Abstract
The BCL6 oncogenic repressor is a master regulator of humoral immunity and B-cell lymphoma survival. Whereas much research has focused on its regulation and function in germinal center B-cells, its role in other mature lymphoid cell compartments is less clear. A novel role for BCL6 in follicular T helper cell development was recently uncovered. The latest discoveries reveal that BCL6 is also an important regulator of other specialized helper T-cell subsets within germinal centers, pre-germinal center events, and peripheral T-cells effector functions. Here, we review newly discovered roles for BCL6 in lymphocyte subsets residing within and outside of germinal centers, and discuss their implications with respect to the molecular mechanisms of BCL6 regulation and potential links to B and T-cell lymphomas.
Introduction
BCL6 (B Cell Lymphoma 6) is a transcriptional repressor and member of the BTB/POZ (bric-á-brac, tramtrack, broad complex/pox virus zinc finger) family of transcription factors. The BCL6 gene was initially cloned by several groups in 1993 from a translocation occurring on chromosome 3q27 in diffuse large B-cell lymphoma (DLBCL) [1]. Targeted disruption of the BCL6 gene revealed a critical role for BCL6 during normal B-cell development, as a master regulator of antibody affinity maturation in germinal centers (GCs) [2]. BCL6 is almost universally expressed in GC-derived B-cell lymphomas, including DLBCL and follicular lymphomas (FLs), regardless of translocations.
Beyond the mature GC B-cell population, novel roles for BCL6 have recently been uncovered in several different T lymphocyte subsets within the GC, including CD4+ follicular helper T (TFH) cells, follicular regulatory T (TFR) cells and follicular helper natural killer T (NKTFH) cells. Newer studies have also revealed a much earlier involvement of BCL6 in pre-GC B and T-cell events. Outside of GCs, new roles for BCL6 have been identified in T regulatory (Treg) cells and in the control of T-cell-dependent inflammation. These new studies point to a broader function of BCL6 both in the GC reaction as well as in T cell-dependent inflammation and autoimmunity.
These discoveries raise many new questions about the full spectrum and context specificity of BCL6 target genes and downstream mechanisms in these various cell types, as well as the molecular mechanisms underlying the regulation and dysregulation of the BCL6 gene in normal and malignant lymphocytes. Recent advances in these topics will be discussed pertinent to the effector functions of normal B- and T-cells and in the development of B- and T-cell lymphomas.
Novel functions of BCL6 within the GC
While the importance of BCL6 in GC B-cell differentiation is well known [3,4], a role for BCL6 in GC T-cells was only recently discovered. Three independent studies identified a critical role for BCL6 in the differentiation of a specialized subset of CD4+ TFH cells in the GC [5–7], which provide help to antigen-specific B-cells and are phenotypically distinguished by high expression of CXCR5, PD-1, IL-21, SAP, ICOS and BCL6 [8]. A primary function of BCL6 in TFH cells appears to be the suppression of genes that are required to drive the differentiation of alternative T helper cell lineages. Competition between BCL6 and T-bet, a driver of T helper type-1 (Th1) cells, is important for determining the commitment of transitional T helper cells toward the TFH phenotype [9,10]. However, the presence of these and other factors, such as IL-21 and IL-2, also enables TFH cells to retain flexibility in their T helper lineage specification [9–11]. Remarkably, unlike GC-derived memory B-cells, which do not express BCL6 [12], BCL6+ effector TFH cells are capable of forming follicular helper-like central memory cells [13–15], implicating BCL6 expression in long-lived T helper cell immunity. BCL6 was also identified recently in a new subset of follicular T regulatory (Treg) cells that localize to GCs and express the TFH markers, CXCR5 and PD-1, and the Treg cell marker, FOXP3 [16,17]. TFR cells, which originate from thymic FOXP3+ precursor cells, require BCL6 for their development and appear to control the magnitude of the GC response [18,19]. Yet another specialized subset of BCL6+ GC T cells, follicular helper natural killer T (NKTFH) cells, were recently shown to provide direct help to antigen-specific B-cells; however, were unable to drive the generation of long-lived plasma cells [20]. Together with its functions in B-cells, BCL6 orchestrates a diverse sequence of T-cell subtypes (Figure 1a) to regulate B cell affinity maturation in the GC.
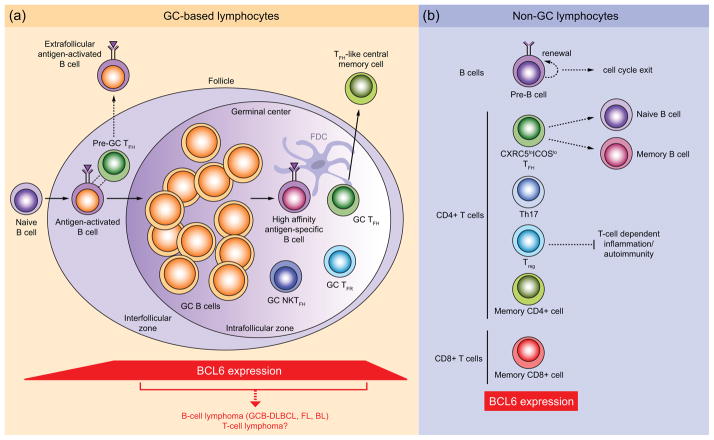
Figure 1.
Lymphocyte subsets that require BCL6 for their development and effector functions. (a) GC-based lymphocyte subsets. Outside of the follicle, naïve B cells that become activated by antigen migrate to the interfollicular zone, where they up-regulate BCL6 and participate in interactions with pre-GC TFH cells, which also express BCL6. BCL6+ pre-GC TFH cells also promote the extrafollicular antibody response and production of short-lived plasma cells. Formation of the GC reaction requires further up-regulation of BCL6 expression in activated B-cells and involves the rapid clonal expansion and somatic hypermutation of GC B cells within the intrafollicular zone. Within the GC, high-affinity antigen-specific B-cells are provided help by BCL6+ GC TFH cells and NKTFH cells (and follicular dendritic cells (FDC) which do not express BCL6), of which GC TFH cells can go on to become BCL6+ TFH-like central memory T cells. The magnitude of the GC reaction appears to be regulated by BCL6+ GC TFR cells. High BCL6 expression in GC B-cells can lead to the development of GC-derived B-cell lymphomas, including GC B-cell (GCB)-type DLBCL, FL and Burkitt’s Lymphoma (BL). It is not yet known whether T-cell lymphomas can also arise from this compartment. (b) Lymphoid subsets that express BCL6 and reside outside of GCs include pre-B cells, CXCR5loICOSlo TFH cells, which provide help to naïve and memory B cells, Th17, Treg and memory CD4+ and CD8+ T-cells. BCL6 expression in CD4+ Treg cells helps to regulate T-cell-dependent inflammatory and autoimmune responses. Solid arrows indicate cell differentiation pathways; dotted lines indicate cell interactions and effector functions.
Pre-GC roles for BCL6 in B and T cells
Recent employment of BCL6-reporter mice and high-resolution intravital cellular imaging, enabling in vivo tracking of specific lymphocyte populations, have alluded to an even earlier role for BCL6 in pre-GC events [21,22] (Figure 1a). In vivo tracking of BCL6+ cells using YFP or RFP reporter systems have shown that BCL6 not only participates in the earliest phase of TFH development [15,23,24], but is also up-regulated in antigen-engaged B cells prior to their clustering to form GC structures [25]. Using intravital imaging in mice, Kerfoot and colleagues independently demonstrated an early role for sustained BCL6 expression during B and TFH cell interactions within the interfollicular zone prior to GC entry [26]. Early B:T-cell interactions that lead to extrafollicular antibody responses, have also been shown to be dependent on SAP and IL-21 and on BCL6+ pre-GC TFH cells (Figure 1a) that express low levels of PD-1 and are phenotypically distinct from TFH cells in the GC [27,28]. What causes BCL6 to become up-regulated in B and T cells at this early stage of GC formation is not fully understood.
New roles for BCL6 outside of GCs
Several new roles for BCL6 in lymphocyte lineages that exist outside of GCs have been recently described (Figure 1b). BCL6 is up-regulated upon pre-B cell receptor signaling in pre-B cells undergoing immunoglobulin V(D)J recombination, protecting cells from DNA-damage-induced apoptosis [29] and, subsequently, enabling cell cycle exit through repression of MYC and CCND2 [30]. This late pre-B cell checkpoint function of BCL6 [30] could help to prevent malignant transformation of pre-B cells to acute lymphoblastic leukemia (ALL). However, BCL6 can also sustain the proliferation of established B-cell ALL (B-ALL), as well as offer a protective role against DNA-damaging classes of chemotherapeutics during B-ALL treatment [31,32]. Within the T-cell compartment, BCL6 was previously shown to be important for the generation and maintenance of effector and memory CD8+ T cells and memory CD4+ T cells [33]. Peripheral CD4+ TFH cells (CXCR5loICOSloIL-7R+) that express BCL6 and are functionally similar to GC TFH cells have recently been identified and provide help to B-cells outside of GCs [34]. Consistent with the severe T helper type-2 (Th2)-type inflammatory disease that develops in BCL6-deficient mice [35], BCL6 has been shown to control the effector functions of peripheral Treg cells [36]. BCL6 also regulates the differentiation of T helper IL-17-expressing (Th17) cells, via both T-cell intrinsic and extrinsic mechanisms [5,6,37]; however, in the BCL6−/− model of Th2 inflammation, increased Th17 cell activity appears to be mediated largely through the T-cell extrinsic function of BCL6 in macrophages [37]. Furthermore, ex vivo over-expression of BCL6 in CD8+ T-cells enables conversion to CD8+ T cells with suppressor activity, which protect against T-cell-dependent autoimmunity [38]. These studies suggest that in addition to controlling chemokine expression in macrophages [39,40], BCL6 could help to control inflammation and autoimmunity by directly regulating the differentiation of Th17, Treg and CD8+ T suppressor cells.
Transcriptional mechanism of action of BCL6 in normal and malignant lymphocytes
BCL6 mediates its actions in GC B-cells through the repression of genes (e.g. ATR, TP53, CDKN1A, EP300) to enable proliferation and survival during affinity maturation, and by repression of terminal differentiation factors (e.g. IRF4, PRDM1) [4,41]. The BCL6-driven gene expression programs of T-cells are less well characterized. In general, it appears that in CD4+ T-cells, BCL6 promotes the expression of genes that confer the GC TFH cell phenotype (CXCR5, CXCR4, PD1) by repressing the transcription factors, Tbx21 (T-bet), Gata3 and Rorγt, to block interferon-gamma (IFNγ-, IL-4- and IL-17-mediated differentiation to the Th1, Th2 and Th17 cell lineages [5,6]. These events appear to be dependent on the DNA binding activity of BCL6 [6]. BCL6 also represses GATA-3 expression in Treg cells and during Th17 cell differentiation [36,37]. As seen in GC B-cells, BCL6-mediated repression of Blimp-1 is required to maintain the TFH phenotype [7,10]. In addition to suppressing T lineage-specific transcription factors, BCL6 directly represses several microRNAs, including miR-17-92, which down-regulates expression of CXCR5 in TFH cells [5] and miR-21, which, via up-regulation of a MAPK pathway inhibitor, decreases GATA-3 expression in Treg cells [42]. Using BCL6-RFP reporter mice, Liu and colleagues showed that CD4+ T cells that express high (but not low) levels of BCL6 are distinguished by a TFH cell-specific gene signature (CXCR5, CXCR4, SAP), but not by genes that are characteristic of Th1, Th2, Th17, or Treg cell subsets [15]. Genome-wide maps of BCL6 binding in GC TFH cells and other non-GC T-cell subsets, together with perturbation of BCL6 and gene expression profiling, will be critical to elucidate the programs of BCL6-mediated repression that specifically direct T-cell (versus B-cell) development and effector functions.
From the biochemical standpoint, BCL6 is believed to mediate its repressive effects largely by recruiting the transcriptional corepressors, silencing mediator for retinoid or thyroid-hormone receptors (SMRT), nuclear receptor corepressor-1 (NCOR) and BCL6-interacting corepressor (BCOR), through its N-terminal BTB domain [3]. SMRT and NCOR, in particular, are ubiquitously expressed and so would be expected to enable BCL6 functions regardless of cell context. However, mice engineered to express a BCL6 mutant that cannot bind to SMRT, NCOR and BCOR only manifest the GC B-cell portion of the BCL6-null phenotype [43]. TFH function was entirely normal in these animals, as were Th2, Th17 and macrophage functions. Accordingly, BCL6 BTB domain-mutant mice lived normal, healthy lives and displayed no evidence of the lethal inflammatory syndrome that is characteristic of BCL6 null mice [43]. Instead, the function of BCL6 in macrophages was apparently linked to competition for DNA binding sites with STAT transcription factors. The molecular function of BCL6 in T-cells remains unknown and must be mediated through distinct and possibly novel biochemical mechanisms. For example, BCL6 regulation of the TFH transcriptional program was recently shown to require cooperation with Maf [44]. In differentiating Th1 cells, BCL6 is recruited by T-bet to repress specific sets of target genes [45]. Altogether, these data indicate that BCL6 regulates cells of the immune system at least in part through distinct and specific biochemical mechanisms.
Mechanisms of BCL6 regulation in normal lymphocytes
Transcriptional and post-transcriptional mechanisms are both critically important for regulation of BCL6 mRNA expression and protein stability in normal lymphocytes [4]. Induction of BCL6 expression during B-cell activation is mediated through IRF8, the only factor that has so far been shown to bind and directly induce expression of BCL6 in B-cells entering and transiting the GC reaction [46] (Figure 2a). Signaling through p110δ phosphatidylinositide 3-kinase (PI3K) was shown to enhance BCL6 expression during class switch recombination, potentially through counter-regulation of NF-κB and/or IRF4 levels [47]. In the GC, proliferating B-cells require IL-21 receptor (IL-21R) signaling to directly up-regulate and maintain expression of BCL6 [48]. In vivo regulation of BCL6 expression in GC TFH cells appears to be independent of IL-21 and IL-6 [48,49], and absence of both factors, while modestly compromising early GC TFH development and expansion, does not affect BCL6 mRNA levels in these cells [50]. Instead, inducible costimulator (ICOS) was shown to be an important initiator of BCL6 expression during early TFH cell differentiation [24]. In vitro induction of BCL6 expression in polarized Th1 cells appears to require the direct binding of the Foxo1 and Foxo3a transcription factors, which direct Th1 cells toward the TFH lineage [10]. What drives BCL6 expression in B and TFH cells that reside in the inter- and outer-follicular regions and participate in pre-GC B:T cell interactions is not currently known, but is proposed to involve strength of antigen stimulation, signaling through toll-like receptors, (potentially via IRF8), IL-21 or SLAM/SAP in B-cells, and in TFH cells, signaling via the T-cell receptor (TCR) and costimulatory molecules, CD28 and ICOS [22].
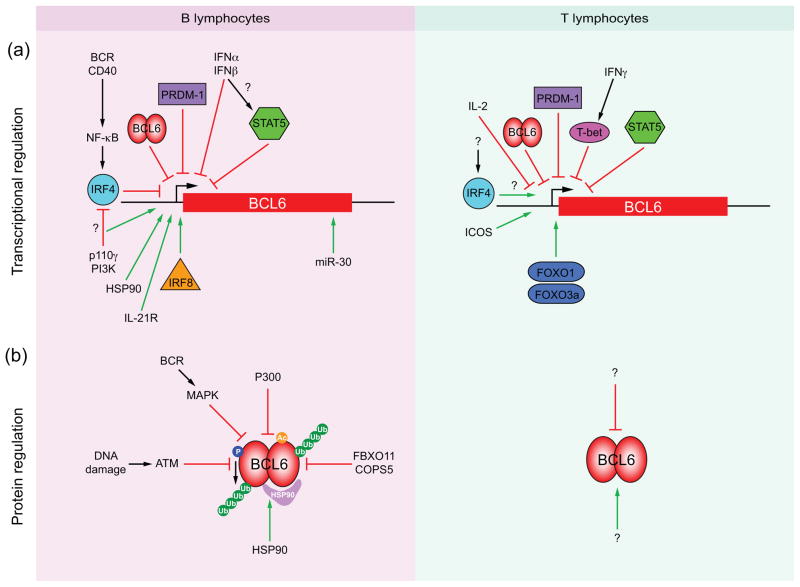
Figure 2.
Molecular mechanisms of BCL6 regulation in B and T lymphocytes. (a) Transcriptional regulators of BCL6 induction (green arrows) and down-regulation (red bars) in B and T lymphocytes. Transcription factors, of which IRF4, IRF8, STAT5, T-bet and Foxo1 and Foxo3a bind directly to the BCL6 gene control region, are indicated by colored shapes. Question marks signify unknown mediators and putative effects. (b) Post-translational regulation of BCL6 protein stabilization (green arrows) and degradation (red bars) in B and T lymphocytes. Acetylation (Ac) of BCL6 by P300 mediates BCL6 protein degradation. Phosphorylation (P) of BCL6 by MAPK or ATM results in BCL6 ubiquitylation (Ub) and proteasome-mediated degradation. FBXO11 and COPS5 also mediate ubiquitin-dependent degradation of BCL6. The HSP90 chaperone stabilizes the BCL6 protein half-life. Question marks signify unknown mediators of BCL6 protein stabilization and degradation in T-cells.
Down-regulation of BCL6 in exiting GC B-cells is mediated by the transcription factors IRF4 and PRDM1 [2,4], and by miR-30 [51] (Figure 2a). In TFH cells, BCL6 expression is also down-regulated by PRDM1 [8,52]; however, IRF-4, which is required for TFH cell differentiation, appears to promote BCL6 expression in CD4+ T cells [53]. Type 1 interferons (IFN-α, -β) directly down-regulate BCL6 mRNA levels in GC B-cells [54], likely through STAT5, which is a negative regulator of BCL6 expression in B and TFH cells [55–57]. Interferon signaling also negatively regulates BCL6 via T-bet, to direct Th1 cell commitment [9,10]. Similarly, IL-2 appears to play a role in the regulation of BCL6 and PRDM1 levels in T helper cells [10], repressing early commitment to the TFH lineage by reducing BCL6 expression [58]. Finally, BCL6 can regulate its own expression in B-cells, which is also thought to occur in GC TFH cells [6].
Post-transcriptional mechanisms that regulate BCL6 protein levels, include functional inactivation of BCL6 via p300-mediated acetylation [59] and degradation through the ubiquitin-proteosome pathway by ATM- and MAPK-mediated phosphorylation [60,61] (Figure 2b). Degradation of BCL6 in B-cells is mediated though FBXO11-CUL1 E3 ligase complexes and the COPS5 protein of the COP9 signalosome [62,63]. In contrast, BCL6 is protected from degradation by Hsp90, which extends the half-life both of BCL6 protein and its mRNA [64]. At BCL6 target genes, BCL6 corepressor complexes are disrupted through CD40 signaling to SMRT and NCoR [65].
Dysregulation of BCL6 in B and T cell lymphomas
BCL6 plays a fundamental role in maintaining the proliferation and survival of B-cell lymphomas that arise from GCs, as documented most clearly in the case of DLBCLs [64,66] (Figure 1a). This function arises from its role as a cell cycle and DNA damage checkpoint suppressor in GC B-cells. Indeed, the ability of BCL6 to impair DNA damage sensing [65] may explain in part the heavy burden of mutations observed in recent genomic re-sequencing studies of DLBCL [67–69]. The oncogenic function of BCL6 is counterbalanced in normal GC B-cells by its repression of oncogenes such as BCL2, MYC and BMI1 [41,70]; however, BCL6 loses control of these targets in B-cell lymphomas at least in part through genetic lesions of these loci. Although BCL6 is expressed almost universally in DLBCLs, its constitutive expression is often dependent on translocations in activated B-cell (ABC)-type DLBCLs, which originate from B-cells that would normally be down-regulating BCL6 [71]. Somatic mutations of genes encoding regulators of BCL6 may also enhance or maintain its functions in lymphomas. For example, somatic loss of function mutations of EP300 that disable its lysine acetyl-transferase activity may stabilize BCL6 by maintaining Hsp90-BCL6 complexes, as well as reducing acetylation of BCL6 [72,73]. BCL6 stability may also be maintained by an inactivating mutation of FBXO11, which reduces the rate of ubiquitin-mediated BCL6 degradation [62].
The discovery of a role for BCL6 in GC TFH cells and other T-cell subsets has raised the question of whether dysregulation of BCL6 and BCL6-mediated pathways could help to drive lymphomas derived from T-cells. Indeed, BCL6 over-expression was previously detected in aberrant CD30+ T-cells that constitute ALK+ anaplastic large cell lymphomas (ALCLs) [74]. Angioimmunoblastic T cell lymphomas (AITLs) appear to express TFH cell-type genes, including BCL6 [75,76]. Cutaneous T-cell lymphomas (CTCLs) also express BCL6 and a TFH-like profile, including PD-1 [77]. These studies suggest that TFH cells could become malignant, or alternatively, that transformed T-cells acquire a TFH-like phenotype and might be addicted to BCL6. It is possible that BCL6+ T cell subsets could support the tumor microenvironment of transformed B or T-cells, as has been shown for Treg cells present in AITL and extranodal NKT-cell lymphomas [78]. Malignant cells that express FOXP3 and have similar functions to Treg cells have also been identified in CTCLs and adult T cell leukemia/lymphomas [78]; however, it is not known whether BCL6 is expressed or genetically altered in these cells, nor how these cells might contribute directly to the disease. Further research is needed to understand whether T-cell lymphomas are addicted to BCL6. Peptidomimetic and small molecule inhibitors that disrupt binding of the SMRT, NCoR and BCoR corepressors to the BCL6 BTB domain potently kill B-cell lymphoma cells [66,79]. However, given that BCL6 does not function in T-cells through this mechanism [43], it seems unlikely that these inhibitors would disrupt putative BCL6 functions in T-cell lymphomas.
Conclusions
The discovery of novel roles for BCL6 in the development and effector functions of a wide variety of mature T-cell subsets, suggest that BCL6, in addition to being a true “master regulator” of the GC reaction, is also an important regulator of T-cell-dependent inflammatory, autoimmune and memory responses in the periphery. Quite remarkably, BCL6 controls the GC reaction by mediating its effects on multiple different cell types, via distinct biochemical mechanisms of action. Advances in cell imaging have also uncovered a critical and early role for BCL6 expression in the events leading up to GC formation. In addition to controlling the effector functions of mature lymphoid cells, BCL6 also regulates the survival of early developing pre-B-cells. Altogether, these breakthrough studies have implicated BCL6 as a central component of the wider adaptive immune system.
Precisely how BCL6 mediates its repressive effects on the gene expression programs of specialized subtypes of T helper and T regulatory cells remains to be determined, but, based on the functional effects of a mutant form of BCL6 in mice, is likely to involve distinct and potentially novel combinations of co-repressor complexes and co-factors that are T cell lineage-specific. Of critical importance to the advancement of new therapies for BCL6+ T-cell lymphomas, is an understanding of the molecular regulatory mechanisms that underlie BCL6 expression in T cells. For example, it is not known whether blockade of the pathways that regulate BCL6 stability in B cells that successfully kill DLBCLs could also be utilized to treat BCL6-dependent T-cell lymphomas. However, it is plausible that BCL6 could simply be a “bystander” differentiation marker of TFH or Treg-like T-cell lymphomas. A direct link between BCL6 and the development, maintenance and survival of T-cell lymphomas requires further investigation.
Highlights.
BCL6 is important for pre-GC events and for TFH, TFR and NKTFH function in GCs.
BCL6 is a regulator of T-cell-dependent inflammation and autoimmune responses.
BCL6 is likely to regulate B and T-cells via cell-specific biochemical mechanisms.
Dysregulation of BCL6 could contribute to BCL6+ T-cell lymphomas.
Acknowledgments
A.M.M. is a Burroughs-Wellcome Clinical Translational Scientist and is funded by the Chemotherapy Foundation and NCI. K.L.B. is supported by a Scholar Award from the American Society of Hematology.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Ohno H. Pathogenetic role of BCL6 translocation in B-cell non-Hodgkin’s lymphoma. Histol Histopathol. 2004;19:637–650. doi: 10.14670/HH-19.637. [DOI] [PubMed] [Google Scholar]
- 2.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 3.Ci W, Polo J, Melnick A. B-cell lymphoma 6 and the molecular pathogenesis of diffuse large B-cell lymphoma. Curr Opin Hematol. 2008;15:381–390. doi: 10.1097/MOH.0b013e328302c7df. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basso K, Dalla-Favera R. Roles of BCL6 in normal and transformed germinal center B cells. Immunol Rev. 2012;247:172–183. doi: 10.1111/j.1600-065X.2012.01112.x. [DOI] [PubMed] [Google Scholar]
- 5.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8••.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. This paper provides an extensive review of the immunological characteristics, functions and molecular regulation of TFH cells. [DOI] [PubMed] [Google Scholar]
- 9•.Nakayamada S, Kanno Y, Takahashi H, Jankovic D, Lu KT, Johnson TA, Sun H, Vahedi G, Hakim O, Handon R, et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity. 2011;35:919–931. doi: 10.1016/j.immuni.2011.11.012. This paper describes the plasticity of T helper cells during differentiation and the transcriptional requirements for Th1 cell development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10•.Oestreich K, Mohn S, Weinmann A. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol. 2012;13:405–411. doi: 10.1038/ni.2242. Along with Ref. [9•], this paper describes the plasticity of T helper cells during differentiation and the molecular requirements for opposing levels of BCL6 and T-bet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11•.Luthje K, Kallies A, Shimohakamada Y, Belz GT, Light A, Tarlinton DM, Nutt SL. The development and fate of follicular helper T cells defined by an IL-21 reporter mouse. Nat Immunol. 2012;13:491–498. doi: 10.1038/ni.2261. Along with Ref. [9•] and Ref. [10•], this paper describes the plasticity of T helper cells during differentiation and the contribution of IL-21 to TFH development. [DOI] [PubMed] [Google Scholar]
- 12.Kuo TC, Shaffer AL, Haddad J, Choi YS, Staudt L, Calame K. Repression of BCL-6 is required for the formation of human memory B cells in vitro. J Exp Med. 2007;204:819–830. doi: 10.1084/jem.20062104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13••.Pepper M, Pagán AJ, Igyártó BZ, Taylor JJ, Jenkins MK. Opposing signals from the bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. See annotation to Ref. [14••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14••.Marshall HD, Chandele A, Jung YW, Meng H, Poholek AC, Parish IA, Rutishauser R, Cui W, Kleinstein SH, Craft J, et al. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 2011;35:633–646. doi: 10.1016/j.immuni.2011.08.016. Along with Ref. [13••], this study was the first to identify BCL6+ CXCR5loICOSlo TFH memory cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Liu X, Yan X, Zhong B, Nurieva RI, Wang A, Wang X, Martin-Orozco N, Wang Y, Chang SH, Esplugues E, et al. Bcl6 expression specifies the T follicular helper cell program in vivo. J Exp Med. 2012;209:1841–1852. S1841–1824. doi: 10.1084/jem.20120219. This paper utilizes a BCL6-RFP reporter mouse to interrogate the gene expression signatures of T-cell subsets expressing variable levels of BCL6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16••.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang Y, Lim H, Reynolds JM, Zhou X, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. See annotation to Ref. [17••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17••.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. Foxp3(+) follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. Along with Ref. [16••], this study was the first to identify BCL6+/FOXP3+ TFR cells, which play a regulatory role in the GC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campbell DJ, Koch MA. T(reg) cells: patrolling a dangerous neighborhood. Nat Med. 2011;17:929–930. doi: 10.1038/nm.2433. [DOI] [PubMed] [Google Scholar]
- 19.Papatriantafyllou M. Regulatory T cells: Pursuing a germinal centre career. Nat Rev Immunol. 2011;11:572. doi: 10.1038/nri3053. [DOI] [PubMed] [Google Scholar]
- 20••.Chang P, Barral P, Fitch J, Pratama A, Ma C, Kallies A, Hogan J, Cerundolo V, Tangye S, Bittman R, et al. Identification of Bcl-6-dependent follicular helper NKT cells that provide cognate help for B cell responses. Nat Immunol. 2012;13:35–43. doi: 10.1038/ni.2166. This was the first study to identify BCL6-dependent NKTFH-cells within the GC. [DOI] [PubMed] [Google Scholar]
- 21.King C. A fine romance: T follicular helper cells and B cells. Immunity. 2011;34:827–829. doi: 10.1016/j.immuni.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 22•.Okada T, Moriyama S, Kitano M. Differentiation of germinal center B cells and follicular helper T cells as viewed by tracking Bcl6 expression dynamics. Immunol Rev. 2012;247:120–132. doi: 10.1111/j.1600-065X.2012.01120.x. This review summarizes in detail the localization, migration, commitment and determinants of BCL6-expressing B and T-cells prior to and during GC formation. [DOI] [PubMed] [Google Scholar]
- 23.Baumjohann D, Okada T, Ansel KM. Cutting Edge: Distinct Waves of BCL6 Expression during T Follicular Helper Cell Development. J Immunol. 2011;187:2089–2092. doi: 10.4049/jimmunol.1101393. [DOI] [PubMed] [Google Scholar]
- 24••.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS Receptor Instructs T Follicular Helper Cell versus Effector Cell Differentiation via Induction of the Transcriptional Repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. See annotation to Ref. [26••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, Okada T. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–972. doi: 10.1016/j.immuni.2011.03.025. See annotation to Ref. [26••] [DOI] [PubMed] [Google Scholar]
- 26••.Kerfoot SM, Yaari G, Patel JR, Johnson KL, Gonzalez DG, Kleinstein SH, Haberman AM. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–960. doi: 10.1016/j.immuni.2011.03.024. Along with Ref. [24••] and Ref. [25••], this study was the first to describe a requirement for BCL6 during the dynamic interactions between B and T-cells prior to GC formation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi H, Cannons J, Klauschen F, Schwartzberg P, Germain R. SAP-controlled T-B cell interactions underlie germinal centre formation. Nature. 2008;455:764–769. doi: 10.1038/nature07345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28••.Lee SK, Rigby RJ, Zotos D, Tsai LM, Kawamoto S, Marshall JL, Ramiscal RR, Chan TD, Gatto D, Brink R, et al. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J Exp Med. 2011;208:1377–1388. doi: 10.1084/jem.20102065. This study identified an important role for BCL6 in non-GC T-cell-dependent extrafollicular antibody responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29••.Duy C, Yu JJ, Nahar R, Swaminathan S, Kweon SM, Polo JM, Valls E, Klemm L, Shojaee S, Cerchietti L, et al. BCL6 is critical for the development of a diverse primary B cell repertoire. J Exp Med. 2010;207:1209–1221. doi: 10.1084/jem.20091299. See annotation to Ref. [30••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30••.Nahar R, Ramezani-Rad P, Mossner M, Duy C, Cerchietti L, Geng H, Dovat S, Jumaa H, Ye BH, Melnick A, et al. Pre-B cell receptor-mediated activation of BCL6 induces pre-B cell quiescence through transcriptional repression of MYC. Blood. 2011;118:4174–4178. doi: 10.1182/blood-2011-01-331181. Along with Ref. [29••], this study identified an important role for BCL6 in the development and survival of early precursor B-cells, suggesting that BCL6 function is not exclusive to mature lymphoid cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Duy C, Hurtz C, Shojaee S, Cerchietti L, Geng H, Swaminathan S, Klemm L, Kweon S, Nahar R, Braig M, et al. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 2011;473:384–388. doi: 10.1038/nature09883. See annotation to Ref. [32••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Geng H, Brennan S, Milne TA, Chen WY, Li Y, Hurtz C, Kweon SM, Zickl L, Shojaee S, Neuberg D, et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012;2:1004–1023. doi: 10.1158/2159-8290.CD-12-0208. Along with Ref. [31••], this study shows a critical link between B-ALL survival and resistance to therapy and the presence of BCL6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–120. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34••.Bentebibel SE, Schmitt N, Banchereau J, Ueno H. Human tonsil B-cell lymphoma 6 (BCL6)-expressing CD4+ T-cell subset specialized for B-cell help outside germinal centers. Proc Natl Acad Sci USA. 2011;108:E488–497. doi: 10.1073/pnas.1100898108. This study identified BCL6-expressing TFH-like cells in the periphery, which provide help to naïve and memory B cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Canfield S, Rothman P. TH2 inflammation repressed by chemokines. Nat Immunol. 2000;1:189–190. doi: 10.1038/79720. [DOI] [PubMed] [Google Scholar]
- 36••.Sawant DV, Sehra S, Nguyen ET, Jadhav R, Englert K, Shinnakasu R, Hangoc G, Broxmeyer HE, Nakayama T, Perumal NB, et al. Bcl6 controls the Th2 inflammatory activity of regulatory T cells by repressing Gata3 function. J Immunol. 2012;189:4759–4769. doi: 10.4049/jimmunol.1201794. This study identified an important role for BCL6 in the differentiation of Treg cells, and thus, in the control of Th2 inflammatory responses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mondal A, Sawant D, Dent AL. Transcriptional repressor BCL6 controls Th17 responses by controlling gene expression in both T cells and macrophages. J Immunol. 2010;184:4123–4132. doi: 10.4049/jimmunol.0901242. [DOI] [PubMed] [Google Scholar]
- 38.Chang CC, Vlad G, D’Agati VD, Liu Z, Zhang QY, Witkowski P, Torkamani AA, Stokes MB, Ho EK, Cortesini R, et al. BCL6 is required for differentiation of Ig-like transcript 3-Fc-induced CD8+ T suppressor cells. J Immunol. 2010;185:5714–5722. doi: 10.4049/jimmunol.1001732. [DOI] [PubMed] [Google Scholar]
- 39.Toney LM, Cattoretti G, Graf JA, Merghoub T, Pandolfi PP, Dalla-Favera R, Ye BH, Dent AL. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 40.Barish GD, Yu RT, Karunasiri M, Ocampo CB, Dixon J, Benner C, Dent AL, Tangirala RK, Evans RM. Bcl-6 and NF-{kappa}B cistromes mediate opposing regulation of the innate immune response. Genes & Development. 2010;24:2760–2765. doi: 10.1101/gad.1998010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ci W, Polo JM, Cerchietti L, Shaknovich R, Wang L, Yang SN, Ye K, Farinha P, Horsman DE, Gascoyne RD, et al. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 2009;113:5536–5548. doi: 10.1182/blood-2008-12-193037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawant DV, Wu H, Kaplan MH, Dent AL. The Bcl6 target gene microRNA-21 promotes Th2 differentiation by a T cell intrinsic pathway. Mol Immunol. 2013;54:435–442. doi: 10.1016/j.molimm.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Huang C, Hatzi K, Melnick A. Lineage-specific functions of Bcl-6 in immunity and inflammation are mediated by distinct biochemical mechanisms. Nat Immunol. 2013;14:380–388. doi: 10.1038/ni.2543. This study generated a BCL6 BTB-mutant mouse and was the first to demonstrate that the BTB domain of BCL6 is required for the development of GC B-cells, but not GC T-cells or macrophages. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44•.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. J Immunol. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. This study was the first to identify a cooperating transcriptional partner of BCL6, Maf, which helps to orchestrate the gene expression program of TFH cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oestreich KJ, Huang AC, Weinmann AS. The lineage-defining factors T-bet and Bcl-6 collaborate to regulate Th1 gene expression patterns. J Exp Med. 2011;208:1001–1013. doi: 10.1084/jem.20102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee CH, Melchers M, Wang H, Torrey TA, Slota R, Qi CF, Kim JY, Lugar P, Kong HJ, Farrington L, et al. Regulation of the germinal center gene program by interferon (IFN) regulatory factor 8/IFN consensus sequence-binding protein. J Exp Med. 2006;203:63–72. doi: 10.1084/jem.20051450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang TT, Makondo KJ, Marshall AJ. p110δ phosphoinositide 3-kinase represses IgE switch by potentiating BCL6 expression. J Immunol. 2012;188:3700–3708. doi: 10.4049/jimmunol.1103302. [DOI] [PubMed] [Google Scholar]
- 48.Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, Corcoran LM, Godfrey DI, Toellner KM, Smyth MJ, et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. 2010;207:365–378. doi: 10.1084/jem.20091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karnowski A, Chevrier S, Belz GT, Mount A, Emslie D, D’Costa K, Tarlinton DM, Kallies A, Corcoran LM. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J Exp Med. 2012;209:2049–2064. doi: 10.1084/jem.20111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lin J, Lwin T, Zhao JJ, Tam W, Choi YS, Moscinski LC, Dalton WS, Sotomayor EM, Wright KL, Tao J. Follicular dendritic cell-induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin’s B-cell lymphomas. Leukemia. 2011;25:145–152. doi: 10.1038/leu.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nutt SL, Tarlinton DM. Germinal center B and follicular helper T cells: siblings, cousins or just good friends? Nat Immunol. 2011;131:472–477. doi: 10.1038/ni.2019. [DOI] [PubMed] [Google Scholar]
- 53.Bollig N, Brustle A, Kellner K, Ackermann W, Abass E, Raifer H, Camara B, Brendel C, Giel G, Bothur E, et al. Transcription factor IRF4 determines germinal center formation through follicular T-helper cell differentiation. Proc Natl Acad Sci U S A. 2012;109:8664–8669. doi: 10.1073/pnas.1205834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Salamon D, Adori M, He M, Bönelt P, Severinson E, Kis LL, Wu L, Ujvari D, Leveau B, Nagy N, et al. Type I interferons directly down-regulate BCL-6 in primary and transformed germinal center B cells: differential regulation in B cell lines derived from endemic or sporadic Burkitt’s lymphoma. Cytokine. 2012;57:360–371. doi: 10.1016/j.cyto.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 55.Walker SR, Nelson EA, Frank DA. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 2007;26:224–233. doi: 10.1038/sj.onc.1209775. [DOI] [PubMed] [Google Scholar]
- 56••.Johnston RJ, Choi YS, Diamond JA, Yang JA, Crotty S. STAT5 is a potent negative regulator of TFH cell differentiation. J Exp Med. 2012;209:243–250. doi: 10.1084/jem.20111174. See annotation to Ref. [57••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57••.Nurieva RI, Podd A, Chen Y, Alekseev AM, Yu M, Qi X, Huang H, Wen R, Wang J, Li HS, et al. STAT5 protein negatively regulates T follicular helper (Tfh) cell generation and function. J Biol Chem. 2012;287:11234–11239. doi: 10.1074/jbc.M111.324046. Along with Ref. [56••], this study was the first to identify a direct role for STAT5 in the regulation of BCL6 expression in TFH cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ballesteros-Tato A, León B, Graf BA, Moquin A, Adams PS, Lund FE, Randall TD. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity. 2012;36:847–856. doi: 10.1016/j.immuni.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 60.Phan R, Saito M, Kitagawa Y, Means A, Dalla-Favera R. Genotoxic stress regulates expression of the proto-oncogene Bcl6 in germinal center B cells. Nat Immunol. 2007;8:1132–1139. doi: 10.1038/ni1508. [DOI] [PubMed] [Google Scholar]
- 61.Niu H, Ye B, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes & Development. 1998;12:1953–1961. doi: 10.1101/gad.12.13.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62••.Duan S, Cermak L, Pagan JK, Rossi M, Martinengo C, di Celle PF, Chapuy B, Shipp M, Chiarle R, Pagano M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature. 2011;481:90–93. doi: 10.1038/nature10688. This study identifies an alternative pathway of ubiquitin-mediated BCL6 protein degradation mediated by FBXO11, which is inactivated in DLBCL and thus provides a mechanistic link to constitutive BCL6 expression in lymphomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63•.Sitte S, Glasner J, Jellusova J, Weisel F, Panattoni M, Pardi R, Gessner A. JAB1 Is Essential for B Cell Development and Germinal Center Formation and Inversely Regulates Fas Ligand and Bcl6 Expression. The Journal of Immunology. 2012:1–11. doi: 10.4049/jimmunol.1101455. Using a BCL6-specific deletion of COPS5, this study identifies a role for the COP9 signalosome in BCL6 expression and stability in GC B-cells. [DOI] [PubMed] [Google Scholar]
- 64.Cerchietti LC, Lopes EC, Yang SN, Hatzi K, Bunting KL, Tsikitas LA, Mallik A, Robles AI, Walling J, Varticovski L, et al. A purine scaffold Hsp90 inhibitor destabilizes BCL-6 and has specific antitumor activity in BCL-6-dependent B cell lymphomas. Nat Med. 2009;15:1369–1376. doi: 10.1038/nm.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ranuncolo S, Polo J, Dierov J, Singer M, Kuo T, Greally J, Green R, Carroll M, Melnick A. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 66.Cerchietti LC, Ghetu AF, Zhu X, Da Silva GF, Zhong S, Matthews M, Bunting KL, Polo JM, Farès C, Arrowsmith CH, et al. A small-molecule inhibitor of BCL6 kills DLBCL cells in vitro and in vivo. Cancer Cell. 2010;17:400–411. doi: 10.1016/j.ccr.2009.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67••.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. See annotation to Ref. [69••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68••.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. See annotation to Ref. [69••] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69••.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, Cruz-Gordillo P, Knoechel B, Asmann YW, Slager SL, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. Along with Ref. [67••] and Ref. [68••], this study uses targeted re-sequencing of DLBCL and FL patients to identify novel and recurring mutations in a variety of important cell regulatory proteins, including chromatin modifying complexes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saito M, Novak U, Piovan E, Basso K, Sumazin P, Schneider C, Crespo M, Shen Q, Bhagat G, Califano A, et al. BCL6 suppression of BCL2 via Miz1 and its disruption in diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2009;106:11294–11299. doi: 10.1073/pnas.0903854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71••.Shaffer AL, Young RM, Staudt L. Pathogenesis of human B cell lymphomas. Annu Rev Immunol. 2012;30:565–610. doi: 10.1146/annurev-immunol-020711-075027. This paper provides an extensive review of the cellular and molecular mechanisms leading to the development of B-cell lymphomas. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72••.Cerchietti LC, Hatzi K, Caldas-Lopes E, Yang SN, Figueroa ME, Morin RD, Hirst M, Mendez L, Shaknovich R, Cole PA, et al. BCL6 repression of EP300 in human diffuse large B cell lymphoma cells provides a basis for rational combinatorial therapy. J Clin Invest. 2010;120:4569–4582. doi: 10.1172/JCI42869. This study was the first to identify a critical role for BCL6 in the repression of P300 and identify P300 mutations in DLBCL, thus providing a mechanistic link to constitutive BCL6 expression and a new combinatorial therapy for DLBCL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73•.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, Kasper LH, Lerach S, Tang H, Ma J, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. This study identifies recurring CBP/P300 mutations within a large cohort of DLBCL and FL patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lamant L, de Reynies A, Duplantier MM, Rickman DS, Sabourdy F, Giuriato S, Brugieres L, Gaulard P, Espinos E, Delsol G. Gene-expression profiling of systemic anaplastic large-cell lymphoma reveals differences based on ALK status and two distinct morphologic ALK+ subtypes. Blood. 2007;109:2156–2164. doi: 10.1182/blood-2006-06-028969. [DOI] [PubMed] [Google Scholar]
- 75.de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, Lamant L, Leroy K, Brière J, Molina T, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007;109:4952–4963. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 76.Piccaluga PP, Agostinelli C, Califano A, Carbone A, Fantoni L, Ferrari S, Gazzola A, Gloghini A, Righi S, Rossi M, et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Research. 2007;67:10703–10710. doi: 10.1158/0008-5472.CAN-07-1708. [DOI] [PubMed] [Google Scholar]
- 77.Cetinözman F, Jansen PM, Willemze R. Expression of programmed death-1 in primary cutaneous CD4-positive small/medium-sized pleomorphic T-cell lymphoma, cutaneous pseudo-T-cell lymphoma, and other types of cutaneous T-cell lymphoma. Am J Surg Pathol. 2012;36:109–116. doi: 10.1097/PAS.0b013e318230df87. [DOI] [PubMed] [Google Scholar]
- 78.Wang J, Ke XY. The four types of Tregs in malignant lymphomas. J Hematol Oncol. 2011;4:50. doi: 10.1186/1756-8722-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cerchietti LC, Yang SN, Shaknovich R, Hatzi K, Polo JM, Chadburn A, Dowdy SF, Melnick A. A peptomimetic inhibitor of BCL6 with potent anti-lymphoma effects in vitro and in vivo. Blood. 2008;113:3397–3405. doi: 10.1182/blood-2008-07-168773. [DOI] [PMC free article] [PubMed] [Google Scholar]