Abstract
West Nile virus (WNV) employs several different strategies to escape the innate immune response. We have previously demonstrated that the WNV NS1 protein interferes with signal transduction from Toll-like receptor 3 (TLR3). NS1 is a glycoprotein that can be found intracellularly or associated with the plasma membrane. In addition, NS1 is secreted to high levels during flavivirus infections. We investigated whether the secreted form of NS1 inhibits innate immune signaling pathways in uninfected cells. Secreted NS1 (sNS1) was purified from supernatants of cells engineered to express the protein. Purified sNS1 associated with and repressed TLR3-induced cytokine production by HeLa cells, and inhibited signaling from TLR3 and other TLRs in bone marrow-derived macrophages and dendritic cells. Footpad administration of sNS1 showed the protein associated predominantly with macrophages and dendritic cells in the draining lymph node. Additionally, sNS1 significantly reduced TLR3 signaling and WNV replicon particle-mediated cytokine transcription in popliteal lymph nodes.
Keywords: West Nile virus, Flaviviridae, nonstructural protein, toll like receptor, innate immunity
Introduction
West Nile virus is a member of the family Flaviviridae whose clinically important members include yellow fever virus, dengue virus, and Japanese encephalitis virus. WNV exists in a transmission cycle between birds and mosquitoes, where humans are incidental hosts. Early WNV replication in mouse models of disease occurs in keratinocytes (Brown et al., 2007; Lim et al., 2011) and skin-resident dendritic cells (DCs), including Langerhans DCs (Wu et al., 2000). Infection initiates migration of Langerhans DCs to draining lymph nodes where further viral expansion occurs concurrently with activation of the immune response (Byrne et al., 2001; Johnston, Halliday, and King, 2000). Upon entry into the bloodstream, WNV infects peripheral tissues such as the spleen and the kidneys. In certain animals, the virus is able to invade the central nervous system and infect neurons of the brain stem, hippocampus, and spinal cord.
The innate immune response is the first line of defense against invading pathogens and can significantly influence viral pathogenesis as well as shape the ensuing adaptive immune response. In recent years, significant progress has been made in identifying virus interactions with the innate immune system.
One arm of the innate immune response involves the recognition of pathogen-associated molecular patterns (PAMPs), eliciting proinflammatory cytokine responses and the production of type I interferon. Several different pattern recognition receptors (PRRs) have been implicated in the recognition of flavivirus infections such as the RNA helicases RIG-I, Mda-5, and a variety of different TLRs (Daffis et al., 2008; Diebold et al., 2004; Fredericksen et al., 2008; Loo et al., 2008; Lund et al., 2004; Nasirudeen et al., 2011; Silva et al., 2007; Town et al., 2009; Tsai et al., 2009; Wang et al., 2006; Wang et al., 2004; Welte et al., 2009).
Our previous work has demonstrated that TLR3 signaling is inhibited in WNV infected cells (Scholle and Mason, 2005) and this inhibition is due to expression of the NS1 protein (Wilson et al., 2008). NS1 is a glycoprotein that is required for RNA replication where it participates in early RNA synthesis (Khromykh et al., 1999; Lindenbach and Rice, 1997; Westaway et al., 1997; Youn et al., 2012). In the infected cell, NS1 is translocated into the lumen of the ER and forms detergent stable, but heat labile, dimers. Additionally, NS1 is secreted from infected cells to high levels (Chung and Diamond, 2008; Macdonald et al., 2005), and this soluble form is detectable as a hexamer (Flamand et al., 1999). Secreted NS1 (sNS1) is known to associate with a number of different cell types in vitro (Avirutnan et al., 2007) and in vivo (Alcon-LePoder et al., 2005) and for both WNV sNS1 and dengue virus sNS1, binding to uninfected endothelial cells is dependent on interactions with sulfated glycosaminoglycans (Avirutnan et al., 2007; Youn et al., 2010).
Given the documented interactions of sNS1 with uninfected cells and our previous data showing NS1-mediated inhibition of TLR3 signaling, we hypothesized that sNS1 can modulate innate immune responses in naïve cells. Our data shows sNS1 purified from cell culture supernatants can inhibit TLR signaling in different cell types of both human and murine origin and impairs cytokine production in response to WNV and replicon particle infection. Importantly, sNS1 was also able to modulate cytokine secretion in response to both TLR3-stimulation and WNV VRP infection in vivo. This is the first evidence of a secreted RNA virus protein capable of modulating TLR signaling.
Results
Secreted NS1 associates with naïve cells and inhibits TLR3 signaling
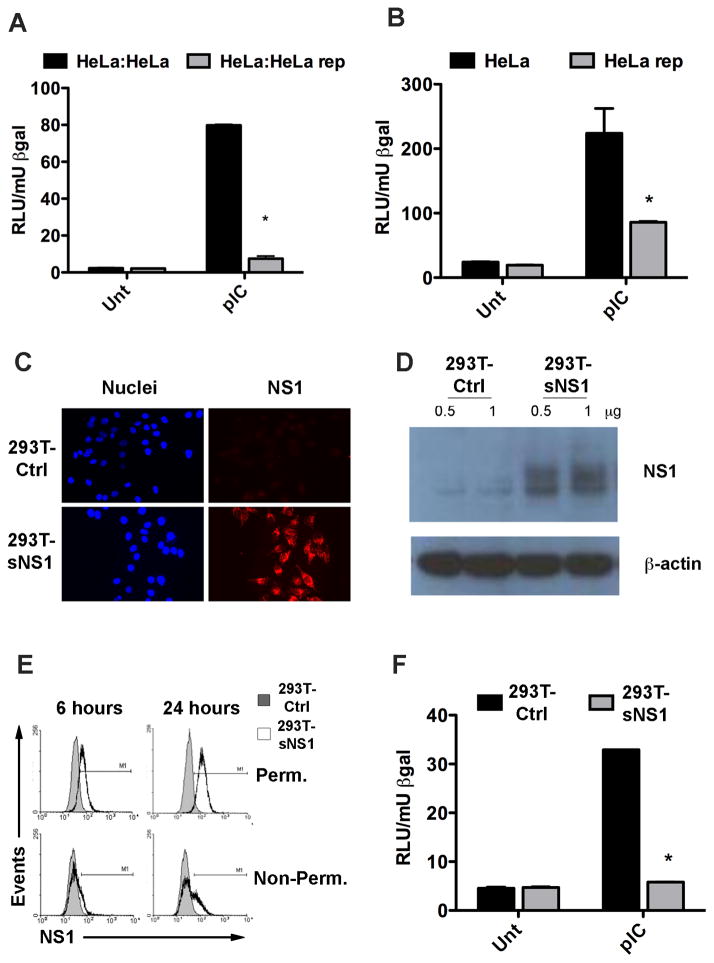
Initial studies were aimed at determining the ability of HeLa cells to associate with NS1 secreted from HeLa cells, which stably express the WNV replicon. (HeLa rep cells) (Scholle and Mason, 2005). Uninfected, i.e., replicon-free HeLa cells were seeded onto chamber slides and then transfected to express a red fluorescent protein (RFP). After transfection the cells were washed extensively to remove all traces of transfection reagent. At this point the HeLa rep cells were added to the uninfected HeLa cells and co-cultured for 16 hours. After incubation, the slides were fixed and permeabilized before being stained with a WNV-specific antibody and an Alexa-488-tagged secondary antibody. Results showed association of a WNV protein (green) with the uninfected cells (red) (Supplemental Figure 1). Therefore, within 16 hours, uninfected cells associated with a potentially secreted WNV non-structural protein. To next address whether WNV infected cells are able to influence TLR3 signaling in neighboring uninfected cells, IFNβ promoter induction was measured by reporter assay in HeLa cells that were incubated in the presence of HeLa rep cells. Uninfected HeLa cells were transfected with a reporter construct in which luciferase (Luc) expression is controlled by the IFNβ promoter, as described previously (Wilson et al., 2008). The transfected cells were then incubated for 16 hours with other uninfected HeLa cells or with HeLa rep cells.. Poly(I:C) was then added directly to the co-cultures to stimulate TLR3 signaling and 4 hours later cell lysates were harvested and analyzed for Luc activity. Strong IFNβ promoter-driven Luc expression was detected in the HeLa cells co-cultured with uninfected HeLa cells, whereas reporter-transfected HeLa cells cultured with HeLa rep cells showed a significantly diminished response to TLR3 stimulation (Figure 1A). These data show WNV replicon infected cells can modulate TLR3 responses in neighboring uninfected cells. To investigate whether this phenomenon was cell contact dependent or mediated by a soluble factor, the experiments were repeated by incubating reporter-transfected HeLa cells with clarified supernatants harvested from HeLa rep cells. Poly(I:C)-induced Luc expression was greatly reduced in HeLa cells incubated with HeLa rep supernatants, whereas Luc expression was unabated in the HeLa cells incubated with supernatants from uninfected HeLa cells. These data confirm that a soluble factor is sufficient for TLR3 inhibition (Fig. 1B).
Figure 1.
NS1 containing cell culture supernatants inhibit the TLR3 response in HeLa cells. (A) HeLa cells were transfected with an IFNβ-Luc reporter construct and a β-gal expressing plasmid and subsequently co-cultured with either regular HeLa cells or HeLa cells harboring a WNV replicon (HeLa rep). Luc activity was determined 4h after the addition of pIC to the culture media and normalized to β-gal activity. Asterisks represent statistically significant differences, determined by student’s t-test, comparing the pIC response of HeLa:HeLa rep samples to that of HeLa:HeLa samples. (B) HeLa cells transfected with IFNβ reporter and β-gal as above were incubated for 16h with cell culture supernatants from either regular HeLa cells or HeLa rep cells then stimulated with pIC before Luc activity was determined. Asterisks represent statistically significant differences, determined by student’s t-test comparing the pIC response of HeLa rep cells to that of HeLa cells. (C) HeLa cells were incubated with cell culture supernatants from either 293T-Ctrl or 293T-NS1 cells for 16h. NS1 association with the cells was detected by immunofluorescence of permeabilized cells using WNV MHIAF. (D) HeLa cells were incubated with control or sNS1-containing supernatants for 16h and extensively washed before lysis. Whole cell lysates were analyzed by immunoblot with WNV MHIAF followed by HRP-labeled mouse antibody. (E) NS1 association with naïve cells by flow cytometry. HeLa cells were incubated with control or sNS1-containing supernatants for 6h and 24h either left unpermeabilized to detect NS1 on the cell surface or permeabilized prior to staining to detect internalized NS1. NS1 was detected with an NS1-specific antibody. (F) Secreted NS1-containing cell culture supernatants inhibit the TLR3 response in HeLa cells. HeLa cells transfected with IFNβ reporter construct and β-gal were incubated with supernatants from either 293T-Ctrl or 293T-NS1 cells for 16h and stimulated as above. Asterisks represent statistically significant differences, determined by student’s t-test of sNS1-containing supernatant treated samples compared to control treated samples
Though NS1 is the only non-structural protein known that is secreted from WNV-infected cells, the HeLa rep cell line expresses all non-structural (NS) proteins. In order to demonstrate that the observed inhibitory effect was sNS1-dependent, a 293T cell line that expresses NS1 from a stably integrated lentivirus vector (293T-NS1) was generated. Western blots of clarified supernatants harvested from 293T-NS1 cells confirmed NS1 secretion from these cells (data not shown). Previous studies have reported the binding and rapid endocytosis of sNS1 by naïve cells (Alcon-LePoder et al., 2005; Avirutnan et al., 2007). In agreement with these studies, the initial characterization of 293T-NS1 supernatants also showed sNS1 association with naïve HeLa cells. Clarified supernatants from the 293T-NS1 cell line or from the 293T lentivirus vector control cell line (293T-Ctrl) were added to HeLa cells and 16 hours later, the cells were analyzed by immunofluorescence using an NS1-specific antibody. Microscopic analysis revealed punctate, NS1-specific staining in cells incubated with sNS1-containing supernatants (Fig. 1C). Additionally, an NS1-specific band was detected by Western blot analysis of whole cell extracts of HeLa cells incubated with 293T-NS1 supernatants (Fig. 1D). To determine whether sNS1 simply binds to the cells or becomes internalized, permeabilized and non-permeabilized cells, incubated with supernatants from either 293T-NS1 or 293T-Ctrl, were compared by flow cytometry. Within 6 hours of sNS1 addition, NS1 positive staining was stronger in permeabilized cells than in non-permeabilized cells, indicating that internalization of sNS1 occurred rapidly (Fig. 1E). NS1 positive staining was still detectable after 24 hours and showed an even higher level of internalized sNS1, suggesting no appreciable degradation of the protein by HeLa cells was occurring. Next, sNS1 containing supernatants from 293T-NS1 cells were examined for their ability to inhibit TLR3 signaling in the absence of other WNV NS proteins. HeLa cells were transfected with the IFNβ Luc reporter construct and incubated with supernatants from either 293T-NS1 cells or 293T-Ctrl cells for 16 hours before stimulation with poly(I:C). IFNβ promoter activation was significantly diminished in cells incubated with sNS1-containing supernatants as compared to cells incubated with 293T-Ctrl supernatants (Fig. 1F). Incubation with sNS1 containing supernatants was not toxic to the cells as confirmed by MTT assay (Supplemental Figure S2). These results confirm that the inhibitory effect is dependent on NS1 expression, however, they do not explicitly demonstrate that sNS1 is directly responsible for the inhibition.
Association of sNS1 with naïve cells is required for inhibition of TLR3 signaling
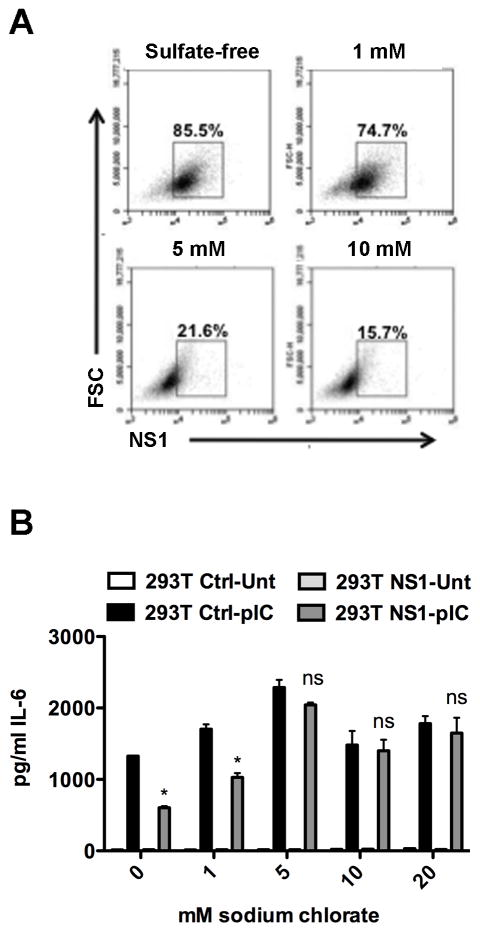
Although our data strongly suggest the inhibition of TLR3 signaling is sNS1-dependent, the possibility remained that this inhibitory effect is attributable to another secreted factor induced by intracellular NS1 rather than sNS1 directly. To address this question, we investigated whether direct association of sNS1 with naïve cells is required for TLR3 signaling inhibition. Secreted NS1 of both dengue and WNV are known to associate with cells via glycosaminoglycans (GAGs), such as heparan sulfate and chondroitin sulfate E (Avirutnan et al., 2007; Youn et al., 2010). Cell binding by dengue or WNV sNS1 is dependent on the sulfation of GAG residues, as pretreatment of cells with sodium chlorate, a chemical used to inhibit GAG sulfation, ablates sNS1-cell binding (Avirutnan et al., 2007; Youn et al., 2010). To determine if GAG sulfation was also required for the binding of sNS1 derived from the 293T-NS1 cell line, supernatants containing sNS1 were collected from 293T-NS1 cells grown in sulfate-free media. HeLa cells were pretreated with increasing concentrations of sodium chlorate for 24 hours, followed by incubation with sulfate-free, sNS1-containing supernatants. The cultures were examined periodically during the sodium chlorate treatment and no cytotoxic effect was observed. Association of sNS1 with HeLa cells was determined by flow cytometry and showed sodium chlorate pretreatment inhibited sNS1 binding to HeLa cells in a concentration dependent manner (Fig. 2A). To determine how the sodium chlorate treatment would affect TLR3 signaling inhibition by sNS1-containing supernatants, HeLa cells were treated with sodium chlorate before incubation with sulfate-free supernatants. After 8 hours of poly(I:C) stimulation, TLR3 signaling was assessed by measuring IL-6 secretion, as previously reported (Wilson et al., 2008). Though TLR3 responses were slightly affected by sodium chlorate alone, the signaling capacity of the cells was retained and treatment with increasing doses of sodium chlorate alleviated sNS1-mediated signaling inhibition (Fig. 2B). Therefore, the inability of sNS1 to bind directly, correlated with the restoration of TLR3 signaling by HeLa cells. Since sNS1-cell binding is required for inhibition of TLR3 signaling, these data strongly suggest that the inhibitory effect is, in fact, directly attributable to sNS1.
Figure 2.
Inhibition of GAG sulfation prevents sNS1 association and restores TLR3 signaling. HeLa cells were incubated for 24h in either DMEM or sulfate-free medium (SF) in the absence or presence of increasing concentrations of sodium chlorate before addition of sNS1-containing or control supernatant under the same conditions. (A) Permeabilized cells were analyzed for sNS1 association by flow cytometry. The initial gate was set on live cells using FSC vs SSC. Cells not incubated with sNS1 served to set the gate shown. (B) Loss of sNS1-cell interaction restores TLR3 signaling. IL-6 was measured from HeLa cells treated as described in A and stimulated with 20μg/mL pIC for 8h. Asterisks denote statistically significant differences between 293T-Ctrl and 293T-NS1 pretreated pIC-stimulated cells.
Purified sNS1 inhibits TLR3 signaling
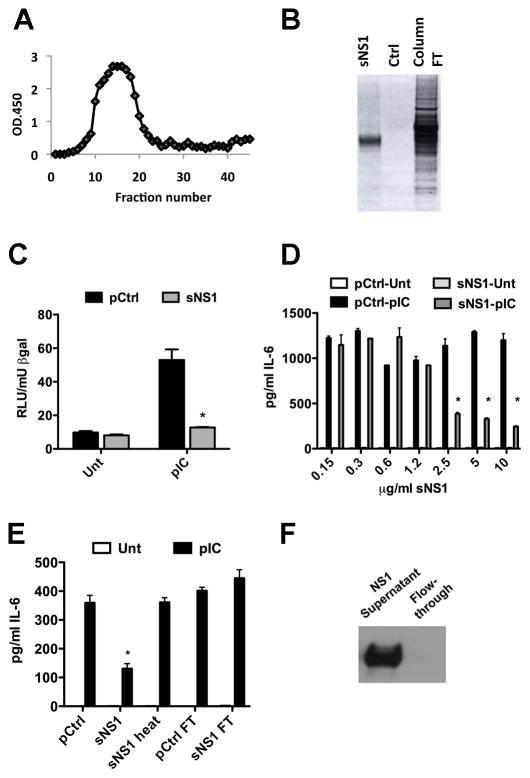
To further establish that the observed effects on TLR3 signaling were sNS1-dependent, sNS1 was purified from the supernatants of the 293T-NS1 cell line by immunoaffinity chromatography using an NS1-specific antibody. Figure 3A depicts a typical elution curve for sNS1 purification and purity was analyzed by silver stain of an SDS-PAGE gel (Figure 3B). Supernatants collected from the 293T-Ctrl cell line were subjected to the same purification procedure; the end product served as a purification control (pCtrl) for purified sNS1 and was used at the same volume as purified sNS1. The effect of purified sNS1 on TLR3-mediated IFNβ promoter activation was determined by reporter assay. After 16 hours of treatment with purified sNS1 or pCtrl, HeLa cells were stimulated with poly(I:C). Cells pretreated with pCtrl strongly induced IFNβ promoter activity. In contrast, cells pretreated with purified sNS1, showed a significantly diminished response to poly(I:C) stimulation (Fig. 3C). TLR3-mediated IL-6 secretion by HeLa cells was also significantly inhibited by pretreatment with purified sNS1. The inhibition was dose-dependent and statistically significant at concentrations greater than 1.25 μg/mL (Fig 3D). Figure 3E shows that signaling inhibition was lost when sNS1 was heated to 95°C (sNS1-heat) or upon antibody depletion of sNS1 from supernatants (NS1 FT), demonstrated in Figure 3F. These data confirm that the observed inhibitory effects are sNS1-dependent.
Figure 3.
Incubation with purified sNS1 inhibits TLR3 signaling in HeLa cells. (A) NS1-specific ELISA on immunoaffinity purification fractions. (B) Silver stain on SDS PAGE gel of purified sNS1, the purification control (pCtrl) and the flow through fraction during the purification. (C) HeLa cells were transfected with IFNβ reporter construct and β-gal expression construct and incubated with 4μg/mL purified sNS1 or an equal volume of pCtrl for 16h prior to stimulation with pIC. Control for these experiments was supernatant from 293T vector control cells that underwent a purification process identical to that of the sNS1-containing supernatants (pCtrl). Asterisks denote statistically significant differences between pCtrl and sNS1 pretreated pIC-stimulated cells.
(D) Dose response curve. Varying concentrations of purified sNS1 or equal volume of pCtrl was added to HeLa cells in full serum media and incubated for 16h before 8h of pIC treatment and subsequent IL-6 ELISA analysis of supernatants. Asterisks denote statistically significant differences between pCtrl and sNS1 pretreated pIC-stimulated cells.
(E) IL-6 secretion was measured from pIC treated HeLa cells following incubation with pCtrl, 5μg/mL sNS1, 5μg/mL sNS1-heat or equal volumes of the flow through fraction generated during the purification process for either control cell supernatant or NS1-expressing cell supernatant. (F) Western blot confirming depletion of sNS1 in the flow through fraction used in Fig. 3E. Asterisks denote statistically significant differences between pCtrl and NS1 pretreated cells, stimulated with pIC.
Secreted NS1 inhibits TLR signaling in bone marrow derived primary cells
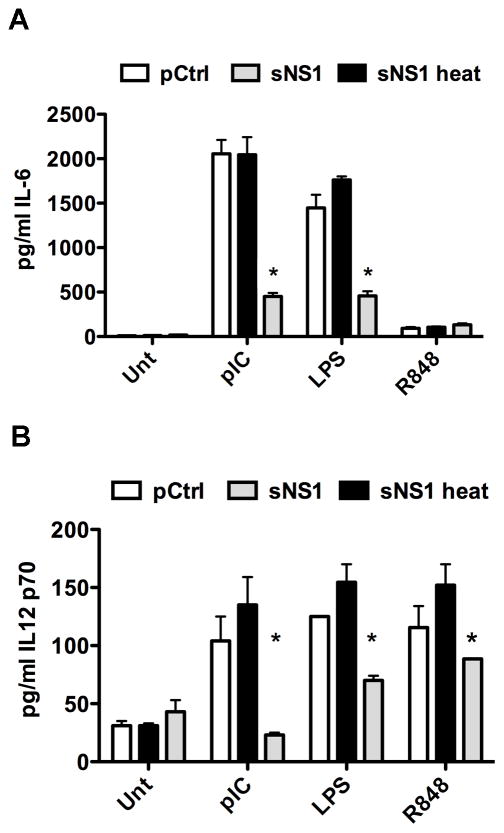
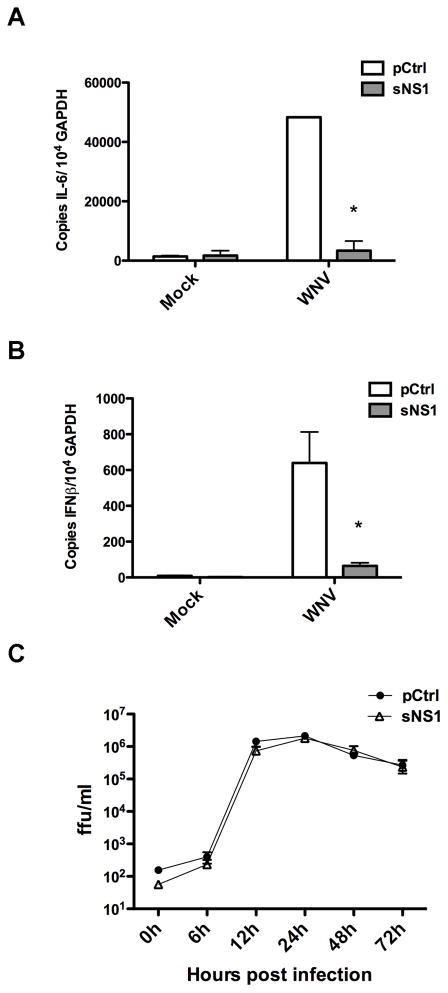
It was of interest to determine whether sNS1 inhibits Toll-like receptor signaling pathways other than those originating from TLR3. We investigated the effect of sNS1 on bone marrow-derived mouse macrophages (BMDMs) and bone marrow-derived myeloid dendritic cells (BMDCs), two immunologically relevant cell types that endogenously express several different TLRs. Because protein degradation is a function of BMDMs and BMDCs, initial experiments were performed to determine the pretreatment times resulting in the highest level of intracellular sNS1 accumulation before protein degradation is observed (data not shown). BMDCs or BMDMs were pretreated with sNS1, heat-inactivated sNS1 (sNS1-heat), or pCtrl for 16 hours. TLR3 was stimulated with poly(I:C), TLR4 with lipopolysaccharide (LPS), and TLR7 with resiquimod (R848). After 8 hours of TLR stimulation, IL-6 secretion by BMDMs was measured by ELISA. The pCtrl-treated cells and cells treated with heat-inactivated sNS1 did secrete high levels of IL-6 in response to stimulation by poly(I:C) and LPS. In contrast, treatment with sNS1 significantly inhibited IL-6 production after stimulation with both poly(I:C) and LPS (Fig. 4A). BMDMs did not respond to R848. BMDCs produced only low levels of IL-6, but did secrete significant levels of IL-12 in response to TLR stimulation (Kawai and Akira, 2008). Therefore, TLR signaling by BMDCs was assessed by measuring IL-12p70 secretion after 12 hours of TLR stimulation. Treatment with sNS1 resulted in diminished IL-12 production by BMDCs stimulated with poly(I:C) or LPS, and also inhibited IL-12 secretion in response to R848 stimulation (Fig. 4B). While sNS1-mediated signaling inhibition of all the TLRs tested in BMDCs was statistically significant, it should be noted that TLR3 signaling was affected more than TLR4 and TLR7 signaling. We next examined how sNS1 treatment would affect the cytokine response to WNV infection in primary cells. BMDMs were pretreated with sNS1 or pCtrl for 16 hours and subsequently infected with WNV at an MOI of 0.1. RNA was isolated 24h after infection and analyzed by real-time RT-PCR. Compared to pretreatment with pCtrl, pretreatment of BMDMs with sNS1 resulted in a significant reduction in WNV-induced IL-6 and IFNβ transcript levels (Fig. 5A and B). Growth of WNV was monitored on sNS1-pretreated cells to determine if sNS1 pretreatment would influence viral replication. Interestingly, growth of WNV on BMDMs was not affected despite decreased cytokine transcription (Fig. 5C). The reasons for these results are not entirely clear, however, it is possible that even the reduced amount of cytokines induced after sNS1 pretreatment was sufficient to impair virus production to levels similar to those in pCtrl-treated cells. Together, these results demonstrate that sNS1 not only inhibits TLR3 signaling, but also inhibits signaling from other TLRs, although the extent of inhibition varied by TLR and by cell type. Additionally, sNS1 pretreatment resulted in a diminished cytokine response to virus infection.
Figure 4.
Purified sNS1 inhibits TLR signaling in BMDMs and BMDCs. (A) BMDMs derived from C57BL/6 mice were incubated for 16h with 5μg/mL sNS1 or sNS1-heat, or pCtrl followed by 8h pIC treatment at 5μg/mL. IL-6 release was measured by ELISA. (B) Myeloid DCs derived from WT C57BL/6 mice were incubated with 5μg/mL sNS1 or controls for 16h and stimulated with 10μg/mL pIC for 12h. IL-12p70 release into supernatants was determined by ELISA. Control for these experiments was supernatant from 293T vector control cells that underwent an identical purification process as NS1 supernatants and was added at the same volume as sNS1. Asterisks denote statistically significant differences between sNS1-heat and sNS1 pretreated -stimulated cells.
Figure 5.
Secreted NS1 modulates cytokine production during WNV infection. (A) BMDMs were pretreated with either purification control or purified sNS1 for 16h, followed by infection with WNV at an MOI of 0.1 for 24h. IL-6 mRNA levels were measured by real-time qRT-PCR. (B) Cells were treated and infected as in (A) and IFNβ mRNA levels were measured by real-time qRT-PCR. Asterisks denote statistically significant differences between Ctrl and NS1 pretreated WNV-stimulated cells as determined by student’s t-test.
(C) BMDMs pretreated with either the purification control or purified sNS1 for 16h were infected with WNV at an MOI of 0.01 and virus production was monitored at the indicated timepoints over 72h by titration of virus containing supernatants on Vero cells.
Secreted NS1 interacts predominantly with macrophages and dendritic cells in vivo
We were interested in examining the possibility of immune modulation by sNS1 in vivo but wanted to first determine the fate of sNS1 upon introduction into mice. Secreted NS1 was delivered by subcutaneous footpad inoculation because this is the most commonly used model of mosquito-delivered WNV infection. Footpad inoculation would also allow monitoring sNS1 migration into the popliteal lymph node (pLN), the draining lymph node of the footpad. Thus, either 5 μg of Alexa 488 (A-488)-labeled sNS1, or an equal volume of unincorporated A-488 dye, were injected into the rear footpads of C57BL/6 mice. The amount of sNS1 injected was within the range of NS1 serum concentrations previously reported for WNV sNS1 in a mouse model (Macdonald et al., 2005) as well as the range reported for sNS1 in dengue patients (Young et al., 2000). The pLNs were harvested 6 hours after sNS1 injection and a single cell suspension of the lymph nodes was analyzed by flow cytometry (Fig. 6A). Analysis of the pLNs showed the A-488 unincorporated dye did not stain any cell type appreciably, however approximately 80% of macrophages and 60% of DCs were positive for A-488 labeled sNS1. Secreted NS1 only associated with 2% of either CD4+ or CD8+ T cells, despite a significantly greater number of these cells in the draining lymph node, suggesting sNS1 preferentially binds to macrophages and DCs.
Figure 6.
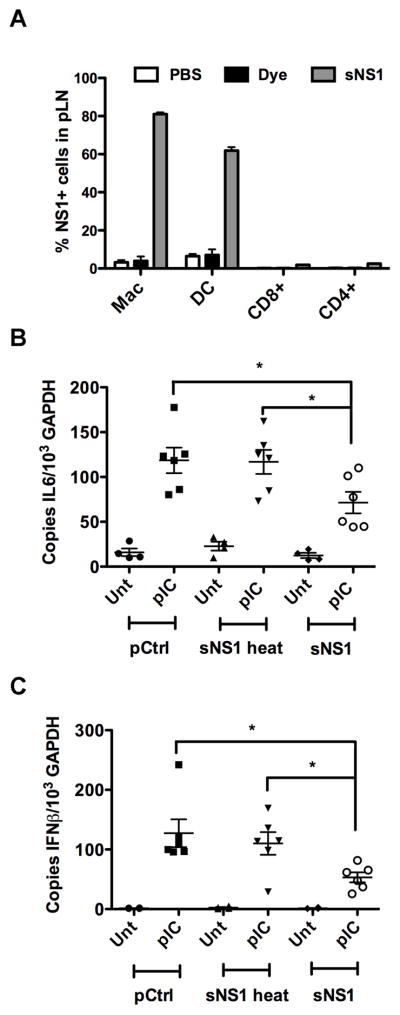
Secreted NS1 associates with innate cells and inhibits PRR signaling in vivo. (A) 5μg of Alexa 488 fluorescently labeled purified sNS1 protein were injected into mice via the rear footpad, After 6h draining lymph nodes were harvested and analyzed for cell type specific markers and NS1 fluorescence. The percentage of NS1 positive cells in the entire cell type population is shown. (B and C) C57BL/6 mice were injected with 5μg of purified sNS1 into each rear footpad. After 6 hours the mice were stimulated by injection of 20μg of Poly(IC:LC) per footpad. After 16h pLNs were harvested and pooled per mouse before RNA extraction and cDNA synthesis. IFNβ mRNA levels (B) and IL-6 mRNA levels (C) were determined by real-time qRT-PCR. Controls for the experiments in (B) and (C) were heat-inactivated sNS1 and the purification control.
Secreted NS1 inhibits the in vivo response to poly(IC:LC) stimulation and VRP infection
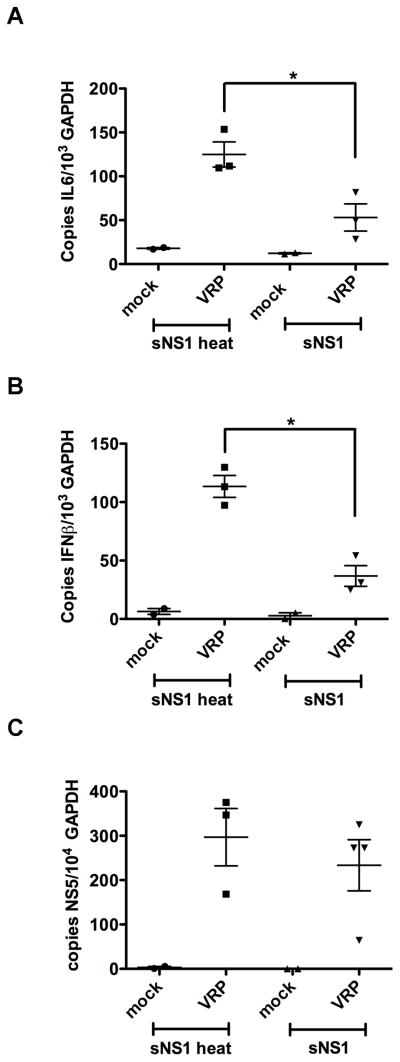
Based on our findings that sNS1 associates with macrophages and DCs, it was of interest to determine whether sNS1 would also inhibit TLR responses in vivo. C57BL/6 mice were injected into the rear footpad with 5 μg of either purified sNS1 or heat-inactivated sNS1, or an equal volume of pCtrl. Six hours later, the mice were stimulated by injection of 20 μg of poly(IC:LC) into the rear footpad. Poly(IC:LC) (Hiltonol) is a polylysine stabilized form of poly(I:C) reported to induce potent innate immune responses in vivo (Caskey et al., 2011). After 16 hours of poly(IC:LC) stimulation, the pLNs were harvested and processed for total RNA extraction. Expression of IL-6 (Fig. 6B), and IFNβ (Fig. 6C) mRNA was determined by real-time qRT-PCR. Mice pretreated with pCtrl and heat-inactivated sNS1 responded to poly(IC:LC) stimulation with upregulation of IL-6 and IFNβ transcription. In contrast, mice pretreated with sNS1 showed significantly reduced cytokine transcription.
To determine the effects of sNS1 in the context of in vivo infection we chose to use virus replicon particles (VRPs). Infection by VRPs offers the unique opportunity to test local responses to the initial infection without resulting in a spreading infection, the response to which might override early inhibitory effects of sNS1. A previous study showed upregulation of IFNβ and IL-6 transcription in the pLN 24 hours after footpad inoculation with WNV VRPs (Bourne et al., 2007). These data indicate that lymphoid tissues are a significant source of IFN production early in WNV infection and that the footpad inoculation model is suitable for monitoring the early innate responses to VRP infection. Six hours after footpad injection of sNS1 or heat-inactivated sNS1, mice were inoculated with WNV VRPs via the footpad and 16 hours later, pLNs were harvested and processed for real-time qRT-PCR analysis. WNV VRP-induced transcriptional upregulation of both IL-6 and IFNβ was significantly diminished in mice pretreated with sNS1 as compared to mice pretreated with heat-inactivated sNS1 (Fig. 7A and 7B). Analysis of genome copy numbers shows replication was not affected by the sNS1 pretreatment, in agreement with our in vitro replication data (Fig. 7C).
Figure 7.
Secreted NS1 diminishes the response to WNV VRP infection in vivo. C57BL/6 mice were pre-injected with purified sNS1 or sNS1-heat as described above and stimulated by injection of 1x105 iu WNV VRPs into each footpad. PLNs were collected and processed as described in Figure 6. IL-6 mRNA (A) and IFNβ mRNA (B) and WNV RNA levels (C) were analyzed by real-time qRT-PCR. A representative experiment out of three independent studies is shown.
Discussion
Based on our previous studies regarding the ability of intracellularly-expressed WNV NS1 to inhibit TLR3 signaling, we explored the possibility that the secreted form of NS1 would also inhibit signal transduction in naïve cells. This hypothesis was initially tested using sNS1-containing supernatants from WNV replicon-bearing cells (HeLa rep) or cells engineered to express NS1 (293T-NS1 cells). Our data show the supernatants from these cells lines were able to inhibit TLR3 signaling in naïve HeLa cells. Additionally, purified sNS1 inhibited TLR3 signaling in a dose-dependent manner and depletion of sNS1 from the culture supernatants or heat-inactivation restored TLR3 signaling in naïve HeLa cells. Our data shows sNS1 explicitly associates with naïve cells by binding to the cell surface and being internalized. It was previously demonstrated that cell-binding by dengue sNS1 and WNV sNS1 can be prevented by sodium chlorate pretreatment to inhibit the sulfation of glycosaminoglycans expressed on the cell surface (Avirutnan et al., 2007; Youn et al., 2010). In agreement with these reports, our data show that binding of sNS1 can be inhibited by sodium chlorate pretreatment of HeLa cells. We show that by preventing binding of sNS1 to naïve cells, TLR3 signaling is restored. Collectively, our data clearly indicate that the inhibitory effect is mediated directly by sNS1.
Baronti et al. have reported that the NS1 proteins of several different flaviviruses, including WNV, do not inhibit TLR3 signaling (Baronti et al., 2010). It is not clear why the results of these authors differ so significantly from ours, however in their study the authors resorted to overexpressing IRF3 in order to detect TLR3-induced transcriptional activation of IFNβ and RT-PCR was used as the main readout for TLR signaling. We determined previously that HeLa cells fail to produce significant levels of IFN-β transcript and protein following pIC stimulation alone, which may explain these conflicting results (Wilson et al., 2008). For our past and present studies, TLR3 signaling was determined using a combination of reporter assays, real-time qRT-PCR, and endogenous cytokine production and overexpression of signaling intermediates was not required to detect activation [(Scholle and Mason, 2005; Wilson et al., 2008) and this report]. We also demonstrate that sNS1-mediated inhibition of TLR3 and other TLRs is not limited to HeLa cells, but extends to more immunologically relevant cell types, such as macrophages and dendritic cells, and can be demonstrated in vivo.
Flaviviruses are recognized by different pattern recognition receptors during infection. Using primary cells, our data demonstrate the ability of sNS1 to inhibit signaling by multiple TLRs as well as reduce the IL-6 and IFNβ responses to WNV infection. In vivo we show a diminished response to both poly(IC:LC) stimulation and VRP infection after sNS1 pretreatment. Several studies have demonstrated that RIG-I and Mda-5 are important in early the recognition and response to flavivirus infection (Fredericksen and Gale, 2006; Fredericksen et al., 2004; Loo et al., 2008). Our data showed treatment with sNS1 diminished the early IL-6 and IFNβ responses to WNV infection in BMDMs and to VRP infection in mice. Therefore, our results suggest that sNS1 may have additional functions in inhibiting signaling events originating from PRRs other than TLRs.
Although our data show a strong inhibitory effect on the immune response to WNV infection, the sNS1-mediated decrease in cytokine production did not translate to increased viral growth. At this point, it is unclear why no enhancement of virus growth was observed after sNS1 pretreatment. However, the sNS1 treatment did not completely ablate IFNβ production in response to WNV infection, and as we have no evidence to suggest that sNS1 inhibits signaling through the type I IFN receptor, and the autocrine effects of residual IFNβ production may have been sufficient to stimulate the IFN amplification loop, limiting spread of the virus. To directly study the impact of sNS1 on virus replication and viral spread in an animal model would require infection with live WNV. However, these studies are complicated by the multifunctional nature of NS1. Secreted NS1 has a number of well-described immunomodulatory effects on the lectin and alternative pathways of the complement system (Avirutnan et al., 2010; Avirutnan et al., 2011; Chung et al., 2006; Fuchs et al., 2010; Fuchs et al., 2011). Therefore, any observed effects of sNS1 on the pathogenesis of live virus infection could not be attributed directly to PRR inhibition.
The cytokine response to infection is important in aspects of viral pathogenesis beyond the direct control of pathogen proliferation, including cell recruitment and the orchestration of B cell and T cell responses. In the pLN, sNS1 was found to associate predominantly with macrophages and DCs, the primary cell types involved in innate immune responses and antigen presentation. This suggests that sNS1 could potentially influence the development of an adaptive immune response to WNV infection. Indeed, a role for TLR3 in the development of the humoral response to WNV single-cycle infectious particles, called RepliVAX WN was recently described (Xia et al., 2013). Using a mouse model, the authors show TLR3-deficiency results in a short lived WNV-specific antibody response after inoculation with RepliVAX WN, and report that TLR3 is important in the maintenance of germinal centers (GCs), the sites of antigen-specific B cell proliferation and maturation. This study also shows impaired B cell activation and hindered development of GCs as well as a diminished memory B cell response in MyD88−/− mice. MyD88 is the adaptor molecule in the TLR7 signaling cascade. TLR7 is also important in the immune response to WNV, and in fact, plays a role in the recognition of flaviviruses by plasmacytoid dendritic cells, triggering the early IFNα response and contributing to immune cell homing to sites of infection. (Diebold et al., 2004; Lund et al., 2004; Silva et al., 2007; Town et al., 2009; Wang et al., 2006; Welte et al., 2009).
To our knowledge, this report is the first to describe PRR signaling inhibition by a secreted protein from an RNA virus. PRR inhibition has been described for other WNV proteins. Whether the sNS1 proteins of other flaviviruses have immunomodulatory properties similar to that of WNV sNS1 remains an important question. Another WNV protein, the structural protein, E, is able to subvert dsRNA-mediated signaling, however this is only true of the E protein of mosquito cell origin and not true of E protein generated in mammalian cells (Arjona et al., 2007). Therefore, TLR3 signaling inhibition by the E protein would occur only during the very first infection cycle. The NS1 protein used for our studies was secreted by mammalian cells and therefore our in vitro and in vivo data indicate immune modulation by sNS1 likely occurs throughout infection, suggesting sNS1 could influence the adaptive immune response to infection.
In summary, our findings provide novel evidence of PRR signaling inhibition by the secreted form of WNV NS1. Our data show targeting of DCs and macrophages by sNS1 in vivo and our in vitro studies show active repression of signaling by these cell types. As DCs and macrophages link the innate and adaptive immune responses, additional studies aimed at characterizing the effect of sNS1 on the adaptive immune response are essential. Our data show immune suppression by sNS1 in vivo, however, further research into the mechanisms used by WNV sNS1 to undermine the host antiviral response and how these immune evasion strategies influence viral pathogenesis is necessary. These studies will provide information critical to the development of treatment strategies and vaccination programs that control WNV infection. The use of vaccine adjuvants can enhance immunity to virus infection and there is evidence that TLR ligands can function as effective vaccine adjuvants (Duthie et al., 2011). Information as to the mechanisms of NS1-mediated TLR-signaling inhibition would contribute to the design of adjuvants that will successfully bolster immunity to WNV and suppress infection. Our data describes a novel function of WNV NS1, a protein with a diverse array of immunomodulatory actions, that contribute to viral pathogenesis and thus the multifunctional nature of this protein necessitates further study.
Materials and methods
Cell Lines
HeLa cells were grown in DMEM (Cellgro) supplemented with 10% HyClone fetal bovine serum (Cellgro),1% antibiotics, and 20μg/ml gentamicin. Human Embryonic Kidney (HEK 293T) cells were transduced with an NS1-His encoding recombinant lentivirus (pLEX-NS1) or an empty vector control lentivirus (pLEX-Ctrl). The pLEX-NS1 and pLEX-Ctrl were packaged into VSV-G pseudotyped, replication defective, lentivirus particles through co-transfection of HEK 293T cells with vector, packaging (pREV and pMDLg), and envelope (pVSV-G) plasmids. Lentivirus-containing supernatants were harvested and clarified 40h post-transfection. The 293T cells were transduced with pLEX-NS1 or pLEX-Ctrl lentivirus for 24h in the presence of 8μg/mL polybrene. Transduced cells were selected by puromycin treatment at 4μg/ml as a pool. The resulting cell lines are termed 293T-NS1 and 293T-Ctrl. 293T-NS1 cells were used in the production of sNS1 containing supernatants used for experiments and for sNS1 purification. 293T-Ctrl cell supernatants were treated identically to NS1-containing supernatants during collection and purification.
Primary Cells
Wild-type C57BL/6 mouse femurs were collected and bone marrow was harvested per standard protocols. Macrophages were grown for 7 days in DMEM containing 10% fetal bovine serum, 1% non-essential amino acids, 1% antibiotics and differentiated in the presence of M-CSF. Myeloid dendritic cells were grown for 10 days in RPMI containing 10% fetal bovine serum, 1% antibiotics, 55μM beta-mercaptoethanol and differentiated in the presence of IL-4.
Purification of NS1
A mouse monoclonal antibody to JEV NS1 (clone 6H4, a kind gift from Dr. Eji Konishi, Osaka University) was coupled to CnBr beads overnight at 4° Celsius. The beads were packed into a column and prepared with oscillating pH washes followed by two additional low pH washes and two high pH washes. Clarified NS1 containing cell supernatant was applied to the column. The beads were then washed at neutral pH and protein was eluted in 0.5mL fractions with a high pH solution. The fractions were immediately neutralized with 1M Tris buffer and assayed by ELISA for NS1 content. NS1 ELISA was performed by coating Immulon 2HB plates (Thermo) with the eluted fractions at 4°C for 12–16 hours followed by detection with the anti-JE NS1 antibody and HRP-labeled mouse secondary antibody. Color was developed with 3,3′,5,5′-Tetramethylbenzidine (TMB) substrate and the reaction was stopped with 2N H2SO4. Fractions positive for NS1 by ELISA were run on 4–12% NuPage protein gels and subjected to silver stain (BioRad) to ensure purity. Pure NS1 fractions were then pooled and dialyzed against PBS overnight at 4°C. The amount of protein present in the samples after dialysis was determined using a protein assay kit (Bio-Rad). Supernatants from the 293T-Ctrl cell line were subjected to the same purification process and the fractions collected were dialyzed against PBS and used as a purification control (pCtrl).
Immunodepletion of NS1
During sNS1 purification, flow-through samples were collected from the immunoaffinity column after the application of the NS1-containing supernatants. Depleted supernatant samples were analyzed by Western blot to confirm the absence of NS1. Samples were dialyzed against PBS before use in cell culture assays.
Reporter Assays
Reporter assays using the luciferase (Luc) reporter gene under control of the IFN-β promoter (IFN-β pGL3; a gift from J. Hiscott) have been described previously (Scholle and Mason, 2005; Wilson et al., 2008). Briefly, 150ng each of IFN-β pGL3 and pCMV β-galactosidase (β-Gal) (Invitrogen) were co-transfected into HeLa cells, using the TransIT-LT1transfection reagent (Mirus). At 4h post-transfection, clarified NS1-containing supernatant or clarified control supernatant was added to the cells. For experiments using purified sNS1, the appropriate concentration of sNS1, or an equal volume of pCtrl, was prepared in full serum media. Heat-inactivation of sNS1 was achieved by heating the sample at 95°C for 1h. After 16h cells were either left untreated or were stimulated by adding 20 μg/ml poly(I:C) (pIC; Calbiochem) to the culture media and incubating for 4 hours. Following treatment, cells were lysed in reporter lysis buffer (0.1% TritonX-100, 10% glycerol, 2mM Dithiothreitol, 2mM trans 1,2 diaminocyclohexane-NNNN acetic acid, and 25mM Tris phosphate) and assayed for Luc and β-Gal activities using a Promega Luc assay system and an ONPG (o-nitrophenyl-β-D-galactopyranoside)-based β-Gal assay. β-Gal activity was used to normalize the Luciferase data for all experiments. All data are expressed as relative light units/mU of β-Gal activity.
Western blot analysis
Protein extracts were prepared in cell lysis buffer (300 mM NaCl, 50 mM Tris-HCl, 0.1% Triton X-100, pH 7.6). Followinga10-min incubation on ice, lysates were clarified by centrifugation, and protein concentrations were determined using a detergent-compatible protein assay kit (Bio-Rad). Equal amounts of protein were electrophoretically separated on 4 to 12% Nu-PAGE gels (Invitrogen) and electroblotted onto polyvinylidenedifluoride membranes (Immobilon-P transfer membrane; Millipore). Following a blocking step with Tris-buffered saline containing 0.1% Tween-20 and 5% dry milk, NS1 was detected using a WNV-specific mouse hyperimmune ascitic fluid [MHIAF] as the primary antibody (Scholle and Mason, 2005), followed by a horseradish peroxidase (HRP)-conjugated secondary antibody (KPL). Bound HRP was visualized with an Immunocruz ECL kit (Santa Cruz). Membranes were stripped and re-probed with a mouse anti-β-actin antibody (Sigma-Aldrich) as a loading control.
Immunofluorescence
HeLa cells were seeded onto LabTek (Nunc) chamber slides and incubated with supernatants from NS1-expressing cells or control cells for 16h at which time the slide was prepared for indirect immunofluorescence analysis (IFA) for identification of NS1 protein. Briefly, cells were washed with PBS, fixed in 4%paraformaldehyde, permeabilized with 0.1% Triton X-100, and blocked (2% bovine serum albumin, 5% normal horse serum, and 10 mM glycine in PBS). The cells were incubated with WNV MHIAF as the primary antibody, followed by incubation with AlexaFluor 568-conjugated anti-mouse antibody (Invitrogen) and then analyzed for presence of NS1 by immunofluorescence using a Zeiss Axioskop 2 microscope.
ELISA
Levels of IL-6 or IL-12p70 in cell culture supernatants were determined by enzyme-linked immunosorbent assay (ELISA) (e-Bioscience) according to the manufacturer’s specifications.
Analysis of sNS1-cell binding to HeLa cells
HeLa cells were incubated in the presence of sNS1 for indicated times, the cells were then washed with PBS before fixing and permeabilizing on ice for 20 minutes (BD Cytofix/Cytoperm kit) or left unpermeabilized and fixed with 4% paraformaldehyde for five minutes on ice. Cells were incubated with a primary antibody generated against the NS1 protein of Japanese Encephalitis virus and then an Alexafluor A488-tagged secondary antibody. Analysis was performed on the Accuri C6 flow cytometer according to standard protocol.
Real-time qRT-PCR
Total RNA was extracted using a Qiagen RNeasy kit with DNase treatment according to the manufacturer’s specifications. Reverse transcription (RT) was carried out with the ImProm II RT kit (Promega) using oligodT as primer. Random hexamer was used as primer for NS5 experiments. Real-time PCR analysis was carried out using the SensiMix SYBR & Fluorescein kit (BioLine) and the following primers; mGAPDH 5′-TGCCCAGAACATCATCCCTG-3′ and 5′-ATCCACGACGGACACATTGG-3′, mIL-6 5′-GACTTCACAGAGGATACCACTCC-3′ and 5′-TTCTGCAAGTGCATCATCGGT-3′,mIFNβ 5′-GGAGATGACGGAGAAGATGC-3′ and 5′-CCCAGTGCTGGAGAAATTGT-3′, Kunjin NS5 5′-GAGTCCAAGAAGTCAGAGGGTACA-3′ and 5′-CCACTCTTCATGGTGACAATGTTCC-3′. Reactions were set up in 96-well PCR plates (Genesee). Amplifications were carried out for 50 cycles, followed by a melt curve analysis of resulting products to confirm the specificity of the reactions. To construct standard curves, total RNA was isolated from the cells, and 300- to 600-bp fragments of the gene of interest encompassing the real-time PCR primer binding sites, were amplified by RT-PCR using the appropriate primer sets. PCR fragments were gel purified and quantified, and the copy number was calculated. Serial 10-fold dilutions were prepared for to create standard curves for real-time qPCR. All data are expressed as the ratio of copy numbers of target gene per 103 or 104copies of GAPDH as indicated.
VRP production and purification
RepliVAX WN contains a deletion of the capsid protein and is only able to replicate for a single infection cycle. RepliVAX can be packaged into infectious particles by supplying the missing capsid protein in trans (Mason, Shustov, and Frolov, 2006). To produce high titer stocks for in vivo studies, BHK (VEErep/Pac-Ubi-C*) cells expressing the WNV capsid protein from a Venezuelan encephalitis virus replicon were inoculated with RepliVAX WN VRPs(Mason, Shustov, and Frolov, 2006) at an MOI of 0.05 and supernatants were harvested 96 hpi. The supernatants were clarified and then concentrated over a 100,000 kilodalton (Kda) cutoff centrifugal filter at 1500g. The concentrate was applied to a 10–40% sucrose gradient and centrifuged at 35,000rpm at 4°C for 2.5hr. The fractions containing infectious particles were collected and washed three times with L15 medium over a centrifugal filter to remove residual sucrose. The final concentrate was resuspended in L15 medium containing 10mM HEPES and 0.1% FBS. VRP titers were determined on Vero cells by immunostain against WNV-specific antigen and reported as infectious units per ml (iu/ml).
In vivo experiments
C57BL/6 mice were obtained from Charles River laboratories and housed under specific-pathogen-free conditions in the Biological Resources Facility at NCSU. All procedures were conducted in accordance with the NCSU Institutional Animal Care and Use Committee. Eight to twelve week old female C57/BL6 mice were injected with 5μg of purified sNS1 into both rear footpads. After 6h, mice were stimulated by injection of 20μg poly(IC:LC) (Hiltonol, Oncovir), PBS control, or 1x105iu purified RepliVAX VRPs into each footpad. Draining lymph nodes were harvested at 16h post stimulation and processed for qRT-PCR analysis.
Secreted NS1-cell association in draining lymph nodes
Purified sNS1 was labeled with Alexafluor 488 using an Invitrogen labeling kit according to the manufacturer’s instructions. Briefly, sNS1 (1mg/ml) was incubated with fluorescent dye for 1hr at RT. The labeled sNS1 was separated from the unincorporated dye by running it through a column packed with the provided purification resin. The final purified protein was resuspended in PBS and quantified prior to use. The unincorporated dye was used as a negative control for the injection experiments. Mice were injected with 5 mg/ml of Alexa488 labeled sNS1 into the rear footpad. After 6hr, pLN from pairs of mice were pooled, homogenized, and passed through a cell strainer to generate single cell suspensions. Cells were washed and resuspended in PBS + 1% FBS before incubation with anti-CD16/CD32 (BD Pharmingen) to block Fc receptors. Cells were then stained with antibodies specific to CD3, CD8, CD4 (T cells), CD11c (DCs), CD335 (NK cells), and F4/80 (macrophages). Antibodies were purchased from eBiosciences, BD Pharmingen, or Biolegend. After cell surface staining, cells were fixed and permeabilized to detect sNS1-488. The cells were washed three times and resuspended in 300 uL of PBS + 1% FBS and samples were subsequently analyzed by flow cytometry on a C6 Accuri cytometer using Accuri CFlow software.
Statistical analysis
Statistical analysis was performed using student’s t-test. P values below 0.05 were considered significant.
Supplementary Material
Highlights.
The secreted form of the WNV NS1 protein (sNS1) inhibits TLR signaling.
sNS1 inhibits TLR3 in HeLa cells, TLR4 and TLR7 in macrophages and dendritic cells
sNS1 inhibits the cytokine response to infection in mouse macrophages.
sNS1 associates with macrophages and dendritic cells in vivo
sNS1 inhibits the cytokine response to WNV replicon particle infection vivo
Acknowledgments
This work was supported by a grant from the National Institutes of Health (NIH U54 AI 057157-07(SERCEB), project SE-RP-012. We thank Lindsey Stevenson for supplying the MTT data in Supplemental Figure S2.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Alcon-LePoder S, Drouet MT, Roux P, Frenkiel MP, Arborio M, Durand-Schneider AM, Maurice M, Le Blanc I, Gruenberg J, Flamand M. The secreted form of dengue virus nonstructural protein NS1 is endocytosed by hepatocytes and accumulates in late endosomes: implications for viral infectivity. J Virol. 2005;79(17):11403–11. doi: 10.1128/JVI.79.17.11403-11411.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjona A, Ledizet M, Anthony K, Bonafe N, Modis Y, Town T, Fikrig E. West Nile virus envelope protein inhibits dsRNA-induced innate immune responses. J Immunol. 2007;179(12):8403–9. doi: 10.4049/jimmunol.179.12.8403. [DOI] [PubMed] [Google Scholar]
- Avirutnan P, Fuchs A, Hauhart RE, Somnuke P, Youn S, Diamond MS, Atkinson JP. Antagonism of the complement component C4 by flavivirus nonstructural protein NS1. J Exp Med. 2010;207(4):793–806. doi: 10.1084/jem.20092545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avirutnan P, Hauhart RE, Somnuke P, Blom AM, Diamond MS, Atkinson JP. Binding of flavivirus nonstructural protein NS1 to C4b binding protein modulates complement activation. J Immunol. 2011;187(1):424–33. doi: 10.4049/jimmunol.1100750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avirutnan P, Zhang L, Punyadee N, Manuyakorn A, Puttikhunt C, Kasinrerk W, Malasit P, Atkinson JP, Diamond MS. Secreted NS1 of dengue virus attaches to the surface of cells via interactions with heparan sulfate and chondroitin sulfate E. PLoS Pathog. 2007;3(11):e183. doi: 10.1371/journal.ppat.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baronti C, Sire J, de Lamballerie X, Querat G. Nonstructural NS1 proteins of several mosquito-borne Flavivirus do not inhibit TLR3 signaling. Virology. 2010;404(2):319–30. doi: 10.1016/j.virol.2010.05.020. [DOI] [PubMed] [Google Scholar]
- Bourne N, Scholle F, Silva MC, Rossi SL, Dewsbury N, Judy B, De Aguiar JB, Leon MA, Estes DM, Fayzulin R, Mason PW. Early production of type I interferon during West Nile virus infection: role for lymphoid tissues in IRF3-independent interferon production. J Virol. 2007;81(17):9100–8. doi: 10.1128/JVI.00316-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown AN, Kent KA, Bennett CJ, Bernard KA. Tissue tropism and neuroinvasion of West Nile virus do not differ for two mouse strains with different survival rates. Virology. 2007;368(2):422–30. doi: 10.1016/j.virol.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne SN, Halliday GM, Johnston LJ, King NJC. Interleukin-1beta but not tumor necrosis factor is involved in West Nile Virus-induced Langerhans cell migration from the skin in C57BL/6 mice. Journal of Investigative Dermatology. 2001;117(3):702–709. doi: 10.1046/j.0022-202x.2001.01454.x. [DOI] [PubMed] [Google Scholar]
- Caskey M, Lefebvre F, Filali-Mouhim A, Cameron MJ, Goulet JP, Haddad EK, Breton G, Trumpfheller C, Pollak S, Shimeliovich I, Duque-Alarcon A, Pan L, Nelkenbaum A, Salazar AM, Schlesinger SJ, Steinman RM, Sekaly RP. Synthetic double-stranded RNA induces innate immune responses similar to a live viral vaccine in humans. J Exp Med. 2011;208(12):2357–66. doi: 10.1084/jem.20111171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KM, Diamond MS. Defining the levels of secreted non-structural protein NS1 after West Nile virus infection in cell culture and mice. J Med Virol. 2008;80(3):547–56. doi: 10.1002/jmv.21091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KM, Liszewski MK, Nybakken G, Davis AE, Townsend RR, Fremont DH, Atkinson JP, Diamond MS. West Nile virus nonstructural protein NS1 inhibits complement activation by binding the regulatory protein factor H. Proc Natl Acad Sci U S A. 2006;103(50):19111–6. doi: 10.1073/pnas.0605668103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Samuel MA, Suthar MS, Gale M, Jr, Diamond MS. Toll-like receptor 3 has a protective role against West Nile virus infection. J Virol. 2008;82(21):10349–58. doi: 10.1128/JVI.00935-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–31. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- Duthie MS, Windish HP, Fox CB, Reed SG. Use of defined TLR ligands as adjuvants within human vaccines. Immunol Rev. 2011;239(1):178–96. doi: 10.1111/j.1600-065X.2010.00978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flamand M, Megret F, Mathieu M, Lepault J, Rey FA, Deubel V. Dengue virus type 1 nonstructural glycoprotein NS1 is secreted from mammalian cells as a soluble hexamer in a glycosylation-dependent fashion. J Virol. 1999;73(7):6104–10. doi: 10.1128/jvi.73.7.6104-6110.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Gale M., Jr West Nile virus evades activation of interferon regulatory factor 3 through RIG-I-dependent and -independent pathways without antagonizing host defense signaling. J Virol. 2006;80(6):2913–23. doi: 10.1128/JVI.80.6.2913-2923.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Keller BC, Fornek J, Katze MG, Gale M., Jr Establishment and maintenance of the innate antiviral response to West Nile Virus involves both RIG-I and MDA5 signaling through IPS-1. J Virol. 2008;82(2):609–16. doi: 10.1128/JVI.01305-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen BL, Smith M, Katze MG, Shi PY, Gale M., Jr The host response to West Nile Virus infection limits viral spread through the activation of the interferon regulatory factor 3 pathway. J Virol. 2004;78(14):7737–47. doi: 10.1128/JVI.78.14.7737-7747.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Lin TY, Beasley DW, Stover CM, Schwaeble WJ, Pierson TC, Diamond MS. Direct complement restriction of flavivirus infection requires glycan recognition by mannose-binding lectin. Cell Host Microbe. 2010;8(2):186–95. doi: 10.1016/j.chom.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Pinto AK, Schwaeble WJ, Diamond MS. The lectin pathway of complement activation contributes to protection from West Nile virus infection. Virology. 2011;412(1):101–9. doi: 10.1016/j.virol.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston LJ, Halliday GM, King NJ. Langerhans cells migrate to local lymph nodes following cutaneous infection with an arbovirus. J Invest Dermatol. 2000;114(3):560–8. doi: 10.1046/j.1523-1747.2000.00904.x. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Toll-like receptor and RIG-I-like receptor signaling. Ann N Y Acad Sci. 2008;1143:1–20. doi: 10.1196/annals.1443.020. [DOI] [PubMed] [Google Scholar]
- Khromykh AA, Sedlak PL, Guyatt KJ, Hall RA, Westaway EG. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. Journal of Virology. 1999;73(12):10272–10280. doi: 10.1128/jvi.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PY, Behr MJ, Chadwick CM, Shi PY, Bernard KA. Keratinocytes are cell targets of West Nile virus in vivo. J Virol. 2011;85(10):5197–201. doi: 10.1128/JVI.02692-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Rice CM. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71(12):9608–17. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, Garcia-Sastre A, Katze MG, Gale M., Jr Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol. 2008;82(1):335–45. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund JM, Alexopoulou L, Sato A, Karow M, Adams NC, Gale NW, Iwasaki A, Flavell RA. Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc Natl Acad Sci U S A. 2004;101(15):5598–603. doi: 10.1073/pnas.0400937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald J, Tonry J, Hall RA, Williams B, Palacios G, Ashok MS, Jabado O, Clark D, Tesh RB, Briese T, Lipkin WI. NS1 protein secretion during the acute phase of West Nile virus infection. J Virol. 2005;79(22):13924–33. doi: 10.1128/JVI.79.22.13924-13933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason PW, Shustov AV, Frolov I. Production and characterization of vaccines based on flaviviruses defective in replication. Virology. 2006;351(2):432–443. doi: 10.1016/j.virol.2006.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasirudeen AM, Wong HH, Thien P, Xu S, Lam KP, Liu DX. RIG-I, MDA5 and TLR3 synergistically play an important role in restriction of dengue virus infection. PLoS Negl Trop Dis. 2011;5(1):e926. doi: 10.1371/journal.pntd.0000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle F, Mason PW. West Nile virus replication interferes with both poly(I:C)-induced interferon gene transcription and response to interferon treatment. Virology. 2005;342(1):77–87. doi: 10.1016/j.virol.2005.07.021. [DOI] [PubMed] [Google Scholar]
- Silva MC, Guerrero-Plata A, Gilfoy FD, Garofalo RP, Mason PW. Differential activation of human monocyte-derived and plasmacytoid dendritic cells by West Nile virus generated in different host cells. J Virol. 2007;81(24):13640–8. doi: 10.1128/JVI.00857-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Town T, Bai F, Wang T, Kaplan AT, Qian F, Montgomery RR, Anderson JF, Flavell RA, Fikrig E. Toll-like receptor 7 mitigates lethal West Nile encephalitis via interleukin 23-dependent immune cell infiltration and homing. Immunity. 2009;30(2):242–53. doi: 10.1016/j.immuni.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai YT, Chang SY, Lee CN, Kao CL. Human TLR3 recognizes dengue virus and modulates viral replication in vitro. Cell Microbiol. 2009;11(4):604–15. doi: 10.1111/j.1462-5822.2008.01277.x. [DOI] [PubMed] [Google Scholar]
- Wang JP, Liu P, Latz E, Golenbock DT, Finberg RW, Libraty DH. Flavivirus activation of plasmacytoid dendritic cells delineates key elements of TLR7 signaling beyond endosomal recognition. J Immunol. 2006;177(10):7114–21. doi: 10.4049/jimmunol.177.10.7114. [DOI] [PubMed] [Google Scholar]
- Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10(12):1366–73. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- Welte T, Reagan K, Fang H, Machain-Williams C, Zheng X, Mendell N, Chang GJ, Wu P, Blair CD, Wang T. Toll-like receptor 7-induced immune response to cutaneous West Nile virus infection. J Gen Virol. 2009;90(Pt 11):2660–8. doi: 10.1099/vir.0.011783-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westaway EG, Mackenzie JM, Kenney MT, Jones MK, Khromykh AA. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71(9):6650–61. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JR, de Sessions PF, Leon MA, Scholle F. West Nile virus nonstructural protein 1 inhibits TLR3 signal transduction. J Virol. 2008;82(17):8262–71. doi: 10.1128/JVI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SJL, Grouard VG, Sun W, Mascola JR, Brachtel E, Putvatana R, Louder MK, Filgueira L, Marovich MA, Wong HK, Blauvelt A, Murphy GS, Robb ML, Innes BL, Birx DL, Hayes CG, Schlesinger FS. Human skin Langerhans cells are targets of dengue virus infection. Nature Medicine. 2000;6(7):816–820. doi: 10.1038/77553. [DOI] [PubMed] [Google Scholar]
- Xia J, Winkelmann ER, Gorder SR, Mason PW, Milligan GN. TLR3- and MyD88-dependent signaling differentially influences the development of West Nile virus-specific B cell responses in mice following immunization with RepliVAX WN, a single-cycle flavivirus vaccine candidate. J Virol. 2013;87(22):12090–101. doi: 10.1128/JVI.01469-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn S, Cho H, Fremont DH, Diamond MS. A short N-terminal peptide motif on flavivirus nonstructural protein NS1 modulates cellular targeting and immune recognition. J Virol. 2010;84(18):9516–32. doi: 10.1128/JVI.00775-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn S, Li T, McCune BT, Edeling MA, Fremont DH, Cristea IM, Diamond MS. Evidence for a Genetic and Physical Interaction between Nonstructural Proteins NS1 and NS4B That Modulates Replication of West Nile Virus. J Virol. 2012;86(13):7360–71. doi: 10.1128/JVI.00157-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PR, Hilditch PA, Bletchly C, Halloran W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. Journal of Clinical Microbiology. 2000;38(3):1053–1057. doi: 10.1128/jcm.38.3.1053-1057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.