Abstract
Illegitimate V(D)J recombination at oncogenes and tumor suppressor genes is implicated in formation of several T cell malignancies. Notch1 and Bcl11b, genes involved in developing T cell specification, selection, proliferation, and survival, were previously shown to contain hotspots for deletional illegitimate V(D)J recombination associated with radiation-induced thymic lymphoma. Interestingly, these deletions were also observed in wild-type animals. In this study, we conducted frequency, clonality, and junctional processing analyses of Notch1 and Bcl11b deletions during mouse development and compared results to published analyses of authentic V(D)J rearrangements at the T cell receptor beta (TCRβ) locus and illegitimate V(D)J deletions observed at the human, nonimmune HPRT1 locus not involved in T cell malignancies. We detect deletions in Notch1 and Bcl11b in thymic and splenic T cell populations, consistent with cells bearing deletions in the circulating lymphocyte pool. Deletions in thymus can occur in utero, increase in frequency between fetal and postnatal stages, are detected at all ages examined between fetal and 7 months, exhibit only limited clonality (contrasting with previous results in radiation-sensitive mouse strains), and consistent with previous reports are more frequent in Bcl11b, partially explained by relatively high Recombination Signal Information Content (RIC) scores. Deletion junctions in Bcl11b exhibit greater germline nucleotide loss, while in Notch1 palindromic (P) nucleotides are more abundant, although average P nucleotide length is similar for both genes and consistent with results at the TCRβ locus. Non-templated (N) nucleotide insertions appear to increase between fetal and postnatal stages for Notch1, consistent with normal terminal deoxynucleotidyl transferase (TdT) activity; however, neonatal Bcl11b junctions contain elevated levels of N insertions. Finally, contrasting with results at the HPRT1 locus, we find no obvious age or gender bias in junctional processing, and inverted repeats at recessed coding ends (Pr nucleotides) correspond mostly to single-base additions consistent with normal TdT activity.
Keywords: illegitimate V(D)J recombination, Notch1, Bcl11b, T cell malignancies, cryptic RSS, T cell development
1. Introduction
V(D)J recombination is the site-specific and evolutionarily conserved DNA recombination process in jawed vertebrates that confers novel antigen-binding domains to immunoglobulins and T cell receptors of lymphocytes. Normally, this process occurs only at particular stages of lymphocyte development and consists of the ordered rearrangements of variable (V), joining (J), and in some cases, diversity (D) gene segments. V(D)J recombinase, comprised mainly of Recombination Activating Gene 1 and 2 Proteins (RAG), recognizes Recombination Signal Sequences (RSSs) adjacent to coding gene segments and mediates recombination [1, 2].
Consensus RSSs are composed of a conserved, palindromic G/C rich heptamer (CACAGTG) and an A/T rich nonamer (ACAAAAACC) separated by a 12 or 23 base pair spacer of poorly conserved nucleotides [1, 3]. In the germline configuration prior to rearrangement, heptamer ends are directly adjacent to their respective coding segments. Following the 12/23 Rule, rearrangement normally juxtaposes coding segments with 12RSSs to those with 23RSSs [4]. The current model for RAG binding to RSSs involves “recombination centers” arising from altered locus accessibility that facilitates the formation of a single RAG/RSS complex followed by locus contraction and capture of the second RSS, ultimately resulting in a paired complex where recombination occurs [1, 5, 6].
To initiate recombination, RAG forms a single-strand nick at the junction between the RSS and coding segment and mediates formation of a hairpin structure via a direct transesterification of the newly freed 3’ hydroxyl on the coding segment with the opposite strand [7]. The resulting four DNA ends, two coding end hairpins and two blunt 5’ phosphorylated signal ends, are contained in a post-cleavage synaptic complex also containing RAG [8-10]. Signal ends are ligated to form flush heptamer-to-heptamer fusions (signal joints), while covalently sealed coding end hairpins undergo further processing prior to ligation [11].
Coding ends are nicked open, and three major processing events may occur that ultimately add diversity to antigen-binding domains. Palindromic (P) nucleotides can result from off-center nicking of hairpins, and nucleolytic activity can remove nucleotides from coding ends, both largely attributed to Artemis nuclease [12, 13]. Lastly, non-templated (N) nucleotides may be added to junctions by terminal deoxynucleotidyl transferase (TdT), a DNA polymerase expressed in lymphocyte precursors. Processed coding ends are then ligated together and retained on the chromosome. The processing and ligation of coding and signal ends involve several components largely associated with the ubiquitous classical nonhomologous end-joining (cNHEJ) pathway for double-strand DNA break repair [14-16].
DNA strand breaks and recombination events, such as those occurring during V(D)J recombination, are potential sources of genomic instability, and several control mechanisms exist to prevent aberrant or illegitimate recombination events outside immune loci or in non-lymphoid cells [1, 17]. First, expression of RAG and activators of V(D)J recombination normally occurs only during certain stages of lymphocyte development, and additionally RAG2 is targeted for degradation at the G1/S phase cell cycle transition [18]. Second, RSSs must be recognized and accessible within chromatin for RAG to initiate recombination. Finally, the large-scale organization of chromatin in the nucleus must be altered to allow for locus contraction and recombination of distant gene segments within the genome [19-21].
Despite these control mechanisms, occurrences of aberrant and illegitimate V(D)J recombination events involving lymphoid and/or non-lymphoid loci are detected in humans and mice, are often associated with lymphoid malignancies, and may be due to several factors [22-24]. While the consensus RSS is conserved among several species, only certain nucleotides of the heptamer are required for recombination, and most bona fide RSSs do not conform precisely to consensus [1, 25]. Thus, many pseudo or cryptic RSSs (cRSSs) can exist throughout the genome that may be recognized by RAG [25, 26]. While RAG1 binding appears to be restricted solely to RSSs within the antigen receptor loci, RAG2 is found bound to trimethylated histone 3 (H3K4me3) broadly throughout the genome, which may allow for recruitment and/or activation of other V(D)J recombinase components at non-lymphoid loci [6, 27]. Rearrangement is also typically correlated with several markers of open or “accessible” chromatin, including particular histone modifications associated with germline transcription [28-30]. Additionally, coding segments separated by large genomic distances can be found juxtaposed in mature lymphocytes, indicating that germline separation, even by thousands of nucleotides, does not preclude recombination. Thus, DNA sequences that bear cRSSs in areas of open chromatin within cells concurrently expressing V(D)J recombinase components may be susceptible to illegitimate recombination.
If these illegitimate events occur at oncogenes or tumor suppressor genes, they may lead to pre-malignant states and/or cell transformation. Notch1 and Bcl11b are two such genes commonly found mutated in several murine and human T cell malignancies [24]. In mice, V(D)J recombinase-mediated deletions in these genes were found in radiation-induced and spontaneous thymic lymphomas, but interestingly deletions were also detected in healthy, wild-type animals [31, 32]. Illegitimate V(D)J recombination is implicated due to cRSSs or heptamer-like sequences and RAG2 binding near breakpoints, characteristic processing of coding junctions (i.e., molecular “signatures” of V(D)J recombination), and the absence or qualitative differences in deletions of RAG2−/− and cNHEJ-deficient mice, respectively [6, 31-34].
Notch1 is a widely expressed oncogene encoding a transmembrane receptor that controls several aspects of T cell development, selection, proliferation, survival, and differentiation [35-37]. Notch1 signaling involves ligand binding and proteolytic cleavages that ultimately release the intracellular portion (ICN), which functions in a nuclear transcription factor complex. Importantly, increased Notch1 expression occurs in early stages of T cell development (DN2/DN3/early DP) during which V(D)J-recombinase components are expressed and active [35-39]. Illegitimate V(D)J recombination mediates a deletion that removes the 5’ portion of the gene containing the promoter and canonical translational start codon in exon 1. Consequently, a truncated ligand- and cleavage-independent ICN can result from usage of cryptic promoters within or downstream of exon 25 [31, 34, 39, 40]. Constitutive ICN activity can potentially promote uncontrolled proliferation and oncogenesis.
Bcl11b is a transcription factor that contains several zinc-finger DNA binding motifs that was originally identified as a haploinsufficient tumor suppressor and is also involved in T cell development, commitment, selection, and survival [41-48]. Bcl11b expression is also upregulated during early stages of T cell development (DN2/DN3/DN4/DP), including those with ongoing V(D)J recombination [38, 44, 48]. Illegitimate V(D)J recombination removes exons 2 and 3, resulting in expression of a less common protein variant known at the γ-isoform that is implicated in the formation of radiation-induced thymic lymphomas [32, 41].
Previous studies of illegitimate V(D)J recombination-mediated deletions in Notch1 and Bcl11b focused mainly on radiation-induced deletions in thymic lymphomas of young adult animals [31, 32]. In this study, we conducted an expanded frequency, clonality, and junctional processing analysis of illegitimate V(D)J deletion events at these loci during several stages of mouse development in wild-type animals. Results are discussed in the context of similar analyses of bona fide V(D)J rearrangements at the T cell receptor beta (TCRβ) locus and illegitimate events at the nonimmune HPRT1 locus found in human peripheral T cells that are not associated with T cell malignancies [49, 50].
2. Materials and Methods
2.1 Mice
C57BL/6 mice (Taconic, Hudson, NY) were bred and maintained in the vivarium of Southeastern Louisiana University under conditions approved by the Institutional Animal Care and Use Committee (IACUC). Mice were used at various developmental stages including fetal (gestational day 16), neonatal (2 days and 1 week), juvenile (2 and 3 weeks), young adult (4, 5, 7, and 10 weeks), and older adult (6 and 7 months). Sex determinations in young mice were confirmed as previously described [51].
2.2 Isolation of cells and DNA
Genomic DNA from whole thymus and spleen was purified from mice at a range of ages between fetal and mature adult stages. While older mice were analyzed individually, fetal samples were pooled from 7 mice. Thymic CD4+ and CD8+ T cells were isolated at 7 weeks. Splenic T and CD43− B cells were purified at 5 weeks.
Thymocytes and splenocytes were isolated into RPMI complete medium containing 10% ΔFCS followed by washing with PBS (pH 7.2). Thymic CD4+ and CD8+ T cells were further purified using an EasySep Mouse CD4+ (or CD8+) Positive Selection Kit (STEMCELL Technologies, Vancouver, Canada). Splenic T and B cells were purified by negative selection using a Dynabead Untouched Mouse T (or CD43− B) Cell Kit (Invitrogen, Grand Island, NY).
All samples were digested in SDS/Proteinase K buffer (0.5% SDS, 100μg/mL Proteinase K, 100mM NaCl, 10mM Tris-HCl, 25mM EDTA, pH 8) at 55°C overnight. Lysates were incubated for 1hr each with 1mM PMSF at room temperature followed by 0.1mg/mL RNaseA at 37°C. Genomic DNA was extracted using phenol, chloroform, and isoamyl alcohol (25:24:1), precipitated in ethanol, resuspended in TE8 buffer (10mM Tris-HCl, 1mM EDTA, pH 8), and quantified using a Nanodrop 1000 (Thermo Fisher Scientific, Wilmington, DE).
2.3 Nested PCR amplification of deletion coding junctions
Detection of illegitimate V(D)J recombinase-mediated deletions in genomic DNA was performed using modified versions of nested PCR protocols previously described [31, 32]. All reactions contained 2mM MgCl2, 200μM each dNTP, 0.5μM each primer (Eurofins MWG Operon, Huntsville, AL), 10ng/μL template DNA, and 25U/mL DNA polymerase (Ex Taq, Takara, Mountain View, CA or HotStar Taq, Qiagen, Valencia, CA) and consisted of 30 cycles of 94°C for 30sec, annealing (Table 1) for 30sec, and 72°C for 1min.
Table 1.
Primers for Nested PCR Amplification of Deletion Coding Junctions
| Primer [31, 32] | Sequence (5’→3→) | Annealing |
|---|---|---|
| NF22 | CCATGGTGGAATGCCTACTTTGTATGAGGC | 61°C |
| NR22 | CCCTCAATTTCTCCTTTAGGTTCCTTTGAG | |
| NF3 | CTCCTGCTGCTCTGTGAGTCCCACTTCCAA | 66°C |
| NR3 | TTCCCCAGTCAGGAGTGGTGGATCCCTCTG | |
| F1 | GGCTGAATTTACAGGATGAGG | 55°C |
| R1 | ACTGGAGTTTCCGATGGCC | |
| F1-2 | GTTTGAGCTTGGAATGGCTGC | 57°C |
| R1-2 | ACATCGCCACCATGGAAGAC |
For detection of Notch1 deletions, outer reactions (45μL) used primers NF22 and NR22 with 450ng of DNA (75,000 cell equivalents), and inner reactions used primers NF3 and NR3 (expected product ~250bp). For detection of Bcl11b deletions, outer reactions (22.5μL) used primers F1 and R1 with 225ng of DNA (37,500 cell equivalents), and inner reactions used primers F1-2 and R1-2 (expected product ~440bp). Some outer reactions for Bcl11b were performed using 50ng of DNA. For whole spleen, outer and inner reactions contained both Notch1 and Bcl11b primers, and outer reactions (30μL) used 30-70ng/μL DNA. All inner reactions (20μL) used 1μL of respective outer reactions.
PCR products were visualized on 1.5-2% agarose gels stained with ethidium bromide and extracted using a Qiaquick (or QiEx II) Gel Extraction Kit (Qiagen, Valencia, CA). Due to extensive genomic distances between primer target sequences, products are only formed when illegitimate deletions are present.
2.4 Determination of deletion frequency in thymus
Multiple nested PCR analyses were performed on DNA samples (minimum of 20 replicates) at limiting dilution to calculate the frequency of deletion events per cell using a modified Poisson distribution formula: F=−ln(Po)/n, where F is the frequency of deletions per cell, Po is the number of negative reactions divided by the total number of reactions, and n is the number of cell equivalents per reaction (assuming 6pg of DNA per diploid genome) [52]. Following limiting dilution theory, the frequency was only calculated when the number of negative reactions (i.e., no amplified deletions) was greater than or equal to 37% of the total number of reactions.
The PCR protocol used was previously shown to have a sensitivity of a single deletion event per reaction [31, 32]. The monoclonality of detected deletion products was confirmed by bacterial cloning and sequencing (TOPO TA Cloning Kit, Invitrogen, Grand Island, NY) or direct DNA sequencing following agarose gel purification (Beckman Coulter Genomics, Danvers, MA).
2.5 Analysis of deletion coding junction sequences and clonality
Sequences of deletion coding junctions were aligned using Se-Al Sequence Alignment Editor v2.0a11 (http://tree.bio.ed.ac.uk/software/seal/) and analyzed using a systematic set of rules for nucleotide scoring based on previous studies of V(D)J recombination [49, 53]. First, coding end termini and nucleolytic processing were assigned based on the longest set of nucleotides matching the known germline sequences. Germline nucleotides that could not be unequivocally assigned to a single coding end were scored as junctional microhomology nucleotides. Second, inserted nucleotides were scored as either non-templated (N) or palindromic (P) nucleotides. Finally, non-templated nucleotides palindromic to germline sequences at coding ends with nucleolytic processing were also scored as inverted repeat nucleotides or “recessed palindromes” (Pr). Nucleotides that could not be unequivocally assigned to a single coding end were excluded from individual 5’ and 3’ coding end analyses.
2.6 Statistical analysis
One-way analysis of variance (ANOVA) was used to examine differences in deletion frequency, nucleolytic processing, and insertion of non-templated (N), palindromic (P), and inverted repeat (Pr) nucleotides between age groups. One-tailed Student's (unpaired) t-tests were used for analyses of sex and overall gene differences. Non-templated (N) dinucleotide pairs were analyzed using a Chi square goodness-of-fit test.
2.7 Analysis of cryptic Recombination Signal Sequences
To identify potential cryptic Recombination Signal Sequences (cRSSs), germline sequences near coding end breakpoints were examined by Recombination Signal Information Content (RIC) analysis using the RSSsite web server [54]. Deletion breakpoints, and accordingly deletion coding and signal ends, were designated based on the longest detected full-length coding ends matching the known germline sequences. Identified cRSSs were also compared to those previously observed in Notch1 and to heptamer-like sequences previously identified in Bcl11b [31, 32].
Possible cRSSs were only considered if the following conditions were met: signal end sequences began within five nucleotides of the deletion breakpoint, the minimum required nucleotides of the heptamer were present (CACNNNN) [1], a potential 12 and 23RSS pairing was possible, and RIC scores were equal to or higher than the lowest known for murine immune loci 12 and 23RSSs of −48.16 and −69.69, respectively [55].
3. Results
We screened for illegitimate V(D)J recombination-mediated deletion events in Notch1 and Bcl11b in thymus and spleen at various developmental stages and in separated lymphocyte populations. Thymus was also analyzed for deletion frequency, clonality, and junctional characteristics of V(D)J recombination at different stages.
3.1 Detection, frequency, and clonality of deletions during mouse development
In thymus, V(D)J-mediated deletions in Notch1 and Bcl11b were found in fetuses at gestational day 16 and at all subsequent ages examined (Fig. 1, Tables 2, 3, 4, and 5). In spleen, deletions were detectable by at least 2 weeks of age. Both CD4+ and CD8+ thymocyte populations contained deletions as did splenic T cells, but deletions were not detected in splenic CD43− B cells (data not shown). Thus, T cells bearing deletions are present in the circulating lymphocyte pool, but deletions are not detected in unactivated splenic B cells.
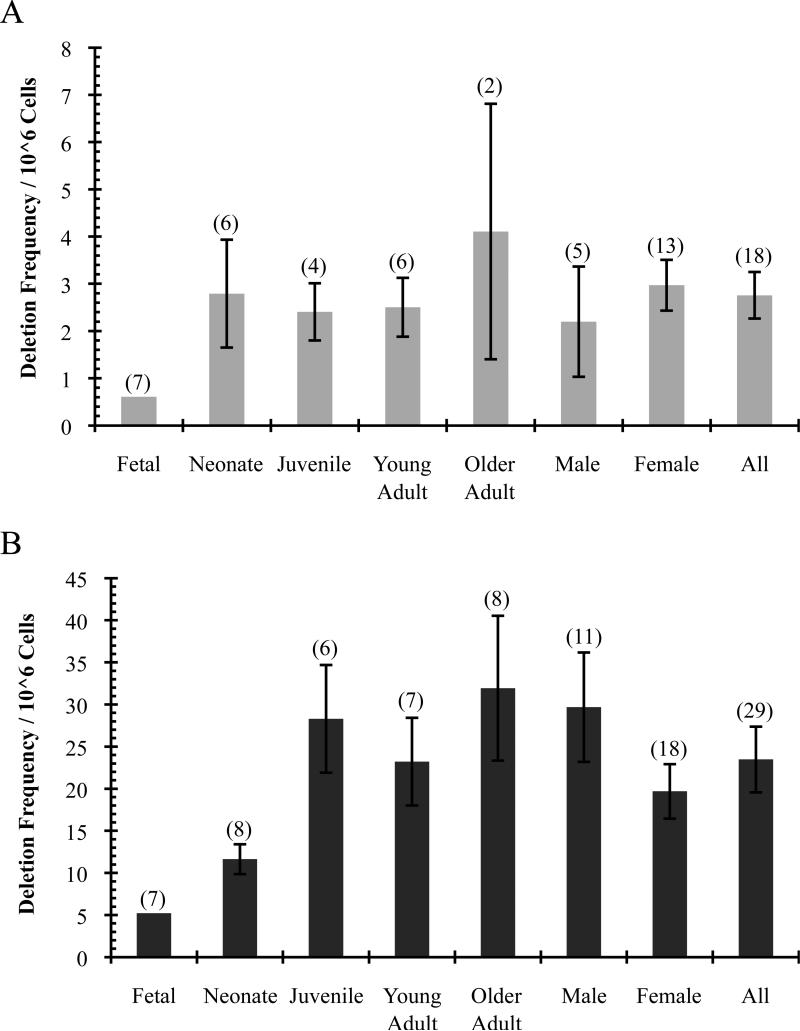
Fig. 1.
Frequency of illegitimate V(D)J deletions detected in thymus. (A) Notch1. (B) Bcl11b. Average frequency of deletions per 106 cells ± SE and number of mice in each group (n) are indicated. Fetal average is not included in Male, Female, or All. Note difference in scale.
Table 2.
Sequences of Notch1 Deletion Coding Junctions in Thymus1
| Group | Age | No. | Sex | 5′ Breakpoint | 5′P | 5′ Pr | N | 3′ Pr | 3′ P | 3′ Breakpoint | Clones (% of total) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Germline Sequence | GAGGATGCCC | TAGGCTTGGG | |||||||||
| Fetal | Fetal | --- | --- | GAGGATGC | TAGGCTTGGG | ||||||
| Neonate | 2 days | 116 | M | GAGGATG | GCTTGGG | ||||||
| 118 | F | GAGGATGCCC | G | AGGCTTGGG | |||||||
| GAGGATGC | G | GGCTTGGG | |||||||||
| 119 | F | GAGGATGC | TA | TAGGCTTGGG | |||||||
| GAGGATGCCC | TA | TAGGCTTGGG | |||||||||
| GAGGATGCC | G | GGCTTGGG | |||||||||
| GAGGATG | ag | GGCTTGGG | 2 (40%) | ||||||||
| 120 | M | GAGGATGCCC | A | TAGGCTTGGG | |||||||
| 1 week | 122 | F | GAGGATGC | AGGCTTGGG | 3 (100%) | ||||||
| 123 | F | GAGGATGCCC | G | GGCTTGGG | 7 (78%) | ||||||
| GAGGATGCCC | GGG | at | GGCTTGGG | ||||||||
| GAGGATGCC | AGGCTTGGG | ||||||||||
| Juvenile | 2 weeks | 113 | F | GAGGATGCCC | G | C | GGCTTGGG | ||||
| 3 weeks | 136 | F | GAGGATG | ggg | A | TAGGCTTGGG | 3 (75%) | ||||
| GAGG | gcg | TAGGCTTGGG | |||||||||
| 137 | M | GAGGATGCCC | cg | GGCTTGGG | |||||||
| GAGGATGCCC | GGG | AGGCTTGGG | |||||||||
| 138 | F | GAGGAT | TA | TAGGCTTGGG | |||||||
| GAGG | g | A | TAGGCTTGGG | ||||||||
| GAGGATGCCC | GG | GGG | |||||||||
| GAGGAT | CCTA | TAGGCTTGGG | |||||||||
| GAGGATGC | G | TAGGCTTGGG | |||||||||
| Young | 4 weeks | 139 | F | GAGGATGCCC | G | GGCTTGGG | 2 (40%) | ||||
| Adult | GAGGATGC | ag | GGCTTGGG | ||||||||
| GAGGATGCCC | G | ag | GGCTTGGG | ||||||||
| GAGGATGCC | tgg | C | GGCTTGGG | ||||||||
| 140 | F | GAGGATG | A | TAGGCTTGGG | 2 (40%) | ||||||
| GAGGAT | tt | GGCTTGGG | |||||||||
| GAGGA | ggat | TA | TAGGCTTGGG | ||||||||
| (−13 total) | ag | GGCTTGGG | |||||||||
| 141 | F | GAGGATGCCC | t | TA | TAGGCTTGGG | ||||||
| 7 weeks | 124 | F | GAGGATGCCC | G | tggg | GGCTTGGG | |||||
| GAGGATG | g | A | TAGGCTTGGG | ||||||||
| GAGGATGCCC | ct | TA | TAGGCTTGGG | ||||||||
| 125 | F | GAGGATGCCC | GG | GGCTTGGG | |||||||
| GAGGATG | att | GCTTGGG | |||||||||
| 10 weeks | 143 | M | GAGGATGCCC | tt | CTTGGG | 2 (100%) | |||||
| Older | 6 months | 101 | F | GAGG | TA | TAGGCTTGGG | |||||
| Adult | GAGGATGCCC | agaga | AGGCTTGGG | ||||||||
| 103 | F | GAGGATG | a | TA | TAGGCTTGGG | 14 (88%) | |||||
| GAGGATGC | t | TAGGCTTGGG | 2 (12%) | ||||||||
| 104 | M | GAGGATGC | a | AGGCTTGGG | |||||||
| 7 months | 144 | M | GAGGATGC | t | GGCTTGGG | 2 (29%) | |||||
| GAGGATG | gg | A | TAGGCTTGGG | 2 (29%) | |||||||
| GAGGAT | c | A | TAGGCTTGGG | ||||||||
| GAGGATGC | G | a | CTTGGG | ||||||||
| GAGG | gtc | A | TAGGCTTGGG | ||||||||
Palindromic (P) nucleotides, bold; Non-templated (N) nucleotides, lower case; inverted repeats (Pr), underlined; junctional microhomology nucleotides, bold-underlined. Italicized nucleotides cannot be unequivocally assigned to one coding end.
Table 3.
Sequences of Bcl11b Deletion Coding Junctions in Thymus1
| Group | Age | No. | Sex | 5′ Breakpoint | 5′ P | 5′ Pr | N | 3′ Pr | 3′ P | 3′ Breakpoint | Clones (% of Total) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Germline Sequence | CAGACACACA | TGAGATTAAT | |||||||||
| Fetal | Fetal | --- | --- | CAGACACAC | GAGATTAAT | 2 (50%) | |||||
| CAGACACACA | TAAT | ||||||||||
| CAGACACA | GATTAAT | ||||||||||
| Neonate | 2 days | 116 | M | CAGACAC | G | GAGATTAAT | |||||
| CAGACACACA | TGT | ATTAAT | |||||||||
| CAGACACACA | TG | ggg | GATTAAT | ||||||||
| CAGACACACA | TG | agg | GATTAAT | ||||||||
| CAGACACA | TG | a | ATTAAT | ||||||||
| 117 | F | CAGAC | G | g | C | GAGATTAAT | |||||
| CAGACACAC | C | GATTAAT | |||||||||
| CAGACACAC | G | A | TAAT | ||||||||
| CAGACACAC | G | aa | ATTAAT | ||||||||
| CAGACA | g | GAGATTAAT | |||||||||
| 118 | F | CAGACACAC | GAGATTAAT | ||||||||
| CAGACACACA | T | C | GAGATTAAT | ||||||||
| CAGACACAC | G | ag | AT | ||||||||
| CAGACACACA | A | TAAT | |||||||||
| 119 | F | CAGACACACA | TG | g | T | ATTAAT | 2 (50%) | ||||
| CAGACACAC | cgggg | TTAAT | |||||||||
| CAGACA | T | ||||||||||
| 120 | M | CAGAC | ccc | C | GAGATTAAT | ||||||
| CAGAC | cc | C | GAGATTAAT | ||||||||
| CAGACACACA | TG | gg | GATTAAT | ||||||||
| 1 week | 121 | F | CAGACAC | ctc | C | GAGATTAAT | |||||
| CAGAC | cct | TGAGATTAAT | 2 (40%) | ||||||||
| CAGAC | G | g | GAGATTAAT | ||||||||
| CAGACACACA | AT | ||||||||||
| 122 | F | CAGACAC | cccccc | (−19 total) | |||||||
| CAGACACACA | TG | g | TTAAT | ||||||||
| 123 | F | CAGACACAC | C | GAGATTAAT | |||||||
| CAGACA | g | TTAAT | |||||||||
| CAGACACAC | G | GATTAAT | |||||||||
| CAGACACAC | G | gg | GATTAAT | ||||||||
| CAGACAC | G | TTAAT | |||||||||
| CAGACACAC | G | g | GATTAAT | ||||||||
| CAGACACAC | G | GAGATTAAT | |||||||||
| CAGAC | GAGATTAAT | ||||||||||
| Juvenile | 2 weeks | 111 | M | CAGACAC | TTAAT | 3 (30%) | |||||
| CAGA | aaat | TTAAT | 2 (20%) | ||||||||
| CAGACACA | ac | (−10 total) | |||||||||
| CAGACACACA | T | agg | GATTAAT | ||||||||
| CA | a | AAT | |||||||||
| CAGAC | cccccgg | C | GATTAAT | ||||||||
| CAGACACAC | c | AGATTAAT | |||||||||
| 112 | F | CAGACA | g | TGAGATTAAT | |||||||
| CAGACA | gggg | AT | |||||||||
| (−23 total) | CCC | ctggggg | A | (−10 total) | |||||||
| CAGAC | cctgg | (−10 total) | |||||||||
| CAGACACAC | G | gg | AAT | ||||||||
| CAGACACAC | G | TTAAT | |||||||||
| CAGACACACA | ga | A | TAAT | ||||||||
| 113 | F | GA | GAGATTAAT | 3 (50%) | |||||||
| CAGACAC | cc | AT | ATTAAT | ||||||||
| CAGAC | TC | GATTAAT | |||||||||
| CAGACAC | TCA | TGAGATTAAT | |||||||||
| 3 weeks | 137 | M | CAGACACAC | G | a | A | TGAGATTAAT | ||||
| CAGACACACA | agg | TTAAT | |||||||||
| CAGACACAC | G | gc | TAAT | ||||||||
| CAG | g | T | ATTAAT | ||||||||
| CAGACACACA | TG | GAGATTAAT | |||||||||
| CAGACACA | AT | ||||||||||
| 138 | F | CAGA | AGATTAAT | ||||||||
| CAGACACAC | G | TAAT | |||||||||
| CAGACACACA | TG | a | C | GATTAAT | |||||||
| CAGACACAC | TTAAT | ||||||||||
| Young | 4 weeks | 139 | F | CAGAC | c | AAT | |||||
| Adult | CAGACACAC | G | g | AT | |||||||
| CAGACA | TGAGATTAAT | ||||||||||
| CAGACACAC | G | GATTAAT | |||||||||
| CAGACACAC | G | (−10 total) | |||||||||
| CAGACACACA | TGAGATTAAT | ||||||||||
| 140 | F | CAGACACAC | C | GAGATTAAT | |||||||
| CAGACACAC | G | agggc | TGAGATTAAT | ||||||||
| CAGACACA | AGATTAAT | ||||||||||
| CAGACACAC | cc | ATTAAT | |||||||||
| 7 weeks | 124 | F | CAGACAC | G | g | GA | (−11 total) | ||||
| CAGACAC | G | gg | TTAAT | ||||||||
| CAGACACAC | c | TTAAT | |||||||||
| CAGACAC | G | gg | GATTAAT | ||||||||
| CAGACACAC | G | gggggg | TAAT | ||||||||
| CAGACAC | TTAAT | ||||||||||
| 125 | F | CAGACACAC | G | gg | TTAAT | ||||||
| CAGACACACA | T | tcg | GATTAAT | ||||||||
| CAGACA | aat | A | TGAGATTAAT | ||||||||
| CAGAC | ccg | C | GAGATTAAT | ||||||||
| CAGACACAC | G | ggg | TAAT | ||||||||
| CAGACACACA | TTAAT | ||||||||||
| CAGACACAC | G | gagaggg | TAAT | ||||||||
| CAGACACACA | T | aggg | TAAT | 2 (15%) | |||||||
| CAGACACAC | G | ggggg | T | ATTAAT | |||||||
| CAGACAC | TAAT | ||||||||||
| CAGACACAC | G | gg | AAT | ||||||||
| (−31 total) | cc | T | AT | ||||||||
| CAGA | (−11 total) | ||||||||||
| 10 weeks | 142 | M | CAGACACAC | tgc | AAT | ||||||
| CAGACACACA | GAGATTAAT | ||||||||||
| CAGACAC | G | act | GATTAAT | ||||||||
| 143 | M | CAG | C | ccctcacgg | GAGATTAAT | ||||||
| CAGACA | AGATTAAT | ||||||||||
| CAGACACACA | TGTG | ac | TAAT | ||||||||
| Older | 6 months | 101 | F | CAGACACAC | G | c | (−10 total) | 2 (40%) | |||
| Adult | CAGACACAC | G | AAT | ||||||||
| CAGACAC | tgg | T | |||||||||
| CAGAC | GT | A | TGAGATTAAT | ||||||||
| 102 | F | CAGACACAC | G | gg | A | TGAGATTAAT | |||||
| CAGACAC | G | ggg | TTAAT | 2 (33%) | |||||||
| (−20 total) | gag | A | TAAT | ||||||||
| CAGACACACA | TG | g | TTAAT | ||||||||
| CAGACAC | tcag | GATTAAT | |||||||||
| 103 | F | CAGACAC | G | ggg | AT | ||||||
| CAGACACAC | GT | GATTAAT | |||||||||
| CAGACACAC | TTAAT | ||||||||||
| (−21 total) | cc | TAAT | |||||||||
| 104 | M | (−23 total) | gc | C | GATTAAT | ||||||
| CAGACAC | G | g | GATTAAT | ||||||||
| 105 | M | CAGACACA | GAGATTAAT | ||||||||
| CAGACACAC | G | TTAAT | |||||||||
| CAGACACAC | GATTAAT | ||||||||||
| CAGACAC | G | gt | GATTAAT | ||||||||
| CAGACAC | ccc | CA | TGAGATTAAT | ||||||||
| CAGAC | ca | A | TAAT | ||||||||
| CAGACACAC | cc | CT | AGATTAAT | ||||||||
| CAGAC | G | gc | (−10 total) | 2 (14%) | |||||||
| CAGACACAC | tcaa | AAT | |||||||||
| CAGACACAC | GATTAAT | ||||||||||
| CAGACACAC | c | AAT | |||||||||
| CAGACAC | t | TGAGATTAAT | |||||||||
| CAGACACAC | cggc | TAAT | |||||||||
| 106 | M | C | tgggg | TAAT | |||||||
| 107 | M | CAGACA | TAAT | ||||||||
| CAGACAC | G | GAGATTAAT | |||||||||
| CAGACACAC | G T | C | GATTAAT | ||||||||
| CAGAC | CT | AGATTAAT | |||||||||
| CAGACAC | cctctc | T | ATTAAT | ||||||||
| CAGACACAC | G | gcggg | TAAT | 2 (22%) | |||||||
| CAGACA | TAAT | ||||||||||
| CAGACAC | G | GATTAAT | |||||||||
| 144 | M | CAGACACAC | G | ag | TAAT | ||||||
| 7 months | (−17 total) | TGAGATTAAT | |||||||||
| CAGACAC | tcaata | AGATTAAT | |||||||||
| 145 | M | CAGACACACA | cggg | AT | |||||||
| CAGACACAC | C | GAGATTAAT | |||||||||
| CAGACAC | G | GAGATTAAT | |||||||||
Palindromic (P) nucleotides, bold; Non-templated (N) nucleotides, lower case; inverted repeats (Pr), underlined; junctional microhomology nucleotides, bold-underlined. Italicized nucleotides cannot be unequivocally assigned to one coding end.
Table 4.
Analysis of Notch1 Deletion Coding Junctions in Thymus
| Deletion Frequency | Fetal | Neonate | Juvenile | Young Adult | Older Adult | Malee | Femalee | All |
|---|---|---|---|---|---|---|---|---|
| No. of mice analyzed | 7 (pool) | 8 | 6 | 7 | 8 | 11 | 18 | 29e |
| No. of mice with detected deletions | --- | 6 (75%) | 4 (67%) | 6 (86%) | 3 (38%) | 5 (45%) | 13 (72%) | 19 (65%)e |
| No. of unique deletions sequenced | 1 | 12 | 10 | 15 | 10 | 11 | 36 | 48 |
| Avg. deletions frequency/106 cells | 0.6 [---] | 2.8 [1.1] | 2.4 [0.6] | 2.5 [0.6] | 4.1 [2.7] | 2.3 [1.2] | 3.0 [0.5] | 2.8 [0.5]e |
| Palindromic (P) nucleotides f | ||||||||
| At full length 5′ coding ends (%)a | --- | 60 | 75 | 57 | 0 | 25 | 69 | 59 |
| At full length 3′ coding ends (%)a | 0 | 100 | 67 | 100 | 83 | 100 | 81 | 81 |
| At all full length coding ends (%)a | 0 | 75 | 70 | 75 | 71 | 63 | 76 | 71 |
| In junctions with full length ends (%)a | 0 | 100 | 70 | 90 | 71 | 71 | 85 | 79 |
| At 5′ coding ends (%) | 0 | 27 | 30 | 27 | 0 | 10 | 25 | 21 |
| At 3′ coding ends (%) | 0 | 27 | 40 | 33 | 50 | 40 | 36 | 36 |
| At all coding ends (%) | 0 | 27 | 35 | 30 | 25 | 25 | 31 | 29 |
| At all coding junctions (%) | 0 | 55 | 70 | 60 | 50 | 50 | 61 | 57 |
| Avg. no. at 5′ coding endsa | --- | 1.7 [0.7] | 2.0 [0.6] | 1.3 [0.3] | --- | 3.0 [---] | 1.4 [0.2] | 1.6 [0.3] |
| Avg. no. at 3′ coding endsa | --- | 1.7 [0.3] | 2.0 [0.7] | 1.6 [0.2] | 1.4 [0.2] | 1.0 [0.0] | 1.8 [0.2] | 1.6 [0.2] |
| Avg. no. at coding endsa | --- | 1.7 [0.3] | 2.0 [0.4] | 1.4 [0.2] | 1.4 [0.2] | 1.4 [0.4] | 1.7 [0.2] | 1.6 [0.2] |
| Avg. no. in deletion junctionsa | --- | 1.7 [0.3] | 2.0 [0.4] | 1.4 [0.2] | 1.4 [0.2] | 1.4 [0.4] | 1.7 [0.2] | 1.6 [0.2] |
| Non-templated (N) nucleotides f | ||||||||
| In deletion junctions (%) | 0 | 36 | 60 | 80 | 100 | 80 | 67 | 68 |
| Avg. no. presentb | --- | 1.5 [0.3] | 2.3 [0.5] | 2.4 [0.3] | 2.0 [0.4] | 1.8 [0.3] | 2.3 [0.3] | 2.2 [0.2] |
| GC content (%)b | --- | 63 [24] | 100 [0] | 42 [10] | 42 [13] | 52 [16] | 57 [8] | 55 [7] |
| Inverted Repeats (Pr) f | ||||||||
| At recessed 5′ coding ends (%)c | 0 | 33 | 17 | 0 | 11 | 17 | 13 | 13 |
| At recessed 3′ coding ends (%)c | --- | 0 | 25 | 10 | 0 | 0 | 10 | 8 |
| At all recessed coding ends (%)c | 0 | 14 | 20 | 6 | 8 | 8 | 12 | 11 |
| In junctions with recessed ends (%)c | 0 | 22 | 20 | 8 | 10 | 11 | 15 | 14 |
| At 5′ coding ends (%) | 0 | 18 | 10 | 0 | 10 | 10 | 8 | 9 |
| At 3′ coding ends (%) | 0 | 0 | 10 | 7 | 0 | 0 | 6 | 4 |
| At all coding ends (%) | 0 | 9 | 10 | 3 | 5 | 5 | 7 | 6 |
| In all deletion junctions (%) | 0 | 18 | 20 | 7 | 10 | 10 | 14 | 13 |
| Avg. no. at 5′ coding endsc | --- | 1.0 [0.0] | 1.0 [---] | --- | 1.0 [---] | 1.0 [---] | 1.0 [0.0] | 1.0 [0.0] |
| Avg. no. at 3′ coding endsc | --- | --- | 1.0 [---] | 1.0 [---] | --- | --- | 1.0 [0.0] | 1.0 [0.0] |
| Avg. no. at coding endsc | --- | 1.0 [0.0] | 1.0 [0.0] | 1.0 [---] | 1.0 [---] | 1.0 [---] | 1.0 [0.0] | 1.0 [0.0] |
| Avg. no. in deletion junctionsc | --- | 1.0 [0.0] | 1.0 [0.0] | 1.0 [---] | 1.0 [---] | 1.0 [---] | 1.0 [0.0] | 1.0 [0.0] |
| Nucleolytic Processing f | ||||||||
| At 5′ coding ends (%) | 100 | 55 | 60 | 53 | 90 | 60 | 64 | 64 |
| At 3′ coding ends (%) | 0 | 73 | 40 | 67 | 40 | 60 | 56 | 55 |
| At all coding ends (%) | 50 | 64 | 50 | 60 | 65 | 60 | 60 | 60 |
| In deletion junctions (%) | 100 | 82 | 100 | 87 | 100 | 90 | 92 | 91 |
| At both coding ends in junctions (%) | 0 | 45 | 0 | 33 | 30 | 30 | 28 | 28 |
| Junctions with no processing (%) | 0 | 18 | 0 | 13 | 0 | 10 | 8 | 9 |
| Avg. nuc. loss at 5′ coding endsd | 2.0 [---] | 1.8 [0.3] | 4.2 [0.7] | 4.3 [1.3] | 3.3 [0.6] | 3.2 [0.7] | 3.5 [0.5] | 3.4 [0.4] |
| Avg. nuc. loss at 3′ coding endsd | --- | 1.6 [0.2] | 3.0 [1.4] | 2.3 [0.2] | 2.0 [0.7] | 2.3 [0.6] | 2.1 [0.3] | 2.2 [0.2] |
| Avg. nuc. loss at coding endsd | 2.0 [---] | 1.7 [0.2] | 3.7 [0.7] | 3.2 [0.6] | 2.9 [0.5] | 2.8 [0.4] | 2.9 [0.3] | 2.8 [0.3] |
| Avg. nuc. loss in deletion junctionsd | 2.0 [---] | 2.7 [0.4] | 3.7 [0.7] | 4.4 [1.0] | 3.8 [0.6] | 3.7 [0.6] | 3.7 [0.5] | 3.7 [0.4] |
1 Dashes indicate no data or not calculable; brackets indicate standard error.
Calculated from full-length coding ends or deletion junctions with palindromic (P) nucleotide insertions.
Calculated from deletions junctions with non-templated (N) nucleotide insertions; includes inverted repeat nucleotides (Pr).
Calculated from recessed coding ends or deletion junctions with inverted repeat (Pr) nucleotide insertions.
Calculated from coding ends or deletion junctions with nucleolytic processing of germline coding ends.
Does not include pooled fetal samples.
Excludes junctions with microhomology nucleotides.
Table 5.
Analysis of Bcl11b Deletion Coding Junctions in Thymus
| Deletion Frequency | Fetal | Neonate | Juvenile | Young Adult | Older Adult | Malee | Femalee | All |
|---|---|---|---|---|---|---|---|---|
| No. of mice analyzed | 7 (pool) | 8 | 6 | 7 | 8 | 11 | 18 | 29e |
| No. of mice with detected deletions | --- | 8 (100%) | 6 (100%) | 7 (100%) | 8 (100%) | 11 (100%) | 18 (100%) | 29 (100%)e |
| No. of unique deletions sequenced | 3 | 34 | 28 | 35 | 43 | 57 | 83 | 143 |
| Avg. deletions frequency/106 cells | 5.2 [---] | 12 [2] | 28 [6] | 23 [5] | 32 [9] | 30 [6] | 20 [3] | 23 [4]e |
| Palindromic (P) nucleotides f | ||||||||
| At full length 5′ coding ends (%)a | 0 | 88 | 60 | 60 | 50 | 70 | 70 | 67 |
| At full length 3′ coding ends (%)a | --- | 0 | 67 | 25 | 60 | 50 | 44 | 46 |
| At all full length coding ends (%)a | 0 | 78 | 63 | 44 | 57 | 64 | 58 | 59 |
| In junctions with full length ends (%)a | 0 | 78 | 63 | 50 | 57 | 64 | 61 | 61 |
| At 5′ coding ends (%) | 0 | 22 | 11 | 9 | 2 | 13 | 9 | 10 |
| At 3′ coding ends (%) | 0 | 0 | 7 | 3 | 7 | 4 | 5 | 4 |
| At all coding ends (%) | 0 | 11 | 9 | 6 | 5 | 8 | 7 | 7 |
| In all deletion junctions (%) | 0 | 22 | 19 | 12 | 9 | 16 | 14 | 14 |
| Avg. no. at 5′ coding endsa | --- | 2.0 [0.2] | 1.7 [0.3] | 2.0 [1.0] | 2.0 [---] | 2.3 [0.4] | 1.6 [0.2] | 1.9 [0.2] |
| Avg. no. at 3′ coding endsa | --- | --- | 2.0 [1.0] | 1.0 [---] | 1.3 [0.3] | 1.5 [0.5] | 1.5 [0.5] | 1.5 [0.3] |
| Avg. no. at coding endsa | --- | 2.0 [0.2] | 1.8 [0.4] | 1.8 [0.8] | 1.5 [0.3] | 2.1 [0.3] | 1.5 [0.2] | 1.8 [0.2] |
| Avg. no. in deletion junctionsa | --- | 2.0 [0.2] | 1.8 [0.4] | 1.8 [0.8] | 1.5 [0.3] | 2.1 [0.3] | 1.5 [0.2] | 1.8 [0.2] |
| Non-templated (N) nucleotides f | ||||||||
| In deletion junctions (%) | 0 | 91 | 78 | 76 | 84 | 80 | 84 | 81 |
| Avg. no. presentb | --- | 2.3 [0.2] | 3.1 [0.5] | 3.6 [0.5] | 2.9 [0.3] | 3.1 [0.3] | 2.9 [0.2] | 2.9 [0.2] |
| GC content (%)b | --- | 84 [5] | 68 [7] | 84 [5] | 78 [5] | 73 [5] | 83 [3] | 79 [3] |
| Inverted Repeats (Pr) f | ||||||||
| At recessed 5′ coding ends (%)c | 0 | 50 | 27 | 52 | 39 | 33 | 49 | 42 |
| At recessed 3′ coding ends (%)c | 0 | 32 | 29 | 17 | 18 | 19 | 27 | 23 |
| At all recessed coding ends (%)c | 0 | 40 | 28 | 34 | 29 | 26 | 38 | 32 |
| In junctions with recessed ends (%)c | 0 | 63 | 44 | 55 | 53 | 45 | 61 | 53 |
| At 5′ coding ends (%) | 0 | 38 | 22 | 44 | 37 | 27 | 43 | 35 |
| At 3′ coding ends (%) | 0 | 31 | 26 | 15 | 16 | 18 | 24 | 21 |
| At all coding ends (%) | 0 | 34 | 24 | 29 | 27 | 22 | 33 | 28 |
| In all deletion junctions (%) | 0 | 63 | 44 | 55 | 53 | 45 | 61 | 53 |
| Avg. no. at 5′ coding endsc | --- | 1.1 [0.1] | 1.3 [0.3] | 1.0 [0.0] | 1.1 [0.1] | 1.1 [0.1] | 1.1 [0.1] | 1.1 [0.1] |
| Avg. no. at 3′ coding endsc | --- | 1.0 [0.0] | 1.3 [0.2] | 1.2 [0.2] | 1.3 [0.2] | 1.2 [0.1] | 1.2 [0.1] | 1.2 [0.1] |
| Avg. no. at coding endsc | --- | 1.0 [0.0] | 1.3 [0.2] | 1.1 [0.1] | 1.2 [0.1] | 1.1 [0.1] | 1.1 [0.1] | 1.1 [0.0] |
| Avg. no. in deletion junctionsc | --- | 1.2 [0.1] | 1.4 [0.3] | 1.2 [0.1] | 1.2 [0.1] | 1.1 [0.1] | 1.3 [0.1] | 1.2 [0.1] |
| Nucleolytic Processing f | ||||||||
| At 5′ coding ends (%) | 50 | 75 | 81 | 85 | 95 | 82 | 88 | 85 |
| At 3′ coding ends (%) | 100 | 97 | 89 | 88 | 88 | 93 | 89 | 91 |
| At all coding ends (%) | 75 | 86 | 85 | 87 | 92 | 88 | 88 | 88 |
| In deletion junctions (%) | 100 | 100 | 100 | 97 | 100 | 100 | 99 | 99 |
| At both coding ends in junctions (%) | 50 | 72 | 70 | 76 | 84 | 75 | 78 | 76 |
| Junctions with no processing (%) | 0 | 0 | 0 | 3 | 0 | 0 | 1 | 1 |
| Avg. nuc. loss at 5′ coding endsd | 1.0 [---] | 2.6 [0.4] | 4.8 [1.1] | 3.4 [1.0] | 4.3 [0.9] | 3.8 [0.6] | 3.8 [0.6] | 3.8 [0.5] |
| Avg. nuc. loss at 3′ coding endsd | 3.5 [2.5] | 3.5 [0.6] | 5.0 [0.5] | 5.1 [0.5] | 4.8 [0.4] | 3.9 [0.3] | 5.0 [0.4] | 4.6 [0.3] |
| Avg. nuc. loss at coding endsd | 2.7 [1.7] | 3.1 [0.4] | 4.9 [0.6] | 4.3 [0.6] | 4.5 [0.5] | 3.9 [0.3] | 4.4 [0.4] | 4.2 [0.3] |
| Avg. nuc. loss in deletion junctionsd | 4.0 [2.0] | 5.3 [0.6] | 8.4 [1.2] | 7.7 [1.1] | 8.3 [0.9] | 6.8 [0.6] | 7.9 [0.7] | 7.4 [0.5] |
1 Dashes indicate no data or not calculable; brackets indicate standard error.
Calculated from full-length coding ends or deletion junctions with palindromic (P) nucleotide insertions.
Calculated from deletions junctions with non-templated (N) nucleotide insertions; includes inverted repeat nucleotides (Pr).
Calculated from recessed coding ends or deletion junctions with inverted repeat (Pr) nucleotide insertions.
Calculated from coding ends or deletion junctions with nucleolytic processing of germline coding ends.
Does not include pooled fetal samples.
Excludes junctions with microhomology nucleotides.
For deletion frequency, the average and standard error for all age groups except fetal was calculated using replicate assays from several individual mice with detected deletions. Fetal samples could not be included in the statistical analysis of deletion frequency due to the pooled nature of the sample. However, the fetal results for each gene represent ~½ of all thymic DNA obtained, and the frequency of Bcl11b deletions appears to increase between fetal and juvenile stages, while Notch1 is similar at all postnatal ages (Fig. 1, Tables 4 and 5).
For all other age groups and both sexes, the average frequency of deletions per 106 cells was not significantly different, but overall was much higher for Bcl11b than Notch1 (23±4 and 2.8±0.5, respectively; p<0.0001; Fig. 1, Tables 4 and 5). Assuming 108 thymocytes per young adult mouse thymus, this yields roughly 280 Notch1 and 2,300 Bcl11b deletions, consistent with previous findings in other mouse strains [31, 32]. While deletion frequency does not appear to significantly increase in older animals, deletions are initiating by day 16 of fetal development and are present through at least 7 months of age (Fig. 1, Tables 4 and 5).
For Notch1, some mice did not contain detectable deletions, and one seemingly unhealthy mouse (#103) exhibited excessive hair loss, and displayed a greatly increased frequency of clonal Notch1 deletions compared to all other mice (Table 2), and neither were included in the frequency analysis.
Clonal populations of deletions were detected in thymus for both genes (Tables 2 and 3), and the percent clones among unique deletion sequences was higher for Notch1 than Bcl11b (23% vs. 8%). This contrasts dramatically with earlier studies in other mouse strains which indicated 75% clonality for Notch1 and no clonality for Bcl11b [31].
3.2 Cryptic Recombination Signal Sequences near deletion breakpoints
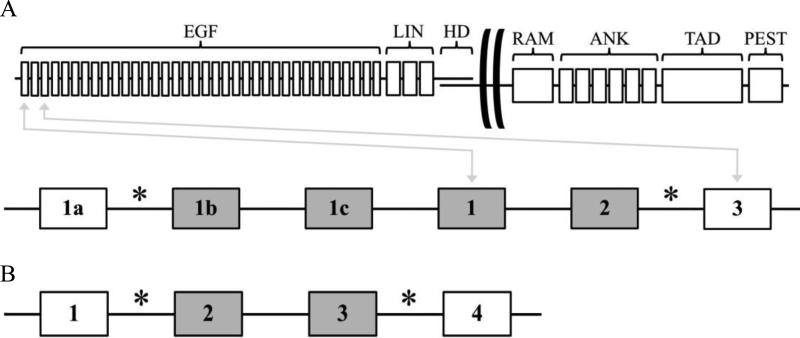
Using Recombination Information Content (RIC) analysis, deletion signal ends were examined for potential cryptic Recombination Signal Sequences (cRSSs) [54]. For Notch1, deletion breakpoints were found at positions 4926 and 16674, resulting in removal of exons 1b through 2 (Fig. 2A). For Bcl11b, removal of exons 2 and 3 were due to breakpoints at positions 9856 and 74128 (Fig. 2B). Breakpoints detected for both genes are consistent with those previously observed in other mouse strains [31, 32].
Fig. 2.
Domain/intron/exon structure and illegitimate V(D)J deletion breakpoint locations. (A) Notch1. (B) Bcl11b. Asterisks denote breakpoints. Exon numbers are indicated. Deleted exons are shown in grey. For Notch1, exons 1a, 1b, and 1c result from alternative transcriptional start site usage.
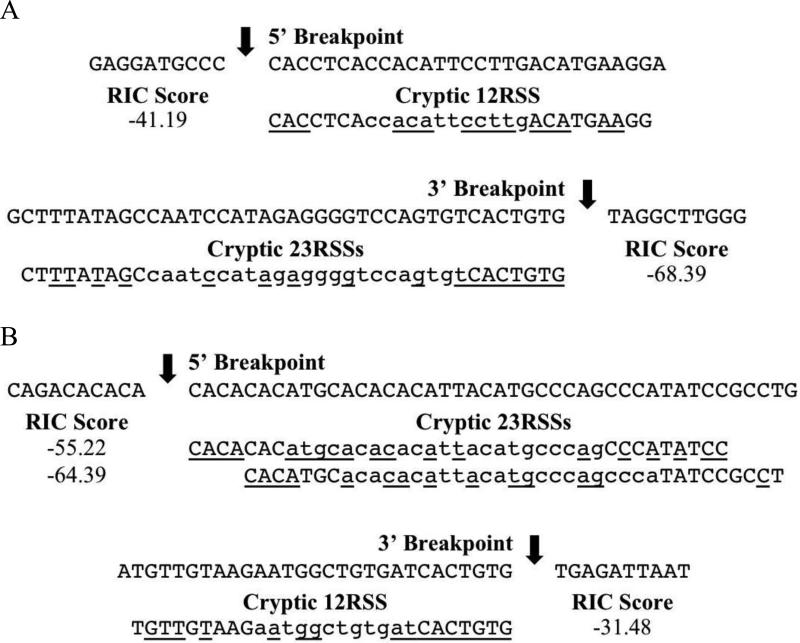
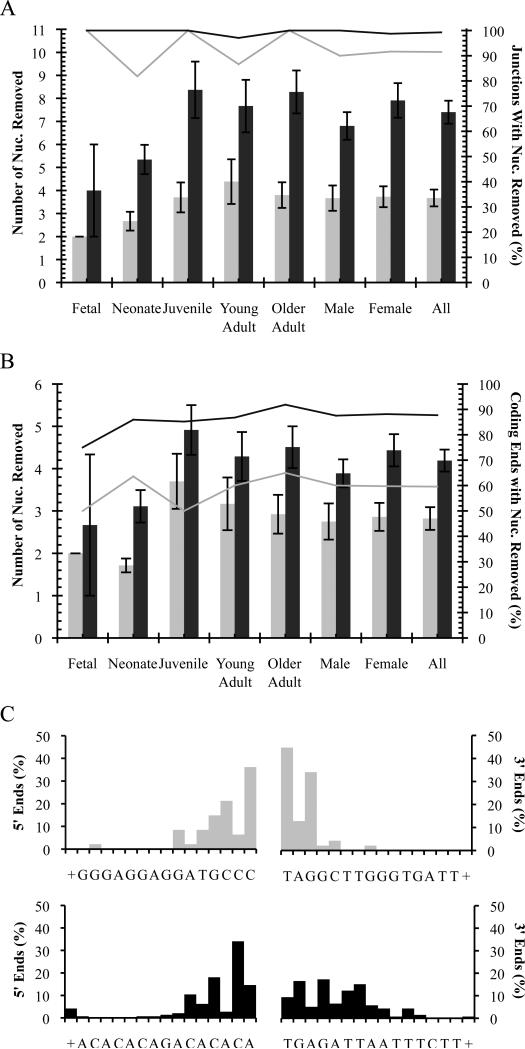
For Notch1, only one plausible set of cRSSs was detected (Fig. 3A). Both were found directly adjacent to the longest deletion coding ends identified and matched those previously observed [31]. The cryptic 12RSS at the 5’ breakpoint is 54% similar to the consensus RSS, and the cryptic 23RSS at the 3’ breakpoint is 44% similar (Fig. 3A). While the cryptic 23RSS has a consensus heptamer, the cryptic 12RSS does not and, interestingly, contains a cytosine at the 4th position of the heptamer shown to destabilize the postcleavage complex and reduce coding joint formation [56], and this may partly explain the lower frequency of deletions detected for Notch1.
Fig. 3.
Cryptic Recombination Signal Sequences (cRSSs) near illegitimate V(D)J deletion breakpoints. (A) Notch1. (B) Bcl11b. Nucleotides are depicted 5’→3’ on the coding strand. Arrows indicate deletion breakpoints. Underlined nucleotides match those of consensus RSSs. RIC scores were calculated using the RSSsite web server [54].
For Bcl11b, two cryptic 23RSSs were found near the 5’ breakpoint with a single cryptic 12RSS partner near the 3’ breakpoint (Fig. 3B). The first cryptic 23RSS is more likely the functional cRSS however, as it lies adjacent to the longest detected 5’ coding end, has the higher RIC score (−55.22), and is a better match to the consensus RSS (49% similarity vs. 36%; Fig. 3B). While the cryptic 23RSS does not contain a consensus heptamer, there are several conserved spacer residues adjacent to the heptamer element that may compensate for lower recombination efficiency due to the lack of consensus nucleotides in positions 5-7 of the heptamer (Fig. 3B) [57]. The cryptic 12RSS at the 3’ breakpoint is 57% similar to consensus, has a high RIC score compared to many immune 12RSSs, and contains a previously identified consensus heptamer [32, 55].
The RIC scores for the cRSSs of Bcl11b are higher than those of Notch1 and many immune RSSs, and this may be a factor in the higher deletion frequency detected for Bcl11b as RIC analysis can effectively predict recombination efficiencies of murine RSSs [25].
3.3 Palindromic (P) nucleotides in deletion coding junctions
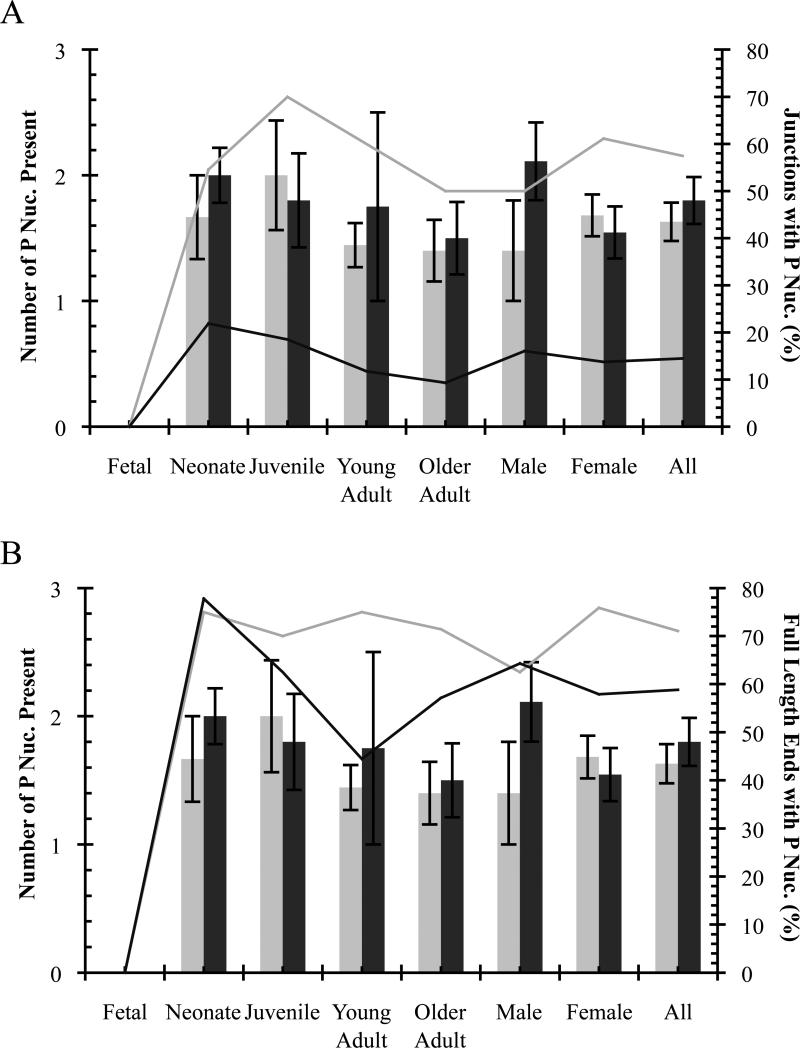
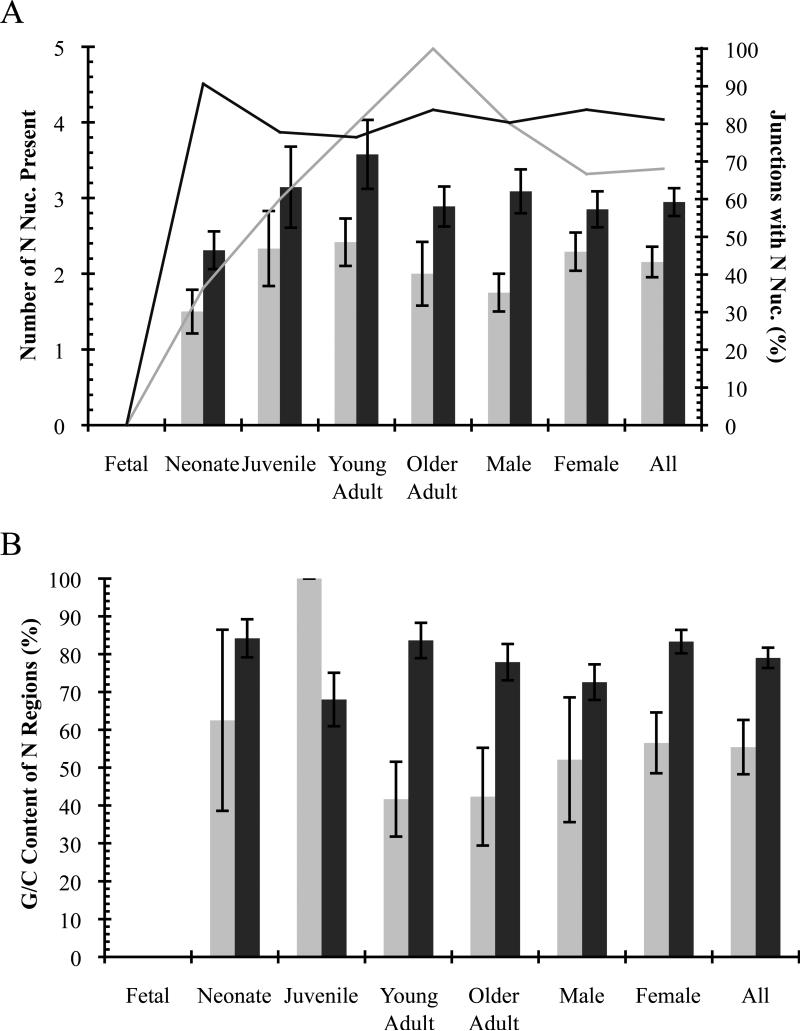
Palindromic (P) nucleotides were found in deletion junctions of both Notch1 and Bcl11b at all ages except fetal (Fig. 4, Tables 2, 3, 4, and 5). The average number present in deletion junctions is not significantly different for any age group or sex, nor overall for Notch1 compared to Bcl11b (1.6±0.2 and 1.8±0.2, respectively; Fig. 4A, Tables 4 and 5). The percentage of all deletion junctions containing P nucleotides is higher for Notch1 than Bcl11b (57% vs. 14%; Fig. 4A, Tables 4 and 5). Full-length coding ends frequently contained P nucleotides with Notch1 ends having slightly more than Bcl11b (71% vs. 59%; Fig. 4B, Tables 4 and 5). The percentage of deletion junctions and full-length ends with P nucleotides is similar for both sexes and all groups except fetal (Fig. 4, Tables 4 and 5). Deletion junctions of fetal mice did not contain P nucleotides, but only a small number of sequences were available for analysis due to low frequency detection of deletions in fetal animals. Of note is that P nucleotides were only found at one coding end in all deletion junctions examined (i.e., either at 5’ or 3’ ends, but not both; Tables 2 and 3).
Fig. 4.
Palindromic (P) nucleotides detected in unique illegitimate V(D)J deletions. Notch1, grey; Bcl11b, black. (A) Deletion coding junctions. (B) Full-length coding ends. Average number of P nucleotides ± SE indicated by columns. Percentage of sequences containing P nucleotides indicated by lines.
3.4 Non-templated (N) nucleotides in deletion coding junctions
Non-templated (N) nucleotides were found in deletion junctions of both Notch1 and Bcl11b at all ages except fetal (Fig. 5A, Tables 2, 3, 4, and 5). The average number present is not significantly different for any age or sex, but is slightly greater for Bcl11b than Notch1 (2.9±0.2 vs. 2.2±0.2; p=0.05; Fig. 5A, Tables 4 and 5). The percentage of deletion junctions containing N nucleotides rises with increasing age for Notch1 but is similar from neonates to older adults for Bcl11b (Fig. 5A, Tables 4 and 5). The overall percentage of deletion junctions with N nucleotides is also higher for Bcl11b than Notch1 (81% vs. 68%; Fig. 5A, Tables 4 and 5).
Fig. 5.
Non-templated (N) nucleotides detected in unique illegitimate V(D)J deletions. Notch1, grey; Bcl11b, black. (A) Average number of N nucleotides ± SE in deletion coding junctions indicated by columns. Percentage of deletion junctions containing N nucleotides indicated by lines. (B) G/C percentage of non-templated (N) nucleotides. Average percentage of G/C nucleotides ± SE in junctions is indicated.
Non-templated nucleotides are inserted by terminal deoxynucleotidyl transferase (TdT), and previous studies of its activity showed that insertion of nucleotides into coding junctions is not random [49, 50, 53]. A preference for G nucleotide insertions was observed during V(D)J recombination of plasmid substrates and in vivo, thus G/C nucleotides are present in coding junctions at higher levels than A/T nucleotides [50, 53]. For Notch1, the percentage of G/C nucleotides was similar for all groups except juvenile, but for Bcl11b all groups were similar (Fig. 5B, Tables 4 and 5). Overall, the G/C content of deletion junctions was higher for Bcl11b than Notch1 (79% vs. 55%; Fig. 5B, Tables 4 and 5).
TdT has also been shown to insert homo-dinucleotide pairs of purines and pyrimidines (RR or YY) at higher frequencies than expected by random insertion of nucleotides and hetero-dinucleotide pairs (RY or YR) at lower than expected levels [53]. Expected frequencies are calculated from the percentage of each nucleotide relative to all detected N nucleotides and the total number of possible dinucleotide pairs (each N region length-1, summed). The product of the percentages of each nucleotide of a particular pair is then multiplied by the total possible pairs. For example, G nucleotides represent 50% of N nucleotides in all Bcl11b deletion junctions, and there are 218 possible dinucleotides. Thus, a GG dinucleotide pair is expected 55 times (0.50*0.50*218). Expected numbers were compared to the observed number of dinucleotide pairs using a Chi square goodness-of-fit test (df=1).
For Notch1, no dinucleotide pairs were present at significantly higher or lower than expected values (data not shown), likely due to the small number of short N regions observed. For Bcl11b, results were consistent with normal TdT activity as four RR/YY dinucleotide pairs were found at significantly higher than expected frequencies (GG, CC, CT, AA), while two RY/YR pairs were found at significantly lower frequencies (GC, CG). All other pairs were not significantly different than expected by random insertion (Table 6).
Table 6.
Dinucleotide Analysis of Non-Templated (N) Nucleotides in Bcl11b Deletion Junctions
| Dinucleotide | No. Observed | No. Expecteda | P-valueb |
|---|---|---|---|
| GG (RR) | 80 | 55 | < 0.001H |
| AA (RR) | 8 | 3 | 0.004H |
| GA (RR) | 15 | 14 | 0.782 |
| AG (RR) | 11 | 14 | 0.407 |
| CC (YY) | 34 | 17 | < 0.001H |
| TT (YY) | 0 | 2 | 0.155 |
| CT (YY) | 12 | 6 | 0.013H |
| TC (YY) | 10 | 6 | 0.098 |
| GC (RY) | 11 | 30 | < 0.001L |
| GT (RY) | 7 | 11 | 0.216 |
| AT (RY) | 4 | 3 | 0.561 |
| AC (RY) | 5 | 7 | 0.442 |
| CG (YR) | 8 | 30 | < 0.001L |
| CA (YR) | 6 | 7 | 0.701 |
| TG (YR) | 6 | 11 | 0.122 |
| TA (YR) | 1 | 3 | 0.245 |
Calculated from the percentage of each nucleotide relative to all N nucleotides in deletion junctions (G=0.500, C=0.276, T=0.100, A=0.124) and the number of possible dinucleotide pairs (n=218).
Calculated with a Chi square goodness of fit test (df=1).
Significantly higher than expected by random insertion.
Significantly lower than expected by random insertion.
3.5 Inverted repeat nucleotides at recessed ends (Pr) in deletion coding junctions
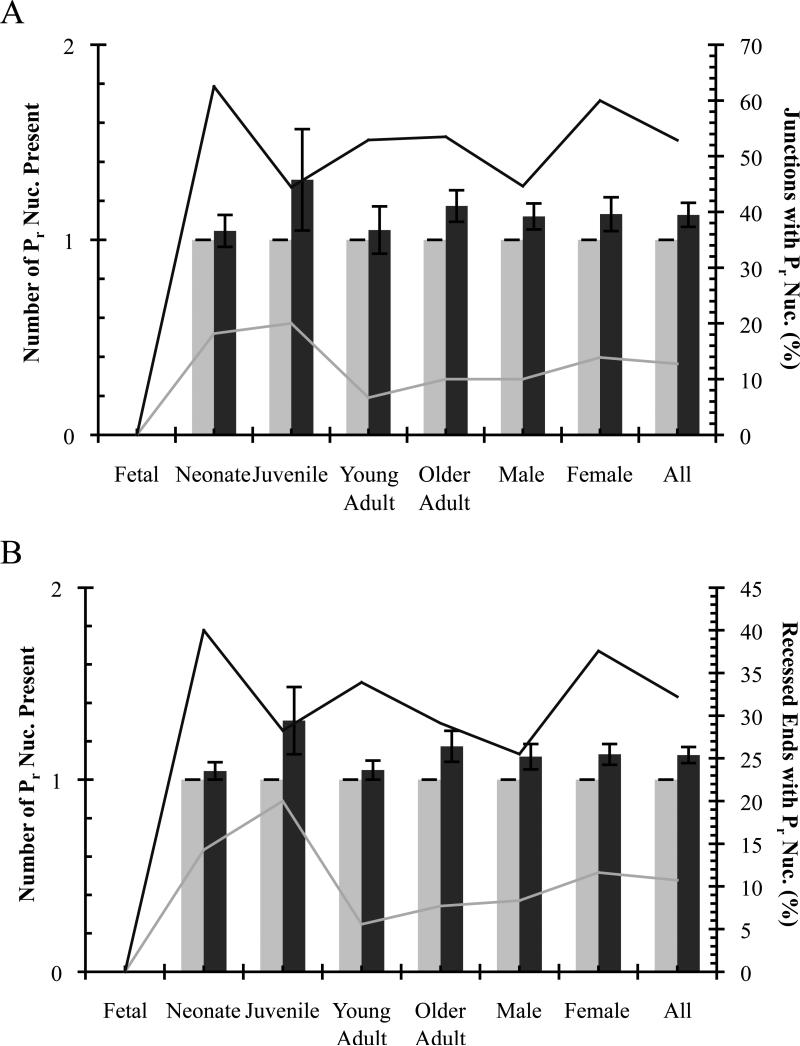
Previously, a novel subset of non-templated (N) nucleotides palindromic to recessed coding ends was described [14, 53]. These inverted repeat nucleotides or “recessed palindromes” (Pr), which ranged from ~1-4 nucleotides in length, were found in V(D)J junctions of plasmid recombination substrates and at some sites of illegitimate V(D)J recombination in vivo (e.g., HPRT1 locus) [49, 53] but not in authentic V(D)J junctions at the antigen receptor loci [58]. Insertion of these nucleotides required TdT, but statistically could not be fully explained by its normal activity. A proposed model for Pr formation involves Artemis processing of stem loops or hairpins formed by complementary N nucleotide addition [12, 14, 50].
We analyzed potential Pr nucleotides in Notch1 and Bcl11b deletion junctions and found their presence to be consistent with normal TdT activity rather than other mechanisms proposed. Pr nucleotides in deletion junctions and at recessed coding ends averaged ~1 base pair, typically were G/C nucleotides, and these did not vary for any age group or sex, nor for Notch1 compared to Bcl11b (Fig. 6, Tables 2, 3, 4, and 5). While the overall percentage of deletion junctions and recessed ends containing Pr nucleotides is higher for Bcl11b than Notch1 (Fig. 6, Tables 4 and 5), this is most likely due to a higher rate of N insertion plus more recessed coding ends for Bcl11b (see below). Pr nucleotides were found associated with the 5’ and 3’ coding ends of each gene and on both ends of some Bcl11b deletion junctions (Tables 2 and 3).
Fig. 6.
Inverted repeat (Pr) nucleotides at recessed coding ends detected in unique illegitimate V(D)J deletions. Notch1, grey; Bcl11b, black. (A) Deletion coding junctions. (B) Recessed coding ends. Average number of Pr nucleotides ± SE indicated by columns. Percentage of sequences containing Pr nucleotides indicated by lines.
3.6 Nucleolytic processing of deletion coding ends
Nucleolytic processing of coding ends was found in deletion junctions of both Notch1 and Bcl11b at all ages examined (Figs. 7A and 7B, Tables 2, 3, 4, and 5). The average number of excised nucleotides was not significantly different for any age group or sex, but overall was more extensive in deletion junctions of Bcl11b compared to Notch1 (7.4±0.5 vs. 3.7±0.4; p<0.0001; Fig. 7A, Tables 4 and 5). For Notch1 deletions, the average number of excised nucleotides was significantly higher for the 5’ coding end compared to the 3’ end (3.4±0.4 vs. 2.2±0.2; p=0.03; Fig. 7C, Table 4).
Fig. 7.
Germline nucleotide loss detected in unique illegitimate V(D)J deletions. Notch1, grey; Bcl11b, black. (A) Deletion coding junctions. (B) Deletion coding ends. In (A) and (B), average number nucleotides removed ± SE indicated by columns. Percentage of sequences with nucleotides removed indicated by lines. Fetal samples are not included in Male and Female groups. (C) Specific nucleotide loss profiles of coding ends of deletion junctions. Percentage of coding ends terminating at a particular germline nucleotide is indicated. Germline nucleotides are depicted 5’–3’ on the coding strand. (+) indicates losses of greater than or equal to 16 nucleotides. Sequences with microhomology nucleotides that cannot be unequivocally assigned to a single coding end are excluded.
While the percentage of all deletion junctions with processing was similar for both genes (91% for Notch1 and 99% for Bcl11b; Fig. 7A, Tables 4 and 5), the percentage of all coding ends with processing was higher for Bcl11b than Notch1 (88% vs. 60%; Fig. 7B, Tables 4 and 5). Most Notch1 deletion junctions contained one full-length and one processed coding end, while both coding ends of most Bcl11b junctions were processed (Tables 2 and 3). These differences in processing are likely due to sequence variations as nucleolytic processing (e.g., by Artemis) is affected by the coding end sequence [13, 53, 59].
3.7 Microhomology mediated end joining usage in deletion coding junctions
In the absence of TdT activity and in deficiencies of cNHEJ, as seen in severe combined immunodeficiency (SCID) mice, microhomology mediated end joining (MMEJ) can be used as an alternative pathway to cNHEJ for double-strand DNA break repair and ligation of DNA ends formed during V(D)J recombination [60]. The use of MMEJ during coding end processing in human peripheral T cells was previously found to be higher for TCRβ junctions with no inserted N nucleotides than at the nonimmune HPRT1 locus in the same sample population [49, 50]. Both Notch1 and Bcl11b contain multiple homologous sites of one or two base pairs near breakpoints, and Notch1 contains several three base pair sites (Tables 2 and 3).
For Notch1, 16 of 48 deletion junctions sequenced did not contain N nucleotides, and one (6%) contained a single base microhomology (Table 2). Likewise for Bcl11b, 31 of 143 deletion junctions contained no N nucleotides, and five (16%) contained a single base microhomology (Table 3). Therefore MMEJ use, while possible, appears to be minimal for both genes in wild-type mice at these loci and was observed previously only for Notch1 deletions in SCID mice with radiation-induced thymic lymphomas [31]. These results support the ideas that junctions formed by MMEJ at immune loci may be preferentially selected while those at nonimmune loci are not or that MMEJ use is less frequent at nonimmune loci.
4. Discussion
We analyzed the frequency, clonality, and junctional characteristics of illegitimate V(D)J recombination-mediated deletion events in Notch1 and Bcl11b in thymocytes at various stages of mouse development. Unexpectedly, we found no statistically significant age- or sex-specific differences, contrasting with previous studies of illegitimate deletions at the human HPRT1 locus and bona fide recombination at the TCRβ locus [49, 50].
While no age- or sex-specific differences in deletion coding end processing in Notch1 and Bcl11b were apparent, gene-specific differences were observed. The number of palindromic (P) nucleotides found in both genes was consistent with V(D)J recombination at the TCRβ locus (~2bp in length) [50, 61], but the percentage of full-length coding ends and all deletion junctions containing P nucleotides was higher for Notch1 deletions than Bcl11b. This is likely explained by Notch1 and Bcl11b coding end differences, which can influence hairpin nicking efficiency and subsequent P nucleotide insertions [13, 59, 61].
Recently, an analysis of P nucleotides at the TCRβlocus in human peripheral T cells showed a greater level of P nucleotide addition in females and especially newborns suggesting a possible selective influence [50]. Another recent deep sequencing study uncovered differences in P nucleotide addition in functional vs. nonfunctional human peripheral TCRβrearrangements and an influence of P nucleotides on reading frame bias, also suggesting impacts on T cell selection [61]. While it is possible to envision junctional sequences of intronic illegitimate V(D)J deletions in Notch1 and Bcl11b influencing regulatory mechanisms of gene expression in a way that impacts cell selection, a direct selection effect seems unlikely.
Non-templated (N) nucleotide addition by TdT is known to increase with age [53, 60, 62, 63], and while deletion junctions of Notch1 follow the typical pattern (very few in fetal, increasing postnatally), those of Bcl11b do not. The percentage of Bcl11b deletion junctions containing N nucleotides is similar across all postnatal ages, including 2 day old mice, while normally TdT activity is reduced in younger animals and their V(D)J junctions contain fewer N nucleotides. Since the cryptic RSSs of Bcl11b are very efficient for recombination, as evidenced by higher RIC scores than many bona fide RSSs, it is possible that the coding end processing complex formed at this illegitimate site is more favorable for TdT activity.
The overall extent of coding end nucleotide loss for Notch1 is ~50% lower than that of Bcl11b, HPRT1, and the TCRβ locus, which may indicate that the coding ends of Notch1 are less efficient substrates for nucleolytic activity [12, 49, 50]. The frequency of nucleolytic processing and extent of nucleotide loss varied with age for HPRT1 (increased in newborns compared to 1-12.5 year olds), but not for Notch1, Bcl11b, or the TCRβ locus, indicating that age-related differences in nucleolytic processing associated with illegitimate V(D)J events at the nonimmune HPRT1 locus in humans may not extend to nonimmune loci in mice [49, 50].
Deletions in Notch1 and Bcl11b were detected at all ages examined, including fetal mice, indicating that the formation of deletions is beginning when mice are acquiring the capacity for V(D)J recombination. Deletions were detected in separated thymic and splenic T cell populations, but not in unactivated splenic B cells nor previously in kidney and tail DNA [31, 32], suggesting that deletions are likely only occurring in T cells. This may relate to accessibility of RSSs during induction of V(D)J recombination, which requires germline transcription (specifically shown at the TCRα locus) and associated epigenetic changes [1, 28, 29]. Notch1 and Bcl11b are not generally expressed in B cells, including those undergoing V(D)J recombination at immunoglobulin loci, and therefore may not be available for recombination.
Normally, Notch1 isoforms are expressed in T cell progenitors in bone marrow and at DN2 through early DP stages of T cell development in thymus [35-37, 39], while Bcl11b expression is upregulated during the DN2 to DP stages [44, 48]. Both genes are important for T cell lineage commitment and maintenance, and their expression overlaps with stages of T cell development during which V(D)J recombination is occurring and RAG proteins are active (DN2, DN3, DP) [38]. The requirement of V(D)J recombination for deletion formation was shown previously as they were not detected in RAG2−/− mice [31, 32]. Interestingly, murine models of T cell acute lymphoblastic leukemia (T-ALL), which can involve Notch1 or Bcl11b mutations [24], suggest that while leukemic precursor cells can originate from both DN and DP thymocytes, enrichment of leukemic potential occurs in DN3 and DN4 stages and requires preTCR signals [64, 65].
The presence of deletions in splenic T cells indicates that deletion-bearing thymocytes leave the thymus and enter the circulating lymphocyte pool or that deletions can also initiate in the periphery. The former is more likely as deletions were not detected in spleen until 2 weeks of age (before which few T cells are present) and re-expression of RAG in peripheral T cells is generally limited to a small number of cells undergoing receptor revision [66, 67]. Additionally, deletions were fairly easily detected while T cells typically represent only ~1/3 of splenocytes. However, the precise origin and specific phenotype of T cells bearing deletions in the spleen remains unresolved as does how long they may survive in the periphery.
The frequency of illegitimate V(D)J recombination mediated deletions per 106 cells is 8-fold higher for Bcl11b than Notch1 in thymocytes of C57BL/6 mice, interestingly does not vary with age or sex, and is similar to results with other mouse strains [31, 32]. Illegitimate HPRT1 deletions in human peripheral T cells was found to vary with age and sex, with the highest levels in 0-5 year old males (0.66 per 106 cells) [68]. The frequency of HPRT1 deletions is at least 4-fold lower than the overall frequency of Notch1 deletions and 35-fold lower than that of Bcl11b. Thus, age/sex frequency bias and overall frequency of deletions, like coding end processing biases, appear to be gene-specific.
The difference in frequency may be due to thymic selection events as HPRT1 deletions were from a post-selection sample of peripheral T cells and most likely do not affect T cell selection, while Notch1 and Bcl11b deletions were from mixed thymocytes and may influence or be influenced by T cell differentiation and selection [69]. Alternatively, it is possible that frequency differences reflect unequal recombination efficiencies of cryptic RSSs at these loci. RSSs directly affect recombination frequency as heptamer sequences influence DNA nicking and hairpin formation and maintain site-specificity of V(D)J recombination, while nonamers provide DNA binding sites for RAG1 [1]. Recombination Information Content (RIC) scores for Bcl11b are higher than those of Notch1, HPRT1, and many bona fide RSSs [55], and this may partly explain the significantly higher deletion frequency for Bcl11b compared to Notch1 and HPRT1 [49].
Notch1 and Bcl11b deletions both showed limited clonality, in contrast with previous reports of extensive deletion clonality for Notch1 and no clonality for Bcl11b [31, 32]. It was proposed that oncogenic Notch1 deletions might escape proliferative controls, while deletions in Bcl11b, previously identified as a tumor suppressor, might be haplosufficient for growth control as cells still contain one wild-type allele [31, 32]. An alternative proposal for extensive Notch1 clonality was that deletion-bearing T cells undergo normal clonal expansion accompanying thymic selection events [31]. The results of this study are not consistent with the proposed proliferative advantage from Notch1 deletions in otherwise normal cells, nor a lack of clonality for Bcl11b deletions.
While extensive Notch1 clonality was shown previously in wild-type mice and increased after irradiation, the mouse strains used (C.B-17 and BALB/c) generally display increased sensitivity to radiation and oncogenesis compared to that used in this study (C57BL/6) [31, 70]. It is thus possible that the increased Notch1 clonality seen previously in the other strains may be due to accumulated mutations that synergistically confer a proliferative advantage to deletion-bearing cells. For example, activation of promoters driving expression of ligand-independent forms of Notch1 at the DP stage in Ikaros deficient mice promotes progression to leukemogenesis [40], and other mouse models of T-ALL also involve collaborating mutations [64, 65].
Functionally, Notch1 is an oncogene that activates transcription promoting proliferation, selection and survival in T cells. Deletions can potentially lead to translation of a truncated, ligand-independent protein, yet healthy, wild-type mice with Notch1 deletions do not regularly develop T cell malignancies as do irradiated mice. In this study, however, one mouse (#103) did appear unhealthy and was found to bear many clonal Notch1 deletions, possibly indicating the development of rare collaborating mutation(s) and T cell malignancy.
In the absence of other mutations, controls on Notch1 (e.g., at the level of transcription and nuclear complex formation) may prevent aberrant activation and/or signaling by deletion-induced, ligand-independent forms [71]. Controls unaffected by deletion events may allow these mutations to exist silently in cells unless other oncogenic events occur. Thus, lower clonality of deletion bearing cells seen in C57BL/6 mice may derive from normal cellular proliferation following T cell selection events, while increased clonality in C.B-17 and BALB/c mice may result from collaborations with additional mutations resulting from DNA repair deficiencies.
Similarly, Bcl11b functions as a transcription factor involved in several aspects of thymic T cell specification, selection, and survival. Although originally discovered in radiation-induced thymic lymphomas and described as a haploinsufficient tumor suppressor gene, recent studies emphasize its importance for suppressing “stem-like” self renewal capacity of progenitor cells and promoting the T cell fate at the DN2 stage of thymocyte development [43-46]. While its precise mechanism of action in T cell oncogenesis is unclear [48], illegitimate V(D)J recombination removes a large portion of the gene which leads to the production of a truncated variant known as the γ-isoform that consists of exons 1 and 4 [32].
Like Notch1, mice bearing Bcl11b deletions do not regularly develop T cell malignancies, possibly indicating that the γ-isoform retains adequate activity or that Bcl11b is a haplosufficient tumor suppressor in wild-type mice. Further clarification is needed as reduced Bcl11b expression has been associated both with promotion and apoptosis of T cell leukemia and lymphoma [72, 73]. Although Bcl11b+/− mice are known to be more susceptible to radiation-induced thymic lymphomas, may undergo modest T cell developmental arrest, and have elevated levels of proliferative βcatenin despite reduced preTCR signaling [48, 74], in the absence of other mutations a single wild-type allele appears sufficient to prevent cell transformation.
The lower level of clonality of Bcl11b deletions observed in this study, and previous lack thereof, but greater frequency of Bcl11b deletions than Notch1, might suggest that Bcl11b deletions are more easily tolerated than deletions in Notch1. Future studies will determine whether this is due to haplosufficiency of Bcl11b for tumor suppressor function, retained activity of the γ-isoform, fewer collaborating pro-proliferative mutations for Bcl11b compared to Notch1, different effects on T cell survival or clonal expansion during development and selection, or as stated above, because deletions simply occur more often in Bcl11b due to better cRSSs present near deletion breakpoints.
Highlights.
Examines illegitimate V(D)J deletion junctions in Notch1 and Bcl11b.
Suggests little influence of deletions alone on clonal outgrowth in wild-type mice.
No age or sex biases in frequency, clonality, or junctional processing observed.
Contrasts with previous results at TCRβ and HPRT1 loci.
Deletions in Bcl11b may be tolerated more easily than those in Notch1.
Acknowledgements
This work was supported by grant AI084023 from National Institutes of Health (NIAID) to P.E.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest statement
The authors declare that there are no conflicts of interest.
References
- 1.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Gen. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 3.Ramsden DA, Baetz K, Wu GE. Conservation of sequence in recombination signal sequence spacers. Nucleic Acids Res. 1994;22:1785–1796. doi: 10.1093/nar/22.10.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Gent DC, Ramsden D, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 5.Curry JD, Geier JK, Schlissel MS. Single-strand recombination signal sequence nicks in vivo: evidence for a capture model of synapsis. Nat Immunol. 2005;6:1272–1279. doi: 10.1038/ni1270. [DOI] [PubMed] [Google Scholar]
- 6.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Gent DC, Mizuuchi K, Gellert M. Similarities between initiation of V(D)J recombination and retroviral integration. Science. 1996;271:1592–1594. doi: 10.1126/science.271.5255.1592. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Schatz DG. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 9.Hiom K, Gellert M. Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell. 1998;1:1011–1019. doi: 10.1016/s1097-2765(00)80101-x. [DOI] [PubMed] [Google Scholar]
- 10.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The rag proteins and V(D)J recombination: Complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 11.Lewis SM. The mechanism of V(D)J joining: lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y, Schwarz K, Lieber MR. The Artemis:DNA-PKcs endonuclease cleaves DNA loops, flaps, and gaps. DNA Repair. 2005;4:845–851. doi: 10.1016/j.dnarep.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Lu H, Schwarz K, Lieber MR. Extent to which hairpin opening by the Artemis:DNA-PKcs complex can contribute to junctional diversity in V(D)J recombination. Nucleic Acids Res. 2007;35:6917–6923. doi: 10.1093/nar/gkm823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, Lu H, Schwarz K, Lieber MR. Repair of double-strand DNA breaks by the human nonhomologous DNA end joining pathway: the iterative processing model. Cell Cycle. 2005;4:1193–1200. doi: 10.4161/cc.4.9.1977. [DOI] [PubMed] [Google Scholar]
- 15.Helmink BA, Sleckman BP. The Response to and Repair of RAG-Mediated DNA Double-Strand Breaks. Annu. Rev. Immunol. 2012;30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malu S, Malshetty V, Francis D, Cortes P. Role of non-homologous end joining in V(D)J recombination. Immunol. Res. 2012;54(1-3):233–246. doi: 10.1007/s12026-012-8329-z. [DOI] [PubMed] [Google Scholar]
- 17.Oettinger MA. How to keep V(D)J recombination under control. Immunol Rev. 2004;200:165–181. doi: 10.1111/j.0105-2896.2004.00172.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L, Reynolds TL, Shan X, Desiderio S. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity. 2011;34:163–174. doi: 10.1016/j.immuni.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, Farrar MA, Sleckman BP, Schatz DG, Busslinger M, Bassing CH, Skok JA. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hewitt SL, Chaumeil J, Skok JA. Chromosome dynamics and the regulation of V(D)J recombination. Immunol Rev. 2010;237:43–54. doi: 10.1111/j.1600-065X.2010.00931.x. [DOI] [PubMed] [Google Scholar]
- 22.Marculescu R, Vanura K, Montpellier B, Roulland S, Le T, Navarro JM, Jäger U, McBlane F, Nadel B. Recombinase, chromosomal translocations and lymphoid neoplasia: targeting mistakes and repair failures. DNA Repair. 2006;5:1246–1258. doi: 10.1016/j.dnarep.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Y, Gostissa M, Hildebrand DG, Becker MS, Boboila C, Chiarle R, Lewis S, Alt FW. The role of mechanistic factors in promoting chromosomal translocations found in lymphoid and other cancers. Adv Immunol. 2010;106:93–133. doi: 10.1016/S0065-2776(10)06004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onozawa M, Aplan PD. Illegitimate V(D)J recombination involving nonantigen receptor loci in lymphoid malignancy. Gene Chromosome Canc. 2012;51:525–535. doi: 10.1002/gcc.21942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowell LG, Davila M, Ramsden D, Kelsoe G. Computational tools for understanding sequence variability in recombination signals. Immunol Rev. 2004;200:57–69. doi: 10.1111/j.0105-2896.2004.00171.x. [DOI] [PubMed] [Google Scholar]
- 26.Lewis SM, Agard E, Suh S, Czyzyk L. Cryptic signals and the fidelity of V(D)J joining. Mol Cell Biol. 1997;17:3125–3136. doi: 10.1128/mcb.17.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimazaki N, Tsai AG, Lieber MR. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell. 2009;34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Corcoran AE. The epigenetic role of non-coding RNA transcription and nuclear organization in immunoglobulin repertoire generation. Sem Immunol. 2010;22:353–361. doi: 10.1016/j.smim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Giallourakis CC, Franklin A, Guo C, Cheng HL, Yoon HS, Gallagher M, Perlot T, Andzelm M, Murphy AJ, Macdonald LE, Yancopoulos GD, Alt FW. Elements between the IgH variable (V) and diversity (D) clusters influence antisense transcription and lineage-specific V(D)J recombination. Proc Natl Acad Sci USA. 2010;107:22207–22212. doi: 10.1073/pnas.1015954107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bossen C, Mansson R, Murre C. Chromatin topology and the regulation of antigen receptor assembly. Annu Rev Immunol. 2012;30:337–356. doi: 10.1146/annurev-immunol-020711-075003. [DOI] [PubMed] [Google Scholar]
- 31.Tsuji H, Ishii-Ohba H, Katsube T, Ukai H, Aizawa S, Doi M, Hioki K, Ogiu T. Involvement of illegitimate V(D)J recombination or microhomology-mediated nonhomologous end-joining in the formation of intragenic deletions of the Notch1 gene in mouse thymic lymphomas. Cancer Res. 2004;64:8882–8890. doi: 10.1158/0008-5472.CAN-03-1163. [DOI] [PubMed] [Google Scholar]
- 32.Sakata J, Inoue J, Ohi H, Kosugi-Okano H, Mishima Y, Hatakeyama K, Niwa O, Kominami R. Involvement of V(D)J recombinase in the generation of intragenic deletions in the Rit1/Bcl11b tumor suppressor gene in gamma-ray-induced thymic lymphomas and in normal thymus of the mouse. Carcinogenesis. 2004;25:1069–1075. doi: 10.1093/carcin/bgh094. [DOI] [PubMed] [Google Scholar]
- 33.Tsuji H, Ishii-Ohba H, Noda Y, Kubo E, Furuse T, Tatsumi K. Rag-dependent and Rag-independent mechanisms of Notch1 rearrangement in thymic lymphomas of Atm(−/−) and scid mice. Mutat Res. 2009;660:22–32. doi: 10.1016/j.mrfmmm.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Ashworth TD, Pear WS, Chiang MY. Deletion-based mechanisms of Notch1 activation in T-ALL: key roles for RAG recombinase and a conserved internal translational start site in Notch1. Blood. 2010;116:5455–5464. doi: 10.1182/blood-2010-05-286328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sandy AR, Maillard I. Notch signaling in the hematopoietic system. Expert Opin Biol Ther. 2009;9:1383–1398. doi: 10.1517/14712590903260777. [DOI] [PubMed] [Google Scholar]
- 36.Radtke F, Fasnacht N, Macdonald HR. Notch signaling in the immune system. Immunity. 2010;32:14–27. doi: 10.1016/j.immuni.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 37.Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ. Functions of notch signaling in the immune system: consensus and controversies. Annu Rev Immunol. 2010;28:343–365. doi: 10.1146/annurev.immunol.021908.132719. [DOI] [PubMed] [Google Scholar]
- 38.Wilson A, Held W, MacDonald HR. Two waves of recombinase gene expression in developing thymocytes. J Exp Med. 1994;179:1355–1360. doi: 10.1084/jem.179.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gómez-del Arco P, Kashiwagi M, Jackson AF, Naito T, Zhang J, Liu F, Kee B, Vooijs M, Radtke F, Redondo JM, Georgopoulos K. Alternative promoter usage at the Notch1 locus supports ligand-independent signaling in T cell development and leukemogenesis. Immunity. 2010;33:685–698. doi: 10.1016/j.immuni.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeannet R, Mastio J, Macias-Garcia A, Oravecz A, Ashworth T, Geimer Le Lay A-S, Jost B, Le Gras S, Ghysdael J, Gridley T, Honjo T, Radtke F, Aster JC, Chan S, Kastner P. Oncogenic activation of the Notch1 gene by deletion of its promoter in Ikaros-deficient T-ALL. Blood. 2010;116:5443–5454. doi: 10.1182/blood-2010-05-286658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakabayashi Y, Inoue J, Takahashi Y, Matsuki A, Kosugi-Okano H, Shinbo T, Mishima Y, Niwa O, Kominami R. Homozygous deletions and point mutations of the Rit1/Bcl11b gene in gamma-ray induced mouse thymic lymphomas. Biochem Biophys Res Comm. 2003;301:598–603. doi: 10.1016/s0006-291x(02)03069-3. [DOI] [PubMed] [Google Scholar]
- 42.Wakabayashi Y, Watanabe H, Inoue J, Takeda N, Sakata J, Mishima Y, Hitomi J, Yamamoto T, Utsuyama M, Niwa O, Aizawa S, Kominami R. Bcl11b is required for differentiation and survival of alphabeta T lymphocytes. Nat Immunol. 2003;4:533–539. doi: 10.1038/ni927. [DOI] [PubMed] [Google Scholar]
- 43.Di Santo JP. Immunology. A guardian of T cell fate. Science. 2010;329:44–45. doi: 10.1126/science.1191664. [DOI] [PubMed] [Google Scholar]
- 44.Li P, Burke S, Wang J, Chen X, Ortiz M, Lee SC, Lu D, Campos L, Goulding D, Ng BL, Dougan G, Huntly B, Gottgens B, Jenkins NA, Copeland NG, Colucci F, Liu P. Reprogramming of T cells to natural killer-like cells upon Bcl11b deletion. Science. 2010;329:85–89. doi: 10.1126/science.1188063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li L, Leid M, Rothenberg EV. An early T cell lineage commitment checkpoint dependent on the transcription factor Bcl11b. Science. 2010;329:89–93. doi: 10.1126/science.1188989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ikawa T, Hirose S, Masuda K, Kakugawa K, Satoh R, Shibano-Satoh A, Kominami R, Katsura Y, Kawamoto H. An essential developmental checkpoint for production of the T cell lineage. Science. 2010;329:93–96. doi: 10.1126/science.1188995. [DOI] [PubMed] [Google Scholar]
- 47.Rothenberg EV, Zhang J, Li L. Multilayered specification of the T-cell lineage fate. Immunol Rev. 2010;238:150–168. doi: 10.1111/j.1600-065X.2010.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kominami R. Role of the transcription factor Bcl11b in development and lymphomagenesis. Pro Jpn Acad Ser B Phys Biol Sci. 2012;88:72–87. doi: 10.2183/pjab.88.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray JM, O'Neill JP, Messier T, Rivers J, Walker VE, McGonagle B, Trombley L, Cowell LG, Kelsoe G, McBlane F, Finette BA. V(D)J recombinase-mediated processing of coding junctions at cryptic recombination signal sequences in peripheral T cells during human development. J Immunol. 2006;177:5393–5404. doi: 10.4049/jimmunol.177.8.5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murray JM, Messier T, Rivers J, O'Neill JP, Walker VE, Vacek PM, Finette BA. VDJ recombinase-mediated TCR β locus gene usage and coding joint processing in peripheral T cells during perinatal and pediatric development. J Immunol. 2012;189:2356–2364. doi: 10.4049/jimmunol.1200382. [DOI] [PubMed] [Google Scholar]
- 51.Lambert JF, Benoit BO, Colvin GA, Carlson J, Delville Y, Quesenberry PJ. Quick sex determination of mouse fetuses. J Neurosci Methods. 2000;95:127–132. doi: 10.1016/s0165-0270(99)00157-0. [DOI] [PubMed] [Google Scholar]
- 52.Luria SE, Delbruck M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gauss GH, Lieber MR. Mechanistic constraints on diversity in human V(D)J recombination. Mol Cell Biol. 1996;16:258–269. doi: 10.1128/mcb.16.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merelli I, Guffanti A, Fabbri M, Cocito A, Furia L, Grazini U, Bonnal RJ, Milanesi L, McBlane F. RSSsite: a reference database and prediction tool for the identification of cryptic Recombination Signal Sequences in human and murine genomes. Nucleic Acids Res. 2010;38:W262–267. doi: 10.1093/nar/gkq391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cowell LG, Davila M, Kepler TB, Kelsoe G. Identification and utilization of arbitrary correlations in models of recombination signal sequences. Genome Biol. 2002;3:RESEARCH0072. doi: 10.1186/gb-2002-3-12-research0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arnal SM, Holub AJ, Salus SS, Roth DB. Non-consensus heptamer sequences destabilize the RAG post-cleavage complex, making ends available to alternative DNA repair pathways. Nucleic Acids Res. 2010;38:2944–2954. doi: 10.1093/nar/gkp1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee AI, Fugmann SD, Cowell LG, Ptaszek LM, Kelsoe G, Schatz DG. A functional analysis of the spacer of V(D)J recombination signal sequences. PLoS Biol. 2003;1:E1. doi: 10.1371/journal.pbio.0000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 59.Nadel B, Feeney AJ. Nucleotide deletion and P addition in V(D)J recombination: a determinant role of the coding-end sequence. Mol Cell Biol. 1997;17:3768–3778. doi: 10.1128/mcb.17.7.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 61.Srivastava SK, Robins HS. Palindromic nucleotide analysis in human T cell receptor rearrangements. PLoS One. 2012;7:e52250. doi: 10.1371/journal.pone.0052250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feeney AJ. Junctional sequences of fetal T cell receptor beta chains have few N regions. J Exp Med. 1991;174:115–124. doi: 10.1084/jem.174.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benedict CL, Gilfillan S, Thai TH, Kearney JF. Terminal deoxynucleotidyl transferase and repertoire development. Immunol Rev. 2000;175:150–157. [PubMed] [Google Scholar]
- 64.Tremblay M, Tremblay CS, Herblot S, Aplan PD, Hebert J, Perreault C, Hoang T. Modeling T-cell acute lymphoblastic leukemia induced by the SCL and LMO1 oncogenes. Genes Dev. 2010;24:1093–1105. doi: 10.1101/gad.1897910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tatarek J, Cullion K, Ashworth T, Gerstein R, Aster JC. M.a. Kelliher, Notch1 inhibition targets the leukemia-initiating cells in a Tal1/Lmo2 mouse model of T-ALL. Blood. 2011;118:1579–1590. doi: 10.1182/blood-2010-08-300343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Velardi A, Cooper MD. An immunofluorescence analysis of the ontogeny of myeloid, T, and B lineage cells in mouse hemopoietic tissues. J Immunol. 1984;133:672–677. [PubMed] [Google Scholar]
- 67.Hale JS, Fink PJ. T-cell receptor revision: friend or foe? Immunol. 2010;129:467–473. doi: 10.1111/j.1365-2567.2010.03250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Finette BA, Kendall H, Vacek PM. Mutational spectral analysis at the HPRT locus in healthy children. Mutat Res. 2002;505:27–41. doi: 10.1016/s0027-5107(02)00119-7. [DOI] [PubMed] [Google Scholar]
- 69.Yuan JS, Tan JB, Visan I, Matei IR, Urbanellis P, Xu K, Danska JS, Egan SE, Guidos CJ. Lunatic Fringe prolongs Delta/Notch-induced self-renewal of committed αβ T-cell progenitors. Blood. 2011;117:1184–1195. doi: 10.1182/blood-2010-07-296616. [DOI] [PubMed] [Google Scholar]
- 70.Okayasu R, Suetomi K, Yu Y, Silver A, Bedford JS, Cox R, Ullrich RL. A deficiency in DNA repair and DNA-PKcs expression in the radiosensitive BALB/c mouse. Cancer Res. 2000;60:4342–4345. [PubMed] [Google Scholar]
- 71.Yatim A, Benne C, Sobhian B, Laurent-Chabalier S, Deas O, Judde JG, Lelievre JD, Levy Y, Benkirane M. NOTCH1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol Cell. 2012;48:445–458. doi: 10.1016/j.molcel.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grabarczyk P, Przybylski GK, Depke M, Völker U, Bahr J, Assmus K, Bröker BM, Walther R, Schmidt C.a. Inhibition of BCL11B expression leads to apoptosis of malignant but not normal mature T cells. Oncogene. 2007;26:3797–3810. doi: 10.1038/sj.onc.1210152. [DOI] [PubMed] [Google Scholar]
- 73.Kurosawa N, Fujimoto R, Ozawa T, Itoyama T, Sadamori N, Isobe M. Reduced level of the BCL11B protein is associated with adult T-cell leukemia/lymphoma. PLoS One. 2013;8:e55147. doi: 10.1371/journal.pone.0055147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Go R, Hirose S, Morita S, Yamamoto T, Katsuragi Y, Mishima Y, Kominami R. Bcl11b heterozygosity promotes clonal expansion and differentiation arrest of thymocytes in gamma-irradiated mice. Canc Sci. 2010;101:1347–1353. doi: 10.1111/j.1349-7006.2010.01546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]