Abstract
The quantification of human cytomegalovirus (HCMV DNA) by real-time PCR is currently a primary option for laboratory diagnosis of HCMV infection. However, the optimal sample material remains controversial due to the use of different PCR assays. To explore the best blood component for HCMV DNA surveillance after liver transplantation, whole blood (WB), serum (SE), and plasma (PL) specimens were collected simultaneously from targeted patients and examined for HCMV DNA using one commercially available assay. The HCMV DNA-positive rate with WB (16.67%) was higher than that with either SE or PL (8.33%, both P<0.01). Quantitative DNA levels in WB were of greater magnitude than those in SE (WB-SE mean log-transformed difference, 0.99; 95%CI=0.74-1.25; P<0.0001) and PL (WB-PL mean log-transformed difference, 1.37; 95%CI=1.07-1.66; P<0.0001). Dynamic monitoring revealed that HCMV DNA in WB was positive sooner and had higher values for a longer period of time during therapy. With earlier positive detection, higher sensitivity, and yield of greater viral loads, WB compared favorably to SE or PL and hence is recommended as the superior material for HCMV DNA surveillance after liver transplantation. In addition, infant recipients require more intensive monitoring and prophylactic care because of their higher susceptibility to primary HCMV infection.
Keywords: HCMV DNA, Blood components, Real-time quantitative PCR, Liver transplantation
Introduction
Human cytomegalovirus (HCMV), a β-herpesvirus, circulates widely in the human population and causes opportunistic infections. HCMV seroprevalence in Chinese adults can be as high as over 90% (1). Although the virus is usually latent in immunocompetent individuals and produces few, obvious lesions, it remains a serious threat to immunocompromised patients, contributing to significant morbidity and mortality among transplant recipients (2,3).
For laboratory diagnosis of HCMV infection, real-time fluorescence quantitative (RTFQ) PCR has been a primary tool to determine quantitative DNA loads. However, the optimal blood component for DNA detection is still under discussion. Peripheral blood leukocytes (PBL) have been cited for their high sensitivity (4-7), but there are limitations in case of leucopenia. EDTA whole blood (WB) has been proposed as the ideal material because of larger yields of DNA loads (8-10); and plasma (PL) or serum (SE) were preferred for their easier sample processing and acceptable analytical performance (11-14). This lack of consensus may be mainly attributed to a variety of PCR assays using different instruments, reagents, DNA extraction methods, and conducted in different centers, leading to interlaboratory variation and creating increasing clinical concern (15-18).
The aim of our study was to explore the best blood component for HCMV DNA surveillance after liver transplantation using one commercially available assay.
Material and Methods
Patient samples
A total of 168 sets of samples (each set consisted of WB, SE, and PL collected simultaneously from 1 patient) were gathered from 140 liver transplant recipients (133 adults and 7 infants) during their admission to the Ren Ji Hospital. The samples included 42 sets of serial specimens obtained during clinical follow-up. SE and PL were obtained from both non-anticoagulated and EDTA-anticoagulated blood after centrifugation at 926 g for 5 min. All samples were stored at -20±3°C before analysis.
Sample analysis
RTFQ-PCR was performed with a commercial HCMV kit (DaAn Gene Co., Ltd., China) using an ABI Prism 7500 Sequence Detection System (Applied Biosystems, USA). Since the kit does not provide reagents for WB DNA extraction, the WB samples were processed with a Tiangen Blood Genome Extraction kit (DP318, Tiangen Biotech Co., Ltd., China). The initial volume for DNA extraction from WB, SE, or PL was 100 µL. According to the manufacturer's protocol, PCR was carried out on 96-well plates with a 45-µL reaction volume containing 2 µL DNA template, 3 µL Taq polymerase, and 40 µL PCR solution. The thermocycler procedure was as follows: initial denaturation at 93°C for 2 min, followed by 10 cycles at 93°C for 45 s and 55°C for 60 s; 30 cycles at 93°C for 30 s and 55°C for 45 s. Four calibrations (4-7 log10 copies/mL), one negative and two positive controls (weakly and highly positive, respectively) provided in the kit were all included in each run. The limit of detection (LOD) was 2.7 log10 copies/mL (i.e., 500 copies/mL) according to the manufacturer's operating manual. Positive results below the LOD were referred to as “weakly positive”. The three comparative specimens (WB, SE, and PL) in each sample set were analyzed in parallel in the same PCR experiment to avoid interassay variation.
Statistical analysis
Chi-square and Fisher exact tests were carried out to compare qualitative results, and paired t-tests were used to analyze quantitative data. All calculations were done with the SPSS 14.0 program (SPSS Inc., USA). The statistical significance was set at P<0.05.
Results
Analysis of sensitivity
As the qualitative results obtained with SE were identical to those for PL, they were combined into one group for comparison with WB. As shown in Table 1, 14 (8.33%) of the sample sets tested positive and 140 (83.33%) tested negative with both WB and SE/PL. The remaining 14 (8.33%) sets were WB-positive and SE/PL-negative. Low DNA levels were observed in these discrepant samples (median, 2.31 log10 copies/mL; range, 1.44-3.06 log10 copies/mL). Therefore, the positivity rate of WB (16.67%) was much higher than that of SE/PL (8.33%) (Χ2 = 76.36, P<0.01).
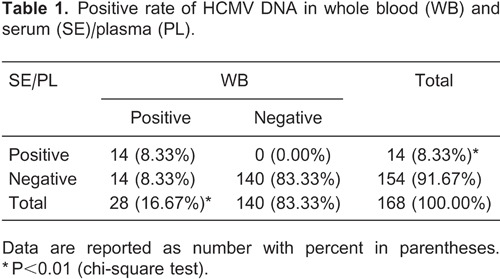
Comparison of HCMV DNA loads of WB, SE and PL samples
In comparing DNA levels quantitatively, the 14 sets with PCR-positive results for all three materials were log-transformed and presented as DNA value ranges, quartile ranges, and median viral load (Figure 1). HCMV DNA loads observed with WB were significantly greater (paired t-test) than those observed in either SE (WB-SE mean log-transformed difference, 0.99; 95%CI=0.74-1.25; P<0.0001) or PL (WB-PL mean log-transformed difference, 1.37; 95%CI=1.07-1.66; P<0.0001). No significant difference was found between the latter two materials (SE-PL mean log-transformed difference, 0.37; 95%CI=0.19-0.55; P=0.6678).
Figure 1. Quantitative comparison of HCMV DNA loads measured with WB, SE and PL. WB: whole blood: SE: serum; PL: plasma. Statistically significant differences (aP<0.0001, bP=0.6678, cP<0.0001, paired t-test).
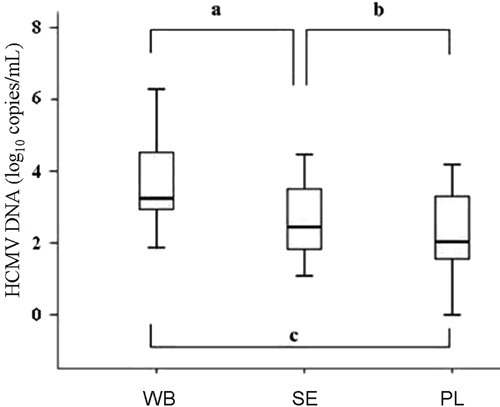
Sequential monitoring of HCMV DNA during clinical follow-up
To study the viral kinetics during the course of active infection and antiviral therapy, we recorded two typical cases of successive DNA values during clinical follow-up.
Case 1A 1-year-old infant, HCMV IgM(-)/IgG(-), received a living donor [HCMV IgM(-)/IgG(+)] liver transplant for congenital bile duct atresia. HCMV DNA was assayed weekly post-transplantation to guide antiviral intervention. As shown in Figure 2A, low-level DNA values were detected in WB at the first (2.76e+1 copies/mL) and second (7.90e+2 copies/mL) weeks after transplantation, while the corresponding SE or PL samples indicated no viral replication. Afterwards, DNA values obtained with WB continued to rise to 3.34e+4 copies/mL at the third week, and high levels persisted (4.16e+4 copies/mL) after the fourth week. The measured viral loads sharply increased to 3.18e+3 and 1.98e+3 copies/mL for SE and PL, respectively, at the third week and persisted at the fourth week of follow-up (5.09e+3 and 3.13e+3 copies/mL, for SE and PL, respectively). These values were both significantly lower than the WB counterparts obtained at the same times. Unfortunately, this young patient died of multiple organ failure on the 27th day after transplantation due to poor response to antiviral therapy.
Figure 2. Monitoring of HCMV DNA for 2 clinical cases. A, 1-year-old infant; B, 44-year-old male adult. Lozenges: whole blood; squares: serum; triangles: plasma. The dashed line indicates the limit of detection (2.7 log10 copies/mL).
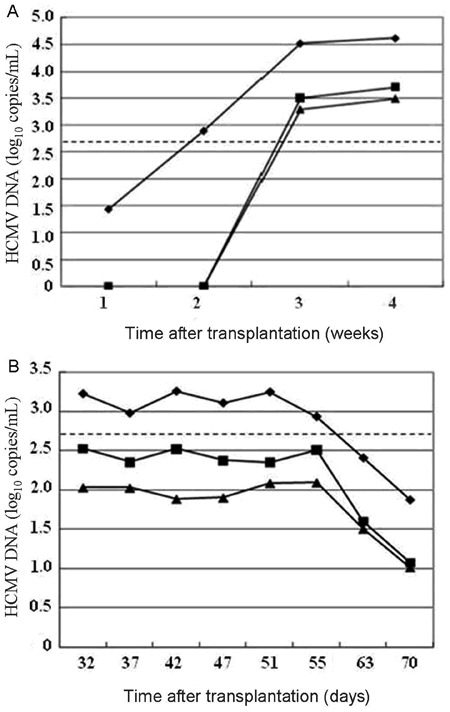
Case 2.A 44-year-old male adult, HCMV IgM(-)/IgG(+), was evaluated for HCMV DNA because of abnormal liver function 1 month after standard orthotopic liver transplantation [donor: HCMV IgM(-)/IgG(+)]. Positive results were obtained in all three samples, and, accordingly, intravenous ganciclovir treatment (250 mg, twice a day) was initiated immediately. Figure 2B summarizes DNA kinetics weekly during the course of treatment. In spite of similar declining rates and degrees, viral loads with WB appeared to persist for a longer period of time than the other two owing to its higher DNA levels at baseline.
The result of HCMV DNA surveillance in these two patients confirmed that the WB sample was positive earlier, yielded higher viral loads, and persisted positive longer during treatment compared with SE or PL.
Discussion
The high risk of HCMV activation after organ transplantation has attracted wide attention in clinical settings. Timely initiation of antiviral therapy and optimal management for infected patients rely mainly on early detection of viral activation as well as effective monitoring of therapeutic response.
Our data indicated that WB is more sensitive and yields higher DNA values than SE or PL (Table 1 and Figure 1). The biological features of HCMV infection may contribute to this finding. PBL is one of the usual sites that harbor HCMV in healthy adults with latent infection. When the virus replicates at a low level, such as at the early stage of reactivation or at the end of certain therapeutic regimens, only a few viruses are released into the serum or plasma. Thus, WB material that contains a large number of PBLs could appear to have higher viral loads than either SE or PL. This result is also consistent with some previously reported data (8-10). As for whether use of different DNA extraction kits for WB introduces technical bias, a previous report also demonstrated the superiority of WB when employing a common extraction method for both WB and PL (9). In this study, each set of comparative samples (i.e., WB, SE, and PL) were measured in parallel in one PCR experiment to maintain the same efficiency of amplification; thus, the PCR results could be reflective of original viral loads in corresponding specimens. Although extra steps required for DNA extraction from WB require additional time and expense, its capability to detect low-level HCMV DNA could provide clinicians with more detailed information about viral replication, which could help guide preemptive protocols as well as predict subsequent relapse following the completion of certain therapeutic regimens.
Since the kinetics of DNA replication is one of the important factors for predicting HCMV disease, monitoring HCMV DNA during follow-up has been proposed as a practical and efficient method to guide individualized therapy (19). Surveillance in clinical Case 1 revealed that WB samples have the advantage of showing earlier DNA positivity and more continuous viral dynamics when monitoring the development of HCMV disease. Moreover, Case 1 also suggests that a sharp increase in viral load during regular monitoring may be a strong signal of a burst of HCMV infection leading to deterioration of the patient. In contrast, Case 2 displayed gradually decreasing HCMV DNA in response to ganciclovir treatment. The downward slope of the DNA curves implied that the patient responded well to antiviral therapy and a good prognosis could be anticipated. Otherwise, persistence of high DNA levels during treatment might indicate drug failure for some reason (e.g., drug resistance or inappropriate dosages). To summarize, WB was superior to SE or PL for HCMV DNA surveillance because of earlier positivity and more continuous and complete recording of viral load dynamics.
With progress in transplant treatment, liver transplantation has been increasingly performed to cure congenital hepatobiliary diseases such as bile duct atresia. However, infant recipients are very susceptible to primary HCMV infection, which may easily be life threatening because of their immature immune system. In our study, infant patients showed significantly higher HCMV DNA and IgM and much lower IgG-positive rates than adults. Thus, more intensive monitoring (e.g., every 2-3 days instead of every week) is recommended, and antiviral intervention should be initiated without any delay in case of primary HCMV infection, even at low DNA levels, as discussed in a previous report by Kalpoe et al. (20). Nevertheless, the number of infant cases in this study was rather small and more cases are needed for further investigation of this issue in the near future.
In summary, this study demonstrated that WB compares favorably to SE or PL for HCMV DNA detection because of higher sensitivity and greater DNA yields. Monitoring DNA loads in WB samples by clinical follow-up offers a practical and effective approach to guide individualized therapy. Infant recipients who are usually at increased risk of primary HCMV infection need closer surveillance and more active intervention to improve survival rates after transplantation.
Acknowledgments
We thank Dr. Xiaosong Chen for reviewing clinical cases and Prof. Hongsheng Zhu for checking the manuscript. The research was supported by the Fund for Young Investigators (2010-2012) of Ren Ji Hospital, School of Medicine, Shanghai Jiao Tong University.
Footnotes
First published online April 7, 2014.
References
- 1.Zhang Y, Ji YH. Surveillance of HCMV DNA in transplant setting. Chin J Lab Med. 2006;29:567–568. [Google Scholar]
- 2.Li M, Zhao WL, Ji YH, Peng YB, Zhi LM, Pan N. Monitoring of HCMV infection in bone marrow transplant recipients by quantitative PCR. Acta Univ Med Sec Shanghai. 2002;22:143–145. [Google Scholar]
- 3.Gao LH, Zheng SS. Cytomegalovirus and chronic allograft rejection in liver transplantation. World J Gastroenterol. 2004;10:1857–1861. doi: 10.3748/wjg.v10.i13.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pellegrin I, Garrigue I, Ekouevi D, Couzi L, Merville P, Merel P, et al. New molecular assays to predict occurrence of cytomegalovirus disease in renal transplant recipients. J Infect Dis. 2000;182:36–42. doi: 10.1086/315688. [DOI] [PubMed] [Google Scholar]
- 5.Boeckh M, Gallez-Hawkins GM, Myerson D, Zaia JA, Bowden RA. Plasma polymerase chain reaction for cytomegalovirus DNA after allogeneic marrow transplantation: comparison with polymerase chain reaction using peripheral blood leukocytes, pp65 antigenemia, and viral culture. Transplantation. 1997;64:108–113. doi: 10.1097/00007890-199707150-00020. [DOI] [PubMed] [Google Scholar]
- 6.Zhang S, Zhou YH, Li L, Hu Y. Monitoring human cytomegalovirus infection with nested PCR: comparison of positive rates in plasma and leukocytes and with quantitative PCR. Virol J. 2010;7:73. doi: 10.1186/1743-422X-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li M, Ji YH, Peng YB, Zhi LM, Liu M, Huang T, et al. Comparison of two methods for earlier detection of active HCMV infection. Chin J Lab Med. 2002;25:220–222. [Google Scholar]
- 8.Garrigue I, Boucher S, Couzi L, Caumont A, Dromer C, Neau-Cransac M, et al. Whole blood real-time quantitative PCR for cytomegalovirus infection follow-up in transplant recipients. J Clin Virol. 2006;36:72–75. doi: 10.1016/j.jcv.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Razonable RR, Brown RA, Wilson J, Groettum C, Kremers W, Espy M, et al. The clinical use of various blood compartments for cytomegalovirus (CMV) DNA quantitation in transplant recipients with CMV disease. Transplantation. 2002;73:968–973. doi: 10.1097/00007890-200203270-00025. [DOI] [PubMed] [Google Scholar]
- 10.Deback C, Fillet AM, Dhedin N, Barrou B, Varnous S, Najioullah F, et al. Monitoring of human cytomegalovirus infection in immunosuppressed patients using real-time PCR on whole blood. J Clin Virol. 2007;40:173–179. doi: 10.1016/j.jcv.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 11.Zhao XT, Zhang Z, Zhang YR, Liu DM, Sun YY. Quantitative detection of CMV DNA by real-time PCR in transplant recipients. Chin J Lab Med. 2006;29:F438–F440. [Google Scholar]
- 12.Kaiser L, Perrin L, Chapuis B, Hadaya K, Kolarova L, Deffernez C, et al. Improved monitoring of cytomegalovirus infection after allogeneic hematopoietic stem cell transplantation by an ultrasensitive plasma DNA PCR assay. J Clin Microbiol. 2002;40:4251–4255. doi: 10.1128/JCM.40.11.4251-4255.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yerly S, Perrin L, Van Delden C, Schaffer S, Thamm S, Wunderli W, et al. Cytomegalovirus quantification in plasma by an automated real-time PCR assay. J Clin Virol. 2007;38:298–303. doi: 10.1016/j.jcv.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Patel R, Smith TF, Espy M, Wiesner RH, Krom RA, Portela D, et al. Detection of cytomegalovirus DNA in sera of liver transplant recipients. J Clin Microbiol. 1994;32:1431–1434. doi: 10.1128/jcm.32.6.1431-1434.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koidl C, Bozic M, Marth E, Kessler HH. Detection of CMV DNA: is EDTA whole blood superior to EDTA plasma? J Virol Methods. 2008;154:210–212. doi: 10.1016/j.jviromet.2008.08.012. [DOI] [PubMed] [Google Scholar]
- 16.Mengelle C, Mansuy JM, Da Silva I, Davrinche C, Izopet J. Comparison of 2 highly automated nucleic acid extraction systems for quantitation of human cytomegalovirus in whole blood. Diagn Microbiol Infect Dis. 2011;69:161–166. doi: 10.1016/j.diagmicrobio.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 17.Gracia-Ahufinger I, Tormo N, Espigado I, Solano C, Urbano-Ispizua A, Clari MA, et al. Differences in cytomegalovirus plasma viral loads measured in allogeneic hematopoietic stem cell transplant recipients using two commercial real-time PCR assays. J Clin Virol. 2010;48:142–146. doi: 10.1016/j.jcv.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Von Muller L, Hampl W, Hinz J, Meisel H, Reip A, Engelmann E, et al. High variability between results of different in-house tests for cytomegalovirus (CMV) monitoring and a standardized quantitative plasma CMV PCR assay. J Clin Microbiol. 2002;40:2285–2287. doi: 10.1128/JCM.40.6.2285-2287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Emery VC, Sabin CA, Cope AV, Gor D, Hassan-Walker AF, Griffiths PD. Application of viral-load kinetics to identify patients who develop cytomegalovirus disease after transplantation. Lancet. 2000;355:2032–2036. doi: 10.1016/S0140-6736(00)02350-3. [DOI] [PubMed] [Google Scholar]
- 20.Kalpoe JS, Kroes AC, de Jong MD, Schinkel J, de Brouwer CS, Beersma MF, et al. Validation of clinical application of cytomegalovirus plasma DNA load measurement and definition of treatment criteria by analysis of correlation to antigen detection. J Clin Microbiol. 2004;42:1498–1504. doi: 10.1128/JCM.42.4.1498-1504.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


