Abstract
Synesthesia is a perceptual condition in which sensory stimulation triggers anomalous sensory experiences. In colored sequence synesthesia (CSS), color experiences are triggered by sequences such as letters or numbers. We performed a family based linkage analysis to identify genetic loci responsible for the increased neural crosstalk underlying CSS. Our results implicate a 23 MB region at 16q12.2-23.1, providing the first step in understanding the molecular basis of CSS.
Keywords: Synesthesia Family, linkage analysis, 16q
1. Introduction
In synesthesia, ordinary stimuli elicit anomalous perceptual experiences [1–3]. For example, in a form we term colored sequence synesthesia (CSS), the visual experience of a specific color is triggered by the viewing of a letter, digit, weekday or month (Fig. 1A) [1,4]. These associations are thought to result from amplified crosstalk between brain areas, such that activity in one area kindles activity in another (Fig. 1B). Although there are several forms of synesthesia, this work will focus exclusively on understanding CSS due to its clustering within families and its straightforward phenotyping, as described below.
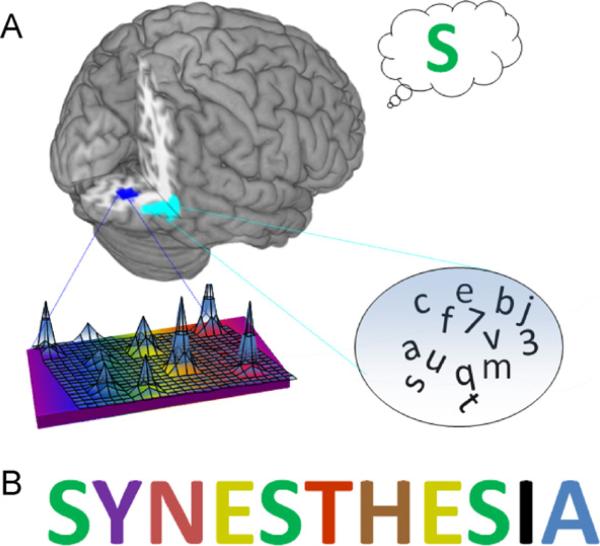
Fig. 1.
(A) In this model of synesthesia, neural populations coding for graphemes (right inferior temporal gyrus, light blue) ramify on those coding for colors (area V4, dark blue). As a result of these connections, activity triggered by a grapheme kindles V4 activity, and (B) synesthetes experience an automatic color association, an example of which is shown here.
Altered levels of crosstalk between brain regions have been found in various disease states, including autism [5,6], Alzheimer's disease [7], and schizophrenia [8,9]. Unfortunately, these studies are merely descriptive, not prescriptive, and give no information about the etiology or modifiability of abnormal patterns. Synesthesia is an ideal model for studying hyperconnectivity at the simplest level because synesthetes are healthy, the condition is easy to phenotype reliably, and it runs in families. The aim of this work is to use genetics to understand how neural crosstalk arises in a healthy brain, in the hopes of ultimately understanding how this mechanism might be modified in a disease state.
One hypothesis of the origin of synesthetic crosstalk suggests that excess connections in the newborn brain are insufficiently pruned during development [10–12], while a second hypothesis posits that synesthesia results from reduced functioning of inhibitory neural networks, either within or across brain regions [1,13,14]. These two hypotheses are indistinguishable by behavior [12,15] and neuroimaging [16,17], both of which demonstrate functional cross-activation but cannot explain the mechanism for the hyperconnectivity. The aim of our genetic study is to find the gene(s) underlying the functional crosstalk. A genetic finding hopes to reveal which of the above hypotheses is correct, and will represent the first genetic mechanism directly linked to increased neural crosstalk.
It has long been noted that synesthesia appears to run in families [18]. Analyses of pedigrees are consistent with the hypothesis of heritability and suggest that the genetic component(s) may be inherited in a dominant fashion with incomplete penetrance [19,20]. Unfortunately, those studies involved no DNA collection.
Asher et al. [29] published the first attempt to link the perceptual traits of a type of synesthesia with a genetic basis by completing a whole-genome scan of synesthetic families. Asher et al. reported linkage to several chromosomal regions, suggesting genetic heterogeneity and several patterns of inheritance; however, it is important to clarify that their study examined a different form of synesthesia (auditory–visual synesthesia) than we examine here (colored weekdays, months, letters, and numbers). Data from our lab indicate that different subtypes of synesthesia fall into clusters, suggesting the possibility that different genetic mechanisms underlie different forms of synesthesia (Novich, Cheng, Eagleman, in press). Thus, previous studies have opened an encouraging line of inquiry but offer no specific genetic conclusions. Our study combines DNA linkage analysis with rigorous phenotyping methods to set the foundation for understanding the genetic mechanism of colored sequence synesthesia.
2. Materials and methods
In an attempt to identify the gene for colored sequence synesthesia, we have performed a family linkage analysis. Although synesthesia has been reported in many forms (e.g., tasting shapes, seeing sounds, etc.) [1], for our analysis we concentrated on the triggering of color perception by letters, numbers, weekdays, or months (colored sequences). This decision was motivated by our analysis of 3194 verified colored sequence synesthetes, which indicated that color synesthesia for one type of sequence (e.g., letters) gives a ~79% likelihood of having color synesthesia for other types of sequences (e.g., weekdays, months or numbers), but gives only chance likelihood of possessing another type of synesthesia (Novich, Cheng, Eagleman, in press). This finding suggests that colored sequences may share a common genetic mechanism, while other forms of synesthesia may have different bases (including possibly non-genetic sources). Therefore, we only pursue the genetic etiology of colored sequence synesthesia here.
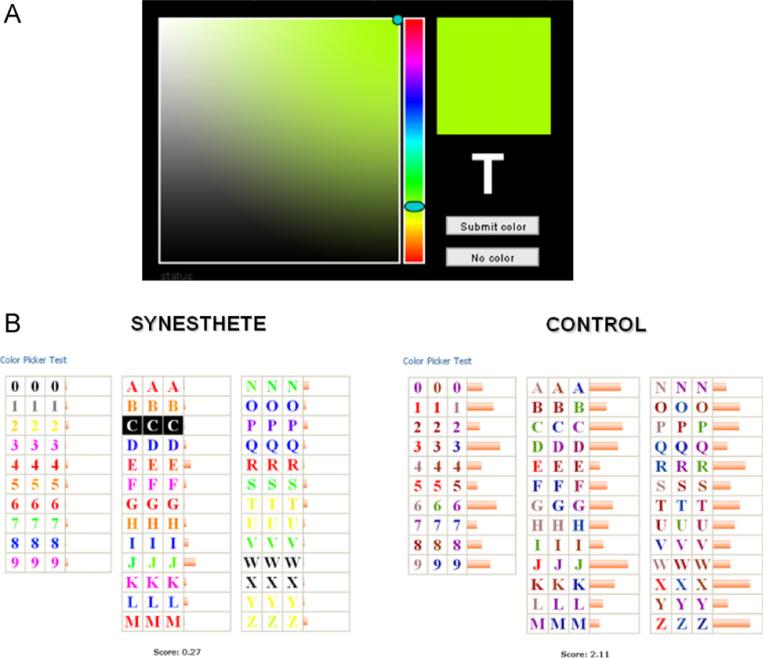
For the purposes of rigorous phenotyping, we have previously developed the Synesthesia Battery (www.synesthete.org), an online battery of tests that sensitively discriminates synesthetes from non-synesthetes [21]. Participants are serially presented with a full set of graphemes (A–Z, 0–9), three times each in a random order. For each grapheme, they are asked to choose the best associated color from an online palette of 16.7 million colors. The palette orientation and the slider bar are randomly rotated with each new grapheme presentation to prevent memorization of a color's location (Fig. 2A). The color choices are then analyzed for consistency within the session (e.g., determining whether the participant chose a similar shade of green upon seeing the letter J). The color variation for each letter (vj) is measured by
| (1) |
which represents the geometric distance in RGB (red, green, and blue) color space. The total variation score for each participant is
| (2) |
where N is the total number of graphemes for which the participant experiences synesthetic colors. Control subjects display low consistency when associating colors to graphemes over 108 trials, while synesthetes perform with high consistency (for a representative example, see Fig. 2B); see Eagleman et al. (2007) for full details of the testing procedure [15,19,21–23].
Fig. 2.
(A) Participants are presented with randomly-ordered graphemes a total of three times each (108 trials). Using a palette with 16 million colors, participants choose the precise color that best matches their synesthetic percept for that grapheme. (B) Representative alphabets and variability scores from one synesthete and one control.
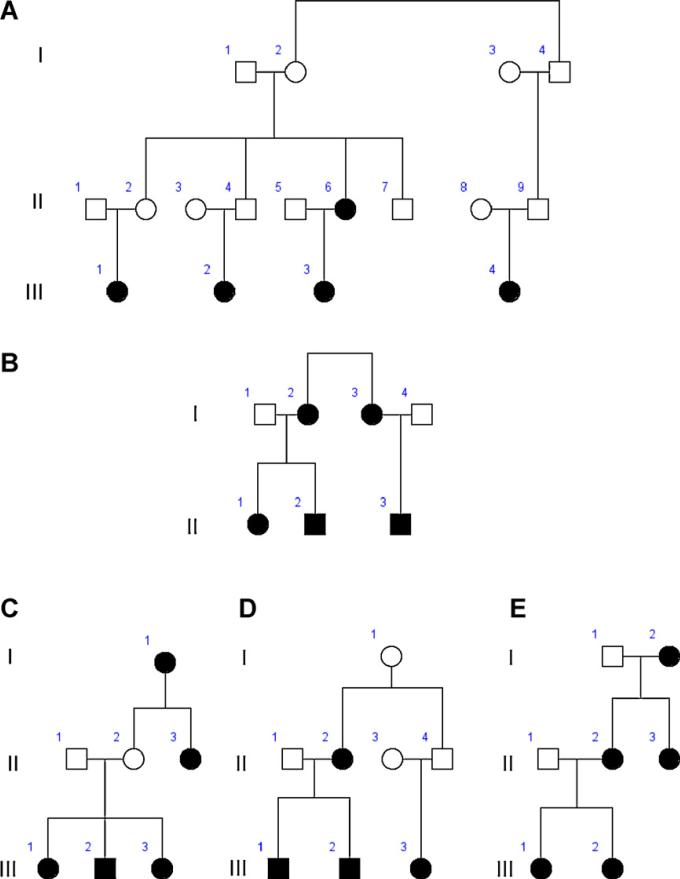
We recruited five families with four or more individuals with colored sequence synesthesia (CSS) (Fig. 3A–E). Once we verified CSS in each proband, we phenotyped CSS in all other members of the family using the tests on the Synesthesia Battery [21]. The pattern in the pedigrees is consistent with an autosomal dominant mode of inheritance with reduced penetrance.
Fig. 3.
Five families with colored sequence synesthesia. Rectangles designate males, circles designate females, and black designates a verified color sequence synesthete.
2.1. Genotyping
After obtaining informed consent, DNA from 48 individuals in five families was obtained. The saliva samples were collected using OraGene sample kits (OraGene, DNA Genotek; [24]). The samples were purified according to Oragene DNA purification protocol and quantified using PicoGreen (Invitrogen, Carlsbad, CA, USA). A single-nucleotide-polymorphism (SNP) array (Linkage-12 BeadChip, Illumina, Inc., San Diego, CA) containing 6090 SNPs was used to genotype 24 affected and 24 unaffected individuals from five large families [25]. The BeadStudio Version 3 genotyping software was used to provide automatic cluster separation, genotype calling, and quality control.
2.2. Genetic linkage analysis
PEDCHECK [26] was used to identify Mendelian inconsistencies, and the MERLIN [27] program was used to detect double recombination events over short genetic distances that were probably due to genotyping errors. Parametric and non-parametric linkage analysis was carried out using the ALLEGRO 1.2c [28] and MERLIN programs. A disease allele frequency of 0.001 was used for the analysis. Allele frequencies for SNP marker loci were obtained from the Hapmap data using the CEU (Caucasian) population. For the parametric linkage analysis, an autosomal dominant mode of inheritance with moderately reduced penetrance 0.8 [29] was used. HLOD scores were also calculated for the parametric linkage analysis allowing for linkage admixture. For the non-parametric analysis, the “all” scoring option was used for the allele sharing and the “exponential” model was used to calculate LOD scores. The Rutgers combined linkage-physical map of the human genome build 36 [30] was used for the multipoint linkage analysis.
Additionally, the non-parametric linkage analysis was carried out since the mode of inheritance of the CSS phenotype has not been established yet. It has been demonstrated that densely distributed SNP marker loci can increase the type-1-error rate due to linkage disequilibrium (LD) between marker loci [31]. Merlin software was used to remove the LD regions, which can result in false positives using the LD cut-off criteria r2 > 0.3. Haplotypes were reconstructed using ALLEGRO1.2c.
3. Results
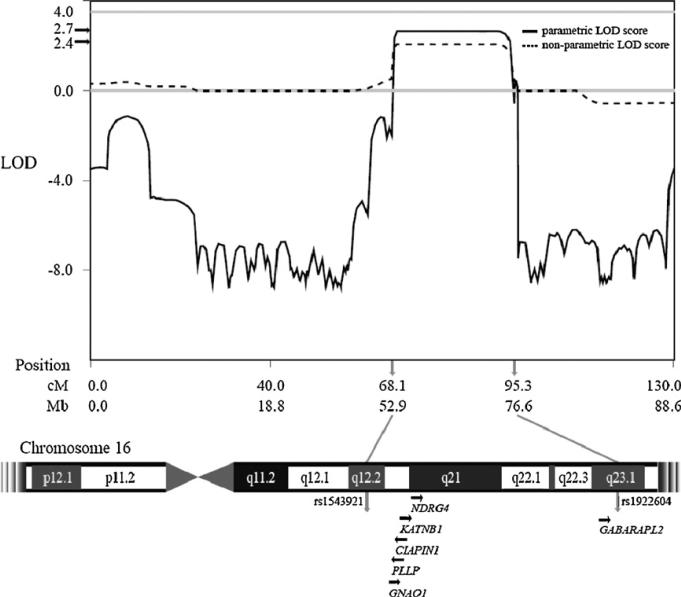
An HLOD score of 2.2 (the proportion of linked families α = 0.36) was observed in the 16q12.2-23.1 region. A close examination of the region revealed that the families A and B had LOD scores of 2.7 (Fig. 4) and 0.9, respectively. The allele sharing linkage analysis provided similar results with the max LOD 2.4 and 0.9 for the families A and B, respectively (Fig. 3). No other significant or suggestive linkages were observed except the 16q12.2-23.1 region from both parametric and allele-sharing linkage analysis.
Fig. 4.
LOD scores from families A and B along chromosome 16. Our data suggest linkage to a 23MB region between rs1543921 and rs1922604 (q12.2-q23.1).
The haplotype analysis revealed that families A and B have very similar haplotype structures on the linked region on 16q12.2-23.1. The reconstructed haplotype of family A was used to define the 23 MB critical interval. This haplotype was not found in families C-E, nor did those families display a haplotype which segregated with the synesthesia phenotype in the 16q12.2-23.1 region, suggesting that families C-E are not linked to this region.
The genomic region between markers rs1543921 and rs1922604 contains 343 genes, many of which are expressed in the brain. Among those genes, six candidate genes were selected to fit the hypothesis of synesthesia (Fig. 4). The genes were: (1) GABARAPL2 (OMIM ID: 607452), a GABA(A) receptor-associated protein that promotes GABAA receptor clustering and has been demonstrated to be an important receptor molecule in intercellular communication; (2) NDRG4, a member of the NDRG gene family that has been implicated in brain development; (3) PLLP (OMIM ID: 600340), a gene involved in myelination and ion transport. (4) KATNB1 (WD repeat containing, OMIM ID: 602703), which produces a microtubule-severing enzyme that may impact neural pruning; (5) Cytokine-induced apoptosis inhibitor 1 (CIAPIN1) (OMIM ID: 608943), a newly-identified anti-apoptotic molecule that is ubiquitously expressed in normal human fetal and adult brains; (6) Guanine nucleotide-binding regulatory protein Go-alpha (GNAO1)(OMIM ID: 139311), a protein with significantly decreased expression in individuals with schizophrenia compared to unaffected family controls.
These genes were sequenced in four synesthetes from families A and B, however none of the sequence variants segregated with all four of the affected individuals, and no novel polymorphisms were discovered. PCR-amplified DNA products were purified with ExoSAP-IT (USB Corp., Cleveland, OH, USA). Sequencing was performed with the BigDye Terminator v3.1 Cycle Sequencing Kit and Applied Biosystems 3730 DNA Analyzer (Applied Biosystems Inc., Foster City, CA, USA). The DNA sequences were assembled and analyzed using the Sequencher software (Gene Codes Corp., Ann Arbor, MI, USA).
4. Discussion
The normal brain utilizes heavy crosstalk between the senses, as evidenced by anatomical tracing [32–35], sensory substitution experiments [36], and cross-sensory illusions [37–40]. The difference between the synesthetic and non-synesthetic brain, therefore, appears to be not whether there is crosstalk, but rather how much there is. The present research may shed light on the way in which abnormal connectivity between brain regions arises.
Our genetic analyses suggest that colored sequence synesthesia is linked to a gene in the 16q12.2-23.1 region. Only two of the five families in this study show linkage to this region, suggesting that CSS is a heterogeneous condition.
Consistent with previous linkage results in auditory–visual synesthesia [29], our data suggest that CSS has locus heterogeneity. Additionally, the condition may be oligogenic and have multiple modes of inheritance. Although we cannot pinpoint a single gene at this time, our future studies include collecting more families to increase the power of the family linkage analysis, and exome sequencing to compare synesthetic and control genomes on a finer scale. These data represent the first in a line of steps to isolate the first gene for colored sequence synesthesia, providing the groundwork for understanding a genetic mechanism for neural hyperconnectivity.
The male–female ratio of synesthetes has been previously examined in several studies, but with widely variable outcomes. Previously, the ratio of male–female synesthetes has been reported between 1:1 [30] and 1:5.5 [41]. Our sample of five families includes nineteen female synesthetes and only five male synesthetes, yielding a male–female ratio of 1:3.8 (even while there is no gender imbalance within the families), similar to a previously reported ratio of 1:3.7 [20]. Larger sample sizes in the future will allow us to determine if the gender bias we find is due to undersampling or a biological factor. It has been previously suggested from pedigree analysis that synesthesia may be X-linked [20], which could possibly explain the gender bias; however, neither the genetic data presented here nor in Asher et al. [29] appear to support the X-linkage hypothesis. Thus, the basis of the possible gender imbalance remains unknown.
Abnormal crosstalk, whether from defects in pruning connections or imbalances in inhibitory networks, is thought to underlie aspects of both autism [30,42–45] and schizophrenia [46–53]. Synesthesia is an example of the same type of crosstalk in healthy brains, and by studying it we hope to elucidate how this type of hyperconnectivity operates in disease states. By finding the gene(s) responsible for this crosstalk, we hope to determine whether the hyperconnectivity is caused by increased anatomical wiring or decreased inhibition. Using synesthesia as a model, we hope to uncover the functional and genetic mechanisms of neural hyper-connectivity.
Acknowledgments
This work was supported by a grant from the Guggenheim Foundation (DME) and the Mind Science Foundation (DME).
Footnotes
Electronic database information
The following URLs were accessed for data in this article: Online Mendelian Inheritance in Man (OMIM), http://www. ncbi.nlm.nih.gov/omim/.
UCSC genome browser, http://genome.ucsc.edu/.
References
- 1.Cytowic RE, Eagleman DM. Wednesday is indigo blue: discovering the brain of synesthesia. MIT Press; Cambridge (MA): 2009. [Google Scholar]
- 2.Hubbard EM, Ramachandran VS. Neurocognitive mechanisms of synesthesia. Neuron. 2005;48:509–20. doi: 10.1016/j.neuron.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Robertson LC, Sagiv N. Synesthesia: perspectives from cognitive neuroscience. Oxford University Press; 2004. [Google Scholar]
- 4.Eagleman DM, Goodale MA. Why color synesthesia involves more than color. Trends Cogn Sci (Regul Ed) 2009;13:288–92. doi: 10.1016/j.tics.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Ebisch SJ, Gallese V, Willems RM, Mantini D, Groen WB, Romani GL, et al. Altered intrinsic functional connectivity of anterior and posterior insula regions in high-functioning participants with autism spectrum disorder. Human Brain Mapping. 2010 doi: 10.1002/hbm.21085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gepner B, Féron F. A world changing too fast for a mis-wired brain? Neurosci Biobehav Rev. 2009;33:1227–42. doi: 10.1016/j.neubiorev.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer's disease. PLoS Comput Biol. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitfield-Gabrieli S, Thermenos HW, Milanovic S, Tsuang MT, Faraone SV, McCarley RW, et al. Hyperactivity and hyperconnectivity of the default network in schizophrenia and in first-degree relatives of persons with schizophrenia. Proc Natl Acad Sci USA. 2009;106:1279–84. doi: 10.1073/pnas.0809141106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lynall ME, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, et al. Functional connectivity and brain networks in schizophrenia. J Neurosci. 2010;30:9477–87. doi: 10.1523/JNEUROSCI.0333-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron-Cohen S, Harrison J, Goldstein LH, Wyke M. Coloured speech perception: is synaesthesia what happens when modularity breaks down? Perception. 1993;22:419–26. doi: 10.1068/p220419. [DOI] [PubMed] [Google Scholar]
- 11.Maurer D. Neonatal synaesthesia: Implications for the processing of speech and faces. Synaesthesia. 1997 [Google Scholar]
- 12.Ramachandran VS, Hubbard EM. Psychophysical investigations into the neural basis of synaesthesia. Proc Biol Sci. 2001;268:979–83. doi: 10.1098/rspb.2001.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grossenbacher PG. Mechanisms of synesthesia: cognitive and physiological constraints. Trends Cognitive Sci. 2001;5(1):36–41. doi: 10.1016/s1364-6613(00)01571-0. [DOI] [PubMed] [Google Scholar]
- 14.Grossenbacher PG. Perception and sensory information in synesthetic experience. Blackwell. 1997 [Google Scholar]
- 15.Dixon MJ, Smilek D, Cudahy C, Merikle PM. Five plus two equals yellow. Nature. 2000;406:365. doi: 10.1038/35019148. [DOI] [PubMed] [Google Scholar]
- 16.Hubbard EM, Arman AC, Ramachandran VS, Boynton GM. Individual differences among grapheme-color synesthetes: brain-behavior correlations. Neuron. 2005;45:975–85. doi: 10.1016/j.neuron.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Nunn JA, Gregory LJ, Brammer M, Williams SC, Parslow DM, Morgan MJ, et al. Functional magnetic resonance imaging of synesthesia: activation of V4/V8 by spoken words. Nat Neurosci. 2002;5:371–5. doi: 10.1038/nn818. [DOI] [PubMed] [Google Scholar]
- 18.Galton F. Visualized numerals. Nature. 1880;21:252–6. [Google Scholar]
- 19.Cytowic RE. Synesthesia: a union of the senses. MIT Press; 2002. [Google Scholar]
- 20.Ward J, Simner J. Is synaesthesia an X-linked dominant trait with lethality in males? Perception. 2005;34:611–23. doi: 10.1068/p5250. [DOI] [PubMed] [Google Scholar]
- 21.Eagleman DM, Kagan AD, Nelson SS, Sagaram D, Sarma AK. A standardized test battery for the study of synesthesia. J Neurosci Methods. 2007;159:139–45. doi: 10.1016/j.jneumeth.2006.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asher JE, Aitken MR, Farooqi N, Kurmani S, Baron-Cohen S. Diagnosing and phenotyping visual synaesthesia: a preliminary evaluation of the revised test of genuineness (TOG-R). Cortex. 2006;42:137–46. doi: 10.1016/s0010-9452(08)70337-x. [DOI] [PubMed] [Google Scholar]
- 23.Mattingley JB, Rich AN, Yelland G, Bradshaw JL. Unconscious priming eliminates automatic binding of colour and alphanumeric form in synaesthesia. Nature. 2001;410:580–2. doi: 10.1038/35069062. [DOI] [PubMed] [Google Scholar]
- 24.Rylander-Rudqvist T, Hakansson N, Tybring G, Wolk A. Quality and quantity of saliva DNA obtained from the self-administrated oragene method–a pilot study on the cohort of Swedish men. Cancer Epidemiol Biomarkers Prev. 2006;15:1742–5. doi: 10.1158/1055-9965.EPI-05-0706. [DOI] [PubMed] [Google Scholar]
- 25.Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat Methods. 2006;3:31–3. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- 26.O'Connell JR, Weeks DE. A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 28.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–3. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 29.Asher JE, Lamb JA, Brocklebank D, Cazier JB, Maestrini E, Addis L, et al. A whole-genome scan and fine-mapping linkage study of auditory-visual synesthesia reveals evidence of linkage to chromosomes 2q24, 5q33, 6p12, and 12p12. Am J Hum Genet. 2009;84:279–85. doi: 10.1016/j.ajhg.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherkassky VL, Kana RK, Keller TA, Just MA. Functional connectivity in a baseline resting-state network in autism. Neuroreport. 2006;17:1687–90. doi: 10.1097/01.wnr.0000239956.45448.4c. [DOI] [PubMed] [Google Scholar]
- 31.Huang Q, Shete S, Amos CI. Ignoring linkage disequilibrium among tightly linked markers induces false-positive evidence of linkage for affected sib pair analysis. Am J Human Genet. 2004;75:1106–12. doi: 10.1086/426000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falchier A, Clavagnier S, Barone P, Kennedy H. Anatomical evidence of multi-modal integration in primate striate cortex. J Neurosci. 2002;22:5749–59. doi: 10.1523/JNEUROSCI.22-13-05749.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockland KS, Ojima H. Multisensory convergence in calcarine visual areas in macaque monkey. Int J Psychophysiol. 2003;50:19–26. doi: 10.1016/s0167-8760(03)00121-1. [DOI] [PubMed] [Google Scholar]
- 34.Fu KM, Johnston TA, Shah AS, Arnold L, Smiley J, Hackett TA, et al. Auditory cortical neurons respond to somatosensory stimulation. J Neurosci. 2003;23:7510–5. doi: 10.1523/JNEUROSCI.23-20-07510.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schroeder CE, Foxe JJ. The timing and laminar profile of converging inputs to multisensory areas of the macaque neocortex. Brain Res Cogn Brain Res. 2002;14:187–98. doi: 10.1016/s0926-6410(02)00073-3. [DOI] [PubMed] [Google Scholar]
- 36.Oslon GC, Lenay CC. Touching for Knowing, Cognitive psychology of haptic manual perception. John Benjamins Publishing Company; 2003. Sensory substitution: limits and perspectives. [Google Scholar]
- 37.Calvert GA, Bullmore ET, Brammer MJ, Campbell R, Williams SC, McGuire PK, et al. Activation of auditory cortex during silent lipreading. Science. 1997;276:593–6. doi: 10.1126/science.276.5312.593. [DOI] [PubMed] [Google Scholar]
- 38.Eagleman DM. Visual illusions neurobiology. Nat Rev Neurosci. 2001;2:920–6. doi: 10.1038/35104092. [DOI] [PubMed] [Google Scholar]
- 39.Macaluso E, Frith CD, Driver J. Modulation of human visual cortex by crossmodal spatial attention. Science. 2000;289:1206–8. doi: 10.1126/science.289.5482.1206. [DOI] [PubMed] [Google Scholar]
- 40.McGurk H, MacDonald J. Hearing lips and seeing voices. Nature. 1976;264:746–8. doi: 10.1038/264746a0. [DOI] [PubMed] [Google Scholar]
- 41.Baron-Cohen S, Burt L, Smith-Laittan F, Harrison J, Bolton P. Prevalence and familiality. Perception. 1996;25:1073–9. doi: 10.1068/p251073. [DOI] [PubMed] [Google Scholar]
- 42.Frith U. Blackwell Science. 1989. Autism: explaining the enigma. [Google Scholar]
- 43.Happe F. Autism: cognitive deficit or cognitive style? Trends Cogn Sci. 1999;3:216–22. doi: 10.1016/s1364-6613(99)01318-2. [DOI] [PubMed] [Google Scholar]
- 44.Just MA, Cherkassky VL, Keller TA, Kana RK, Minshew NJ. Functional and Anatomical Cortical Underconnectivity in Autism: Evidence from an fMRI Study of an Executive Function Task and Corpus Callosum Morphometry. Cereb Cortex. 2006 doi: 10.1093/cercor/bhl006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kana RK, Keller TA, Minshew NJ, Just MA. Inhibitory Control in High-Functioning Autism: Decreased Activation and Underconnectivity in Inhibition Networks. Biol Psychiatry. 2006 doi: 10.1016/j.biopsych.2006.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adler LE, Pachtman E, Franks RD, Pecevich M, Waldo MC, Freedman R. Neuro-physiological evidence for a defect in neuronal mechanisms involved in sensory gating in schizophrenia. Biol Psychiatry. 1982;17:639–54. [PubMed] [Google Scholar]
- 47.Benes FM. Cortical pathology: a new generation of quantitative microscopic studies. Neuropathol Schizophrenia. 2000 [Google Scholar]
- 48.Daskalakis ZJ, Christensen BK, Chen R, Fitzgerald PB, Zipursky RB, Kapur S. Evidence for impaired cortical inhibition in schizophrenia using transcranial magnetic stimulation. Arch Gen Psychiatry. 2002;59:347–54. doi: 10.1001/archpsyc.59.4.347. [DOI] [PubMed] [Google Scholar]
- 49.Freedman R, Adler LE, Myles-Worsley M, Nagamoto HT, Miller C, Kisley M, et al. Inhibitory gating of an evoked response to repeated auditory stimuli in schizophrenic and normal subjects. Human recordings, computer simulation, and an animal model. Arch Gen Psychiatry. 1996;53:1114–21. doi: 10.1001/archpsyc.1996.01830120052009. [DOI] [PubMed] [Google Scholar]
- 50.Griffith J, Hoffer LD, Adler LE, Zerbe GO, Freedman R. Effects of sound intensity on a midlatency evoked response to repeated auditory stimuli in schizophrenic and normal subjects. Psychophysiology. 1995;32:460–6. doi: 10.1111/j.1469-8986.1995.tb02097.x. [DOI] [PubMed] [Google Scholar]
- 51.Rose KN, Swerdlow GF. Schizophrenia, mania, depression: toward a unified hypothesis of cortico-striatal-pallido-thalamic function. Behav Brain Sci. 1987;10:197–245. [Google Scholar]
- 52.Waldo M, Myles-Worsley M, Madison A, Byerley W, Freedman R. Sensory gating deficits in parents of schizophrenics. Am J Med Genet. 1995;60:506–11. doi: 10.1002/ajmg.1320600605. [DOI] [PubMed] [Google Scholar]
- 53.Walker EF. Developmentally moderated expressions of the neuropathology underlying schizophrenia. Schizophr Bull. 1994;20:453–80. doi: 10.1093/schbul/20.3.453. [DOI] [PubMed] [Google Scholar]