controls Foxp3 expression via PI3K, Akt, and mTOR. P Natl Acad Sci USA. 2008;105:7797–7802. doi: 10.1073/pnas.0800928105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, Worley PF, Kozma SC, Powell JD. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K, Gudapati P, Dragovic S, Spencer C, Joyce S, Killeen N, Magnuson MA, Boothby M. Mammalian target of rapamycin protein complex 2 regulates differentiation of Th1 and Th2 cell subsets via distinct signaling pathways. Immunity. 2010;32:743–753. doi: 10.1016/j.immuni.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Battaglia M, Stabilini A, Roncarolo M-G. Rapamycin selectively expands CD4+CD25+FoxP3+ regulatory T cells. Blood. 2005;105:4743–4748. doi: 10.1182/blood-2004-10-3932. [DOI] [PubMed] [Google Scholar]
- 31.Basu S, Golovina T, Mikheeva T, June CH, Riley JL. Cutting Edge: Foxp3-Mediated Induction of Pim 2 Allows Human T Regulatory Cells to Preferentially Expand in Rapamycin. J Immunol. 2008;180:5794–5798. doi: 10.4049/jimmunol.180.9.5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strauss L, Whiteside TL, Knights A, Bergmann C, Knuth A, Zippelius A. Selective survival of naturally occurring human CD4+CD25+Foxp3+ regulatory T cells cultured with rapamycin. J Immunol. 2007;178:320–329. doi: 10.4049/jimmunol.178.1.320. [DOI] [PubMed] [Google Scholar]
- 33.Chakraborty AK, Das J. Pairing computation with experimentation: a powerful coupling for understanding T cell signalling. Nat Rev Immunol. 2010;10:59–71. doi: 10.1038/nri2688. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein B, Faeder JR, Hlavacek WS. Mathematical and computational models of immune-receptor signalling. Nat Rev Immunol. 2004;4:445–456. doi: 10.1038/nri1374. [DOI] [PubMed] [Google Scholar]
- 35.Altan-Bonnet G, Germain RN. Modeling T cell antigen discrimination based on feedback control of digital ERK responses. PLoS Biol. 2005;3:e356. doi: 10.1371/journal.pbio.0030356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faeder JR, Hlavacek WS, Reischl I, Blinov ML, Metzger H, Redondo A, Wofsy C, Goldstein B. Investigation of early events in Fc epsilon RI-mediated signaling using a detailed mathematical model. J Immunol. 2003;170:3769–3781. doi: 10.4049/jimmunol.170.7.3769. [DOI] [PubMed] [Google Scholar]
- 37.Lipniacki T, Hat B, Faeder JR, Hlavacek WS. Stochastic effects and bistability in T cell receptor signaling. J TheorBiol. 2008;254:110–122. doi: 10.1016/j.jtbi.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell. 2009;136:337–351. doi: 10.1016/j.cell.2008.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kauffman SA. Metabolic stability and epigenesis in randomly constructed genetic nets. J Theor Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- 40.Glass L, Kauffman SA. The logical analysis of continuous, nonlinear biochemical control networks. J TheorBiol. 1973;39:103–129. doi: 10.1016/0022-5193(73)90208-7. [DOI] [PubMed] [Google Scholar]
- 41.Morris MK, Saez-Rodriguez J, Sorger PK, Lauffenburger DA. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang R, Shah MV, Yang J, Nyland SB, Liu X, Yun JK, Albert R, Loughran TP., Jr. Network model of survival signaling in large granular lymphocyte leukemia. Proc Natl Acad Sci USA. 2008;105:16308–16313. doi: 10.1073/pnas.0806447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saez-Rodriguez J, Simeoni L, Lindquist JA, Hemenway R, Bommhardt U, Arndt B, Haus UU, Weismantel R, Gilles ED, Klamt S, Schraven B. A logical model provides insights into T cell receptor signaling. Plos Comput Biol. 2007;3:e163. doi: 10.1371/journal.pcbi.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong T, Xing J, Li L, Tyson JJ. A simple theoretical framework for understanding heterogeneous differentiation of CD4+ T cells. BMC Syst Biol. 2012;6:66. doi: 10.1186/1752-0509-6-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mendoza L. A network model for the control of the differentiation process in Th cells. Biosystems. 2006;84:101–114. doi: 10.1016/j.biosystems.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Naldi A, Carneiro J, Chaouiya C, Thieffry D. Diversity and plasticity of Th cell types predicted from regulatory network modelling. Plos Comput Biol. 2010;6:e1000912. doi: 10.1371/journal.pcbi.1000912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song J, Lei FT, Xiong X, Haque R. Intracellular signals of T cell costimulation. CellMolImmunol. 2008;5:239–247. doi: 10.1038/cmi.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim HP, Leonard WJ. The basis for TCR-mediated regulation of the IL-2 receptor alpha chain gene: role of widely separated regulatory elements. EMBOJ. 2002;21:3051–3059. doi: 10.1093/emboj/cdf321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Macian F. NFAT proteins: key regulators of T-cell development and function. Nat Rev Immunol. 2005;5:472–484. doi: 10.1038/nri1632. [DOI] [PubMed] [Google Scholar]
- 50.Passerini L, Allan SE, Battaglia M, Nunzio SDi, Alstad AN, Levings MK, Roncarolo MG, Bacchetta R. STAT5-signaling cytokines regulate the expression of FOXP3 in CD4+CD25+ regulatory T cells and CD4+CD25-effector T cells. IntImmunol. 2008;20:421–431. doi: 10.1093/intimm/dxn002. [DOI] [PubMed] [Google Scholar]
- 51.Yao Z, Kanno Y, Kerenyi M, Stephens G, Durant L, Watford WT, Laurence A, Robinson GW, Shevach EM, Moriggl R, Hennighausen L, Wu C, OShea JJ. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–4375. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 53.Gong D, Malek TR. Cytokine-Dependent Blimp-1 Expression in Activated T Cells Inhibits IL-2 Production. The Journal of Immunology. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 54.Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohkura N, Kitagawa Y, Sakaguchi S. Development and Maintenance of Regulatory T cells. Immunity. 2013;38:414–423. doi: 10.1016/j.immuni.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Buckler JL, Liu X, Turka LA. Regulation of T-cell responses by PTEN. Immunol Rev. 2008;224:239–248. doi: 10.1111/j.1600-065X.2008.00650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma J, Ding Y, Fang X, Wang R, Sun Z. Protein kinase C-theta inhibits inducible regulatory T cell differentiation via an AKT-Foxo1/3a–dependent pathway. J Immunol. 2012;188:5337–5347. doi: 10.4049/jimmunol.1102979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-beta-mediated induction of Foxp3+ T regulatory cells. J Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 60.Albert I, Thakar J, Li S, Zhang R, Albert R. Boolean network simulations for life scientists. Source Code Biol Med. 2008;3:16. doi: 10.1186/1751-0473-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang RS, Saadatpour A, Albert R. Boolean modeling in systems biology: an overview of methodology and applications. Physical biology. 2012;9:055001. doi: 10.1088/1478-3975/9/5/055001. [DOI] [PubMed] [Google Scholar]
- 62.Alon U. Network motifs: theory and experimental approaches. Nature reviews. Genetics. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- 63.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Section 22.3. Gray Codes". New York: Cambridge University Press; 2007. [Google Scholar]
- 64.Tone Y, Furuuchi K, Kojima Y, Tykocinski ML, Greene MI, Tone M. Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat Immunol. 2008;9:194–202. doi: 10.1038/ni1549. [DOI] [PubMed] [Google Scholar]
- 65.Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan-Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science. 2008;321:1081–1084. doi: 10.1126/science.1158013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gottschalk RA, Hathorn MM, Beuneu H, Corse E, Dustin ML, Altan-Bonnet G, Allison JP. Distinct influences of peptide-MHC quality and quantity on in vivo T-cell responses. Proc Natl Acad Sci USA. 2012;109:881–886. doi: 10.1073/pnas.1119763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katzman SD, O’Gorman WE, Villarino AV, Gallo E, Friedman RS, Krummel MF, Nolan GP, Abbas AK. Duration of antigen receptor signaling determines T-cell tolerance or activation. Proc Natl Acad Sci USA. 2010;107:18085–18090. doi: 10.1073/pnas.1010560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J, Ioan-Facsinay A, van der Voort EI, Huizinga TW, Toes RE. Transient expression of FOXP3 in human activated nonregulatory CD4+ T cells. Eur J Immunol. 2007;37:129–138. doi: 10.1002/eji.200636435. [DOI] [PubMed] [Google Scholar]
- 69.Miyao T, Floess S, Setoguchi R, Luche H, Fehling Hans J, Waldmann H, Huehn J, Hori S. Plasticity of Foxp3+ T Cells Reflects Promiscuous Foxp3 Expression in Conventional T Cells but Not Reprogramming of Regulatory T Cells. Immunity. 2012;36:262–275. doi: 10.1016/j.immuni.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 70.Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E, Klein-Hessling S, Serfling E, Hamann A, Huehn J. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klamt S, Saez-Rodriguez J, Lindquist JA, Simeoni L, Gilles ED. A methodology for the structural and functional analysis of signaling and regulatory networks. BMCBioinformatics. 2006;7:56. doi: 10.1186/1471-2105-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tubo NJ, Pagan AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bensinger SJ, Walsh PT, Zhang J, Carroll M, Parsons R, Rathmell JC, Thompson CB, Burchill MA, Farrar MA, Turka LA. Distinct IL-2 Receptor Signaling Pattern in CD4+CD25+ Regulatory T Cells. The Journal of Immunology. 2004;172:5287–5296. doi: 10.4049/jimmunol.172.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.