Abstract
Hepatitis E virus (HEV) is classified within the family Hepeviridae, genus Hepevirus. HEV genotype 3 (Gt3) infections are endemic in pigs in Western Europe and in North and South America and cause zoonotic infections in humans. Several serological assays to detect HEV antibodies in pigs have been developed, at first mainly based on HEV genotype 1 (Gt1) antigens. To develop a sensitive HEV Gt3 ELISA, a recombinant baculovirus expression product of HEV Gt3 open reading frame-2 was produced and coated onto polystyrene ELISA plates. After incubation of porcine sera, bound HEV antibodies were detected with anti-porcine anti-IgG and anti-IgM conjugates. For primary estimation of sensitivity and specificity of the assay, sets of sera were used from pigs experimentally infected with HEV Gt3. For further validation of the assay and to set the cutoff value, a batch of 1100 pig sera was used. All pig sera were tested using the developed HEV Gt3 assay and two other serologic assays based on HEV Gt1 antigens. Since there is no gold standard available for HEV antibody testing, further validation and a definite setting of the cutoff of the developed HEV Gt3 assay were performed using a statistical approach based on Bayes' theorem. The developed and validated HEV antibody assay showed effective detection of HEV-specific antibodies. This assay can contribute to an improved detection of HEV antibodies and enable more reliable estimates of the prevalence of HEV Gt3 in swine in different regions.
Keywords: Hepatitis E virus, Zoonosis, Swine, ELISA
Introduction
Hepatitis E virus (HEV) is a small nonenveloped RNA virus of approximately 7.2 kb, and has been classified within the family Hepeviridae, genus Hepevirus. Four mammalian genotypes (Gt1-4) were identified in the 1990s, and avian HEV had been identified before that (1). More recently, HEVs have also been identified in rats, rabbits, ferrets, bats, and trout (2). Gt1 and Gt2 are found exclusively in humans and are responsible for large waterborne epidemics and sporadic cases in endemic regions (Asia, India, Africa, and Mexico) (1). Gt1 and Gt2 can be divided further into five (a-e) and two (a and b) subtypes, respectively. In contrast, Gt3 and Gt4 are responsible for sporadic cases in humans and are widespread in animals. Especially for HEV Gt3, it is assumed that zoonotic transmission plays an important role in human infections (3). Genotypes 3 and 4 seem more variable and can be divided into 10 (a-j) and 7 (a-g) subgenotypes, respectively (4).
Despite the considerable genetic variability observed between genotypes, to date only one serotype has been identified. HEV Gt3 infections have been reported to be endemic in pigs in Europe (5) and in North and South America (6). HEV infections in humans often run a subclinical course but can also cause serious acute hepatitis. The disease is characterized by a self-limiting jaundice of varying severity, which is hard to distinguish from hepatitis of other viral origin, and is often accompanied by nonspecific symptoms such as fever, headache, and pain in the upper abdomen. In pigs naturally infected with HEV Gt3, microscopic lesions of hepatitis are observed, but overt clinical symptoms in pigs have not been reported. Autochthonous HEV Gt3 and Gt4 infections in humans may be zoonotic and may originate from swine reservoirs. For this reason, it is suggested that the study of HEV dynamics in pig populations be done in Europe and the Americas.
For swine serology, several HEV antibody assays have been developed, mainly based on HEV Gt1 antigens, in particular open reading frame (ORF)-2 proteins or peptides. HEV Gt3 antibodies have been shown to cross-react with Gt1 antigens (7), but, given the high variability of strains, homologous assays may perform better. Since HEV Gt3 is the predominant genotype in pigs in Europe and North America, in this study an HEV antibody assay for pigs was developed based on the HEV Gt3 antigen. Due to the fact that there is no gold standard available, in-house developed or commercially available HEV immunoassays often have not been validated properly. For final validation and a definite choice for cutoff of the developed HEV Gt3 assay in this study, posterior Bayesian statistical inference was used.
A total of 1100 serum samples from fattening pigs were tested using the newly developed HEV Gt3 assay and two other HEV immunoassays for comparison. Test results of the three assays were analyzed simultaneously with a latent class model [see Engel et al. (8) and references therein]. This statistical model accounts for the lack of a gold standard, i.e., no assay or combination of assays was chosen as a reference.
Material and Methods
HEV ELISA development
A recombinant expression product of HEV (ORF-2, Gt3, Subtype 3a) was produced in a baculovirus expression system as previously described (9,10). A synthetic gene encoding 497 amino acids of swine Gt3 HEV ORF-2, Subtype 3a, was engineered using the amino acid sequence published in GenBank AY585859 (Geneart, Germany). The part of the HEV capsid encoded by this gene includes the motifs needed for self-assembly into empty virus-like particles when expressed in Sf9 insect cells. Codon usage and percentage G and C nucleotide content were optimized for expression in Sf9 Spodoptera frugiperda insect cells and a Kozak sequence was introduced upstream of the ATG. The recombinant baculovirus was obtained using the Bac-to-Bac¯ Baculovirus Expression System (Invitrogen, France) according to the manufacturer's instructions. Sf9 insect cells adapted to a serum-free medium (Invitrogen) were grown at 27°C in SF900 serum-free medium supplemented with penicillin, streptomycin, and fungizone (Invitrogen). Viral stocks were prepared on Sf9 cells, after two plaque purifications also on Sf9 cells.
Protein expression in the culture medium and cell lysate was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting using negative and positive sera from swine. Sf9 cells were infected with the recombinant baculovirus at a multiplicity of infection of five. The culture medium was harvested 5 days after infection. Cells and debris were removed by centrifugation at 10,000 g for 30 min. The supernatant was then centrifuged at 11,000 g for 3 h in a Beckman SW28 rotor. The resulting pellet was resuspended in 4 mL PBS and held overnight at 4°C. The sample was then mixed with 0.44 g/mL CsCl and centrifuged at 15,000 g for 18 h at 4°C in a Beckman SW50.1 rotor. Fractions of 0.5 mL were collected from the bottom of each gradient and fraction densities were determined using an Abbe refractometer (Bioblock, France) and stored at a concentration of 0.3 μg/mL.
For the test principle, recombinant protein was coated onto polystyrene ELISA plates. Recombinant protein was dissolved 1:1500 in coating buffer (see ELISA procedures), and incubated with the test serum and HEV-specific swine antibodies were detected using anti-porcine anti-IgG and IgM conjugates. Antibodies present in the test serum bound to the antigen during the first incubation, and during the second incubation, the bound antibodies were detected using anti-swine monoclonal antibodies against IgM and against the L-chain conjugated to the enzyme horseradish peroxidase (HRP, see ELISA procedures). Bound conjugate was visualized by adding a substrate chromogen. Wells containing samples negative for HEV antibodies remained colorless. To optimize the ELISA, different concentrations of the purified recombinant protein were tested using a series of swine sera from an experimental infection. Swine sera were tested in different dilutions to establish the optimal dilution for test sera. A primary cutoff of the assay was established using sera from two cesarean-delivered colostrum-deprived, specific pathogen-free pigs. For primary estimation of sensitivity and specificity of the assay, sets of sera were used from pigs experimentally infected with HEV Gt3 (10,11).
All experiments in this study involving the use of animals were approved by the Ethics Committee for Animal Experiments of Wageningen University, in accordance with legislation of The Netherlands and the European Union.
HEV ELISA comparison assays
Two different HEV immunoassays were used for comparison, a commercially available immunoassay and an HEV immunoassay that was developed in-house. The commercially available HEV Ab-ELISA kit (Axiom, Germany) is a double-antigen sandwich ELISA based on a recombinant Burmese HEV Gt1 capsid protein derivative covering the carboxy-terminal amino acid residues 394-606. Due to its test principle, it can detect HEV-specific antibodies independently of the host species and immunoglobulin class. The manufacturer's instructions were strictly adhered to in the assay, including the recommended thresholds for definition of a positive serum. The in-house developed HEV immunoassay is a previously standardized in-house ELISA based on a purified 55-kDa truncated recombinant capsid protein of the Sar-55 strain of human HEV Gt1 (Gt1 assay). Based on its test principles, this immunoassay detects HEV-specific IgG in pig sera. This immunoassay was performed as described by Martin et al. (12). The absorbance of each sample was read with an ELISA plate reader with a 450-nm filter. Test samples with absorbances equal to or greater than the cutoff value (0.300) were considered to be positive for anti-HEV IgG. In all immunoassays, sera of HEV naive and HEV infected swine were used as negative and positive controls in each plate.
HEV ELISA validation
For further validation of the assay and to definitely set its cutoff value, a batch of 1100 pig sera was used. The sera were obtained at slaughter from pigs from at least 550 pig farms (maximum two pigs per farm) in The Netherlands. Potential HEV infections in these pigs were of Gt3. To date, only HEV Gt3 infections have been found in pigs in The Netherlands. All pig sera were tested using the developed HEV Gt3 assay and the two comparison serologic assays based on HEV Gt1 antigens.
Statistical evaluation of diagnostic test accuracy
The pig sera obtained at slaughter were used for a Bayesian evaluation of the accuracy of the three ELISA tests. In a Bayesian analysis two sources of information are combined: prior information about prevalence, test sensitivity, and test specificity, and new observations, which in this case were the ELISA test results. The prior information attaches a probability to each possible value of a parameter in the model, e.g., an unknown test sensitivity. Consequently, the prior information takes the form of a probability distribution for this sensitivity. This prior distribution is combined with the data into a posterior distribution, which is an up-to-date summary of all available information about the sensitivity. The median of this posterior distribution is presented as an estimate and the 95% range as a Bayesian confidence interval (credible interval) for the sensitivity.
The statistical model distinguishes between the test results and the true antibody status of the sera. Because the tests are fallible, observed test results and unobserved true antibody status are not necessarily the same. The test results of the three ELISA tests were analyzed with a latent class model, where the true-antibody status (seropositive or seronegative) represents the latent classes.
Data were collected in relatively small batches from different producers. In the statistical model, each batch has its own true prevalence, i.e., probability for a true seropositive, and these different prevalences are assumed to be sampled from a distribution of prevalence values. The probabilistic behavior of the tests, in relation to true antibody status, is modeled according to Engel et al. (8). The model accounts for possible dependence between tests for true seropositives and true seronegatives.
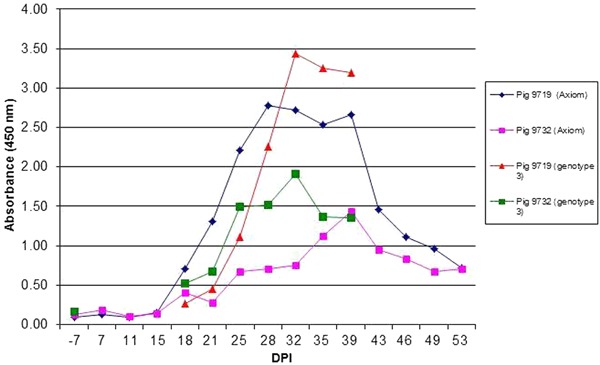
Prior distributions were based on literature and expert opinion. For assays based on a Gt3 antigen, prior emphasis was on a slightly higher specificity. For assays using more isotypes of antibodies in the conjugate, prior emphasis was on slightly higher sensitivity. The prior median and 95% ranges for sensitivity and specificity are shown in Table 1. The prior range for median prevalence was chosen from 0.5 to 0.9. Further (technical) details about model and priors may be found in Supplementary Material.
Because batches of sera from the same producer tended to be small, often as small as two sera from the same producer, parameters for prevalence were hard to identify from the data. In order to obtain more reliable inference about the sensitivity and specificity of the tests, it was assumed that dependence between tests, conditional upon true-negative status, was negligible.
For the CVI (Central Veterinary Institute) test, binary data were generated at three different thresholds (20,23,25) and three separate analyses were performed, one for each threshold.
HEV ELISA procedures
For coating onto polystyrene ELISA plates, the optimal dilution of the recombinant baculoprotein stock (0.3 µg/mL) was 1:1500 using a coating buffer (0.05 M sodium carbonate buffer, pH 9.65). In the final procedure, 100 µL diluted baculoprotein was dispensed to each well of a polystyrene ELISA plate (Costar 9018, Greiner Bio-one, Alphen a/d Rijn, The Netherlands). Plates were covered with adhesive foil (plate sealer) and incubated overnight at room temperature. Before usage, ELISA plate(s) were washed three times using tap water supplemented with 0.05% Tween 80 in an ELISA plate washer. A volume of 100 µL StabilCoat¯ (Diarect AG, Freiburg, Germany) (diluted 1:1 with PBS) was dispensed to all wells and incubated 30 min at room temperature. After removal of StabilCoat¯, ELISA plates were dried for 3 h at 37°C and stored at 4°C until use. In the final test procedure, sera were diluted 1:5 in serum dilution buffer: 10% bovine serum albumin+10% Elk+1% Tergitol (dissolved in PBS) added to low salt buffer (2.5 mL 10% Tween 80 in 500 mL PBS). One hundred microliters of diluted test serum was dispensed to the wells and, after sealing, plates were incubated at room temperature for 1 h. After this first incubation, plates were washed three times using tap water supplemented with 0.05% Tween 80 in an ELISA plate washer. Anti-swine IgM HRP-conjugated monoclonal antibody (CVI 28.4.1.) was diluted 1:2000 and the anti-swine L-chain HRP-conjugated monoclonal antibody (CVI 27.2.1.) was diluted 1:1000 in conjugate buffer (5% fetal calf serum) in high salt buffer (14.5 g NaCl in 500 mL PBS, 2.5 mL 10% Tween 80). For the second incubation, 100 µL of the diluted conjugate solution was dispensed to each well. Plates were covered with a plate sealer and incubated for 1 h at room temperature. Subsequently, plates were washed three times with tap water supplemented with 0.05% Tween 80 using an ELISA plate washer, and 100 µL ready-to-use 3,3′,5,5′-tetramethylbenzidine substrate was dispensed to each well and incubated for 10 min at room temperature. The reaction was stopped by adding 100 µL 0.5 M sulfuric acid to all wells. The color intensity was measured at 450 nm within 15 min after color development had been stopped.
Results
HEV ELISA development
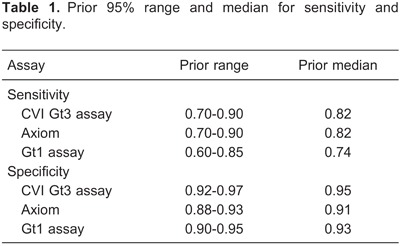
As described in Material and Methods, several protocols were tested to set up an immunoassay based on HEV Gt3 recombinant antigen. Final performance of the developed HEV Gt3 immunoassay was assessed using sera of pigs experimentally infected with HEV Gt3, in comparison with a commercially available HEV Ab-ELISA (Axiom). The results showed slight differences in the kinetics of anti-HEV antibody detection, but seroconversion of HEV-infected pigs was detected with both detection methods (Figure 1).
Figure 1. Hepatitis E virus genotype 3 (Gt3) antibody developments in experimentally HEV Gt3-infected pigs measured by HEV Gt3 ELISA CVI and HEV ELISA Axiom. Test procedures of the developed assay are described in Material and Methods.
HEV ELISA validation
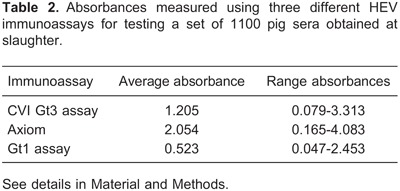
The average absorbance values for the set of 1100 pig sera obtained at slaughter and measured using three different HEV antibody assays are reported in Table 2.
The estimated sensitivity and specificity of each of the three tests are shown in Table 3, together with a 95% confidence interval (95%CI, credible interval). These results correspond to a threshold value of 20 for the CVI Gt3 assay. For threshold values of 23 and 25, similar values for estimated sensitivity and specificity were found for the Axiom and Gt1 assays. For a threshold value of 20, the CVI Gt3 assay showed the highest sensitivity. For threshold values of 20, 23, and 25, the CVI Gt3 assay showed similar specificities. For the CVI Gt3 immunoassay, the best results were obtained for the threshold value of 20; therefore, this value was chosen to be used as the primary choice cutoff value for the test.
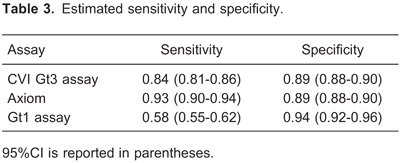
The higher specificity of the Gt1 assay (0.94, 95%CI=0.92-0.96) was apparently achieved at the cost of a modest sensitivity (0.58, 95%CI=0.55-0.62). CVI Gt3 and Axiom had a similar specificity (0.89, 95%CI=0.88-0.90), but the estimated sensitivity of Axiom (0.93, 95%CI=0.90-0.94) was higher than that of CVI Gt3 (0.84, 95%CI=0.81-0.86).
The estimated median prevalence was 0.95 (95%CI=0.90-0.97). The estimate for standard deviation σ is 0.87 (95%CI=0.66-1.03), which implies considerable variation among batches in prevalence: the odds ratio of two batches can be as high as 11, e.g., odds ratio 0.9/(1-0.9) vs 0.45/(1-0.45) for prevalences of 0.90 and 0.45.
Discussion
To establish an HEV antibody assay for pigs, where HEV Gt3 is the predominant genotype in Europe and North America, an HEV antibody assay was developed based on the HEV Gt3 antigen. Since there is increasing evidence that human autochthonous hepatitis E cases are linked to swine reservoirs (5,13), HEV antibody testing in swine is indicated to estimate HEV prevalence. For this purpose, a validated HEV antibody assay for Europe, preferably HEV Gt3 based, is needed. Therefore, a recombinant expression product of HEV (ORF-2, Gt3, Subtype 3a) was chosen as antigen. For sensitivity testing of HEV antibody development in the acute phase of the infection, a conjugate including HEV-specific IgG and IgM was used. The effective performance of this format was tested using a set of serum samples obtained from experimentally infected pigs.
Validation of HEV antibody assays is hampered by the fact that a gold standard is not available. To get around this problem, different approaches have been used including Western blot and Bayesian analyses (9,14). In this study, posterior Bayesian statistical inference was used for final validation and definitive choice of the cutoff value. Through a combined approach of testing, using swine sera obtained from experimental infections and from the field, and comparison of immunoassays using Bayesian evaluation of the accuracy of the assays, an adequate validation of the immunoassay could be established.
In the statistical model, it was assumed that there is an interdependency between assays for true seropositives and independence for true seronegatives. Without the last assumption, i.e., allowing for dependence between assays for true seronegatives as well, quite unrealistic results were found for the specificity of all three assays. Quite likely, this is connected to identification problems that influenced the prevalence structure of batches in the data set. Such identification problems are not specific to the statistical approach used in this article, and are potential problems shared by all methods used for diagnostic test evaluation in the absence of a gold standard. For true positives, dependence is likely, since all three immunoassays are based on more or less the same principle. Moreover, based on previous observations, it has been postulated repeatedly that it is likely that there is just one single serotype of mammalian HEV (1). Assumed independence for true seronegatives was considered reasonable, because in that case there will not be an immunogenic reaction related to the selected antigens in the assays.
To better understand HEV transmission in autochtho-nous HEV infections in humans, HEV surveillance in pigs is needed. Using an accurately validated, sensitive HEV antibody assay based on the predominant antigen will result in more reliable surveillance data. The HEV antibody assay developed and validated in this study is suitable for detection of HEV-specific antibodies in swine reservoirs and can contribute to reliable estimates of HEV prevalence in swine in different regions.
Supplementary Material
Acknowledgments
Research was commissioned by the Dutch Ministry of Economic Affairs (Min EZ) and supported by MEDVETNET, Network of Excellence for the Prevention and Control of Foodborne Zoonoses, WP31 ZOOVIRNET, Food Producing Animals as a Potential Source of Emerging Viral Zoonoses.
Footnotes
First published online March 21, 2014.
References
- 1.Worm HC, van der Poel WH, Brandstatter G. Hepatitis E: an overview. Microbes Infect. 2002;4:657–666. doi: 10.1016/S1286-4579(02)01584-8. [DOI] [PubMed] [Google Scholar]
- 2.Smith DB, Purdy MA, Simmonds P. Genetic variability and the classification of hepatitis E virus. J Virol. 2013;87:4161–4169. doi: 10.1128/JVI.02762-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christou L, Kosmidou M. Hepatitis E virus in the Western world - a pork-related zoonosis. Clin Microbiol Infect. 2013;19:600–604. doi: 10.1111/1469-0691.12214. [DOI] [PubMed] [Google Scholar]
- 4.Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 5.van der Poel WH, Verschoor F, van der Heide R, Herrera MI, Vivo A, Kooreman M, et al. Hepatitis E virus sequences in swine related to sequences in humans, The Netherlands. Emerg Infect Dis. 2001;7:970–976. doi: 10.3201/eid0706.010608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.dos Santos DR, de Paula V, de Oliveira JM, Marchevsky RS, Pinto MA. Hepatitis E virus in swine and effluent samples from slaughterhouses in Brazil. Vet Microbiol. 2011;149:236–241. doi: 10.1016/j.vetmic.2010.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Emerson SU, Clemente-Casares P, Moiduddin N, Arankalle VA, Torian U, Purcell RH. Putative neutralization epitopes and broad cross-genotype neutralization of Hepatitis E virus confirmed by a quantitative cell-culture assay. J Gen Virol. 2006;87:697–704. doi: 10.1099/vir.0.81545-0. [DOI] [PubMed] [Google Scholar]
- 8.Engel B, Swildens B, Stegeman A, Buist W, De Jong MCM. Estimation of sensitivity and specificity of three conditionally dependent diagnostic tests in the absence of a gold standard. J Agric Biol Environ Stat. 2006;11:360–380. doi: 10.1198/108571106X153534. [DOI] [Google Scholar]
- 9.Bouwknegt M, Frankena K, Rutjes S, Wellenberg GJ, De Roda Husman AM, Van der Poel WHM, et al. Basic reproduction ratio for hepatitis E virus transmission in swine by contact-exposure in one-to-one trials. Vet Res. 2008;39:40–50. doi: 10.1051/vetres:2008017. [DOI] [PubMed] [Google Scholar]
- 10.Rose N, Boutrouille A, Fablet C, Madec F, Eloit M, Pavio N. The use of Bayesian methods for evaluating the performance of a virus-like particles-based ELISA for serology of hepatitis E virus infection in swine. J Virol Methods. 2010;163:329–335. doi: 10.1016/j.jviromet.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 11.Bouwknegt M, Rutjes SA, Reusken CB, Stockhofe-Zurwieden N, Frankena K, de Jong MC, et al. The course of hepatitis E virus infection in pigs after contact-infection and intravenous inoculation. BMC Vet Res. 2009;5:7. doi: 10.1186/1746-6148-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin M, Segales J, Huang FF, Guenette DK, Mateu E, de Deus N, et al. Association of hepatitis E virus (HEV) and postweaning multisystemic wasting syndrome (PMWS) with lesions of hepatitis in pigs. Vet Microbiol. 2007;122:16–24. doi: 10.1016/j.vetmic.2006.12.020. [DOI] [PubMed] [Google Scholar]
- 13.Bouquet J, Tesse S, Lunazzi A, Eloit M, Rose N, Nicand E, et al. Close similarity between sequences of hepatitis E virus recovered from humans and swine, France, 2008-2009. Emerg Infect Dis. 2011;17:2018–2025. doi: 10.3201/eid1711.110616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez de Oya N, Galindo I, Girones O, Duizer E, Escribano JM, Saiz JC. Serological immunoassay for detection of hepatitis E virus on the basis of genotype 3 open reading frame 2 recombinant proteins produced in Trichoplusia ni larvae. J Clin Microbiol. 2009;47:3276–3282. doi: 10.1128/JCM.00750-09. [DOI] [PMC free article] [PubMed] [Google Scholar]


