Evaluation of host immune response to infection at the molecular level is a promising avenue to obtain diagnostic and prognostic tools for the clinical management of patients with sepsis. A recent report from Cajander and colleagues [1] has shown the potential of HLA-DR mRNA quantification by real-time PCR as a biomarker of immunosuppression in these patients. IgM is the first immunoglobulin produced in response to infection. In a pilot study, we have employed a next generation quantitative PCR method (nanoliter-sized droplet technology paired with digital PCR (ddPCR)) for detecting the early transcriptomic response of IgM in blood from patients with sepsis. Approval for the study protocol for both scientific and ethical aspects was obtained from the Committee for Clinical Research of Hospital Clínico Universitario, Valladolid, Spain. Written informed consent was obtained directly from each patient or a legal surrogate. The target gene transcript was IGHM, which encodes the constant region of the mu heavy chain, which defines the IgM isotype [2]. In blood, the cells producing IgM transcripts are B lymphocytes expressing CD20 [3], which was employed as housekeeping gene.
Fifty-five patients with sepsis were recruited, 42 of them presenting criteria of septic shock (Additional file 1). Septic patients were predominantly older males (n = 40, 72.7%; mean age 72 years (standard deviation 9.3)). Mean Sepsis-related Organ Failure Assessment score was 8.4 (standard deviation 2.9). Overall ICU mortality was 34%. Emergency surgery was needed in 54% of cases, with cardiac and abdominal surgery the most frequent (45% and 40%, respectively). Respiratory infection was present in 34.5% of the cases. Frequency of abdominal infection was also 34.5%. Gram-negative bacteria were the most frequent isolated (56. 4% of cases). In parallel, we recruited 20 patients with post-surgical systemic inflammatory response syndrome (SIRS) and 15 healthy controls.
Compared to real-time quantitative PCR, ddPCR offers greater precision and reproducibility [4]. ddPCR allowed us to identify the presence of an early molecular response of IgM in the blood of patients with sepsis compared with healthy controls and patients with SIRS. This response was more intense in the most severe patients (Figure 1). When accuracy and the predictive value of the IGHM/CD20 ratio for diagnosing sepsis were analyzed, the area under the receiver operating characteristic curve was 0.72 (95% confidence interval 0.60 to 0.85; P = 0.003; Figure 1). In conclusion, quantification of IgM response at the transcriptomic level by ddPCR represents a promising approach for the early detection of sepsis.
Figure 1.
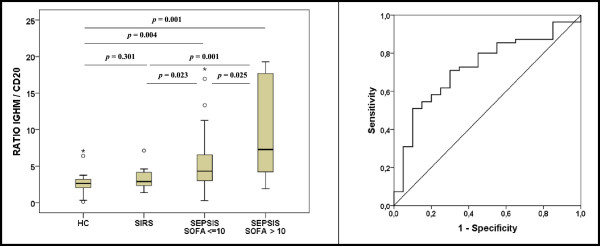
IgM transcriptomic response in the presence/absence of sepsis. Left: comparison of immunoglobulin (Ig)M transcriptomic response between groups. HC, healthy control (n = 15); SIRS, systemic inflammatory response syndrome (n = 20); SEPSIS SOFA < =10, Sepsis with Sepsis-related Organ Failure Assessment score ≤10 (n = 39); SEPSIS SOFA >10, sepsis with SOFA score >10 (n = 16). Results for the ratio are expressed as (Copies IGHM/Copies CD20). Right: receiver operating characteristic curve of IGHM/CD20 predicting presence of sepsis in the comparison (sepsis versus SIRS). For this comparison septic patients were considered as a single group.
Abbreviations
ddPCR: droplet digital PCR; Ig: Immunoglobulin; PCR: Polymerase chain reaction; SIRS: Systemic inflammatory response syndrome.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ET, JW, and JFBM helped with the study design, provided a critical review of the results and participated in article writing; RA, AAA, ARF, DAO, JIGH, and JME analyzed the data and participated in article writing; EC, MH, EGS, SS, and AMM recruited the patients, provided a critical review of the results and participated in article writing; LR and VI performed the molecular works and provided a critical review of the results. All authors read and approved the final manuscript.
Supplementary Material
Additional file 1 is the supplemental methods.
See related research by Cajander et al., http://ccforum.com/content/17/5/R223
Contributor Information
Eduardo Tamayo, Email: tamayo@med.uva.es.
Raquel Almansa, Email: ralmansa@saludcastillayleon.es.
Elena Carrasco, Email: elenacarrascoserrano@gmail.com.
Ana Ávila-Alonso, Email: a.avila.al@gmail.com.
Ana Rodríguez-Fernández, Email: a.rodfer@hotmail.es.
John Wain, Email: J.Wain@uea.ac.uk.
María Heredia, Email: maria_her_05@hotmail.com.
Esther Gomez-Sanchez, Email: esthergzam@hotmail.com.
Susana Soria, Email: susasori@telefonica.net.
Lucia Rico, Email: lrico@saludcastillayleon.es.
Verónica Iglesias, Email: viglesiasl@saludcastillayleon.es.
Ángel Martínez-Martínez, Email: ajmmza@yahoo.es.
David Andaluz-Ojeda, Email: davidandaluz78@yahoo.es.
Jose Ignacio Gómez Herreras, Email: jgomez012001@yahoo.es.
Jose Maria Eiros, Email: eiros@med.uva.es.
Jesús F Bermejo-Martin, Email: jfbermejo@saludcastillayleon.es.
Acknowledgements
The authors deeply thank Eva Obregón, Angel Burgaleta from BioRad Spain and Yann Jouvenot from BioRad USA for their technical support with the ddPCR assay. This work was supported by 'Fondo de Investigaciones Sanitarias, Instituto de Salud Carlos III', Spain, grant number PI13/02110.
References
- Cajander S, Bäckman A, Tina E, Strålin K, Söderquist B, Källman J. Preliminary results in quantitation of HLA-DRA by real-time PCR: a promising approach to identify immunosuppression in sepsis. Crit Care. 2013;17:R223. doi: 10.1186/cc13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefranc M-P, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, Wu Y, Gemrot E, Brochet X, Lane J, Regnier L, Ehrenmann F, Lefranc G, Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemark M, Holmqvist J, Abrahamsson J, Mellgren K. Reconstitution after haematopoietic stem cell transplantation - revelation of B cell developmental pathways and lineage phenotypes. Clin Exp Immunol. 2012;167:15–25. doi: 10.1111/j.1365-2249.2011.04469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindson CM, Chevillet JR, Briggs HA, Gallichotte EN, Ruf IK, Hindson BJ, Vessella RL, Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1 is the supplemental methods.


