Summary
RNA interference (RNAi) is the process of specific gene silencing by the use of dsRNA. In cultured Drosophila cells, RNAi methodologies are well established and easily executed: double stranded RNA (dsRNA), when added to the cell culture medium, is efficiently internalized by the cells and, through the activity of endogenous processing machinery, targets the specified mRNA for degradation resulting in reduced levels of its encoded protein. This technique has proven very useful in studying the role of host genes during Legionella pneumophila infections, as it allows the effect of host factor depletion on intracellular growth of the bacterium to be examined. In this chapter we present the methods commonly used in our laboratory to study intracellular growth of L. pneumophila using dsRNA in Drosophila cells.
Keywords: dsRNA, RNA interference, Kc167, Legionella, Drosophila melanogaster
1. Introduction
During RNA-mediated gene silencing, or RNAi, dsRNAs are internalized by receptor-mediated endocytosis (1). Once inside the cell, dsRNAs are processed into small, single stranded nucleotide sequences of ~21bp by the host protein complex Dicer (2, 3). The resulting ssRNAs then hybridize to their complementary sequences in mRNA transcripts, which leads to cleavage of the mRNA (4, 5). The result is post-transcriptional modulation of gene expression through targeted degradation of specific mRNAs, which results in decreased protein production. The ability to deplete a single host factor, or a combination of factors, using this technique has been instrumental in characterizing the role of individual host proteins and various host cellular processes during infection with bacterial pathogens (6).
Gene silencing in cultured Drosophila cell lines using RNAi has several advantages. First, this method is not technically demanding, as cells efficiently internalize dsRNA when added directly to the cell culture medium in the absence of serum, and they possess all necessary enzymes for dsRNA processing and mRNA-targeted cleavage. This bypasses the need to rely on dominant negative constructions, which depend on transfection or expression efficiencies, which can be low, or the construction of stable cell lines. Second, RNAi alleviates the need to generate mutant cell lines in order to study the effect of loss of host factor function. Third, depletion rather than gene inactivation allows the affect of genes to be analyzed that are normally lethal when disrupted. Finally, the elegant simplicity of the system is highly amenable to high throughput screening strategies allowing the contribution of multiple, individual host factors to be analyzed simultaneously.
The discovery that Drosophila cell lines have macrophage-like properties and can efficiently internalize microorganisms greatly facilitated the identification of host proteins that modulate intracellular growth of pathogens (7). Moreover, because many genes and the biological functions of their encoded proteins are conserved between Drosophila and mammalian systems, phenotypes in Drosophila cells can often be recapitulated in biologically relevant hosts (8–12). As an added benefit, dsRNA treatment of Drosophila cells does not induce a strong immune response, contrary to that observed in mammalians systems, thus avoiding the complication of innate immune activation. In addition, the fruit fly genome encodes components of innate immunity but not those that function in adaptive immunity offering the advantage of exploring innate immune surveillance and signaling during host-pathogen interactions in the absence of the confounding effects of an adaptive immune response (13).
Legionella pneumophila intracellular replication in Drosophila melanogaster Kc167 cultured cells is similar to that in natural hosts of this pathogen, macrophages and amoeba (6), making Drosophila cells an excellent model system to study L. pneumophila pathogenesis. To date, both animals and cultured cells have been used to study L. pneumophila virulence strategies (14). Initial studies in Drosophila cells using RNAi indicated that there was an important role for host cell ubiquitination during intracellular growth, and that there were multiple membrane trafficking pathways that support formation of the Legionella-containing vacuole (LCV) (6). The use of RNAi in Drosophila cells should prove invaluable in expanding our current knowledge of how host factors contribute to L. pneumophila pathogenesis.
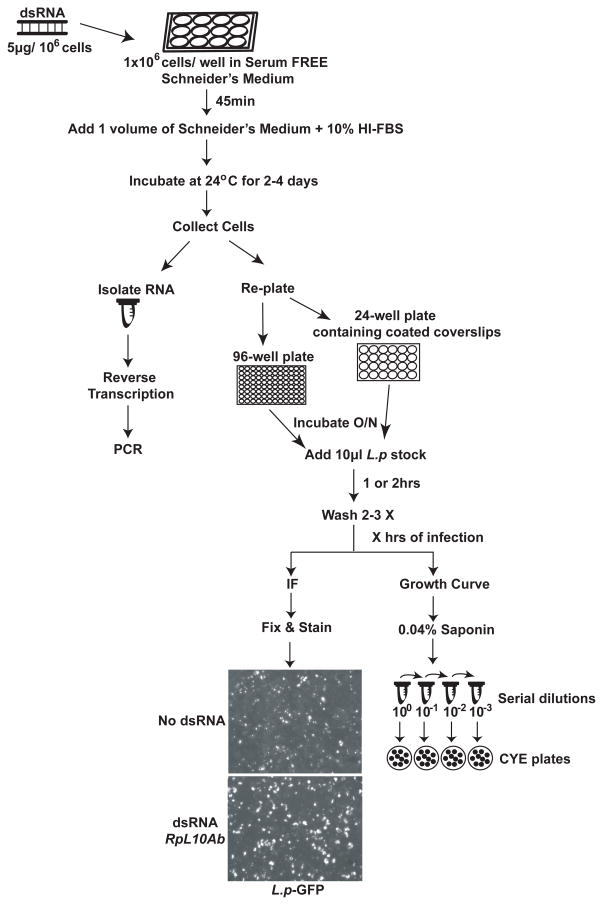
The following sections present a general procedure for the use of RNAi to study L. pneumophila growth within cultured Drosophila cells. The methods in this chapter cover the propagation of Drosophila cells in a laboratory setting, dsRNA design and synthesis, treatment of Drosophila cells with dsRNA, how to test the efficiency of gene silencing and selected assays designed to measure the effects of host factor depletion on L. pneumophila intracellular growth (Figure 1).
Figure 1. Schematic representation of methods used to study L. pneumophila infection in cultured Drosophila cells depleted of host factors using RNA interference.
Drosophila cells are treated with dsRNA against the target gene of interest for 2–4 days. After incubation, cells are re-plated and samples from each condition are used to isolate RNA and test depletion efficiency. Re-plated cells are then infected with L. pneumophila and the infection is allowed to proceed for the length of time desired. Growth curve or immunofluorescence microscopy assays can be performed to test the effects of host factor depletion on L. pneumophila intracellular growth. Pictures were taken at 20X. HS-FBS: Heat-Inactivated Fetal Bovine Serum; O/N: overnight; IF: immunofluorescence; L.p-GFP: L. pneumophila expressing GFP.
2. Materials
2.1. Cell lines and media
Drosophila melanogaster cultured embryonic Kc167 (preferred) or S2 cell lines. Cell lines can be obtained from the Drosophila Genome Resource Center (https://dgrc.cgb.indiana.edu/cells/).
Schneider’s Drosophila Medium (Invitrogen; See Note 1).
Heat Inactivated Fetal Bovine Serum (HI-FBS; Invitrogen).
Schneider’s Drosophila Medium supplemented with 10% Heat-Inactivated Fetal Bovine Serum.
0.05% Trypsin-EDTA (Gibco)
2.2. RNAi reagents
Drosophila genomic DNA.
Oligonucleotides to amplify gene of interest containing a 5′ T7 promoter sequence.
PCR components (dNTPs, DNA polymerase; DNA polymerase buffer; dH20).
Superscript Reverse transcriptase II (Invitrogen) and oligodT primer (T(20)).
Total RNA isolated from dsRNA treated and untreated Drosophila cells (store at −80°C).
F actin-specific primers for RT-PCR: act5F: atgtgtgacgaagaagtt; act5R: agtccagaacgataccg
2.3. Analysis of Legionella growth in dsRNA treated Drosophila cells
2.3.1. Growth Curves
Liquid media: ACES Yeast Extract (AYE). For 1L weight 10g of yeast extract (VWR) and 10g of ACES (Sigma). Add them to a beaker containing ~900mL of dH20. Mix well until everything is dissolved. Adjust pH to 6.9 using 10M KOH. Adjust final volume to 1L. Filter sterilize using 0.22um filters into 500mL bottles.
AYE additions: L-cysteine (Sigma) and Ferric Nitrate (Sigma). Prepare a 5mL solution of 0.08g/mL of L-cysteine in dH2O and another 5mL of 0.027g/mL of Ferric Nitrate in dH2O. Filter solutions individually using 0.22um filters. Add 5mL of each supplement per 1L of AYE previously filter sterilized. Do not mix Ferric Nitrate and AYE before filter sterilizing. Once iron comes into contact with AYE it forms a precipitate, consequently if the solution were subsequently filtered, the iron would be removed. Allow media to stand overnight to clear. Store at room temperature protected from light.
Plates: Charcoal Buffered Yeast Extract (CYE). Prepare 2L AYE (without L-cysteine and Ferric Nitrate additives and do not filter sterilize). Add 4g of activated charcoal (Sigma), 32g of agar (VWR) and a stir bar to a 4L flask then add the 2L of AYE. Cover the mouth of the flask with aluminum foil and autoclave tape. Autoclave for 30 minutes at 121°C. Let it cool to 65°C to prevent media from solidifying.
CYE additions. Prepare just before pouring plates while waiting for media to cool down. Make sure all solutions are completely dissolved before filtering them. See Table 1 for stock solutions guidelines. Once the CYE reaches ~65°C, add the additives and mix well for 5 minutes and pour plates. 1L of CYE is enough for ~50- 150×15mm petri dishes.
2% saponin (Sigma) stock solution in ddH2O, filter sterilized
Sterile water
Table 1.
Recommended stocks for CYE plate’s supplements
| Supplement | Final Concentration | Stocks for CYE plates1,2 | ||
|---|---|---|---|---|
| 2L | 4L | 6L | ||
| L-cysteine* | 0.4g/L | 1.2g/15mL | 2.0g/25mL | 2.8g/35mL |
| Ferric Nitrate | 0.135g/L | 405mg/15mL | 675mg/25mL | 945mg/35mL |
| Thymidine | 0.1g/L | 300mg/15mL | 500mg/25mL | 700mg/35mL |
Prepare in 50mL tubes and filter sterilize using 0.22um filters before adding to media
Stock solutions to be prepare if making 2, 4 or 6 Liter of media. Add 5mL of these stocks per L of CYE made.
L-cysteine might not dissolve in H2O, can be dissolved in 0.5–1.0M KOH if necessary
2.3.2. Infectious Center Assays
Phosphate buffered saline (PBS) (Sigma for solutions; Gibco for cell culture)
Goat Serum (Gibco)
Fixation Buffer: 4% Paraformaldehyde, 40mM HEPES, 6.5% Sucrose, pH 7.4
Blocking Buffer: 4% goat serum (Gibco) in phosphate buffered saline (PBS), pH 7.4. Alternatively, 4% bovine serum albumin (BSA) (Sigma) in PBS can be used.
Permeabilization Buffer: 0.1% TritonX-100 in PBS. Alternatively, 0.2% Saponin can also be used to permeabilize the cells.
Hank’s Balanced Salt Solution (Gibco)
Hoechst (Molecular Probes)
Anti-Legionella primary antibody (6) and fluorophore-conjugated secondary antibody
2.4. Equipment
Incubator at ~24°C (not supplemented with CO2)
PCR thermocycler
Culture tubes
37°C incubator with rotating wheel or platform shaker
Bench-top centrifuge.
12, 24 and 96-well tissue culture treated plates
Agarose gel reagents and electrophoresis equipment
Spectrophotometer
Biosafety cabinet
3. Methods
3.1. Propagation of Drosophila cells
Routine propagation of cultured Drosophila Kc167 cells is performed in a biosafety cabinet to avoid contamination. Cells are typically cultured in tissue cultured treated T75 culture flasks. Cells are passaged when they are roughly 90% confluent (See Note 2).
Warm Schneider’s Drosophila Medium supplemented with 10% HI-FBS to room temperature.
In a biosafety cabinet, aspirate the medium from the flask.
Rinse the cells by adding 1ml of room temperature 1X PBS. Gently tilt the flask so the PBS covers the entire surface area of the cells then carefully remove the PBS.
Add 1ml of 0.05% Trypsin-EDTA, tilt the flask to ensure that all cells are submerged. Incubate at room temperature for 5 minutes.
Add 10mL of Schneider’s Drosophila Medium supplemented with 10% HI-FBS and lift the cells by pipetting the media over the cells along the wall of the flask.
In a new flask, dilute the resuspended Drosophila cells 1:3, 1:4 or 1:5, equivalent to roughly 1×106 cells/ml in fresh Schneider’s Drosophila Medium supplemented with 10% HI-FBS.
Culture cells at 24°C until confluent.
3.2. Synthesis of dsRNA
Design primers for your gene of interest. Using the flybase.org database, retrieve the annotated sequence of your target gene and design primers to target sequences that correspond to gene exon regions that will generate a fragment in the range of 250–600 bp. (Use http://www.flyrnai.org/cgi-bin/RNAi_find_primers.pl to identify regions predicted to have off-targets sequences (15) and avoid these regions when designing your primers (see Note 3)). Alternatively, http://www.flyrnai.org provides a series of primer pairs for generating dsRNA for Drosophila genes that have been tested for off-target effects. These can be accessed through http://www.flyrnai.org/cgi-bin/RNAi_gene_lookup_public.pl by searching with the name of your target gene.
Add the T7 promoter sequence (taatacgactcactataggg) to the 5′-end of each primer. T7 RNA polymerase will be used to transcribe the target DNA fragment, or amplicon, containing your sequence of interest to generate the dsRNA.
Amplify each amplicon using Drosophila genomic DNA by the polymerase chain reaction (PCR) using standard PCR reagents and conditions (see Ramadan et al. (16) for guidance).
Purify the PCR product. We routinely use the PCR purification Kit from Qiagen following the manufacturer’s protocol.
Synthesize dsRNA using the purified amplicon as template. We routinely use the MEGAscript RNAi Kit from Ambion following the manufacturer’s protocol.
Newly synthetized dsRNA can be store at −20°C for up to one year.
3.3. Treatment of Drosophila cells with dsRNA
Harvest cells from a confluent flask, roughly 3–4 days after their last passage, by treating with 0.05% Trypsin-EDTA then collecting in Drosophila Schneider’s medium supplemented with 10% HI-FBS as described above and transfer to a 50 ml Falcon tube (see Note 4).
Pellet the cells by centrifuging at 200 × g for 5 minutes at room temperature in a bench-top centrifuge.
Resuspend the cells in 30ml of room temperature Schneider’s Drosophila medium without HI-FBS.
Determine the cell concentrations and dilute (if necessary) to 1×106 cell/ml.
In a 12 well tissue culture treated plate, aliquot 1 ml of cell suspension, equivalent to 1×106 cells per well for each dsRNA treatment plus 1 well for untreated control sample (see Note 5).
Add 5μg of dsRNA per 1×106 cells (each well) and mix by gently swirling the plate by hand.
Incubate for 45 min at 24°C.
Add one volume (1 ml) of Schneider’s Drosophila medium supplemented with 10% HI-FBS to each well.
Incubate cells at 24°C for the desired treatment time. We usually treat the cells for 3 or 4 days (see Note 6).
3.4. Legionella Infection of dsRNA treated Drosophila cells
3.4.1. 1-Day prior to infection
After Drosophila cells have been treated with dsRNA for the desired time, rinse the cells once with 1 ml of PBS to remove traces of media, treat with 0.05% Trypsin-EDTA and harvest in Schneider’s Drosophila medium containing 10% HI-FBS as described above.
Pellet the cells by centrifuging at 200 × g for 5 minutes at room temperature, then resuspend the cells in fresh, room temperature Schneider’s Drosophila medium containing 10% HI-FBS and quantitate using hemocytometer.
Dilute cells and plate in a tissue culture-treated plate at the appropriate concentration depending on the assay used. See Table 2 for the recommended conditions for each assay. If using a L. pneumophila strain that is a thymidine auxotroph, use Schneider’s Drosophila medium containing 10% HI-FBS supplemented with 0.4 mg/ml thymidine. For growth curves, we perform infections in 96 well plates. For immunofluorescence microscopy studies we performed infections in 24-well plates containing Concanavalin A or poly-L-lysine coated coverslips or in 96-well flat bottom tissue culture treated plates (see Notes 7 and 8).
Save an aliquot of dsRNA treated and untreated cells from each condition (typically 1×106 cells) to isolate RNA and test for target mRNA depletion.
Incubate the cells overnight at 24°C to allow the cells to settle.
Isolate RNA from dsRNA treated and untreated cells. We routinely use QIAshredder (Qiagen) for homogenization of samples and RNeasy Mini Kit (Qiagen) to isolate RNA. RNA samples can be stored temporarily at −80°C. Methods for RT-PCR to measure gene silencing are described below.
Prepare serially diluted cultures of the appropriate L. pneumophila strains in AYE media containing the proper supplements and grow overnight at 37°C with shaking.
Table 2.
Recommended cell densities and MOI to be use per assay
| Assay1 | Plate | Conc. dsRNA- treated Drosophila cell suspension needed (cells/ml) | Volume cells to add per well | Number cells/well | Recommend ed MOI2 | Conc. L.p.3 Stock Culture Suspension (bacteria/ul) | Amount of L.p stock add per well (ul) |
|---|---|---|---|---|---|---|---|
| Growth curve | 96- well plate | 1×106 | 100 ul | 1×105 | 0.05 | 5×102 | 10 |
| IF | 96- well plate | 1×106 | 100 ul | 1×105 | 1 | 1×104 | 10 |
| IF | 24- well plate | 2×105 | 0.5 ml | 1×105 | 1 or 2 | 1×104 2×104 |
10 |
IF = immunofluorescence;
MOI = multiplicity of infection;
L.p. = L. pneumophila
3.4.2. Day of Infection
Identify motile L. pneumophila cultures by light microscopy.
Determine the optical density (A600) for each strain.
Given that 1 OD unit at A600 is equivalent to 1×106 bacteria/ul, dilute selected bacterial cultures in room temperature Schneider’s Drosophila medium containing 10% HI-FBS to a concentration of 0.1x that of the number of cells to be infected based on the desired multiplicity of infection (MOI) to be used (see Table 2). For example, to infect at an MOI=1, having plated 1×105 cells, dilute the bacteria culture to 1×104 bacteria/ul.
Add 10 ul of diluted L. pneumophila stock solution to each well then mix by gently rotating by hand in a circular motion.
Centrifuge at 200 × g for 5 minutes at room temperature to synchronized the infection.
Incubate at ~24°C for 1 hr if MOI≥1 or for 2 hrs if MOI=0.05.
Wash 2–3X with Schneider’s Drosophila medium supplemented with 10% HI-FBS.
Incubate at ~24°C for the desired length of time.
3.4.3. For L. pneumophila growth curves
At 2, 30, 48 and 72 hours after infection lyse the cells by adding 5 ul of 0.04% saponin per well and incubating for 10 minutes at room temperature.
Mix by pipetting and harvest the supernatant.
Rinse each well with 100 ul of sterile water and combine the appropriate fractions (see Note 9).
Serial dilute samples in sterile water and plate 10 ul aliquots on CYE plates containing the appropriate supplements using the guidelines outlined in Table 3.
Incubate plates at 37°C for 4 days then quantitate colony-forming units.
Table 3.
Recommended dilutions for growth curve assays
| Hours post infection | Dilutions |
|---|---|
| 2 | 100 |
| 30 | 100, 10−1 |
| 48 | 10−1, 10−2 |
| 72 | 10−2, 10−3 |
3.4.4. For immunofluorescence assays
At the appropriate time points (see Note 10), aspirate the culture media and add 0.5ml (24 well plate) or 100 ul (96 well plate) of Fixation Buffer and incubate at room temperature for 30 minutes.
Gently remove the Fixation Buffer and discard in the appropriate container.
Add 0.5ml (24 well plate) or 100 ul (96 well plate) of Permeabilization Buffer and incubate for 10 minutes at room temperature.
Wash 2–3X with Hank’s Medium Salt Solution. Always add and remove media very carefully to avoid lifting the cells (see Note 11).
Aspirate the media and add 0.5ml (24 well plate) or 100 ul (96 well plate) of Blocking Buffer and incubate for 15 minutes at room temperature.
Remove the Blocking Buffer and add anti-Legionella primary antibody dilute in Blocking Buffer to the recommended concentration and incubate for 1 hr at room temperature.
Wash 3X with Blocking Buffer, 5 minutes each.
Aspirate the Blocking Buffer and add fluorophore-conjugated secondary antibody diluted in Blocking Buffer to the manufacturer’s specifications and incubate for 1 hr at room temperature.
Wash 3X with Blocking Buffer, 5 minutes each.
For nuclei staining, add 0.5ml (24 well plate) or 100 ul (96 well plate) of freshly made Hoechst stain diluted to 1 ug/ml in Blocking Buffer or PBS and incubate for 10 minutes at room temperature (see Note 12).
Wash 2X with 0.5ml (24 well plate) or 100 ul (96 well plate) Hank’s Medium Salt Solution.
For infections in 24 well tissue culture treated plates on coverslips, mount the coverslips on glass slides in 4 ul of mounting solution. Visualize by fluorescence microscopy (see Note 13)
3.5. Measuring target mRNA depletion by RT-PCR
-
1
Using RNA samples isolated from dsRNA treated and untreated Drosophila cells as template perform a reverse transcription reaction.
Combine 1 ul of oligodT (5 ug/ml) and 1 ug of RNA in a 20 ul volume of ddH2O.
Incubate at 70°C for 10 minutes.
Add dNTPs and reverse transcriptase. We use the SuperScript RT II kit (Invitrogen) following the manufacturer’s instructions.
Incubate at 42°C for 1.5 hours then heat inactivate the enzyme at 65°C for 20 minutes.
-
2
Use 1 ul of the cDNA reaction from Step 1 to perform a partial PCR reaction using the same primers used to generate the dsRNA amplicon from genomic DNA and typically 20–25 cycles. For each dsRNA treatment, the corresponding fragment should be amplified from dsRNA treated and untreated samples for comparison. For normalization, we amplify the F actin gene using similar PCR conditions but with fewer cycles as this transcript is highly abundant (see Note 14).
-
4
Separate the PCR reaction in an agarose gel.
-
5
Quantify the intensity of the PCR product for each gene targeted and calculate fold depletion relative to the untreated control.
Footnotes
All Drosophila media should be filtered sterilized using a 0.22 um filter and stored at 4°C. All media should be pre-warmed to room temperature prior to use.
Drosophila cell densities lower than 1×105 cells/ml or above 1×107 cell/ml are not recommended.
To completely rule out off-target effects, it is important to design at least 2 sets of primers corresponding to different non-overlapping regions of the gene and test these individually.
Never use recently thawed cells. Cells should be passaged at least once prior to use for RNAi. In addition, do not use Kc167 cells that are >90% confluent for dsRNA treatment as we have observed reduced dsRNA uptake in cells that are overgrown.
Typically, one well of 1×106 cells after 3–4 days incubation with dsRNA will yield roughly 3×106 cells, sufficient for 30 individual samples of 1×105 cells each for subsequent assays. Depending on the number of conditions, time points and L. pneumophila strains to be analyzed, additional wells of dsRNA treated cells may be required. We typically add at least 1 additional well per dsRNA to harvest RNA from these cells to test for gene silencing by RT-PCR.
For some host targets, dsRNA treatments for 3–4 days can render the cells very unhealthy, even apoptotic, especially for targets that perform vital functions. In these cases, reducing the treatment time to 2 days often yields sufficient depletion while preserving the cells in a state that allows them to support growth of L. pneumophila upon infection.
We use Corning Costar 3603 96-well plates for immunofluorescence studies. These plates have a flatter thinner surface resulting in better resolution for microscopy. The poor adherence of Drosophila cells can be problematic. If cells do not attach well to the plate surface, wells and/or coverslips can be coated with 25–50 ug/ml of Concanavalin A (17).
To generate poly-L-lysine-coated coverslips, incubate coverslips in 1 M HCl for 4 hrs at room temperature, rinse 3x with ddH2O, and then incubate in 1 mg/ml poly-L-lysine (Sigma) for 2–3 hrs with agitation. We typically treat 100 coverslips in a 20 ml volume in a 50 ml Falcon tube. Rinse the coverslips 7x with ddH2O (poly-L-lysine is toxic to cells so ensure removal of all trace amounts of unused reagent). Rinse the coverslips in 70% ethanol, dry on Whatman paper and UV sterilize for 20 minutes.
Make sure to mix well by pipetting up and down several times at each step to avoid inconsistencies in the number of colony forming units recovered due to the incomplete lysis of the Drosophila cells.
For infectious center assays, time points are typically 1, 6 and 16 hours post infection and any other appropriate time point in between.
As an alternative detection system, L. pneumophila strains expressing the green fluorescent protein (GFP) can be used to circumvent the need for antibody staining and the potential loss of cells during washing.
If DNA staining if too bright, decrease the incubation time to 5 minutes.
For 24 well plate infections on coverslips, use a 100x objective and count the number of bacteria per phagosome of at least 100 phagosomes per coverslip. For 96-well plates we use 20X objectives and take pictures of 4 sites per well for each channel. Using image analysis software, we determine the pixel areas for each LCV present in each picture. The data per site is then averaged to obtain the average size, as determined by the LCV area, per well and/or condition tested. It is recommended to perform microscopy as soon as possible after preparation of slides. However, mounted slides and 96-well plates containing Hank’s Medium Salt Solution can be stored temporarily at 4°C protected from light.
Alternatively, qRT-PCR instead of RT-PCR can be used to determine the relative levels of mRNA transcripts in dsRNA treated and untreated cells.
References
- 1.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O’Farrell PH, Andino R. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- 5.Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- 6.Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 8.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–1253. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 9.Cheng LW, Viala JP, Stuurman N, Wiedemann U, Vale RD, Portnoy DA. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc Natl Acad Sci U S A. 2005;102:13646–13651. doi: 10.1073/pnas.0506461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–1251. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 11.Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5:901–911. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- 12.Akimana C, Al-Khodor S, Abu Kwaik Y. Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS One. 2010;5:e11025. doi: 10.1371/journal.pone.0011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherry S. Genomic RNAi screening in Drosophila S2 cells: what have we learned about host-pathogen interactions? Curr Opin Microbiol. 2008;11:262–270. doi: 10.1016/j.mib.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 16.Ramadan N, Flockhart I, Booker M, Perrimon N, Mathey-Prevot B. Design and implementation of high-throughput RNAi screens in cultured Drosophila cells. Nat Protoc. 2007;2:2245–2264. doi: 10.1038/nprot.2007.250. [DOI] [PubMed] [Google Scholar]
- 17.Cheng LW, Portnoy DA. Drosophila S2 cells: an alternative infection model for Listeria monocytogenes. Cell Microbiol. 2003;5:875–885. doi: 10.1046/j.1462-5822.2003.00327.x. [DOI] [PubMed] [Google Scholar]