Abstract
Purpose of review
The purpose of this review is to describe the recent knowledge gathered from the identification of seven genomic regions that have been linked to the risk of developing malignant glioma.
Recent findings
The recent novel discoveries in fine mapping and genotype-phenotype studies will be highlighted. Through imputation and next generation sequencing a novel genetic variant, rs55705857 with a strong association at 8q24 has been discovered and validated in two studies. This locus is specifically associated with IDH1 and IDH2 mutated tumors and oligodendroglial tumors, albeit the specific mechanism of tumor development is not understood. The genetic variants associated with risk of glioma in the EGFR gene have also been associated with specific somatic aberrations, including loss at the CDKN2A/B locus and allele specific loss of EGFR in the tumors. A specific TP53 low frequency variant has also been associated with glioma risk and validated in a separate data set. The genetic risk in the telomere regulating genes TERT and RTEL appear to be associated with higher grade tumors without IDH mutations.
Summary
The link of genetic loci to specific tumor subtypes may have relevance for understanding glioma biology, for developing new diagnostic tools and targeted therapy for glioma.
Keywords: glioma etiology, genotype, phenotype, glioblastoma, oligodendroglioma
Introduction
Understanding the etiology of a disease is very important to identify strategies for prevention, surveillance and potential targets for treatments. The etiology of glioma has for many years not been very well understood. Rare genetic syndromes with germline mutations are well known to be causal for gliomas, such as the Li-Fraumeni syndrome caused by TP53 mutations, neurofibromatosis type 1 and 2. Common environmental exposures such as smoking, alcohol and diet have not been consistently associated with malignant brain tumors. The only consistent associations of external exposure is higher doses of ionizing radiation, described in cohort studies from the tinea capitis cohort in Israel and atomic bomb survivors in Japan. In addition there is an inverse association of asthma and allergies observed in many case- control studies. However, if the underlying causes of this is due to the host immune system, external agents or medication against asthma and allergies has not been entangled [1]. During the last years a new genetic era has emerged with the technology of screening the whole genome. This review gives an overview in the recent discoveries of low penetrant genetic variants that contribute to our understanding of the iniation and development of gliomas.
Established genetic risk loci
Since the first published genome wide association studies in 2009 [2, 3], several loci associated with glioma risk have been discovered. This includes loci mapped in or near genes such as CCDC26 in 8q24, PHLDB1 at chromosome 11q23.3 , the TP53 polyadenylation site by rs78378222 at 17p13.1 [4], rs11979158 and rs2252586 in EGFR at chromosome 7,[5, 6]and rs4977756 in CDKN2A/B [2, 3]. In addition, there are two genetic variants associated with telomere regulation also associated with glioma risk, rs2736100 in the TERT gene and rs6010620 in RTEL [2, 3]. A recent additional GWAS of 1850 glioma cases confirmed the previously identified loci but did not identify any novel genomic areas [7]. Separating the analyses on cases with family history found support for an association with the RTEL gene in cases with a family history of brain tumors [8]. The lack of identification of additional loci implies that most of the common high risk glioma loci have been discovered and that larger and more pathologically homogeneous data sets will be necessary to delineate additional rare loci. Interestingly, several of the genetic variants are in or near genes that often are found to acquire somatic mutation s in glioma: for example, TP53, CDKN2A/B, EGFR and TERT [9, 10]. One should remember that tagging SNPs identified through GWAS should be seen only as a marker of a genomic area and that there are likely one or more functional genetic variants in linkage disequilibrium with tagging SNP. Therefore additional fine mapping studies have been performed for some of the regions to discover true functional variants and to understand the relationship between the germline variants and the acquired somatic events.
Fine mapping and tagging studies
Several efforts have been made after the initial genome wide association studies to explore the genomic area by resequencing and fine mapping.
8q24
The chromosomal 8q24 region has been associated with risk of several common cancer sites (reviewed in Huppi K et al [11]). However, the loci for the other cancers are approximately 1-2 MB proximal to the 8q24.21glioma locus at. An imputation effort combined with next generation sequencing revealed a genetic variant rs55705857 at 8q24.21 with a strong association to IDH mutated tumors and the histopathological subtype oligodendroglioma with an odds ratio of approximately 6.0, a level of association rarely seen in low penetrant association studies [12]. The association was confirmed by a separate European study that performed fine-mapping of the genomic area using 1000 genomes and a case control set of a total of 4147 glioma cases and 7435 controls. This study found an equally strong association with an odds ratio for low-grade glioma associated with rs55705857 was 4.3 (P = 2.31 × 10(-94) [13] The mechanism by which the 8q24 genetic variant confers an increased risk of glioma is unknown, The SNP is completely conserved throughout mammalian evolution and the surrounding sequence models as a miRNA [12]. The variant resides within the CCDC26 locus which has been implicated in several inflammatory pathways [13]. The association with IDH mutation suggests that the variant may also be involved in the generation of the promoter DNA hypermethylation phenotype. The mechanism of action cannot simply be mediated only through IDH1 or IDH2 mutations, as oligodendroglioma with wild type IDH also are linked to the risk locus. The PHLDB1 locus is likewise association primarily to IDH mutated tumors [14] – thus there is clearly an interaction between germline variants and the acquisition of specific somatic alterations.
Tp53
In 2005 Malmer et al. observed an association of a specific p53 haplotype and glioma [15].More recently, a GWAS –imputation based study was performed in the Icelandic population (with validation in two US populations) showing that a genetic variant in the polyadenylation site of TP53 (rs78378222) is associated with glioma [4]. The genetic variant has recently been validated by a fine mapping study showing association both for glioblastoma and other gliomas [16]. Studies of RNA transcripts suggest that the rs78378222[C] variant gives a impaired termination and polyadenylation of the TP53. Transcript [4], but no studies of correlation to protein expression has yet been published.
RTEL and TERT
Several loci within or near the TERT gene have been associated with multiple kinds of cancer [17].The association has also been observed in the Han Chinese population [18]. A genetic tagging study of the TERT locus analyzed the association between rs2736100 with telomere length and attrition rate at age 50 and 60 in 900 individuals and found an association between rs2736100 and telomere length especially at older age [15]. This result indicates that TERT genotypes might have higher impact of disease at older age [19]. This is supported by a study showing a stronger association with glioma at higher age irrespective of glioma subtype [20].
Genotype- phenotype correlations
Some of the risk variants for glioma have distinctly been correlated to specific acquired tumor alterations (Figure 1). Glioblastoma, the most aggressive types of glioma, are genetically heterogeneous tumors, but a study using micro dissection of several areas within the same tumor has shown that somatic events in the EGFR and CDKN2A/B locus are early events appearing in all areas of the tumors, while later mutations are a stochastic process (See Figure 2; redrawn with permission [21]). The germline EGFR variants have been associated with allele specific loss of EGFR and homozygous loss at the CDKN2A/B locus using SNP array data from a Swedish cohort and TCGA data as validation (See Table 1 from the publication [22]). The EGFR risk genotypes has not been associated with EGFR amplification measured by fluorescence in situ hybridization (FISH) [23], but FISH has not the potential to detect allele specific loss as done by paired samples of blood and tumor with a SNP array using the recently developed ASCAT algorithm [22]. Overall the SNPs in EGFR and CDKN2A/B seem to be associated with all types of glioma and the SNPs in TERT with higher grade and chromosome 10 loss [23].
Figure 1. Pathways for glioma development.
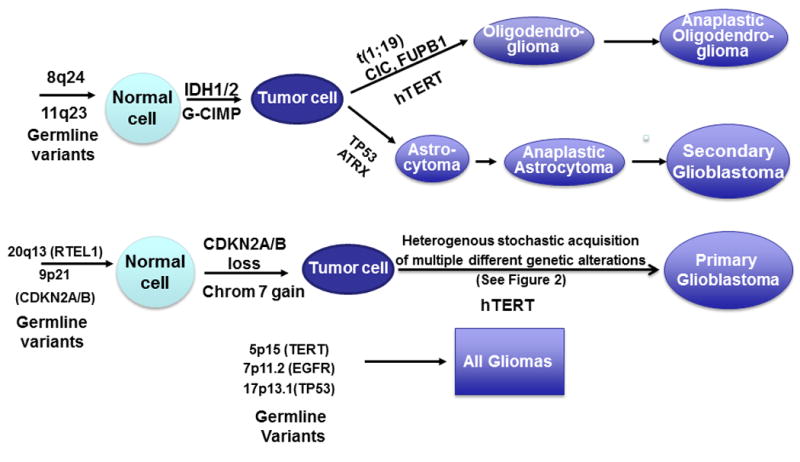
Figure 2.
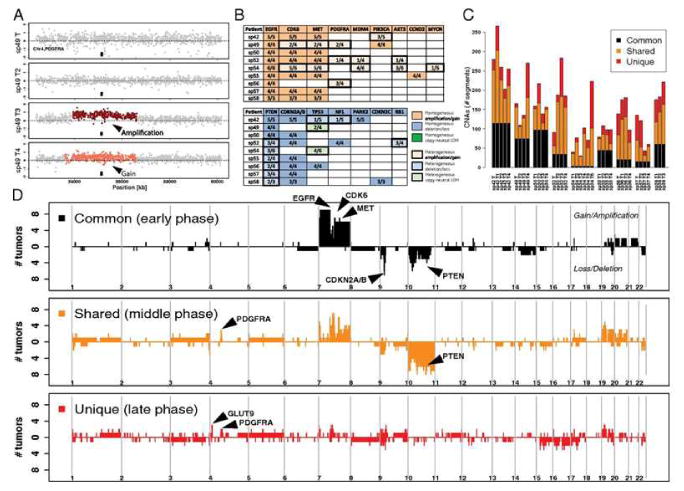
Gliomas are not only many different subtypes but the intratumour heterogeneity on a molecular level is also evident. (From reference 21)
Table 1.
Wibom et al, plos one table 5 redrawn with permission showing a correlation between risk genotypes in EGFR and somatic events in EGFR and CDKN2A/B
| UMU | TCGA | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
||||||||||||||||
| Risk variant | Region | Cytoband | Gene | Event | Variant | n (major)a |
n (major) eventa |
n (rare+hz)a |
n(rare+hz) eventa |
P | Variant | n (major) |
n (major) event |
n(rare+ hz) |
n(rare+ hz) event |
P | Approach |
| rs17172430 | chr7:55054218-55242525 | 7p11.2 | EGFR | LOH | rs1015793 | 60 (41/19) | 19 (15/4) | 21 (14/7) | 2 (2/0) | 0.0385 | rs17172430 | 236 | 57 | 49 | 6 | 0.0455 | GOI |
|
| |||||||||||||||||
| rs11979158 | Chr9:21957750-21965132 | 9p21.3 | CDKN2A | HD | rs10245472 | 57 (39/18) | 33 (26/7) | 24 (16/8) | 8 (7/1) | 0.0374 | rs11979158 | 209 | 51 | 76 | 10 | 0.0267 | GOI |
|
| |||||||||||||||||
| rs11979158 | Chr9:21992901-21999312 | 9p21.3 | CDKN2B | HD | rs10245472 | 57 (39/18) | 32 (25/7) | 24 (16/8) | 7 (6/1) | 0.0233 | rs11979158 | 209 | 122 | 76 | 32 | 0.0107 | GOI |
|
| |||||||||||||||||
| rs17172430 | chr9:21992901-21999312 | 9p21.3 | CDKN2B | HD | rs1015793 | 60 (41/19) | 34 (27/7) | 21 (14/7) | 5 (4/1) | 0.0088 | rs17172430 | 236 | 134 | 49 | 20 | 0.0300 | GOI |
|
| |||||||||||||||||
| rs17172430 | chr9:21961989-21978896 | 9p21.3 | MTAP, CDKN2A | HD | rs1015793 | 60 (41/19) | 35 (28/7) | 21 (14/7) | 5 (4/1) | 0.0062 | rs17172430 | 236 | 135 | 49 | 20 | 0.0264 | global |
|
| |||||||||||||||||
| rs17172430 | chr9:22010493-22055620 | 9p21.3 | MTAP, CDKN2BAS | HD | rs1015793 | 60 (41/19) | 33 (27/6) | 21 (14/7) | 4 (3/1) | 0.0040 | rs17172430 | 236 | 132 | 49 | 19 | 0.0210 | global |
n number, major samples homozygous for the major allele, rare+hz samples homozygous for the rare allele plus heterozygous samples, event samples positive for given event, GOI genes of interest.
total number of samples (glioblastoma samples/non-glioblastoma samples).
doi:10.1371/journal.pone,0047929.t005
The PHLDB1 (11q23.3) and CCDC26 (8q24) genetic variants have been strongly associated with IDH1 and 2 mutation [24]. Thus it is likely that these SNPs are also associated with a hypermethylated phenotype [25]. The results of the US study showed that rs55705857 (8q24.21) is associated with risk of oligodendroglial tumors regardless of tumor 1p/19q and IDH mutation status but IDH mutated and not wild type astrocytoma showed association [12]. In the study by Jenkins et al, approximately 40% of patients of these glioma subtypes carry one or more of the risk alleles for rs55705857 (8q24.21) - compared to 8% of the controls - indicating that this marker could be used to support diagnosis in intracranial tumors when biopsy is difficult. While the sensitivity of such a single germline marker is still not sufficient for diagnosis, in the future such blood tests may be used in combination with other clinical, laboratory and radiologic information such as PET-CT methionine or PET-MR imaging. Somatic mutations have recently been observed in the promoter of TERT in a high frequency in glioblastoma and progressive astrocytoma and oligodendroglioma, giving a clear picture of TERT as an important mediator of glioma progression and malignancy grade [10]. The TERT and RTEL risk genotypes are also correlated with higher grade tumors indicating that there is a genotype- phenotype correlation [23].
Conclusions
Many of the genes that are associated with an increased risk of glioma also have been identified as genes often mutated in gliomas or highly correlated with specific acquired mutations. EGFR and CDKN2A/B are frequently mutated in gliomas and SNPs within or near these genes are associated with the development of gliomas. SNPs in 8q24 and 11q23.3 are associated with gliomas that acquire IDH mutation . Taken together these observations lead to the hypothesis that germline genetic risk variant s interact with acquired alterations to influence glioma development. Furthermore, it is likely that additional risk variant and acquired alteration interactions will be discovered in the future. The coming years will witness the performance and publication of functional studies that will further delineate the mechanisms behind the pathologic and molecular associations that have been discovered.
Bullet points.
Common genetic variants contribute to glioma risk
Fine mapping of the associated regions has discovered stronger associations in 8q24 and TP53
The genetic variants are correlated to specific histologic and molecular subtypes of gliomas
Acknowledgments
This review was funded by Acta Oncologica foundation through the Royal Swedish Academy of Science (BM salary).
References
- 1.Bondy ML, Scheurer ME, Malmer B, et al. Brain tumor epidemiology: consensus from the Brain Tumor Epidemiology Consortium. Cancer. 2008 Oct 1;113(7 Suppl):1953–68. doi: 10.1002/cncr.23741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shete S, Hosking FJ, Robertson LB, et al. Genome-wide association study identifies five susceptibility loci for glioma. Nat Genet. 2009 Aug;41(8):899–U54. doi: 10.1038/ng.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009 Aug;41(8):905–8. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011 Nov;43(11):1098–U85. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andersson U, Schwartzbaum J, Wiklund F, et al. A comprehensive study of the association between the EGFR and ERBB2 genes and glioma risk. Acta Oncol. 2010 Aug;49(6):767–75. doi: 10.3109/0284186X.2010.480980. [DOI] [PubMed] [Google Scholar]
- 6.Sanson M, Hosking FJ, Shete S, et al. Chromosome 7p11.2 (EGFR) variation influences glioma risk. Hum Mol Genet. 2011 Jul 15;20(14):2897–904. doi: 10.1093/hmg/ddr192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajaraman P, Melin BS, Wang ZM, et al. Genome-wide association study of glioma and meta-analysis. Hum Genet. 2012 Dec;131(12):1877–88. doi: 10.1007/s00439-012-1212-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melin B, Dahlin AM, Andersson U, et al. Known glioma risk loci are associated with glioma with a family history of brain tumours - A case-control gene association study. Int J Cancer. 2013 May;132(10):2464–8. doi: 10.1002/ijc.27922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct 23;455(7216):1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Killela PJ, Reitman ZJ, Jiao YC, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013 Apr;110(15):6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huppi K, Pitt JJ, Wahlberg BM, et al. The 8q24 gene desert: an oasis of non-coding transcriptional activity. Frontiers in genetics. 2012;3:69. doi: 10.3389/fgene.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Jenkins RB, Xiao YY, Sicotte H, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet. 2012 Oct;44(10):1122–+. doi: 10.1038/ng.2388. A finemapping resequencing of the 8q24 discovered a locus vith a much stronger association than previous detected. The Lo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enciso-Mora V, Hosking FJ, Kinnersley B, et al. Deciphering the 8q24.21 association for glioma. Hum Mol Genet. 2013 Jun 1;22(11):2293–302. doi: 10.1093/hmg/ddt063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins RB, Wrensch MR, Johnson D, et al. Distinct germ line polymorphisms underlie glioma morphologic heterogeneity. Cancer Genet. 2011 Jan;204(1):13–8. doi: 10.1016/j.cancergencyto.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malmer B, Feychting M, Lonn S, et al. p53 genotypes and risk of glioma and meningioma. Cancer Epidemiology Biomarkers & Prevention. 2005 Sep;14(9):2220–3. doi: 10.1158/1055-9965.EPI-05-0234. [DOI] [PubMed] [Google Scholar]
- 16.Enciso-Mora V, Hosking FJ, Di Stefano AL, et al. Low penetrance susceptibility to glioma is caused by the TP53 variant rs78378222. Br J Cancer. 2013 May 28;108(10):2178–85. doi: 10.1038/bjc.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bojesen SE, Pooley KA, Johnatty SE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet. 2013 Apr;45(4):371–84. 84e1–2. doi: 10.1038/ng.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao Y, Chen G, Zhao Y, et al. Fine-mapping of a region of chromosome 5p15.33 (TERT-CLPTM1L) suggests a novel locus in TERT and a CLPTM1L haplotype are associated with glioma susceptibility in a Chinese population. Int J Cancer. 2012 Oct 1;131(7):1569–76. doi: 10.1002/ijc.27417. [DOI] [PubMed] [Google Scholar]
- 19.Melin BS, Nordfjall K, Andersson U, et al. hTERT Cancer Risk Genotypes Are Associated With Telomere Length. Genet Epidemiol. 2012 May;36(4):368–72. doi: 10.1002/gepi.21630. [DOI] [PubMed] [Google Scholar]
- 20.Walsh KM, Rice T, Decker PA, et al. Genetic variants in telomerase-related genes are associated with an older age at diagnosis in glioma patients: evidence for distinct pathways of gliomagenesis. Neuro Oncol. 2013 Jun 3; doi: 10.1093/neuonc/not051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Sottoriva A, Spiteri I, Piccirillo SGM, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013 Mar;110(10):4009–14. doi: 10.1073/pnas.1219747110. The study shows heterogenity in glioblastoma development by analysing different parts of the tumor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wibom C, Ghasimi S, Van Loo P, et al. EGFR Gene Variants Are Associated with Specific Somatic Aberrations in Glioma. PLoS One. 2012;7(12):e47929. doi: 10.1371/journal.pone.0047929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Di Stefano AL, Enciso-Mora V, Marie Y, et al. Association between glioma susceptibility loci and tumour pathology defines specific molecular etiologies. Neuro Oncol. 2013 May;15(5):542–7. doi: 10.1093/neuonc/nos284. A large data set displaying the clinical characteristics in association to glioma subtypes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rice T, Zheng S, Decker PA, et al. Inherited variant on chromosome 11q23 increases susceptibility to IDH-mutated but not IDH-normal gliomas regardless of grade or histology. Neuro Oncol. 2013 May;15(5):535–41. doi: 10.1093/neuonc/nos324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010 May 18;17(5):510–22. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]


