Abstract
In this review, we consider a selection of recent advances in chloroplast biology. These include new findings concerning chloroplast evolution, such as the identification of Chlamydiae as a third partner in primary endosymbiosis, a second instance of primary endosymbiosis represented by the chromatophores found in amoebae of the genus Paulinella, and a new explanation for the longevity of captured chloroplasts (kleptoplasts) in sacoglossan sea slugs. The controversy surrounding the three-dimensional structure of grana, its recent resolution by tomographic analyses, and the role of the CURVATURE THYLAKOID1 (CURT1) proteins in supporting grana formation are also discussed. We also present an updated inventory of photosynthetic proteins and the factors involved in the assembly of thylakoid multiprotein complexes, and evaluate findings that reveal that cyclic electron flow involves NADPH dehydrogenase (NDH)- and PGRL1/PGR5-dependent pathways, both of which receive electrons from ferredoxin. Other topics covered in this review include new protein components of nucleoids, an updated inventory of the chloroplast proteome, new enzymes in chlorophyll biosynthesis and new candidate messengers in retrograde signaling. Finally, we discuss the first successful synthetic biology approaches that resulted in chloroplasts in which electrons from the photosynthetic light reactions are fed to enzymes derived from secondary metabolism.
General characteristics of chloroplasts
The first photosynthetic eukaryotes originated more than 1000 million years ago through the primary acquisition of a cyanobacterial endosymbiont by a eukaryotic host, which subsequently gave rise to glaucophytes (whose photosynthetic organelles are called “cyanelles”), red algae (containing “rhodoplasts”) and green algae and plants (with “chloroplasts”). Other major photosynthetic eukaryotic lineages arose when eukaryotic hosts engulfed a free-living photosynthetic eukaryote (e.g. red or green alga), initiating secondary and tertiary endosymbioses [1]. Therefore, chloroplasts are organelles that are characteristic of plant and green algal cells, but still exhibit many prokaryotic features.
During evolution, the cyanobacterium-derived genome has undergone a dramatic reduction in size, mainly as a result of outright gene loss and the large-scale transfer of genes to the nuclear genome. Thus, the genomes of modern chloroplasts (plastomes) contain only 120-130 genes, most of which encode components of the organelle's gene expression machinery and its photosynthetic apparatus, and are organized in nucleoids that show both prokaryotic and eukaryotic features. However, chloroplasts contain many more protein species than their plastomes can code for. Hence, the majority of chloroplast proteins are now encoded by the nuclear genome and must be imported post-translationally into the organelle [2].
Apart from photosynthesis, chloroplasts are capable of performing many other specialized functions that are essential for plant growth and development — nitrate and sulphate assimilation, and the synthesis of amino acids and fatty acids, chlorophyll and carotenoids. To carry out these tasks, their membrane systems are equipped with specialized transport functions. The outer and inner envelope membranes mediate the import and sorting of proteins and the exchange of metabolites, while protein complexes in the thylakoid membranes implement the proton and electron transport processes that are an essential part of the photosynthetic light reactions. The thylakoids of land plants, where photosynthesis takes place, display an intricate architecture, with regions of stacked and appressed thylakoid membranes forming so-called grana. Moreover, plastids communicate with the nucleus by retrograde signaling to adjust the expression of nuclear genes according to the metabolic and developmental state of the organelle.
Chloroplast evolution
Primary endosymbiosis: a ménage à trois?
New evidence implies that primary endosymbiosis might have been more complex than has been envisaged hitherto, possibly involving a “ménage à trois”. Members of the genus Chlamydia are obligate intracellular bacteria, which include important pathogens of humans and other animals, and are found as endosymbionts in amoebae and insects. Although Chlamydiae are not found in plants, an unexpected number of chlamydial genes share significant similarities with plant genes [3,4], and these often contain a plastid-targeting signal [5]. In several studies, between 21 and 55 genes were shown to be transferred between Chlamydiae and primary photosynthetic eukaryotes [6-8]. This suggests that a protist lineage that could enter into a symbiosis with a particular cyanobacterium was routinely infected by an ancestor of extant Chlamydia that facilitated the establishment of the cyanobacterial endosymbiont by Chlamydia-to-protist lateral gene transfer [7]. Recent studies suggest that the chlamydial symbiont compartment was probably not the site of any essential biochemical pathway and was maintained only until all possible chlamydial genes had been transferred to the host [6]. Thus, the two critical steps in primary plastid endosymbiosis might have been the secretion of effector proteins into the host cytosol by intracellular chlamydial pathogens, together with the maintenance of the afflicted host by the cyanobiont, which supplied photosynthetic carbon to a chlamydia-controlled assimilation pathway [6]. If such complex interactions were indeed necessary for the establishment of the primary endosymbiotic relationship between plastid and host cytoplasms, this could explain why endosymbiotic relationships between heterotrophs and photoautotrophs were so rarely successful in the long term [7].
A second primary endosymbiosis
Evidence has also emerged for an independent instance of the primary endosymbiotic acquisition of a cyanobacterium — by the rhizarian amoeba Paulinella chromatophora about 60 million years ago [9,10]. This organism contains stably transmitted cyanobacterium-like photosynthetic organelles termed “chromatophores”, the genome of which encodes about a quarter of the protein-coding genes that can be found in its free-living relative Synechococcus WH5701 [10]. Eleven putative pseudogenes were identified, indicating that reductive genome evolution is ongoing. More than 30 expressed genes have been transferred from the chromatophore to the nuclear genome of the host. In the case of three photosynthetic genes that now reside in the nucleus, biochemical evidence indicates that their products are synthesized in the amoeba cytoplasm and delivered to the chromatophores, where they form complexes with chromatophore-encoded subunits [11]. This highlights P. chromatophora as an exceptional model for the study of early events in the generation of an organelle, and suggests that protein import into bacterial endosymbionts might be more widespread than is currently assumed [11].
What is the basis for the longevity of kleptoplasts?
Kleptoplasts are a special case of transient internal photosynthetic symbionts in otherwise non-photosynthetic eukaryotes. In contrast to some lineages, in which the cells of photosynthetic symbionts are retained in their entirety (“photosymbionts”), other eukaryotes collect and retain only the chloroplasts of photosynthetic species, generating structures termed “kleptoplasts” (reviewed in [12]). The most dramatic kleptoplast association known to date occurs in the sacoglossan sea slug Elysia chlorotica, which can maintain photosynthetically active kleptoplasts derived from ingested xanthophyte algae for up to 10 months [13]. This gives these animals their distinctive green colour, which is why they are also called “leaves that crawl”, “solar-powered slugs” or “photosynthetic slugs”. It is widely assumed that the slugs survive starvation by means of kleptoplast photosynthesis, yet direct evidence for this is lacking. Moreover, the inference that kleptoplasts require many proteins in order to support a photosynthetic lifestyle implies that essential genes for photosynthesis have been transferred by lateral gene transfer (LGT) from the alga to the slug, and in fact one instance of a tentative transfer has been reported so far [14]. However, no evidence for massive LGT has been obtained, and genome- and transcriptome-wide approaches actually argue against it [15,16]. Recently, doubts have been raised as to whether these molluscs are actually dependent upon photosynthesis, and the role of light in the survival of the sea slugs was reinvestigated [17]. Surprisingly, photosynthesis was found not to be essential for the slugs to survive months of starvation, which explains the lack of LGT from alga to animal in these species. A possible explanation for the longevity of the sacoglossan kleptoplast was suggested previously: plastids that remain photosynthetically active within slugs for periods of months share the property of encoding FtsH, a D1 quality-control protease that is essential for photosystem II repair [18]. A replenishable supply of chloroplast-encoded FtsH could, in principle, rescue kleptoplasts from D1 photodamage, thereby influencing plastid longevity in sacoglossan slugs.
Chloroplast structure
Nucleoids
A single mesophyll chloroplast can contain up to 300 chromosomes, which are organized into complex structures called “nucleoids”, each consisting of 10-20 copies of the plastid genome, together with RNA and various proteins (for a recent review see [19]). Owing to their endosymbiotic origin and the fact that photosynthetic metabolism goes on all around them, nucleoids have a unique composition and organization, and display features typical of prokaryotic nucleoids, as well as attributes of eukaryotic chromatin. Nucleoids contain all the enzymes necessary for transcription, replication and segregation of the plastid genome (reviewed in [20]). In addition, mRNA processing and editing, as well as ribosome assembly, take place in association with the nucleoid, suggesting that these processes occur co-transcriptionally. However, few nucleoid proteins have been characterized in detail [19,21].
Proteomic analysis of nucleoid preparations has identified new DNA-binding proteins, some of which were not inherited from the prokaryotic ancestors [22,23]. One group of proteins in particular have been described, which contains a so-called SWIB domain that has previously been shown to be part of chromatin remodelling complexes in yeast. This domain is present in 20 proteins in Arabidopsis, and at least four of these are located in the chloroplast [23]. The SWIB-domain proteins in chloroplasts are small proteins with a high isoelectric point and a high lysine content and might serve as functional replacements for the bacterial histone-like, DNA-binding HU proteins known from Escherichia coli [23]. Thus one of them, SWIB-4, has a histone H1-like motif and binds to DNA, and recombinant SWIB-4 has been shown to induce compaction and condensation of nucleoids, and functionally complements an E. coli mutant that lacks the histone-like nucleoid structuring protein H-NS [23].
The two suppressor of variegation 4 (SVR4) proteins (SVR4 and SVR4-like), originally identified in Arabidopsis [24], both have orthologues in all dicot and monocot plants sequenced so far, whereas the moss Physcomitrella patens and spikemoss Selaginella moellendorffii contain only one gene copy, indicating that a gene duplication took place in the progenitor of vascular plants [25]. Inactivation of either SVR4 or SVR4-like in Arabidopsis results in seedling lethality [24,26]. Both proteins are localized in the chloroplast, expressed during early stages of chloroplast development, and contain 20% negatively charged amino acid residues [26]. Given the inherent risk of random aggregation of the negatively charged nucleic acids and basic proteins, such as histones and ribosomal subunits [27-29], SVR4 and SVR4-like could function as negatively charged molecular chaperones that mimic nucleic acids or serve as decoys [28,30] to allow for the establishment of productive DNA/RNA-protein interaction in developing chloroplasts, where dramatic rearrangements in nucleoid organization take place [26].
Thylakoid architecture
A structural hallmark of thylakoid membranes in plants are the so-called “grana” (reviewed in [31]). Grana cylinders are made up of stacks of flat grana membrane discs with a diameter of about 300-600 nm, which are enwrapped in (and interconnected by) the unstacked stroma lamellae. Tightly curved margins form the periphery of each discoid sac. For a typical granum from Arabidopsis thaliana the membrane bilayers are on average 4.0 nm thick, lumen thickness is 4.7 nm and discs are separated by a 3.6 nm gap [32].
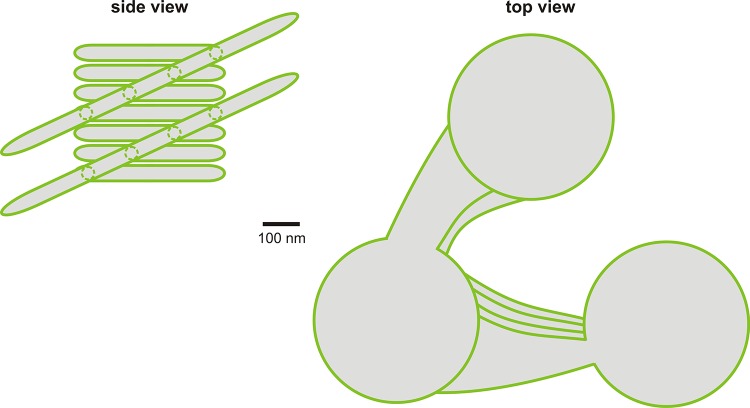
The exact three-dimensional architecture of grana is still under debate, and two quite different types of models have been proposed: the “helical” model and several “fork/bifurcation” models. In the helical model, thylakoids comprise a fretwork of stroma lamellae, which wind around grana stacks as a right-handed helix, connecting individual grana disks via narrow membrane protrusions (Figure 1). The grana are connected to each other solely by the stroma lamella helices, which are tilted at an angle ranging from 10 to 25°, with respect to the grana stacks [33-35], and make multiple contacts with successive layers in the grana through slits located in the rims of the stacked discs. The fork/bifurcation models, on the other hand, postulate that the grana themselves are formed by bifurcations of stroma lamellae. Thus, Arvidsson and Sundby (1999) suggested that a granum contains piles of repeat units, each containing three grana discs, which are formed by symmetrical invaginations of a thylakoid pair caused by the bifurcation of the thylakoid membrane [36] (Figure 1). Shimoni et al. (2005) presented another model in which grana discs are paired units formed by simple bifurcation of stroma thylakoids [37] (Figure 1). Here, the granum-stroma assembly is formed by bifurcations of the stroma lamellar membranes into multiple parallel discs. The stromal membranes form wide lamellar sheets that intersect the granum body roughly perpendicular to the long axis of the granum cylinder [37,38]. In this model, adjacent granum layers are joined not only through the stroma lamellae, but via the bifurcations and through direct membrane bridges. This model can also be used to explain the rearrangements seen in thylakoids during state transitions [39]. The mutual incompatibility of the helical and bifurcation models has led to a great deal of debate [33,35,38,40,41], but recent tomographic data clearly support the helical model [33,40].
Figure 1. The helical model of thylakoid architecture.
A fretwork of stroma lamellae, which wind around the ascending grana stacks as a right-handed helix connects to individual grana discs via narrow membrane protrusions (indicated by dotted circles in the side view). Adapted with permission [31] 2014. Journal of Experimental Botany doi:10.1093/jxb/eru090.
Lateral heterogeneity of thylakoids
The term “lateral heterogeneity” refers to the observation that grana and stroma lamellae differ in their protein composition. Photosystem II and light-harvesting complex (LHC)II are concentrated in the grana, while photosystem I with its LHCI and the chloroplast ATP synthase are localized in the unstacked thylakoid regions, that is the stroma lamellae and grana end membranes. The cytochrome b6f complex (Cyt b6f) can be found in both appressed and non-appressed regions of thylakoids (reviewed in [41,42]). The NDH complex and the PGRL1-PGR5 heterodimer – the two thylakoid complexes specifically involved in cyclic electron flow – are less abundant than the aforementioned four major thylakoid complexes and are located in the stroma lamellae [43–45], where they can functionally interact with photosystem I as an electron donor. While the bulkiness of the NDH complex precludes its location in grana, PGRL1 homodimers have been detected in grana [43].
Detection of several of the major thylakoid multiprotein complexes in margin-enriched fractions of thylakoids by biochemical methods has been reported in some experiments. However, the marked curvature of thylakoid membranes at the grana margins is essentially incompatible with the presence of the larger multiprotein complexes at these sites. Therefore, grana margins have been thought to be essentially protein-free (reviewed in [42]). However, following the recent demonstration, by immunogold labelling, that CURT1 proteins — small polypeptides with two transmembrane regions and a putative N-terminal amphipathic helix — are localized to grana margins [46], this view must be revised. Interestingly, the CURT1 proteins appear to control the level of grana stacking, which points to an unsuspected role of grana margins in regulating the fraction of thylakoid membranes incorporated into the appressed regions that make up grana.
Chloroplast functions
Chloroplast proteins: mutants and proteomes
Estimates for the size of the chloroplast proteome in Arabidopsis range from 2000 [47] to 4400 (http://www.plastid.msu.edu/) different proteins. In the course of the Chloroplast 2010 Project (http://www.plastid.msu.edu/), homozygous mutants for several thousand nuclear genes with chloroplast functions were identified and phenotypically characterized. Despite extensive screening, for several hundred genes no homozygous mutant alleles were discovered, suggesting that these might represent genes with essential functions. More recently, lines that had failed to yield any homozygotes when grown in soil were tested for homozygous lethality owing to defects either in seed or seedling development [48,49]. Mutants arrested at various stages of seed development (and with defects in seedling development that responded to supplementation with sucrose, amino acids or to CO2 enrichment) were indeed uncovered. This resulted in an annotation of more than 200 publically available Arabidopsis mutants, including 36 and 33 genes with one and two, respectively, independent seed- or seedling-development-defective mutant alleles. The study also resulted in the submission of 521 homozygous mutants and 128 seed stocks segregating for lethal alleles to the Arabidopsis Biological Resource Center (ABRC; http://www.arabidopsis.org).
Proteomics usefully complement the reverse genetics approach to chloroplast function outlined above. Driven by recent advances in bioanalytical and computational technologies, the strategy allows for identification, and reasonably accurate quantification, of thousands of proteins in complex mixtures, as well as the ability to characterize post-translational modifications, such as acetylation, glycosylation and phosphorylation (reviewed in [50]). An attempt to obtain a high-quality inventory of the plastid proteome has led to the identification of 1564 and 1559 proteins for maize and Arabidopsis, respectively [51]. These estimates were based on both manual curation of published experimental information, including more than 150 proteomics studies devoted to different subcellular fractions, and new quantitative proteomics experiments on plastid subfractions. These figures correspond to an estimated 40% and 50% of all plastid proteins in maize and Arabidopsis, respectively — the most comprehensive inventory assembled so far. Recently, members of the Arabidopsis proteomics community decided to develop a summary aggregation portal that is capable of retrieving proteomics data from a series of online resources on the fly [52]. The web portal is known as the MASCP Gator and can be accessed at the following address: http://gator.masc-proteomics.org/.
Chlorophyll biosynthesis
Biosynthesis of chlorophyll takes place in the plastid, and the initial steps in the pathway leading to protoporphyrin IX are common to the biosynthesis of other tetrapyrroles, such as heme. Important discoveries in chlorophyll biosynthesis include the demonstration that plastid glutamyl-transfer RNA is involved in the formation of glutamate-1-semialdehyde [53], which is subsequently converted into 5-aminolevulinic acid — the universal precursor of tetrapyrrole biosynthesis in all organisms (reviewed in [54]), and the finding that the enzyme Mg-chelatase (which catalyses the insertion of the Mg2+ ion into protoporphyrin IX) contains three different protein subunits: ChlH, ChlI, and ChlD [55,56]. Later, the GENOME UNCOUPLER4 (GUN4) was found to bind both the substrate and the product of the Mg-chelatase, thereby dramatically enhancing the activity of the enzyme [57]. GUN4 also reduces the threshold Mg2+ concentration required for activity at low porphyrin concentrations [58], and it was proposed to have a protective function in tetrapyrrole trafficking [59] and to control Mg-chelatase activity at physiologically significant Mg2+ concentrations [58]. More recently, it was shown that GUN4 interacts with the ChlH subunit of the enzyme [60,61].
One of the least understood steps in chlorophyll biosynthesis is the formation of the isocyclic “fifth” ring (ring-E), which is catalysed in plants by the aerobic cyclase system (ACS). The overall cyclase reaction is a six-electron oxidation proposed to occur in three sequential steps: (a) hydroxylation of the methyl-esterified ring-C propionate by incorporation of atmospheric oxygen; (b) oxidation of the resulting alcohol to the corresponding ketone; (c) reaction of the activated methylene group with the γ -mesocarbon of the porphyrin nucleus in an oxidative reaction involving the removal of two protons to yield the “fifth” ring [62]. At the biochemical level, the ACS requires both soluble and membrane-bound chloroplast fractions and, in barley, at least two mutants exist (xantha-l and viridis-k) which are defective in the membrane components [63]. Thus, the ACS may be composed of three gene products: a soluble protein and two membrane-bound components — one encoded in barley by Xantha-l and the other by Viridis-k. So far, only AcsF (which corresponds to Xantha-l in barley, CRD1 in Chlamydomonas or CHL27 in Arabidopsis) has been unambiguously identified [63–65]. As diiron enzymes are known to perform hydroxylation and cyclization of keto intermediates, AcsF could be involved in one or more of the proposed cyclase steps. Recent progress has come from pull-down experiments using FLAG-tagged versions of the two AcsF-like gene products in Synechocystis in combination with protein mass spectrometry, which have identified the soluble YCF54 protein as a new putative subunit of ACS [66]. Inactivation of the Synechocystis ycf54 gene resulted in significantly reduced chlorophyll levels, marked accumulation of the substrate of the cyclase, Mg-protoporphyrin IX methyl ester, and only traces of its product, protochlorophyllide, indicating that YCF54 is essential for the activity and/or stability of the oxidative cyclase. Future experiments must clarify whether YCF54 is the long-sought soluble component of the cyclase system, or whether it functions in AcsF synthesis/maturation or in cyclase assembly. Low chlorophyll accumulation A (LCAA), the tobacco homologue of YCF54, might have an additional role in the feedback-control of 5-aminolevulinic acid biosynthesis [67]. Because the structure of YCF54 is similar to that of the photosystem II assembly factor Psb28 (see Table 1), YCF54 might also be involved in coordinating chlorophyll biosynthesis and photosystem biogenesis.
Table 1. Accessory factors involved in the assembly of thylakoid multiprotein complexes in plants and cyanobacteria.
| PSII | PSI | Cyt b6f | cpATPase | NDH |
|---|---|---|---|---|
| HCF136A[103]/ YCF48S[104] | YCF3C,T[85,86] | CCS1C[105] | ALB4A[106] | AtCYP20-2A[107] |
| ALB3A,C[108,109]/ Slr1471S[110] | YCF4C,T[87,88] | CCB1C,A[111,112] | AtCGL160A[113] | CRR1A[114] |
| YCF39S[115] | YCF37S[116]/Pyg7[117] | CCB2C,A[111,112] | CRR6A[118]/Slr1097S[119] | |
| LPA1A[120]/REP27C[121] | PPD1T[122] | CCB4C,A[111,112] | CRR7A[123] | |
| LPA2A[124] | Y3IP1T,A[125] | DACA[126] | CRR41A[127] | |
| LPA3A[128] | PBF1T[129] | CRR42A[127] | ||
| Slr2013S[130] | HCF101A[131–133] | NDF5A[134] | ||
| Psb27S[135–138]/ LPA19A[139] | RubAS[140,141] | PAM68LA[75] | ||
| Psb28S[142] | ||||
| Psb29/THF1S,A[143–145] | ||||
| PratAS[146] | ||||
| PittS[147] | ||||
| AtCYP38A[148] | ||||
| PAM68A,S[149] |
Organism in which the assembly factor was functionally characterized: A, Arabidopsis, C, Chlamydomonas, S, Synechocystis or Synechococcus, T, tobacco. Abbreviations: cpATPase, chloroplast ATP synthase; Cyt, cytochrome; PSI, photosystem I; PSII, photosystem II.
Photosynthesis: new proteins and new functions
It comes as a surprise to learn that some proteins that are directly involved in the light reactions of photosynthesis have remained unidentified until very recently. Thus, although antimycin A-sensitive cyclic electron flow (AA-sensitive CEF), which serves to recycle electrons from ferredoxin to plastoquinone, was discovered by Arnon and co-workers more than 50 years ago, it is only a few years since the proteins responsible were identified. A role in AA-sensitive CEF has been attributed to the two thylakoid proteins PGR5 [68] and PGRL1 [69] ever since their identification, but this assignment has remained controversial. Indeed, current technical limitations still preclude unequivocal clarification of their precise function in CEF in vivo, but recent biochemical experiments have shown that PGRL1/PGR5 complexes possess ferredoxin-plastoquinone reductase (FQR) activity in vitro [43]. Consequently, PGRL1-PGR5 complexes in flowering plants appear to shuttle between photosystem I and the cytochrome (Cyt) b6f complex, whereas in the green alga Chlamydomonas PGRL1 (but not PGR5) has been detected in a photosystem I cytochrome b6f supercomplex that has intrinsic CEF activity [70].
The second pathway mediating CEF involves the so-called “NAD(P)H dehydrogenase complex” or “NDH complex”. Although the plant NDH complex is related to the NADH dehydrogenase complexes of bacteria and mitochondria, its function and composition are enigmatic. Recently, the Shikanai group has identified three novel subunits of plant NDH (CRR-31, -J and –L) and their functional characterization clearly indicated that CRR-31 supplies a docking site for ferredoxin [71,72]. Therefore it can be concluded that the plant NDH complex accepts electrons from ferredoxin rather than NAD(P)H. Consequently, the authors of the first study proposed that the term “NDH” be retained, but used to mean “NADH dehydrogenase-like complex” rather than “NAD(P)H dehydrogenase complex” [71]. In a strict sense, the NDH complex is also an FQR like the PGRL1/PGR5 complex (Figure 2). With respect to its physical interaction with other thylakoid complexes, the NDH complex has been shown, on the basis of genetic and biochemical experiments [73], as well as by electron microscopy analyses [74], to form super-complexes with photosystem I, such that two photosystem I complexes bind to one NDH complex, with Lhca5 and 6 acting as linkers [73,74]. Interestingly, the association of photosystem I with light-harvesting 1 proteins and the NDH complex had evolved before the emergence of vascular plants, as evidenced by analyses of photosystem I in the moss P. patens [75,76].
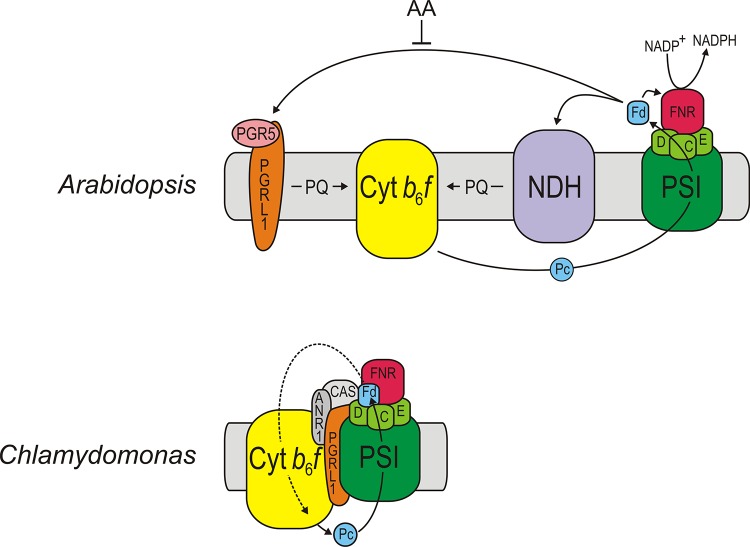
Figure 2. Model for the different roles of PGRL1 in cyclic electron flow in vascular plants and green algae.
In the vascular plant Arabidopsis, cyclic electron flow (CEF) around photosystem I (PSI) operates via two partially redundant pathways, an NDH-dependent and the PGRL1/PGR5-dependent pathway. Only the latter is inhibited by AA. Note that both the PGRL1/PGR5 and the NDH complex (via Lhca5 and 6) can physically interact with PSI in Arabidopsis and accept electrons only from ferredoxin (Fd) [43,71]. In the green alga Chlamydomonas, CEF can be mediated by a PSI-Cyt b6f-PGRL1-ANR1-CAS supercomplex [70,150]. FNR, ferredoxin NADP+ oxidoreductase; Pc, plastocyanin; PQ, plastoquinone; C, D and E, stromal subunits of PSI. Adapted with permission 2013 Journal of Experimental Botany doi:10.1093/jxb/eru090.
The major thylakoid protein complexes require not only structural proteins that serve as subunits but also accessory proteins that mediate the correct assembly of the complexes. Here, a plethora of factors have been identified during the last few years and a picture emerged in which such accessory factors constitute an integrative network mediating the stepwise assembly of multiprotein complex components (reviewed in [77-84]). Thus, it appears that besides the set of actual subunits of the photosystems, even more proteins are required for the expression of the chloroplast-encoded subunits and their assembly. In Table 1, we provide a list of the current inventory of assembly factors identified in Arabidopsis, Chlamydomonas, tobacco or cyanobacteria (Synechocystis or Synechococcus) (Table 1). Interestingly, two photosystem I assembly factors are encoded by chloroplast genes: ycf3 and ycf4 [85-88]. An important function in the chloroplast protein import machinery has been recently assigned to another chloroplast open reading frame, ycf1 [89]. However, such a tentative assignment of an important function of the encoded Tic214 protein is somehow at variance with the observation that chloroplast genomes of Poaceae species lack the ycf1 gene.
Novel retrograde signals
The term “retrograde signalling” refers to the idea that signals emanating from chloroplasts or mitochondria can modulate nuclear gene expression. Proposed almost 30 years ago, the initial notion that a single plastid signal might regulate the expression of nuclear genes involved in plastid biogenesis has since expanded to accommodate the insight that multiple signals are produced by plastids. While the ultimate effects of retrograde signalling on nuclear gene expression have now been clearly defined, many aspects of the initiation and transmission of the signals, and their mode of action, remain unresolved, speculative or controversial [90,91]. Relevant signals are thought to be derived from various sources, including (a) the pool of reactive oxygen species (ROS), (b) the reduction/oxidation (redox) state of the organelle, (c) organellar gene expression, and (d) the tetrapyrrole pathway. More recently, “brand-new” retrograde signaling pathways have been described that involve (e) metabolites — particularly 3'-phosphoadenosine 5'-phosphate (PAP) [92] and methylerythritol cyclodiphosphate (MEcPP) [93] — and (f) a carotenoid derivative (β-cyclocitral [β-CC]) [94].
Synthetic biology
Synthetic biology can broadly be defined as “the deliberate (re)design and construction of novel biological and biologically based systems to perform new functions for useful purposes, that draws on principles elucidated from biology and engineering” (http://www.erasynbio.eu/index.php?index=32).
The plastome, at least in some species, such as tobacco, tomato and Chlamydomonas, can be manipulated by genetic transformation with large constructs made up of foreign or synthetic DNA segments [95]. In fact, due to its prokaryotic origin, the chloroplast genome offers many advantages for genetic engineering because its genes are organized in operons and many are co-expressed from a single promoter as a polycistronic transcript that may subsequently be processed further into monocistronic mRNAs. Moreover, no position effects or epigenetic gene-silencing mechanisms, like those observed with nuclear transgenes, have been reported in chloroplasts [96]. These features make the chloroplast compartment especially amenable to the application of synthetic biology to goals such as the sustainable synthesis of chemicals and high-value products. Lu et al. [97] successfully demonstrated this by expressing the tocochromanol pathway (which produces tocopherols and tocotrienols, collectively called “vitamin E”) in the chloroplasts of tobacco and tomato and achieving up to a tenfold increase in total tocochromanol accumulation. This represents a prime example of how overexpression of enzymes in the chloroplast can redirect photosynthetically generated carbon skeletons from the endogenous isoprenoid biosynthetic pathway into the production of higher levels of tocopherols and tocotrienols.
It is highly desirable that novel pathways introduced into the chloroplast should be able to tap directly the chemical energy derived from sunlight in the form of ATP, NADPH or even photo-reduced ferredoxin. One group of enzymes which could potentially be used for this purpose are the cytochrome P450 mono-oxygenases (P450s), which are represented in all biological kingdoms and constitute one of the largest superfamilies of enzymes known [98]. Most P450s are located in the endoplasmatic reticulum, where they act as key enzymes in the biosynthesis of a large number of high-value bioactive natural compounds. Many of these compounds are normally made in very small quantities and are difficult to produce by chemical synthesis due to their often complex structures [99]. P450s generally obtain the electrons needed for their catalytic reactions from NADPH or NADH, but bacterial and mitochondrial P450s are also known to accept electrons from ferredoxin. Therefore, a direct link between photoreduced ferredoxin and P450s is possible if the evolutionary compartmentalization of the photosystems in the chloroplasts and of the majority of the P450 pathways in the endoplasmatic reticulum can be broken down.
The potential value of combining P450-mediated mono-oxygenation reactions with photosynthesis was first demonstrated in vitro when spinach chloroplasts were brought together with microsomes from yeast expressing a fusion between a P450 from rat (CYP1A1) and a reductase. This mixture supported the light-driven conversion of the P450 substrate 7-ethoxycoumarin into 7-hydroxycoumarin [100]. More recently, it was shown in vitro that electrons supplied by photosystem I purified from barley could be transferred with high efficiency to a P450 (CYP79A1) from Sorghum bicolor via ferredoxin, thus eliminating the need for an NADPH recycling system and a reductase [101]. Subsequently, it was shown that the P450-catalysed pathway for the biosynthesis of dhurrin (a cyanogenic glycoside) can be transferred from the cytosolic endoplasmatic reticulum of S. bicolor into the tobacco chloroplast [102]. To this end, fusion proteins between a chloroplast transit peptide and the coding regions of two P450 enzymes and a uridine 5'-diphosphate (UDP) glucosyltransferase, which together constitute the route to dhurrin biosynthesis, were successfully expressed in the chloroplasts of transiently transformed tobacco leaves. Interestingly, the chloroplast was able to provide the heme cofactor for the proper assembly of the P450s, the tyrosine and UDP-glucose substrates. The electron-demanding P450-catalysed synthesis of dhurrin was driven by directly tapping into light-driven reduction of ferredoxin by photosystem I (Figure 3). Thus, this example demonstrates that P450s that normally reside in the endoplasmic reticulum membranes can be targeted to the chloroplast and inserted into the thylakoids and can act as receptors for electrons from the light reactions of photosynthesis for use in the biosynthesis of dhurrin.
Figure 3. Schematic representation of a light-driven metabolon introduced into the thylakoids.

Photosystem I (PSI) receives electrons from photosystem II via plastocyanin (PC) and directs them to ferredoxin (Fd), which give them either to the ferredoxin NADP+ oxidoreductase (FNR) for NADPH production or directly to the two P450 enzymes (P450s). The two membrane-bound P450s hydroxylate the substrate in two consecutive steps, and this is followed by glycosylation by a soluble glucosyltransferase (GT) to form the final stable product. The novel aspect of this approach is that photosynthetic reducing power, in the form of reduced ferredoxin, is used directly by a novel biosynthetic pathway to produce the product without the need for numerous energy consuming metabolic conversions.
Acknowledgments
Dario Leister is grateful to the Deutsche Forschungsgemeinschaft for funding (FOR 804, FOR 2092, LE1265/20, /21 and /28). Poul Erik Jensen gratefully acknowledges financial support (a) from the VILLUM Center of Excellence “Plant Plasticity”, (b) from the “Center of Synthetic Biology” funded by the UNIK research initiative of the Danish Ministry of Science, Technology and Innovation, (c) from “bioSYNergy” funded by the UCPH Excellence Programme for Interdisciplinary Research, and (d) from “Plant Power: Light-Driven Synthesis of Complex Terpenoids Using Cytochrome P450s” (12-131834) funded by the Danish Council for Strategic Research, Programme Commission on Strategic Growth Technologies.
Abbreviations
- AA-sensitive CEF
antimycin A-sensitive cyclic electron
- ACS
aerobic cyclase system
- Cyt
cytochrome
- FNR
ferredoxin NADP+ oxidoreductase
- GUN4
GENOME UNCOUPLER4
- FQR
ferredoxin plastoquinone reductase
- LHC
light-harvesting complex
- MEcPP
methylerythritol cyclodiphosphate
- NDH
NAD(P)H dehydrogenase
- PAP
3'-phosphoadenosine 5'-phosphate
- SVR4
suppressor of variegation 4
- UDP
uridine 5'-diphosphate.
Disclosures
The authors declare that they have no disclosures.
The electronic version of this article is the complete one and can be found at:http://f1000.com/prime/reports/b/6/40
REFERENCES
- 1.Kleine T, Maier UG, Leister D. DNA transfer from organelles to the nucleus: the idiosyncratic genetics of endosymbiosis. Annu Rev Plant Biol. 2009;60:115–38. doi: 10.1146/annurev.arplant.043008.092119. [DOI] [PubMed] [Google Scholar]
- 2.Jarvis P, López-Juez E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat Rev Mol Cell Biol. 2013;14:787–802. doi: 10.1038/nrm3702. [DOI] [PubMed] [Google Scholar]
- 3.Horn M, Collingro A, Schmitz-Esser S, Beier CL, Purkhold U, Fartmann B, Brandt P, Nyakatura GJ, Droege M, Frishman D, Rattei T, Mewes H, Wagner M. Illuminating the evolutionary history of chlamydiae. Science. 2004;304:728–30. doi: 10.1126/science.1096330. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/1017232
- 4.Stephens RS, Kalman S, Lammel C, Fan J, Marathe R, Aravind L, Mitchell W, Olinger L, Tatusov RL, Zhao Q, Koonin EV, Davis RW. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science. 1998;282:754–9. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- 5.Brinkman, Fiona SL, Blanchard JL, Cherkasov A, Av-Gay Y, Brunham RC, Fernandez RC, Finlay BB, Otto SP, Ouellette, Francis BF, Keeling PJ, Rose AM, Hancock Robert EW, Jones Steven JM, Greberg H. Evidence that plant-like genes in Chlamydia species reflect an ancestral relationship between Chlamydiaceae, cyanobacteria, and the chloroplast. Genome Res. 2002;12:1159–67. doi: 10.1101/gr.341802. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1009412
- 6.Ball SG, Subtil A, Bhattacharya D, Moustafa A, Weber Andreas PM, Gehre L, Colleoni C, Arias M, Cenci U, Dauvillée D. Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell. 2013;25:7–21. doi: 10.1105/tpc.112.101329. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718386056
- 7.Huang J, Gogarten JP. Did an ancient chlamydial endosymbiosis facilitate the establishment of primary plastids? Genome Biol. 2007;8:R99. doi: 10.1186/gb-2007-8-6-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1098074
- 8.Moustafa A, Reyes-Prieto A, Bhattacharya D. Chlamydiae has contributed at least 55 genes to Plantae with predominantly plastid functions. PLoS ONE. 2008;3:e2205. doi: 10.1371/journal.pone.0002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marin B, Nowack Eva C M, Melkonian M. A plastid in the making: evidence for a second primary endosymbiosis. Protist. 2005;156:425–32. doi: 10.1016/j.protis.2005.09.001. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718386061
- 10.Nowack, Eva C M, Melkonian M, Glöckner G. Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes. Curr Biol. 2008;18:410–8. doi: 10.1016/j.cub.2008.02.051. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718386136
- 11.Nowack EC, Grossman AR. Trafficking of protein into the recently established photosynthetic organelles of Paulinella chromatophora. Proc Natl Acad Sci USA. 2012;109:5340–45. doi: 10.1073/pnas.1118800109. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717697959
- 12.Dorrell RG, Howe CJ. What makes a chloroplast? Reconstructing the establishment of photosynthetic symbioses. J Cell Sci. 2012;125:1865–75. doi: 10.1242/jcs.102285. [DOI] [PubMed] [Google Scholar]
- 13.Rumpho ME, Pelletreau KN, Moustafa A, Bhattacharya D. The making of a photosynthetic animal. J Exp Biol. 2011;214:303–11. doi: 10.1242/jeb.046540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR. Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elyia chlorotica. Proc Natl Acad Sci USA. 2008;105:17867–71. doi: 10.1073/pnas.0804968105. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/1127814
- 15.Bhattacharya D, Pelletreau KN, Price DC, Sarver KE, Rumpho ME. Genome analysis of Elysia chlorotica egg DNA provides no evidence for horizontal gene transfer into the germ line of this kleptoplastic mollusc. Mol Biol Evol. 2013;30:1843–52. doi: 10.1093/molbev/mst084. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718025758
- 16.Wägele H, Deusch O, Händeler K, Martin R, Schmitt V, Christa G, Pinzger B, Gould SB, Dagan T, Klussmann-Kolb A, Martin W. Transcriptomic evidence that longevity of acquired plastids in the photosynthetic slugs Elysia timida and Plakobranchus ocellatus does not entail lateral transfer of algal nuclear genes. Mol Biol Evol. 2011;28:699–706. doi: 10.1093/molbev/msq239. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13398967
- 17.Christa G, Zimorski V, Woehle C, Tielens Aloysius G M, Wägele H, Martin WF, Gould SB. Plastid-bearing sea slugs fix CO2 in the light but do not require photosynthesis to survive. Proc Biol Sci. 2014;281:20132493. doi: 10.1098/rspb.2013.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718184017
- 18.Vries J de, Habicht J, Woehle C, Huang C, Christa G, Wägele H, Nickelsen J, Martin WF, Gould SB. Is ftsH the key to plastid longevity in sacoglossan slugs? Genome Biol Evol. 2013;5:2540–8. doi: 10.1093/gbe/evt205. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718201168
- 19.Krupinska K, Melonek J, Krause K. New insights into plastid nucleoid structure and functionality. Planta. 2013;237:653–64. doi: 10.1007/s00425-012-1817-5. [DOI] [PubMed] [Google Scholar]
- 20.Sakai A, Takano H, Kuroiwa T. Organelle nuclei in higher plants: structure, composition, function, and evolution. Int Rev Cytol. 2004;238:59–118. doi: 10.1016/S0074-7696(04)38002-2. [DOI] [PubMed] [Google Scholar]
- 21.Pfalz J, Pfannschmidt T. Essential nucleoid proteins in early chloroplast development. Trends Plant Sci. 2013;18:186–94. doi: 10.1016/j.tplants.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Majeran W, Friso G, Asakura Y, Qu X, Huang M, Ponnala L, Watkins KP, Barkan A, van Wijk Klaas J. Nucleoid-enriched proteomes in developing plastids and chloroplasts from maize leaves: a new conceptual framework for nucleoid functions. Plant Physiol. 2012;158:156–89. doi: 10.1104/pp.111.188474. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718386137
- 23.Melonek J, Matros A, Trösch M, Mock H, Krupinska K. The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell. 2012;24:3060–73. doi: 10.1105/tpc.112.099721. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718386138
- 24.Yu F, Park SS, Liu XY, Foudree A, Fu AG, Powikrowska M, Khrouchtchova A, Jensen PE, Kriger JN, Gray GR, Rodermel SR. SUPPRESSOR OF VARIEGATION4, a new var2 suppressor locus, encodes a pioneer protein that is required for chloroplast biogenesis. Mol Plant. 2011;4:229–40. doi: 10.1093/mp/ssq074. [DOI] [PubMed] [Google Scholar]
- 25.Qiao J, Ma C, Wimmelbacher M, Börnke F, Luo M. Two novel proteins, MRL7 and its paralog MRL7-L, have essential but functionally distinct roles in chloroplast development and are involved in plastid gene expression regulation in Arabidopsis. Plant Cell Physiol. 2011;52:1017–30. doi: 10.1093/pcp/pcr054. [DOI] [PubMed] [Google Scholar]
- 26.Powikrowska M, Khrouchtchova A, Martens HJ, Zygadlo-Nielsen A, Melonek J, Schulz A, Krupinska K, Rodermel S, Jensen PE. SVR4 (suppressor of variegation 4) and SVR4-like: two proteins with a role in proper organization of the chloroplast genetic machinery. Physiol Plant. 2014;150:477–92. doi: 10.1111/ppl.12108. [DOI] [PubMed] [Google Scholar]
- 27.Frehlick LJ, Eirín-López JM, Ausió J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- 28.Jäkel S, Mingot J, Schwarzmaier P, Hartmann E, Görlich D. Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 2002;21:377–86. doi: 10.1093/emboj/21.3.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindström MS. NPM1/B23: A multifunctional chaperone in ribosome biogenesis and chromatin remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koch B, Mitterer V, Niederhauser J, Stanborough T, Murat G, Rechberger G, Bergler H, Kressler D, Pertschy B. Yar1 protects the ribosomal protein Rps3 from aggregation. J Biol Chem. 2012;287:21806–15. doi: 10.1074/jbc.M112.365791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pribil M, Labs M, Leister D. Structure and dynamics of thylakoids in land plants. J Exp Bot. 2014 doi: 10.1093/jxb/eru090. in press. [DOI] [PubMed] [Google Scholar]
- 32.Kirchhoff H, Hall C, Wood M, Herbstová M, Tsabari O, Nevo R, Charuvi D, Shimoni E, Reich Z. Dynamic control of protein diffusion within the granal thylakoid lumen. Proc Natl Acad Sci USA. 2011;108:20248–53. doi: 10.1073/pnas.1104141109. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13418956
- 33.Austin JR, Staehelin LA. Three-dimensional architecture of grana and stroma thylakoids of higher plants as determined by electron tomography. Plant Physiol. 2011;155:1601–11. doi: 10.1104/pp.110.170647. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718386139
- 34.Daum B, Nicastro D, Austin J, McIntosh JR, Kühlbrandt W. Arrangement of photosystem II and ATP synthase in chloroplast membranes of spinach and pea. Plant Cell. 2010;22:1299–312. doi: 10.1105/tpc.109.071431. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/3427957
- 35.Mustárdy L, Buttle K, Steinbach G, Garab G. The three-dimensional network of the thylakoid membranes in plants: quasihelical model of the granum-stroma assembly. Plant Cell. 2008;20:2552–7. doi: 10.1105/tpc.108.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arvidsson PO, Sundby C. A model for the topology of the chloroplast thylakoid membrane. Austr J Plant Physiol. 1999;26:687–94. doi: 10.1071/PP99072. [DOI] [Google Scholar]
- 37.Shimoni E, Rav-Hon O, Ohad I, Brumfeld V, Reich Z. Three-dimensional organization of higher-plant chloroplast thylakoid membranes revealed by electron tomography. Plant Cell. 2005;17:2580–6. doi: 10.1105/tpc.105.035030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brumfeld V, Charuvi D, Nevo R, Chuartzman S, Tsabari O, Ohad I, Shimoni E, Reich Z. A note on three-dimensional models of higher-plant thylakoid networks. Plant Cell. 2008;20:2546–9. doi: 10.1105/tpc.108.062299. author reply 2549-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chuartzman SG, Nevo R, Shimoni E, Charuvi D, Kiss V, Ohad I, Brumfeld V, Reich Z. Thylakoid membrane remodeling during state transitions in Arabidopsis. Plant Cell. 2008;20:1029–39. doi: 10.1105/tpc.107.055830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daum B, Kühlbrandt W. Electron tomography of plant thylakoid membranes. J Exp Bot. 2011;62:2393–402. doi: 10.1093/jxb/err034. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718386140
- 41.Nevo R, Charuvi D, Tsabari O, Reich Z. Composition, architecture and dynamics of the photosynthetic apparatus in higher plants. Plant J. 2012;70:157–76. doi: 10.1111/j.1365-313X.2011.04876.x. [DOI] [PubMed] [Google Scholar]
- 42.Dekker JP, Boekema EJ. Supramolecular organization of thylakoid membrane proteins in green plants. Biochim Biophys Acta. 2005;1706:12–39. doi: 10.1016/j.bbabio.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 43.Hertle AP, Blunder T, Wunder T, Pesaresi P, Pribil M, Armbruster U, Leister D. PGRL1 is the elusive ferredoxin-plastoquinone reductase in photosynthetic cyclic electron flow. Mol Cell. 2013;49:511–23. doi: 10.1016/j.molcel.2012.11.030. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/717970512
- 44.Lennon AM, Prommeenate P, Nixon PJ. Location, expression and orientation of the putative chlororespiratory enzymes, Ndh and IMMUTANS, in higher-plant plastids. Planta. 2003;218:254–60. doi: 10.1007/s00425-003-1111-7. [DOI] [PubMed] [Google Scholar]
- 45.Rumeau D, Peltier G, Cournac L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007;30:1041–51. doi: 10.1111/j.1365-3040.2007.01675.x. [DOI] [PubMed] [Google Scholar]
- 46.Armbruster U, Labs M, Pribil M, Viola S, Xu W, Scharfenberg M, Hertle AP, Rojahn U, Jensen PE, Rappaport F, Joliot P, Dörmann P, Wanner G, Leister D. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell. 2013;25:2661–78. doi: 10.1105/tpc.113.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/718029019
- 47.Richly E, Leister D. An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene. 2004;329:11–6. doi: 10.1016/j.gene.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 48.Lu Y, Savage LJ, Last RL. Chloroplast phenomics: systematic phenotypic screening of chloroplast protein mutants in Arabidopsis. Methods Mol Biol. 2011;775:161–85. doi: 10.1007/978-1-61779-237-3_9. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718386141
- 49.Savage LJ, Imre KM, Hall DA, Last RL. Analysis of essential Arabidopsis nuclear genes encoding plastid-targeted proteins. PLoS ONE. 2013;8:e73291. doi: 10.1371/journal.pone.0073291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petersen J, Rogowska-Wrzesinska A, Jensen ON. Functional proteomics of barley and barley chloroplasts - strategies, methods and perspectives. Front Plant Sci. 2013;4:52. doi: 10.3389/fpls.2013.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang M, Friso G, Nishimura K, Qu X, Olinares, Paul Dominic B, Majeran W, Sun Q, van Wijk Klaas J. Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships; the concept of orthoproteomics. J Proteome Res. 2013;12:491–504. doi: 10.1021/pr300952g. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718386142
- 52.Joshi HJ, Hirsch-Hoffmann M, Baerenfaller K, Gruissem W, Baginsky S, Schmidt R, Schulze WX, Sun Q, van Wijk, Klaas J, Egelhofer V, Wienkoop S, Weckwerth W, Bruley C, Rolland N, Toyoda T, Nakagami H, Jones AM, Briggs SP, Castleden I, Tanz SK, Millar AH, Heazlewood JL. MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 2011;155:259–70. doi: 10.1104/pp.110.168195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schön A, Krupp G, Gough S, Berry-Lowe S, Kannangara CG, Söll D. The RNA required in the first step of chlorophyll biosynthesis is a chloroplast glutamate tRNA. Nature. 1986;322:281–4. doi: 10.1038/322281a0. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka R, Tanaka A. Tetrapyrrole biosynthesis in higher plants. Annu Rev Plant Biol. 2007;58:321–46. doi: 10.1146/annurev.arplant.57.032905.105448. [DOI] [PubMed] [Google Scholar]
- 55.Gibson LC, Willows RD, Kannangara CG, Wettstein D von, Hunter CN. Magnesium-protoporphyrin chelatase of Rhodobacter sphaeroides: reconstitution of activity by combining the products of the bchH, -I, and -D genes expressed in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1941–4. doi: 10.1073/pnas.92.6.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jensen PE, Gibson LC, Henningsen KW, Hunter CN. Expression of the chlI, chlD, and chlH genes from the cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J Biol Chem. 1996;271:16662–7. doi: 10.1074/jbc.271.28.16662. [DOI] [PubMed] [Google Scholar]
- 57.Larkin RM, Alonso JM, Ecker JR, Chory J. GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science. 2003;299:902–6. doi: 10.1126/science.1079978. [DOI] [PubMed] [Google Scholar]
- 58.Davison PA, Schubert HL, Reid JD, Iorg CD, Heroux A, Hill CP, Hunter CN. Structural and biochemical characterization of Gun4 suggests a mechanism for its role in chlorophyll biosynthesis. Biochemistry. 2005;44:7603–12. doi: 10.1021/bi050240x. [DOI] [PubMed] [Google Scholar]
- 59.Verdecia MA, Larkin RM, Ferrer J, Riek R, Chory J, Noel JP. Structure of the Mg-chelatase cofactor GUN4 reveals a novel hand-shaped fold for porphyrin binding. PLoS Biol. 2005;3:e151. doi: 10.1371/journal.pbio.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sobotka R, Dühring U, Komenda J, Peter E, Gardian Z, Tichy M, Grimm B, Wilde A. Importance of the cyanobacterial Gun4 protein for chlorophyll metabolism and assembly of photosynthetic complexes. J Biol Chem. 2008;283:25794–802. doi: 10.1074/jbc.M803787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou S, Sawicki A, Willows RD, Luo M. C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett. 2012;586:205–10. doi: 10.1016/j.febslet.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 62.Porra RJ, Schäfer W, Gad'on N, Katheder I, Drews G, Scheer H. Origin of the two carbonyl oxygens of bacteriochlorophyll a. Demonstration of two different pathways for the formation of ring E in Rhodobacter sphaeroides and Roseobacter denitrificans, and a common hydratase mechanism for 3-acetyl group formation. Eur J Biochem. 1996;239:85–92. doi: 10.1111/j.1432-1033.1996.0085u.x. [DOI] [PubMed] [Google Scholar]
- 63.Rzeznicka K, Walker CJ, Westergren T, Kannangara CG, Wettstein D von, Merchant S, Gough SP, Hansson M. Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc Natl Acad Sci USA. 2005;102:5886–91. doi: 10.1073/pnas.0501784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pinta V, Picaud M, Reiss-Husson F, Astier C. Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J Bacteriol. 2002;184:746–53. doi: 10.1128/JB.184.3.746-753.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tottey S, Block MA, Allen M, Westergren T, Albrieux C, Scheller HV, Merchant S, Jensen PE. Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc Natl Acad Sci USA. 2003;100:16119–24. doi: 10.1073/pnas.2136793100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hollingshead S, Kopecná J, Jackson PJ, Canniffe DP, Davison PA, Dickman MJ, Sobotka R, Hunter CN. Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J Biol Chem. 2012;287:27823–33. doi: 10.1074/jbc.M112.352526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Albus CA, Salinas A, Czarnecki O, Kahlau S, Rothbart M, Thiele W, Lein W, Bock R, Grimm B, Schöttler MA. LCAA, a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 2012;160:1923–39. doi: 10.1104/pp.112.206045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell. 2002;110:361–71. doi: 10.1016/S0092-8674(02)00867-X. [DOI] [PubMed] [Google Scholar]
- 69.DalCorso G, Pesaresi P, Masiero S, Aseeva E, Schünemann D, Finazzi G, Joliot P, Barbato R, Leister D. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell. 2008;132:273–85. doi: 10.1016/j.cell.2007.12.028. [DOI] [PubMed] [Google Scholar]
- 70.Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature. 2010;464:1210–3. doi: 10.1038/nature08885. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/2990959
- 71.Yamamoto H, Peng L, Fukao Y, Shikanai T. An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell. 2011;23:1480–93. doi: 10.1105/tpc.110.080291. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/10134958
- 72.Yamamoto H, Shikanai T. In planta mutagenesis of Src homology 3 domain-like fold of NdhS, a ferredoxin-binding subunit of the chloroplast NADH dehydrogenase-like complex in Arabidopsis: a conserved Arg-193 plays a critical role in ferredoxin binding. J Biol Chem. 2013;288:36328–37. doi: 10.1074/jbc.M113.511584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng L, Fukao Y, Fujiwara M, Takami T, Shikanai T. Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell. 2009;21:3623–40. doi: 10.1105/tpc.109.068791. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/3698957
- 74.Kouřil R, Strouhal O, Nosek L, Lenobel R, Chamrád I, Boekema EJ, Sebela M, Ilík P. Structural characterization of a plant photosystem I and NAD(P)H dehydrogenase supercomplex. Plant J. 2014;77:568–76. doi: 10.1111/tpj.12402. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718201652
- 75.Armbruster U, Rühle T, Kreller R, Strotbek C, Zühlke J, Tadini L, Blunder T, Hertle AP, Qi Y, Rengstl B, Nickelsen J, Frank W, Leister D. The PHOTOSYNTHESIS AFFECTED MUTANT68-LIKE protein evolved from a PSII assembly factor to mediate assembly of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Cell. 2013;25:3926–43. doi: 10.1105/tpc.113.114785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Busch A, Petersen J, Webber-Birungi MT, Powikrowska M, Lassen, Lærke Marie Münter, Naumann-Busch B, Nielsen AZ, Ye J, Boekema EJ, Jensen ON, Lunde C, Jensen PE. Composition and structure of photosystem I in the moss Physcomitrella patens. J Exp Bot. 2013;64:2689–99. doi: 10.1093/jxb/ert126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nickelsen J, Rengstl B. Photosystem II assembly: from cyanobacteria to plants. Annu Rev Plant Biol. 2013;64:609–35. doi: 10.1146/annurev-arplant-050312-120124. [DOI] [PubMed] [Google Scholar]
- 78.Komenda J, Sobotka R, Nixon PJ. Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr Opin Plant Biol. 2012;15:245–51. doi: 10.1016/j.pbi.2012.01.017. [DOI] [PubMed] [Google Scholar]
- 79.Chi W, Ma J, Zhang L. Regulatory factors for the assembly of thylakoid membrane protein complexes. Philos Trans R Soc Lond, B, Biol Sci. 2012;367:3420–9. doi: 10.1098/rstb.2012.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schöttler MA, Albus CA, Bock R. Photosystem I: its biogenesis and function in higher plants. J Plant Physiol. 2011;168:1452–61. doi: 10.1016/j.jplph.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 81.Rochaix J. Assembly of the photosynthetic apparatus. Plant Physiol. 2011;155:1493–500. doi: 10.1104/pp.110.169839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Peng L, Yamamoto H, Shikanai T. Structure and biogenesis of the chloroplast NAD(P)H dehydrogenase complex. Biochim Biophys Acta. 2011;1807:945–53. doi: 10.1016/j.bbabio.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 83.Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J. Recent advances in understanding the assembly and repair of photosystem II. Ann Bot. 2010;106:1–16. doi: 10.1093/aob/mcq059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mulo P, Sirpiö S, Suorsa M, Aro E. Auxiliary proteins involved in the assembly and sustenance of photosystem II. Photosyn Res. 2008;98:489–501. doi: 10.1007/s11120-008-9320-3. [DOI] [PubMed] [Google Scholar]
- 85.Boudreau E, Takahashi Y, Lemieux C, Turmel M, Rochaix JD. The chloroplast ycf3 and ycf4 open reading frames of Chlamydomonas reinhardtii are required for the accumulation of the photosystem I complex. EMBO J. 1997;16:6095–104. doi: 10.1093/emboj/16.20.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ruf S, Kössel H, Bock R. Targeted inactivation of a tobacco intron-containing open reading frame reveals a novel chloroplast-encoded photosystem I-related gene. J Cell Biol. 1997;139:95–102. doi: 10.1083/jcb.139.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Krech K, Ruf S, Masduki FF, Thiele W, Bednarczyk D, Albus CA, Tiller N, Hasse C, Schöttler MA, Bock R. The plastid genome-encoded Ycf4 protein functions as a nonessential assembly factor for photosystem I in higher plants. Plant Physiol. 2012;159:579–91. doi: 10.1104/pp.112.196642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ozawa S, Nield J, Terao A, Stauber EJ, Hippler M, Koike H, Rochaix J, Takahashi Y. Biochemical and structural studies of the large Ycf4-photosystem I assembly complex of the green alga Chlamydomonas reinhardtii. Plant Cell. 2009;21:2424–42. doi: 10.1105/tpc.108.063313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M. Uncovering the protein translocon at the chloroplast inner envelope membrane. Science. 2013;339:571–4. doi: 10.1126/science.1229262. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718003600
- 90.Estavillo GM, Chan KX, Phua SY, Pogson BJ. Reconsidering the nature and mode of action of metabolite retrograde signals from the chloroplast. Front Plant Sci. 2012;3:300. doi: 10.3389/fpls.2012.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Leister D. Retrograde signaling in plants: from simple to complex scenarios. Front Plant Sci. 2012;3:135. doi: 10.3389/fpls.2012.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, Brearley C, Hell R, Marin E, Pogson BJ. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23:3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/13543956
- 93.Xiao Y, Savchenko T, Baidoo Edward E K, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149:1525–35. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/717648074
- 94.Ramel F, Birtic S, Ginies C, Soubigou-Taconnat L, Triantaphylidès C, Havaux M. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109:5535–40. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/14263016
- 95.Scharff LB, Bock R. Emerging tools for synthetic biology in plants. Plant J. 2013 doi: 10.1111/tpj.12356. [DOI] [PubMed] [Google Scholar]
- 96.Bock R. Strategies for metabolic pathway engineering with multiple transgenes. Plant Mol Biol. 2013;83:21–31. doi: 10.1007/s11103-013-0045-0. [DOI] [PubMed] [Google Scholar]
- 97.Lu Y, Rijzaani H, Karcher D, Ruf S, Bock R. Efficient metabolic pathway engineering in transgenic tobacco and tomato plastids with synthetic multigene operons. Proc Natl Acad Sci USA. 2013;110:E623–32. doi: 10.1073/pnas.1216898110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems--biological variations of electron transport chains. Biochim Biophys Acta. 2007;1770:330–44. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 99.Lassen LM, Zygadlo Nielsen A, Friis Ziersen BE, Gnanasekaran T, Moller BL, Jensen PE. Redirecting photosynthetic electron flow into light-driven synthesis of alternative products including high-value bioactive natural products. ACS Synth Biol. 2014;3:1–12. doi: 10.1021/sb400136f. [DOI] [PubMed] [Google Scholar]
- 100.Kim YS, Hara M, Ikebukuro K, Miyake J, Ohkawa H, Karube I. Photo-induced activation of cytochrome P450/reductase fusion enzyme coupled with spinach chloroplasts. Biotechnol Techniques. 1996;10:717–20. doi: 10.1007/BF00222553. [DOI] [Google Scholar]
- 101.Jensen K, Jensen PE, Møller BL. Light-driven cytochrome P450 hydroxylations. ACS Chem Biol. 2011;6:533–9. doi: 10.1021/cb100393j. [DOI] [PubMed] [Google Scholar]
- 102.Zygadlo Nielsen A, Ziersen B, Jensen K, Lassen LM, Olsen CE, Møller BL, Jensen PE. Redirecting photosynthetic reducing power toward bioactive natural product synthesis. ACS Synth Biol. 2013;2:308–15. doi: 10.1021/sb300128r. [DOI] [PubMed] [Google Scholar]
- 103.Meurer J, Plücken H, Kowallik KV, Westhoff P. A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J. 1998;17:5286–97. doi: 10.1093/emboj/17.18.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Komenda J, Nickelsen J, Tichý M, Prásil O, Eichacker LA, Nixon PJ. The cyanobacterial homologue of HCF136/YCF48 is a component of an early photosystem II assembly complex and is important for both the efficient assembly and repair of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem. 2008;283:22390–9. doi: 10.1074/jbc.M801917200. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/7466956
- 105.Inoue K, Dreyfuss BW, Kindle KL, Stern DB, Merchant S, Sodeinde OA. Ccs1, a nuclear gene required for the post-translational assembly of chloroplast c-type cytochromes. J Biol Chem. 1997;272:31747–54. doi: 10.1074/jbc.272.50.31747. [DOI] [PubMed] [Google Scholar]
- 106.Benz M, Bals T, Gügel IL, Piotrowski M, Kuhn A, Schünemann D, Soll J, Ankele E. Alb4 of Arabidopsis promotes assembly and stabilization of a non chlorophyll-binding photosynthetic complex, the CF1CF0-ATP synthase. Mol Plant. 2009;2:1410–24. doi: 10.1093/mp/ssp095. [DOI] [PubMed] [Google Scholar]
- 107.Sirpiö S, Holmström M, Battchikova N, Aro E. AtCYP20-2 is an auxiliary protein of the chloroplast NAD(P)H dehydrogenase complex. FEBS Lett. 2009;583:2355–8. doi: 10.1016/j.febslet.2009.06.031. [DOI] [PubMed] [Google Scholar]
- 108.Bellafiore S, Ferris P, Naver H, Göhre V, Rochaix J. Loss of Albino3 leads to the specific depletion of the light-harvesting system. Plant Cell. 2002;14:2303–14. doi: 10.1105/tpc.003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G. ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell. 1997;9:717–30. doi: 10.1105/tpc.9.5.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Spence E, Bailey S, Nenninger A, Møller SG, Robinson C. A homolog of Albino3/OxaI is essential for thylakoid biogenesis in the cyanobacterium Synechocystis sp. PCC6803. J Biol Chem. 2004;279:55792–800. doi: 10.1074/jbc.M411041200. [DOI] [PubMed] [Google Scholar]
- 111.Kuras R, Vitry C de, Choquet Y, Girard-Bascou J, Culler D, Büschlen S, Merchant S, Wollman FA. Molecular genetic identification of a pathway for heme binding to cytochrome b6. J Biol Chem. 1997;272:32427–35. doi: 10.1074/jbc.272.51.32427. [DOI] [PubMed] [Google Scholar]
- 112.Lezhneva L, Kuras R, Ephritikhine G, de Vitry C. A novel pathway of cytochrome c biogenesis is involved in the assembly of the cytochrome b6f complex in Arabidopsis chloroplasts. J Biol Chem. 2008;283:24608–16. doi: 10.1074/jbc.M803869200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rühle T, Razeghi JA, Vamvaka E, Viola S, Gandini C, Kleine T, Schünemann D, Barbato R, Jahns P, Leister D. The Arabidopsis protein CGL160 promotes assembly of the CFo part of the chloroplast ATP synthase. Plant Physiol. 2014 doi: 10.1104/pp.114.237883. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shimizu H, Shikanai T. Dihydrodipicolinate reductase-like protein, CRR1, is essential for chloroplast NAD(P)H dehydrogenase in Arabidopsis. Plant J. 2007;52:539–47. doi: 10.1111/j.1365-313X.2007.03256.x. [DOI] [PubMed] [Google Scholar]
- 115.Ermakova-Gerdes S, Vermaas W. Inactivation of the open reading frame slr0399 in Synechocystis sp. PCC 6803 functionally complements mutations near the QA niche of photosystem II. A possible role of Slr0399 as a chaperone for quinone binding. J Biol Chem. 1999;274:30540–9. doi: 10.1074/jbc.274.43.30540. [DOI] [PubMed] [Google Scholar]
- 116.Wilde A, Lünser K, Ossenbühl F, Nickelsen J, Börner T. Characterization of the cyanobacterial ycf37: mutation decreases the photosystem I content. Biochem J. 2001;357:211–6. doi: 10.1042/0264-6021:3570211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stöckel J1, Bennewitz S, Hein P, Oelmüller R. The evolutionarily conserved tetratrico peptide repeat protein pale yellow green7 is required for photosystem I accumulation in Arabidopsis and copurifies with the complex. Plant Physiol. 2006;141:870–8. doi: 10.1104/pp.106.078147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Munshi MK, Kobayashi Y, Shikanai T. Chlororespiratory reduction 6 is a novel factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant Physiol. 2006;141:737–44. doi: 10.1104/pp.106.080267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dai H, Zhang L, Zhang J, Mi H, Ogawa T, Ma W. Identification of a cyanobacterial CRR6 protein, Slr1097, required for efficient assembly of NDH-1 complexes in Synechocystis sp. PCC 6803. Plant J. 2013;75:858–66. doi: 10.1111/tpj.12251. [DOI] [PubMed] [Google Scholar]
- 120.Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell. 2006;18:955–69. doi: 10.1105/tpc.105.037689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Park S, Khamai P, Garcia-Cerdan JG, Melis A. REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast-localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol. 2007;143:1547–60. doi: 10.1104/pp.107.096396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu J, Yang H, Lu Q, Wen X, Chen F, Peng L, Zhang L, Lu C. PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell. 2012;24:4992–5006. doi: 10.1105/tpc.112.106542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kamruzzaman Munshi M, Kobayashi Y, Shikanai T. Identification of a novel protein, CRR7, required for the stabilization of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis. Plant J. 2005;44:1036–44. doi: 10.1111/j.1365-313X.2005.02604.x. [DOI] [PubMed] [Google Scholar]
- 124.Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L. LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell. 2007;19:1980–93. doi: 10.1105/tpc.107.050526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 125.Albus CA, Ruf S, Schöttler MA, Lein W, Kehr J, Bock R. Y3IP1, a nucleus-encoded thylakoid protein, cooperates with the plastid-encoded Ycf3 protein in photosystem I assembly of tobacco and Arabidopsis. Plant Cell. 2010;22:2838–55. doi: 10.1105/tpc.110.073908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Xiao J, Li J, Ouyang M, Yun T, He B, Ji D, Ma J, Chi W, Lu C, Zhang L. DAC is involved in the accumulation of the cytochrome b6/f complex in Arabidopsis. Plant Physiol. 2012;160:1911–22. doi: 10.1104/pp.112.204891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Peng L, Fukao Y, Fujiwara M, Shikanai T. Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell. 2012;24:202–14. doi: 10.1105/tpc.111.090597. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717949800
- 128.Cai W, Ma J, Chi W, Zou M, Guo J, Lu C, Zhang L. Cooperation of LPA3 and LPA2 is essential for photosystem II assembly in Arabidopsis. Plant Physiol. 2010;154:109–20. doi: 10.1104/pp.110.159558. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]; http://f1000.com/prime/7445956
- 129.Krech K, Fu H, Thiele W, Ruf S, Schöttler MA, Bock R. Reverse genetics in complex multigene operons by co-transformation of the plastid genome and its application to the open reading frame previously designated psbN. Plant J. 2013;75:1062–74. doi: 10.1111/tpj.12256. [DOI] [PubMed] [Google Scholar]
- 130.Kufryk GI, Vermaas Wim F J. Slr2013 is a novel protein regulating functional assembly of photosystem II in Synechocystis sp. strain PCC 6803. J Bacteriol. 2003;185:6615–23. doi: 10.1128/JB.185.22.6615-6623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schwenkert S, Netz Daili J A, Frazzon J, Pierik AJ, Bill E, Gross J, Lill R, Meurer J. Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem J. 2010;425:207–14. doi: 10.1042/BJ20091290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lezhneva L, Amann K, Meurer J. The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 2004;37:174–85. doi: 10.1046/j.1365-313X.2003.01952.x. [DOI] [PubMed] [Google Scholar]
- 133.Stöckel J, Oelmüller R. A novel protein for photosystem I biogenesis. J Biol Chem. 2004;279:10243–51. doi: 10.1074/jbc.M309246200. [DOI] [PubMed] [Google Scholar]
- 134.Ishida S, Takabayashi A, Ishikawa N, Hano Y, Endo T, Sato F. A novel nuclear-encoded protein, NDH-dependent cyclic electron flow 5, is essential for the accumulation of chloroplast NAD(P)H dehydrogenase complexes. Plant Cell Physiol. 2009;50:383–93. doi: 10.1093/pcp/pcn205. [DOI] [PubMed] [Google Scholar]
- 135.Roose JL, Pakrasi HB. Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J Biol Chem. 2004;279:45417–22. doi: 10.1074/jbc.M408458200. [DOI] [PubMed] [Google Scholar]
- 136.Roose JL, Pakrasi HB. The Psb27 protein facilitates manganese cluster assembly in photosystem II. J Biol Chem. 2008;283:4044–50. doi: 10.1074/jbc.M708960200. [DOI] [PubMed] [Google Scholar]
- 137.Komenda J, Knoppová J, Kopečná J, Sobotka R, Halada P, Yu J, Nickelsen J, Boehm M, Nixon PJ. The Psb27 assembly factor binds to the CP43 complex of photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2012;158:476–86. doi: 10.1104/pp.111.184184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Liu H, Huang RY, Chen J, Gross ML, Pakrasi HB. Psb27, a transiently associated protein, binds to the chlorophyll binding protein CP43 in photosystem II assembly intermediates. Proc Natl Acad Sci USA. 2011;108:18536–41. doi: 10.1073/pnas.1111597108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wei L, Guo J, Ouyang M, Sun X, Ma J, Chi W, Lu C, Zhang L. LPA19, a Psb27 homolog in Arabidopsis thaliana, facilitates D1 protein precursor processing during PSII biogenesis. J Biol Chem. 2010;285:21391–8. doi: 10.1074/jbc.M110.105064. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/7461956
- 140.Shen G, Zhao J, Reimer SK, Antonkine ML, Cai Q, Weiland SM, Golbeck JH, Bryant DA. Assembly of photosystem I. I. Inactivation of the rubA gene encoding a membrane-associated rubredoxin in the cyanobacterium Synechococcus sp. PCC 7002 causes a loss of photosystem I activity. J Biol Chem. 2002;277:20343–54. doi: 10.1074/jbc.M201103200. [DOI] [PubMed] [Google Scholar]
- 141.Shen G, Antonkine ML, van der Est A, Vassiliev IR, Brettel K, Bittl R, Zech SG, Zhao J, Stehlik D, Bryant DA, Golbeck JH. Assembly of photosystem I. II. Rubredoxin is required for the in vivo assembly of FX in Synechococcus sp. PCC 7002 as shown by optical and EPR spectroscopy. J Biol Chem. 2002;277:20355–66. doi: 10.1074/jbc.M201104200. [DOI] [PubMed] [Google Scholar]
- 142.Dobáková M, Sobotka R, Tichý M, Komenda J. Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 2009;149:1076–86. doi: 10.1104/pp.108.130039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Huang W, Chen Q, Zhu Y, Hu F, Zhang L, Ma Z, He Z, Huang J. Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol Plant. 2013;6:1673–91. doi: 10.1093/mp/sst069. [DOI] [PubMed] [Google Scholar]; http://f1000.com/prime/718009864
- 144.Keren N, Ohkawa H, Welsh EA, Liberton M, Pakrasi HB. Psb29, a conserved 22-kD protein, functions in the biogenesis of Photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell. 2005;17:2768–81. doi: 10.1105/tpc.105.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL. Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 2004;136:3594–604. doi: 10.1104/pp.104.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Klinkert B, Ossenbühl F, Sikorski M, Berry S, Eichacker L, Nickelsen J. PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J Biol Chem. 2004;279:44639–44. doi: 10.1074/jbc.M405393200. [DOI] [PubMed] [Google Scholar]
- 147.Schottkowski M, Ratke J, Oster U, Nowaczyk M, Nickelsen J. Pitt, a novel tetratricopeptide repeat protein involved in light-dependent chlorophyll biosynthesis and thylakoid membrane biogenesis in Synechocystis sp. PCC 6803. Mol Plant. 2009;2:1289–97. doi: 10.1093/mp/ssp075. [DOI] [PubMed] [Google Scholar]
- 148.Sirpiö S, Khrouchtchova A, Allahverdiyeva Y, Hansson M, Fristedt R, Vener AV, Scheller HV, Jensen PE, Haldrup A, Aro E. AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J. 2008;55:639–51. doi: 10.1111/j.1365-313X.2008.03532.x. [DOI] [PubMed] [Google Scholar]
- 149.Armbruster U, Zühlke J, Rengstl B, Kreller R, Makarenko E, Rühle T, Schünemann D, Jahns P, Weisshaar B, Nickelsen J, Leister D. The Arabidopsis thylakoid protein PAM68 is required for efficient D1 biogenesis and photosystem II assembly. Plant Cell. 2010;22:3439–60. doi: 10.1105/tpc.110.077453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Terashima M, Petroutsos D, Hüdig M, Tolstygina I, Trompelt K, Gäbelein P, Fufezan C, Kudla J, Weinl S, Finazzi G, Hippler M. Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc Natl Acad Sci USA. 2012;109:17717–22. doi: 10.1073/pnas.1207118109. [DOI] [PMC free article] [PubMed] [Google Scholar]; http://f1000.com/prime/717959855