Summary
Zygotic genome activation (ZGA) is a major genome programming event whereby the cells of the embryo begin to adopt specified fates. Experiments in Drosophila and zebrafish revealed that ZGA depends on transcription factors that provide large-scale control of gene expression by direct and specific binding to gene regulatory sequences [1–5]. Zelda (Zld) plays such a role in the Drosophila embryo, where it was shown to control the action of patterning signals [1, 2], however the mechanisms underlying this effect remain largely unclear. A recent model proposed that Zld binding sites act as quantitative regulators of the spatiotemporal expression of genes activated by Dorsal (Dl), the morphogen that patterns the dorsoventral (DV) axis [6]. We test this model experimentally, using enhancers of brinker (brk) and short gastrulation (sog), both of which are directly activated by Dl, but at different concentration thresholds [7–9]. In agreement with the model, we show that there is a clear positive correlation between the number of Zld binding sites and the spatial domain of enhancer activity. Likewise, the timing of expression could be advanced or delayed. We present evidence that Zld facilitates binding of Dl to regulatory DNA, and that this is associated with increased chromatin accessibility. Importantly, the change in chromatin accessibility is strongly correlated with the change in Zld binding, but not Dl. We propose that the ability of genome activators to facilitate read-out of transcriptional input is key to widespread transcriptional induction during ZGA.
Results and Discussion
In blastoderm embryos, brk is activated in an 8–10 cell-wide domain that develops into the ventral neurogenic ectoderm (vNE), while sog is expressed in a broader band of 16–18 cells encompassing the entire neurogenic ectoderm (NE) (see Figures 1A and 1I). Both genes have the same ventral expression boundary due to repression by Snail (Sna) in the presumptive mesoderm [10–14]. The dorsal borders of their domains lie in regions of the Dl gradient where amounts are low and change little, raising the question of how their enhancers can interpret small differences in Dl concentrations.
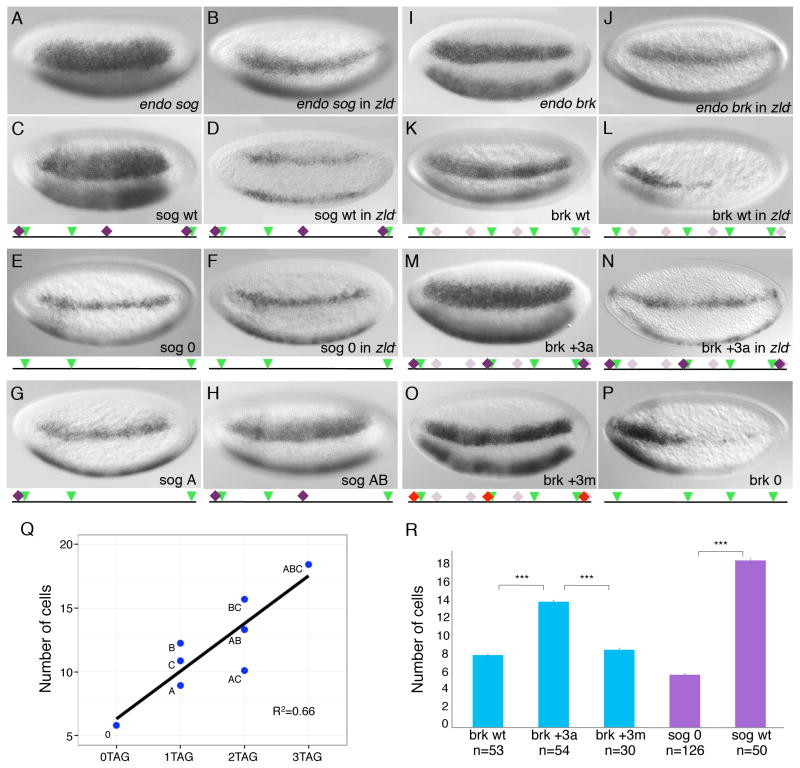
Figure 1. The number of Zld binding sites determines the spatial extent of Dl target gene expression.
Wild-type (A, C, E, G, H, I, K, M, O, P) and zld− (B, D, F, J, L, N) embryos in nc 14 were hybridized with RNA probes synthesized against cDNA sequences for sog (A, B), brk (I, J) or lacZ (C–H, K–P) for transgenic embryos. Embryos are oriented anterior to the left and dorsal up here and in subsequent figures. A schematic representation of the enhancer that drives lacZ expression is shown below transgenic embryos (C–H, K–P). Green triangle = Dl site; Dark purple diamond = canonical Zld site; Light purple diamond = non-canonical Zld site; Red diamond = mutagenized Zld site. (C–F) Mutation of all three Zld sites in the sog NEE caused a reduction in the expression domain it drives. (G, H) Elimination of one (H) or two (G) Zld sites in sog NEE resulted in a step-wise narrowing of expression domain. (K–N) Addition of 3 Zld sites to the brk5′ enhancer led to a Zld -dependent expansion in expression. (O) Mutation of the 3 added Zld sites gave an expression similar to that driven by brk wt enhancer. (P) Removal of all Zld sites in brk5′ enhancer led to sporadic and thin expression pattern. Anterior-posterior modulation seems to be in play for the expression of brk, which could be explained by the presence of two Bcd sites in this enhancer [15]. (Q) Scatter plot showing the width of expression domain (in the number of cells it spans) driven by different forms of the sog NEE that contains 0 (0TAG), 1 (1TAG), 2 (2TAG) or 3 (3TAG) Zld sites. Each dot represents the average from at least 20 embryos. The width of expression domain correlates with number of Zld sites (linear regression R2=0.66). (R) Bar chart showing the width of expression domain driven by brk wt, brk +3a, brk +3m, sog 0 and sog wt enhancers. Data are represented as mean ± standard error of the mean (SEM) (*** means t-test p-value < 0.005).
See also Figures S1–S3.
sog and brk each have two reported cis-regulatory modules (enhancers) that are active in early embryos [15–20]. The sog intronic Lateral Stripe Enhancer (LSE) [16] is less well-conserved and drives a slightly narrower stripe of expression relative to the sog shadow enhancer [17], also known as the Neurogenic Ectoderm Enhancer (NEE), which recapitulates the broad endogenous sog pattern [18]. The brk5′ and 3′ enhancers both support lateral stripes similar to endogenous brk [15, 17], however the 3′ enhancer drives a more dynamic pattern that broadens at cellularization [19], thus we focused on the brk5′ enhancer to avoid confounding dynamic change of width.
The sog 426bp NEE contains 3 CAGGTAG heptamer sites for optimal Zld binding. However, the brk 498bp 5′ enhancer does not have any canonical Zld binding sites (also known as TAGteam sites [21]). To explain its Zld dependence, we used EMSA to look for Zld binding sites in the brk5′ enhancer. We identified 3 CAGGTCA sequences and a tandem GAGGCACAGGCAC sequence that promote very weak Zld binding, which was abolished upon mutation of the sites (Figure S1 in Supplemental Information).
To test whether altering the number of Zld binding sites in the NE enhancers can affect the expression they drive, we created mutant forms of the brk and sog enhancers. The sog NEE (sog wt, Figure 1C) drives a lacZ reporter expression pattern identical to endogenous sog (Figure 1A). Mutation of all 3 CAGGTAG sites dramatically reduced the expression width (sog 0, Figures 1E and 1R). Similar changes were also observed by Liberman et al. (2009) when they mutated the CAGGTAG sites in the sog LSE [20]. Co-staining of lacZ and endogenous sog illustrates that the narrowed lacZ domain resulted from a collapse of the dorsal, not the ventral border (data not shown). We infer that without Zld, sog is unable to be activated by the lower levels of Dl in the dorsal neuroectoderm region. In embryos lacking maternal Zld [1] (referred to herein as zld−), both the endogenous sog and sog wt domains shrink and become sporadic (Figures 1B and 1D). This is not due to an indirect effect on the Dl concentration gradient because it is unchanged in zld− (Figure S2). Thus, loss of Zld in trans, or Zld binding sites in cis, has the same effect on NEE activity, indicating a direct modulation of Zld on sog.
Next we did the opposite experiment by introducing 3 CAGGTAG sites into the brk5′ enhancer. This modified enhancer (brk +3a) drives a considerably expanded expression domain (Figure 1M) compared to brk wt (Figures 1K and 1R). A second form of the brk enhancer with CAGGTAG sites added to different locations (brk +3b) also drives the same expanded expression domain (Figure S3), arguing against the requirement of precise motif grammar in Zld’s regulation of NE genes.
To rule out the possibility that the expansion in domain width of brk +3 is caused by inadvertent disruption of a repressor binding site rather than addition of Zld binding sites, we mutated the 3 added CAGGTAG sequences in brk +3a into 7-mers that are neither the original sequence, nor Zld binding sites (Figure 1O, brk +3m). Mutation of these sites reduced the expanded domain of brk +3a back to a width similar to brk wt (Figure 1R). When each of the brk +3a, brk +3b, and brk +3m transgenic enhancers was placed into a zld− background, narrow and sporadic expression resulted resembling that of endogenous brk in zld− (Figures 1J, 1N and data not shown), supporting again, that the CAGGTAG driven broadened expression is Zld-dependent. Moreover, mutation of the newly found weak Zld binding sites led to a narrowed and weakened stripe of expression, identical to the pattern of brk wt in zld− (Figures 1L and 1P).
To better correlate the number of Zld sites with the extent of reporter expression, we constructed six different forms of the sog NEE containing either one or two of the three CAGGTAG sites (see Figure 1G for a 1-site line (sog A) and Figure 1H for a 2-site line (sog AB)). The width of expression correlated moderately to the number of Zld sites in the enhancer (Figure 1Q; R2=0.66). However, some sites appear to be more important than the others in contributing to the expression width, indicating a context dependency for Zld binding sites. From our results and others’ work demonstrating weakened NE gene expression upon removal of Zld or Zld sites [1, 2, 20, 22, 23], it is evident that Zld is indispensable for the proper expression of NE genes.
We next asked if the number of Zld binding sites also influences the timing of Dl target expression, since previous reports have implicated Zld as a developmental timer. Harrison et al. (2011) observed a correlation between the onset of zygotic gene expression and strength of Zld binding at nuclear cycle (nc) 8 [3]. Besides that, when the enhancer region of zen, which contains four Zld binding sites was multimerized, it drove precocious activation of reporterexpression [21]. And finally, Nien et al., (2011) showed that the expression of many patterning genes is delayed in zld− embryos [2], including sog and brk. We reasoned that since Dl nuclear concentrations increase from nc 10 to 14 [24–26], the lower levels of Dl present in earlier cycles would no longer be adequate to activate target genes without Zld’s input, resulting in delayed activation of sog and brk [6, 27].
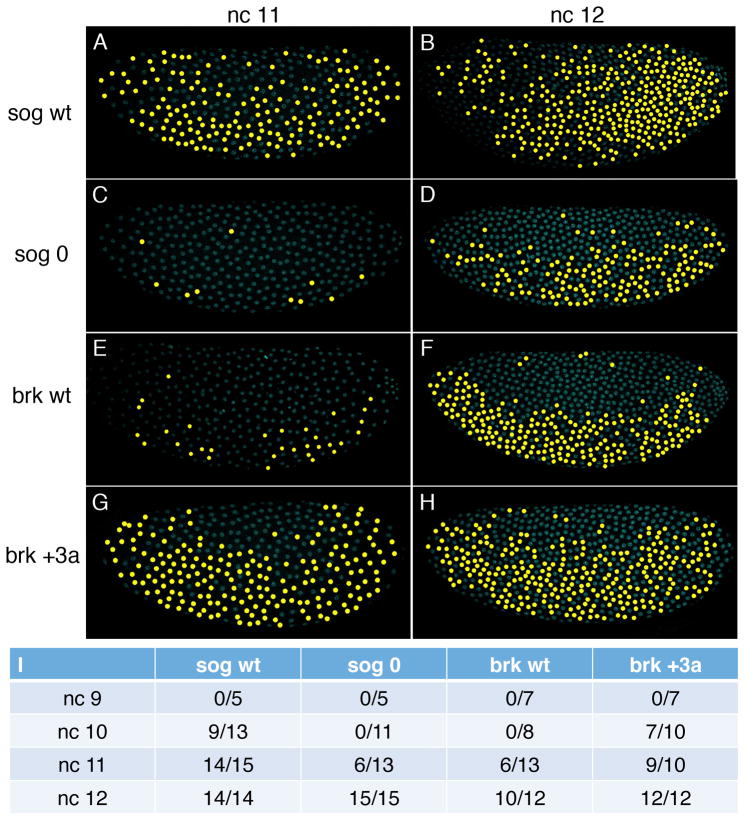
To measure the onset of transcription, we determined when the four transgenic enhancers (sog wt, sog 0, brk wt and brk +3a) could activate an intron-containing yellow reporter gene [28], which allows us to detect nascent transcripts. Reporter expression driven by the sog wt enhancer was first detectable in nc 10 embryos, while no reporter activity was observed for sog 0 enhancer until nc 11 (Figure 2I; see false color of FISH signal). Even in nc 12, the expression driven by sog 0 is more sporadic compared to sog wt (Figures 2A–2D). Unlike in nc 14 embryos, reporter expression can be seen in ventral nuclei of nc 11 and nc 12 embryos because the Sna repressor has not yet accumulated to high levels [14]. Adding 3 Zld sites to the brk enhancer resulted in advanced initiation of reporter activity from nc 11 to nc 10 (Figure 2I), and reporter expression also became more robust, in terms of both the proportion of nuclei showing expression and the ratio of embryos with expressing nuclei (Figures 2E–2I). Our results clearly illustrate that by manipulating Zld binding sites, the timing of NE gene activation can be altered. Temporal regulation by transcription factor binding sites has also been shown inCiona where the number of Brachyury binding sites governs the timing of notochord gene expression [29].
Figure 2. The number of Zld binding sites determines the timing of Dl target gene activation.
nc 11 (A, C, E, G) and 12 (B, D, F, H) embryos carrying sog(A–D) or brk(E–H) transgenes were hybridized with RNA probes synthesized against intronic sequences of the yellow reporter gene. Dl antibody staining (not shown) was used to orient embryos. DAPI stained nuclei expressing yellow reporter gene are pseudo-colored in yellow. Compared to the sog wt enhancer (A, B), mutation of all Zld sites in the sog NEE (sog 0; C, D) results in delayed and sporadic expression. (E–H) Embryos carrying the brk enhancer with added Zld sites (brk +3a; G, H) have advanced initiation of transcription compared to embryos carrying the brk wt enhancer (brk wt; E, F). (I) Table showing the number of embryos carrying the four transgenic lines that display any expression from nc 9 to nc 12.
We believe that Zld regulates the temporal and spatial expression of NE genes by promoting Dl activity, rather than acting independently, because nuclear Dl is absolutely required for the activation of brk and sog, which exhibit no expression in genetic backgrounds lacking nuclear Dl [20, 30]. One possible mechanism may involve cooperativity at the level of DNA binding [6]. To test the hypothesis that the extent of Zld binding impacts Dl binding at target enhancers, we performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) to measure Zld and Dl binding to the different transgenic enhancers.
The sog 0 enhancer without Zld sites has diminished Zld binding when compared to sog wt (t-test p-value = 0.004; Figure 3A). Dl binding is also much reduced (p-value = 0.002; Figure 3B). As an internal control, Zld and Dl binding to the endogenous sog locus showed no significant difference between the lines (p-value = 0.464 and 0.288, respectively; Figures 3A and 3B). On the other hand, introduction of Zld sites into the brk transgenic enhancer led to higher Zld binding (p-value = 0.0047; Figure 3D) and Dl binding (p-value = 0.004; Figure 3E), while Zld and Dl binding to the endogenous locus remained similar between lines (p-value = 0.221 and 0.452, respectively; Figures 3D and 3E). These results illustrate that changing the number of Zld sites, and therefore changing the amount of Zld binding to the NE enhancers, influences the level of Dl binding to its target sites in vivo.
Figure 3. Zld promotes Dl binding to target enhancers and increases chromatin accessibility.
Bar charts showing Zld (A, D, G) and Dl (B, E, H) ChIP-qPCR results and DNase I digestion-qPCR results (C, F, I) performed on 1.5–3 hr embryos carrying transgenic enhancers. (A, B, C) Embryos carrying the sog enhancer with mutated Zld sites (sog 0) has reduced Zld (A) and Dl (B) binding, as well as lower sensitivity to DNase I digestion (C) on the reporter region compared to embryos carrying the sog wt enhancer. (D, E, F) the brk transgenic enhancer with added Zld sites (brk +3a) has higher Zld (D) and Dl (E) binding, and higher sensitivity to DNase I digestion (F) than the brk wt enhancer. (G, H, I) the brk transgenic enhancer with mutated Dl sites (brk 0Dl) has reduced Zld (G) and Dl (H) binding, and slightly decreased sensitivity to DNase I digestion (I) than the brk wt enhancer. Shown are ChIP enrichment or DNase I hypersensitivity relative to an unrelated genomic region (see Experimental Procedures). Error bars indicate SEM from three biological replicates (*: p<0.05, **: p<0.01 and ***: p < 0.005).
Our results from reporter expression analyses and ChIP experiments suggest that Zld promotes transcriptional output by facilitating Dl DNA binding. Zld might directly interact with Dl, leading to cooperative DNA binding as in the Dl-Twist (Twi) interaction [13, 31–33]. Alternatively, Zld might assist factor binding by interacting with common co-activators, or by changing the local chromatin accessibility [34, 35]. We favor the latter possibility for several reasons. 1) Zld binding greatly overlaps with that of many other transcription factors such as Bicoid (Bcd), Hunchback, Dl, Twi, Sna, and Mother against Dpp (Mad) [2]; 2) Zld helps the binding of Twi and Bcd to target DNA [23, 36]; 3) the presence of Zld binding sites is associated with high levels of transcription factor binding [37]; 4) and the Zld site (CAGGTA; [2]) is the most enriched motif in transcription factor binding “HOT regions”, which were seen to correlate with decreased nucleosome density [37–39]. Hence, it is more likely that Zld plays a more general role, such as “opening” the underlying chromatin, than interacting specifically with multiple other factors.
We therefore went on to address the hypothesis that Zld facilitates the binding of Dl by making the local chromatin more accessible. DNase I’s preferential digestion of nucleosome-depleted DNA in the genome can be used to map active regulatory regions accessible for transcription factor binding [40, 41]. We performed DNase I hypersensitivity assays followed by qPCR (DNase I-qPCR) to measure the chromatin “openness” of transgenic enhancers carrying varying numbers of Zld sites. The sog transgenic enhancer region had significant reduction of chromatin accessibility when Zld sites were mutated (~1.6 fold, p-value = 0.002; Figure 3C), while adding Zld sites to the brk transgenic enhancer increased sensitivity to DNase I digestion (~1.6 fold, p-value = 0.002; Figure 3F). The DNase I hypersensitivity assessed on endogenous brk and sog loci were comparable between transgenic lines (p-value = 0.118 and 0.114, respectively; Figures 3C and 3F), serving as a control for embryo staging between transgenic lines and the DNase I digestion procedure.
These results suggest that the presence of Zld sites, and thus Zld binding, makes the local chromatin more accessible for Dl, and potentially other transcription factors. However, it is feasible that the total number of factor binding sites influences chromatin accessibility rather than the number of Zld sites in particular. Therefore, we assayed the DNase I hypersensitivity of a transgenic brk enhancer that lacks all Dl binding sites and shows no reporter expression (brk 0Dl, Figure S3). Dl binding decreased nearly to background levels (~2.3 fold, p-value = 0.001; Figure 3H) compared to brk wt, but the Zld binding and DNase I hypersensitivity showed only slight decreases (~1.5 fold, p-value = 0.012 and ~1.2 fold, p-value = 0.0002, respectively; Figures 3G and 3I), which is not comparable to the effects seen upon manipulation of Zld sites on the brk and sog enhancers (Figures 3A–F). We reason that the binding of each transcription factor may contribute to the DNase I hypersensitivity to a certain extent, but the major influence comes from Zld binding. To further evaluate the contribution of Zld vs. Dl sites to chromatin accessibility, we calculated the fold change in Zld and Dl binding for sog 0, brk +3a, and brk 0Dl relative to their corresponding wt transgenic enhancers, then correlated the fold change in factor binding with the change in DNase I hypersensitivity (Figure 4). We found a strong correlation between the change in Zld binding and DNase I hypersensitivity (R2 = 0.98), while the change in Dl binding and DNase I hypersensitivity do not correlate (R2 = 0.02). These results support the idea that the number of Zld sites rather than Dl sites is important in determining chromatin accessibility.
Figure 4. The change in chromatin accessibility correlates with the change in Zld binding on target enhancers.

Zld and Dl ChIP enrichment and DNase I hypersensitivity on the transgenic region were normalized to endogenous enhancer locus, then the fold change was calculated for the two lines under comparison (sog 0 vs. sog wt, brk +3a vs. brk wt, and brk 0Dl vs. brk wt). Blue dots and green dots represent Zld and Dl, respectively. The change in Zld binding between lines strongly correlates with the change in DNase I hypersensitivity (linear regression R2=0.98) while the change in Dl binding does not (R2=0.02).
Using Zld’s co-regulation of NE genes as a case in point, we have shed light on how Zld functions as a zygotic genome activator. Our data revealed that Zld works in combination with Dl and regulates Dl target genes by binding differentially to their regulatory sequences. Changing the number of Zld sites on Dl target gene enhancers had a pronounced effect on their expression both temporally and spatially. As a uniformly distributed factor, Zld supplies positional information by promoting Dl binding to target enhancers, thereby increasing the “apparent dosage” of Dl. Zld’s input is especially important where the level of morphogen is low, and likely plays a similar role for other key factors in the blastoderm embryo, such as Twi, Bcd, and Mad. Uniform factors have been found to act in combination with Sonic-hedgehog in neural tube differentiation [42], and our findings on how Zld potentiates morphogen activity will be relevant to vertebrate systems.
While our results do not rule out other possible mechanisms, they strongly support the idea that Zld binding increases chromatin accessibility, which we believe contributes greatly to how it activates such a wide range of targets. In this model, the amount of Zld binding on a region would determine how open and therefore how active it is. At the center of this property is Zld’s ability to occupy a large fraction of its recognition sites in early embryos [3]. Besides that, Zld is present in nuclei as early as nc 2 [2], which is considerably earlier than other factors (for example, Bcd: nc 6, Dl: nc 10) [25, 26, 43]. Therefore, Zld may indeed act as a pioneer factor as previously suggested [3, 27], but whether Zld binds to its sites in nucleosomes and repositions them, or recruits histone modifiers that in turn affect binding of other factors like Dl, awaits further investigation. Interestingly, this idea may extend beyond flies since newly discovered genome activators in zebrafish ZGA were seen to cooperate with developmental regulators and prime the genome for subsequent activation [4, 5], thus it seems that developmental control of ZGA is highly similar in flies and fish.
Experimental Procedures
Transgenic reporter analysis
Mutant forms of the 426bp sog NEE and the 498bp brk5′ enhancer were created via site-specific mutagenesis or by direct synthesis using Integrated DNA Technologies custom gene synthesis service. Enhancer and primer sequences can be found in Supplemental Experimental Procedures.
In situ hybridization and antibody staining were performed as previously described [1, 44].
Zld and Dl ChIP-qPCR
ChIP was performed on 1.5–3 hr embryos, using a modified protocol from the Zeitlinger lab [45]. Three biological replicates were performed for each ChIP experiment. Three primer sets (see Supplemental Experimental Procedures for primer sequences) were used to probe the reporter locus (targetOut), the endogenous enhancer (targetEn), and an unrelated genomic region on chr2L (control), respectively. ChIP enrichment was then calculated as (ChIPtarget/ChIPcontrol)/(inputtarget/inputcontrol) for both reporter and endogenous loci.
DNase I digestion-qPCR
DNase I digestion was performed on 1.5–3 hr embryos, as previously described [41] with some modifications. Three biological replicates were performed for each DNase I digestion experiment. The same primer sets as in the ChIP-qPCR experiments were used. We first calculated the percent remaining DNA at target loci relative to the control region, which did not show DNase I hypersensitivity [41], then the percent remaining DNA after 15 min digestion was normalized to that without DNase I digestion, giving rise to normalized percent remaining DNA ([target15min/control15min]/[target0min/control0min]). DNase I hypersensitivity was finally presented as 1/(normalized percent remaining DNA) for both reporter and endogenous loci.
Supplementary Material
Highlights.
Zelda “potentiates” Dorsal morphogenetic activity both temporally and spatially.
Zelda binding to target enhancers leads to earlier and more robust expression.
Zelda increases chromatin accessibility and facilitates Dorsal binding to DNA.
Acknowledgments
We thank Randy Hui for help with DNA binding assays. We are indebted to Kai Chen and Julia Zeitlinger for guidance on ChIP-qPCR, providing anti-Dl antibodies, and many insightful discussions. This study was supported by grants from the National Institutes of Health (GM63024 to C. Rushlow) and National Science Foundation (EFRI-MIKS 1136913 to S. Shvartsman).
Footnotes
Author Contributions
Conceived and designed the experiments: SMF YS NK SYS CAR. Performed the experiments: SMF YS BL RZ KOB NK. Analyzed the data: all authors. Wrote the paper: SMF YS SYS CAR.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011;7:e1002339. doi: 10.1371/journal.pgen.1002339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison MM, Li XY, Kaplan T, Botchan MR, Eisen MB. Zelda binding in the early Drosophila melanogaster embryo marks regions subsequently activated at the maternal-to-zygotic transition. PLoS Genet. 2011;7:e1002266. doi: 10.1371/journal.pgen.1002266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature. 2013;503:360–364. doi: 10.1038/nature12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leichsenring M, Maes J, Mossner R, Driever W, Onichtchouk D. Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science. 2013;341:1005–1009. doi: 10.1126/science.1242527. [DOI] [PubMed] [Google Scholar]
- 6.Kanodia JS, Liang HL, Kim Y, Lim B, Zhan M, Lu H, Rushlow CA, Shvartsman SY. Pattern formation by graded and uniform signals in the early Drosophila embryo. Biophys J. 2012;102:427–433. doi: 10.1016/j.bpj.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stathopoulos A, Levine M. Genomic regulatory networks and animal development. Dev Cell. 2005;9:449–462. doi: 10.1016/j.devcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 8.Hong JW, Hendrix DA, Papatsenko D, Levine MS. How the Dorsal gradient works: insights from postgenome technologies. Proc Natl Acad Sci USA. 2008;105:20072–20076. doi: 10.1073/pnas.0806476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves GT, Stathopoulos A. Graded dorsal and differential gene regulation in the Drosophila embryo. Cold Spring Harb Perspect Biol. 2009;1:a000836. doi: 10.1101/cshperspect.a000836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosman D, Ip YT, Levine M, Arora K. Establishment of the mesoderm-neuroectoderm boundary in the Drosophila embryo. Science. 1991;254:118–122. doi: 10.1126/science.1925551. [DOI] [PubMed] [Google Scholar]
- 11.Leptin M. twist and snail as positive and negative regulators during Drosophila mesoderm development. Genes Dev. 1991;5:1568–1576. doi: 10.1101/gad.5.9.1568. [DOI] [PubMed] [Google Scholar]
- 12.Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- 13.Zinzen RP, Senger K, Levine M, Papatsenko D. Computational models for neurogenic gene expression in the Drosophila embryo. Curr Biol. 2006;16:1358–1365. doi: 10.1016/j.cub.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 14.Bothma JP, Magliocco J, Levine M. The snail repressor inhibits release, not elongation, of paused Pol II in the Drosophila embryo. Curr Biol. 2011;21:1571–1577. doi: 10.1016/j.cub.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Markstein M, Zinzen R, Markstein P, Yee KP, Erives A, Stathopoulos A, Levine M. A regulatory code for neurogenic gene expression in the Drosophila embryo. Development. 2004;131:2387–2394. doi: 10.1242/dev.01124. [DOI] [PubMed] [Google Scholar]
- 16.Markstein M, Markstein P, Markstein V, Levine MS. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc Natl Acad Sci USA. 2002;99:763–768. doi: 10.1073/pnas.012591199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crocker J, Tamori Y, Erives A. Evolution acts on enhancer organization to fine-tune gradient threshold readouts. PLoS Biol. 2008;6:e263. doi: 10.1371/journal.pbio.0060263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunipace L, Saunders A, Ashe HL, Stathopoulos A. Autoregulatory feedback controls sequential action of cis-regulatory modules at the brinker locus. Dev Cell. 2013;26:536–543. doi: 10.1016/j.devcel.2013.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liberman LM, Stathopoulos A. Design flexibility in cis-regulatory control of gene expression: synthetic and comparative evidence. Dev Biol. 2009;327:578–589. doi: 10.1016/j.ydbio.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- 22.Wunderlich Z, Bragdon MD, Depace AH. Comparing mRNA levels using in situ hybridization of a target gene and co-stain. Methods. 2014 doi: 10.1016/j.ymeth.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yanez-Cuna JO, Dinh HQ, Kvon EZ, Shlyueva D, Stark A. Uncovering cis-regulatory sequence requirements for context-specific transcription factor binding. Genome Res. 2012;22:2018–2030. doi: 10.1101/gr.132811.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reeves GT, Trisnadi N, Truong TV, Nahmad M, Katz S, Stathopoulos A. Dorsal-ventral gene expression in the Drosophila embryo reflects the dynamics and precision of the dorsal nuclear gradient. Dev Cell. 2012;22:544–557. doi: 10.1016/j.devcel.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liberman LM, Reeves GT, Stathopoulos A. Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila. Proc Natl Acad Sci USA. 2009;106:22317–22322. doi: 10.1073/pnas.0906227106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanodia JS, Rikhy R, Kim Y, Lund VK, DeLotto R, Lippincott-Schwartz J, Shvartsman SY. Dynamics of the Dorsal morphogen gradient. Proc Natl Acad Sci USA. 2009;106:21707–21712. doi: 10.1073/pnas.0912395106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rushlow CA, Shvartsman SY. Temporal dynamics, spatial range, and transcriptional interpretation of the Dorsal morphogen gradient. Curr Opin Genet Dev. 2012;22:542–546. doi: 10.1016/j.gde.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 28.Perry MW, Boettiger AN, Bothma JP, Levine M. Shadow enhancers foster robustness of Drosophila gastrulation. Curr Biol. 2010;20:1562–1567. doi: 10.1016/j.cub.2010.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Katikala L, Aihara H, Passamaneck YJ, Gazdoiu S, Jose-Edwards DS, Kugler JE, Oda-Ishii I, Imai JH, Nibu Y, Di Gregorio A. Functional Brachyury binding sites establish a temporal read-out of gene expression in the Ciona notochord. PLoS Biol. 2013;11:e1001697. doi: 10.1371/journal.pbio.1001697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jazwinska A, Rushlow C, Roth S. The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development. 1999;126:3323–3334. doi: 10.1242/dev.126.15.3323. [DOI] [PubMed] [Google Scholar]
- 31.Szymanski P, Levine M. Multiple modes of dorsal-bHLH transcriptional synergy in the Drosophila embryo. EMBO J. 1995;14:2229–2238. doi: 10.1002/j.1460-2075.1995.tb07217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiang J, Levine M. Binding affinities and cooperative interactions with bHLH activators delimit threshold responses to the dorsal gradient morphogen. Cell. 1993;72:741–752. doi: 10.1016/0092-8674(93)90402-c. [DOI] [PubMed] [Google Scholar]
- 33.Ip YT, Park RE, Kosman D, Yazdanbakhsh K, Levine M. dorsal-twist interactions establish snail expression in the presumptive mesoderm of the Drosophila embryo. Genes Dev. 1992;6:1518–1530. doi: 10.1101/gad.6.8.1518. [DOI] [PubMed] [Google Scholar]
- 34.Spitz F, Furlong EE. Transcription factors: from enhancer binding to developmental control. Nat Rev Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 35.Lelli KM, Slattery M, Mann RS. Disentangling the many layers of eukaryotic transcriptional regulation. Annu Rev Genet. 2012;46:43–68. doi: 10.1146/annurev-genet-110711-155437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Z, Chen H, Ling J, Yu D, Struffi P, Small S. Impacts of the ubiquitous factor Zelda on Bicoid-dependent DNA binding and transcription in Drosophila. Genes Dev. 2014;28:608–621. doi: 10.1101/gad.234534.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Satija R, Bradley RK. The TAGteam motif facilitates binding of 21 sequence-specific transcription factors in the Drosophila embryo. Genome Res. 2012;22:656–665. doi: 10.1101/gr.130682.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.mod EC, Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kvon EZ, Stampfel G, Yanez-Cuna JO, Dickson BJ, Stark A. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev. 2012;26:908–913. doi: 10.1101/gad.188052.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell O, Tiwari VK, Thoma NH, Schubeler D. Determinants and dynamics of genome accessibility. Nat Rev Genet. 2011;12:554–564. doi: 10.1038/nrg3017. [DOI] [PubMed] [Google Scholar]
- 41.Thomas S, Li XY, Sabo PJ, Sandstrom R, Thurman RE, Canfield TK, Giste E, Fisher W, Hammonds A, Celniker SE, et al. Dynamic reprogramming of chromatin accessibility during Drosophila embryo development. Genome biology. 2011;12:R43. doi: 10.1186/gb-2011-12-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cohen M, Briscoe J, Blassberg R. Morphogen interpretation: the transcriptional logic of neural tube patterning. Curr Opin Genet Dev. 2013;23:423–428. doi: 10.1016/j.gde.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 43.Little SC, Tkacik G, Kneeland TB, Wieschaus EF, Gregor T. The formation of the Bicoid morphogen gradient requires protein movement from anteriorly localized mRNA. PLoS Biol. 2011;9:e1000596. doi: 10.1371/journal.pbio.1000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanodia JS, Kim Y, Tomer R, Khan Z, Chung K, Storey JD, Lu H, Keller PJ, Shvartsman SY. A computational statistics approach for estimating the spatial range of morphogen gradients. Development. 2011;138:4867–4874. doi: 10.1242/dev.071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.