Abstract
Mitochondrial dysfunction is an important part of the decline in cardiac function in heart failure.
Aims
We hypothesize that there would be specific abnormalities in mitochondrial function and proteome with the progression of ischemic heart failure (HF).
Materials and Methods
We used a high left anterior descending artery (LAD)ligation in 3–4 month old male rats to generate HF. Rats were studied 9 weeks post ligation.
Key findings
Electron microscopy of left ventricle samples showed, mitochondrial changes including decreased size, increased number, abnormal distribution, and cristae loss. Mitochondria in ischemic HF exhibited decreased total ATP, impaired mitochondrial respiration, as well as reduced complex I activity. Analysis of LV mitochondrial proteins by mass spectrometry was performed, and 31 differentially expressed proteins (p < 0.05) of more than 500 total proteins were identified. Of these proteins, 15 were up-regulated and 16 down-regulated in the failing heart. A set of complex I proteins were significantly decreased, consistent with the impairment of complex I activity. There were distinct changes in mitochondrial function and proteome in ischemic HF. Although there were similarities, the distinction between the reported proteomic changed with TAC pressure overload induced HF and ischemic HF in the current study suggested different pathological mechanisms.
Significance
Specific changes in mitochondrial protein expression, which correlate with changes in mitochondrial function, have been identified in ischemic HF for the first time.
Keywords: proteome remodeling, mitochondria, ischemic heart failure, NADH dehydrogenase, complex I, NDUFA5, NDUFV1, heart, heart failure, complex II, complex IV
Introduction
Impaired mitochondrial function is a key feature in the decline in cardiac function in heart failure (Bayeva et al., 2013, Chen and Knowlton, 2011, Rosca and Hoppel, 2012). A number of cardiomyopathies are associated with changes in expression of mitochondrial proteins (Hollander et al., 2011, Sterba et al., 2011, Zhang et al., 2012). Proteomic changes differ amongst different types of heart failure HF. Studies have been done on pressure-overload induced HF (Bugger et al., 2010, Dai et al., 2012), diabetes related HF, and tachycardia-induced HF (Birner et al., 2012). However, the mitochondrial proteome changes in response to ischemic heart failure (HF) have not been investigated.
In the present study, we hypothesized that there would be specific abnormalities in mitochondrial structure, function and proteome with the progression of ischemic heart failure. We used our established rat model of ischemic HF with high ligation of the left anterior descending artery (LAD) as previously reported (Lin et al., 2007). We investigated mitochondrial function, including individual complex function and respiration. We used mass spectrometry to analyze the mitochondrial proteome, and Ingenuity Pathway Analysis software (IPA) to algorithmically generate protein connections. This allowed us to identify the most plausible signaling pathway abnormalities related to mitochondria in ischemic HF. This is the first study, to our knowledge, to investigate mitochondrial proteome changes in ischemic HF. The mitochondrial proteome had multiple changes, and some of these may be distinct to ischemic HF. Although end-stage systolic HF appears very similar regardless of etiology, the underlying molecular and cellular changes may differ. Thus, investigation of fundamental cell and molecular changes in HF has the potential to provide new insights into HF and to lead to the development of new treatments.
Material and Methods
Animal Model
2–3 month old male Sprague- Dawley rats (weight approximately 300g) were obtained from Charles River (Wilmington, MD). Rats were anesthetized with an intraperitoneal (IP) injection of Ketamine (45 mg/Kg) and Xylazine (2.28 mg/Kg). Prior to thoracotomy and during surgery, deep anesthesia was confirmed by the absence of the corneal and toe pinch reflexes. A high ligation of the LAD was performed, as previously described (Lin, Kim, 2007). Mortality was approximately 40% in the first 24h, which is consistent with the literature for this model, and zero thereafter. Post-operative rats were treated with buprenorphine 0.05 mg/Kg every 12 h for 48h by intramuscular (IM) injection. The animal protocol was approved by the University of California, Davis, Animal Research Committee in accordance with the NIH Guide for the Care and Use of Laboratory Animals.
Heart Failure Model
A rat coronary ligation model of heart failure was used as previously described (Lin, Kim, 2007). Decreased fractional shortening was confirmed by echocardiography. Sham rats underwent thoracotomy, but no ligation. Rats were studied at 9 weeks post-surgery, at age 4–5 months (weight approximately 350g) at which time HF had developed.
Echocardiography
was done 9 weeks post ligation, as previously described (Chen et al., 2012, Lin, Kim, 2007). The rats were anesthetized with Ketamine/Xylazine 45/2.28 mg/kg IP. Two-dimensional imaging was used to identify the short-axis position. M-mode images were recorded in the short-axis view and saved for analysis of chamber size and fractional shortening (FS %). A minimum of three images were analyzed for each rat, and the average values each rat for the two groups were pooled and compared.
Following echocardiography, the rats were overdosed with Ketamine/Xylazine 75/3.9 mg/kg IP and the hearts were collected for mitochondrial isolation. Additional measurements performed at this time including tibia length, heart weight and body weight. A small piece of the left ventricle free wall was fixed for electron microscopy (EM).
Mitochondrial Isolation
Freshly isolated left ventricle was minced immediately, and functional mitochondria were isolated using the method of Frezza et al. (Frezza et al.,2007). Instead of using ultracentrifugation with sucrose gradients, isolating mitochondria with higher purity, the method used isolated mitochondria with higher activity and preserved electron transport integrity. The minced LV was homogenized with a glass pestle in isolation buffer (IB) 1 (1 M sucrose, 1 M Tris-HCl, 1 M KCl, 1 M EDTA, and 10% BSA, pH 7.4). The homogenized tissue was centrifuged at 700 g for 10 minutes at 4 °C. The pellet was re-suspended in IB2 (1 M sucrose, 0.1 M EGTA-Tris, 1 M Tris-HCl, pH 7.4), centrifuged at 8000 g for 10 minutes, and then washed (Chen, Liu, 2012). Protein levels, measured by the BCA assay, were used to assess the mitochondrial concentration. Mitochondria not used for functional studies were stored in −80°C in IB2, and saved for proteomic analysis.
Mitochondrial Respiration Measurement
Mitochondrial oxygen consumption rate (OCR) was measured with a Seahorse XF24 Analyzer, as previously described (Chen, Liu, 2012). Fresh KCl based mitochondrial assay solution (MAS, 115 mM KCl, 10 mM KH2PO4, 2 mM MgCl2, 3 mM HEPES, 1 mM EGTA and 0.2% BSA) was prepared to match in vivo physiologic conditions. Pilot experiments were done to determine optimal conditions to reach plateau states. Baseline OCR (state II respiration) was measured after adding 5 mmol/L succinate in the presence of 2 µmol/L rotenone as substrates. ADP OCR (State III respiration) was measured by the addition of ADP to a final concentration of 2.0 mmol/L. FCCP (mesoxalonitrile 4-trifluoromethoxyphenylhydrazone) stimulated OCR was measured after adding FCCP to a final concentration of 4.0 µmol/L to determine maximal uncoupled respiration. Respiration due to proton leak was measured using 2.0 µmol/L oligomycin and non-mitochondrial OCR was measured by addition of antimycin A (AA). The mitochondrial plate was prepared at 37 °C and read as previously described (Chen, Liu, 2012).
LC-MS/MS
Samples were separated briefly on a standard 10% SDS PAGE. Gel pieces, containing all the proteins in the sample, were cut out and digested using the Shevchenko protocol (Shevchenko et al., 2002). Digested peptides were analyzed using a QExactive mass spectrometer (Thermo Fisher Scientific) coupled with an Easy-LC (Thermo Fisher Scientific) and a nanospray ionization source. The peptides were loaded onto a trap (100micron, C18 100Å 5U) and desalted online before being separated using a reverse phase column (75micon, C18 200Å 3U). The gradient duration for separation of peptides was 60 minutes using 0.1% formic acid and 100% acetonitrile for solvents A and B, respectively. Data were acquired using a data dependent MS/MS method which had a full scan range of 300–1600Da and a resolution of 70,000. The MS/MS method’s resolution is 17,500 and an isolation width of 2 m/z with normalized collision energy of 27. The nanospray source was operated using 2.2 kV spray voltage and a heated transfer capillary temperature of 250°C.
Database Searching
Tandem mass spectra were extracted by Mass Matrix File converter version 3. Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (The GPM, thegpm.org; version CYCLONE (2011.05.01.1)). X! Tandem was set up to search the uniprot rat complete proteome (August 2012, 57,276 entries) plus an equal number of reverse sequences and 113 common laboratory contaminants from the Common Repository of Adventitious Proteins database (http://www.thegpm.org/crap/) assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 20 PPM and a parent ion tolerance of 20 PPM. Iodoacetamide derivative of cysteine was specified in X! Tandem as a fixed modification. Deamidation of asparagine and glutamine, oxidation of methionine and tryptophan, sulphone of methionine and tryptophan oxidation to formylkynurenin of tryptophan were specified in X! Tandem as variable modifications.
Criteria for Protein Identification
Scaffold (version Scaffold_3.6.2, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 80.0% probability as specified by the Peptide Prophet algorithm (Keller et al., 2002). Protein identifications were accepted if they could be established at greater than 80.0% probability and contained at least 2 identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Using the above criteria, the False Discovery rate was calculated as 0% on the peptide level and 0.2% on the protein level (Tabb, 2008).
Statistics and Spectral Counting
Differentially expressed proteins were determined using spectral counting. Two groups of comparisons were done with each group comprised of 5 biological replicates. Two approaches were used to analyze the spectral counting data. In the first approach Spectral counts were assigned to each protein within scaffold and analyzed using scaffold F-Test (Zhang et al., 2006). In the second approach, assigned spectral counts were exported from scaffold and corrected for shared peptides (Zhang et al., 2010) and then analyzed using Q-spec (Keller, Nesvizhskii, 2002). Proteins with a p-value ≤ 0.05 were considered significant and analyzed using Ingenuity Pathway Analysis software (IPA) (Qiagen, Redwood City, California). Networks of these proteins were algorithmically generated based on their direct connectivity from IPA database.
Complex Activities Measurement
Complex I, II, IV activity were measured with kits from Mitosciences (Eugene, Oregon). 2.5 ug of mitochondrial, based on protein concentration, were used for each assay based on recommendations of manufacturer. Assays were performed following the directions of the manufacturer.
Electron microscopy (EM)
was done in the core Electron Microscopy Laboratory, Department of Pathology and Laboratory Medicine, School of Medicine, University of California at Davis. Left ventricular samples were taken from freshly excised hearts, washed, fixed in 4% paraformaldehyde, post-fixed in 2% osmium tetroxide, embedded in resin, and sectioned (Chen et al., 2009). Standard transmission EM with a Philips CM120 microscope was used for all studies. The EM images were analyzed with Photoshop CS3, using the counting and area analysis function as we have reported before (Chen, Gong, 2009).
Western Blotting
Western blotting was done as previously described (Stice et al., 2011). The membrane was stained with Ponceau after transfer to verify the proteins were equally loaded. An internal control was used to allow comparison among different membranes. Values were normalized to the internal control to correct for variations amongst blots as previously described (Voss et al., 2003). The same mitochondrial samples were analyzed for ATP-synthase (ab109867, Abcam, Cambridge, MA), NADH dehydrogenase flavoprotein 1 (NDUFV1) (ab55535, Abcam, Cambridge, MA), Tripartite motif-containing protein 72 (TRIM72) (ab68061, Abcam, Cambridge, MA), Heat shock cognate 70 (HSC 70) (PA5–29241, Thermo Scientific, Rockford, I.L.), and Voltage- dependent anion channel-1 (VDAC-1) (ab15895, Abcam, Cambridge, MA) with 1:1000 dilution, and NADH dehydrogenase 1 alpha subcomplex subunit 5 (NDUFA5) (ab119308, Abcam, Cambridge, MA) with 1:100 dilution. Anti-mouse secondary antibody and anti-rabbit secondary antibody (respectively NA931V, NA934V GE Healthcare, Pittsburgh, PA) were used at a 1:1000 dilution.
After washing, the membrane was developed with a chemiluminescent system and analyzed as previously described (Stice, Chen, 2011).
ELISA
A commercial ELISA (StressGen/Enzo, Farmingdale, NY) was used to measure HSP60 levels following the instructions of the manufacturer. Samples were diluted 1:25 and measured in duplicate.
Statistics
Unless specified, all results are expressed as mean ± SEM. Control and HF data were compared using a Student t-test (Sigma Stat, Version 4, Aspire Software International, Ashburn, VA). A p<0.05 was considered significant.
Results
Cardiac Function
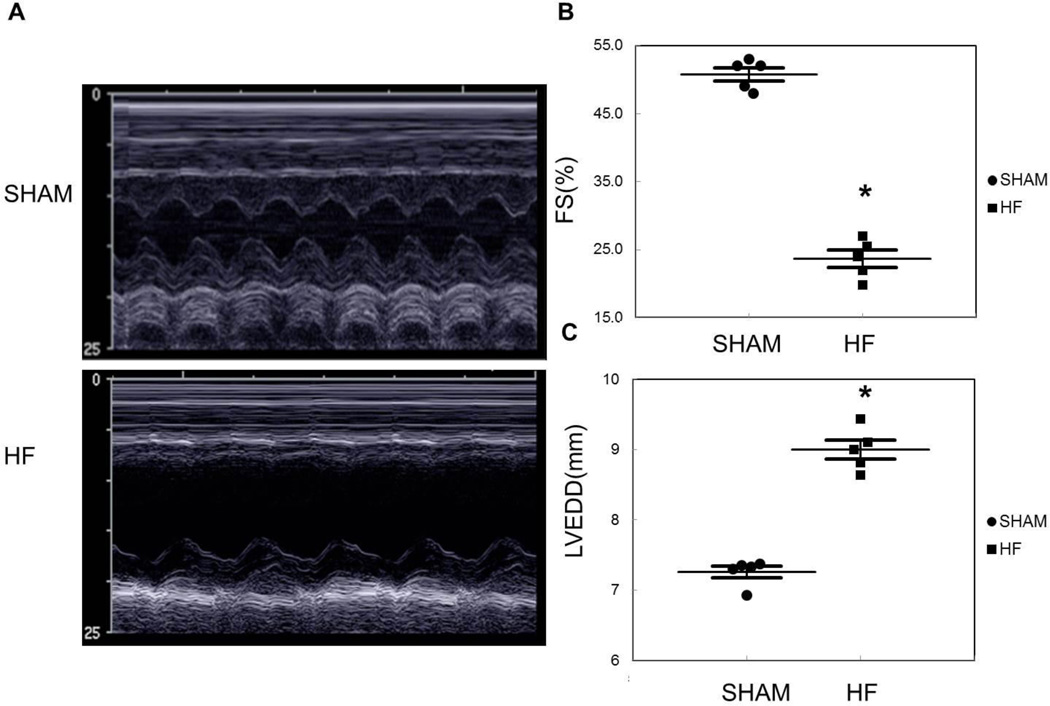
High LAD ligation was performed to generate ischemic heart failure in 2–3 month old male rats (weight 300g approximately), labeled HF group. Controls underwent the same surgery without coronary ligation, and are designated as Shams. Cardiac echocardiography was used to follow changes in function and chamber size. Nine weeks post coronary ligation, heart weight, heart weight/ tibia length ratio, and lung weight/ body weight ratio were all significantly increased in the HF group (p < 0.05, Table 1). Representative M-Mode images are shown in Figure 1A. Fractional shortening dropped from 50.8 ± 1.0% in Sham to 23.7 ± 1.3% in HF (p < 0.05, Figure 1B). The left ventricular was dilated in the HF group with a left ventricular end-diastolic dimension (LVEDD) of 9.0 ± 0.1 mm in the HF vs. 7.3 ± 0.1 mm in the Shams (p < 0.05) (Figure 1C).
Table 1.
Measurements of cardiac mass in rats with LAD ligation compared with SHAM. N= 5/group.
| Sham | Heart Failure | |
|---|---|---|
| Heart Weight (g) | 1.12±0.04 | 2.21±0.08* |
| Tibia Length (mm) | 38.52±0.04 | 38.50±0.04 |
| Heart weight-tibia length ratio (mg/mm) | 29.13±1.12 | 57.51±2.00* |
| Body Weight (g) | 305.64±1.71 | 313.98±2.77 |
| Lung Weight-Body Weight Ratio (mg/g) | 4.90±0.05 | 11.78±0.24* |
| Heart weight-Body Weight Ratio (mg/g) | 3.67±0.12 | 7.05±0.24* |
p < 0.05 vs. Sham
Figure 1.
A) Representative M mode echocardiography images from Sham and HF rats. B) Graph summarizes differences in fractional shortening (FS %). C) Left ventricular end-diastolic diameter (LVEDD) for Sham vs. HF 9 weeks after surgery. N=5/group, *p < 0.05 vs. Sham.
Electron microscopy (EM)
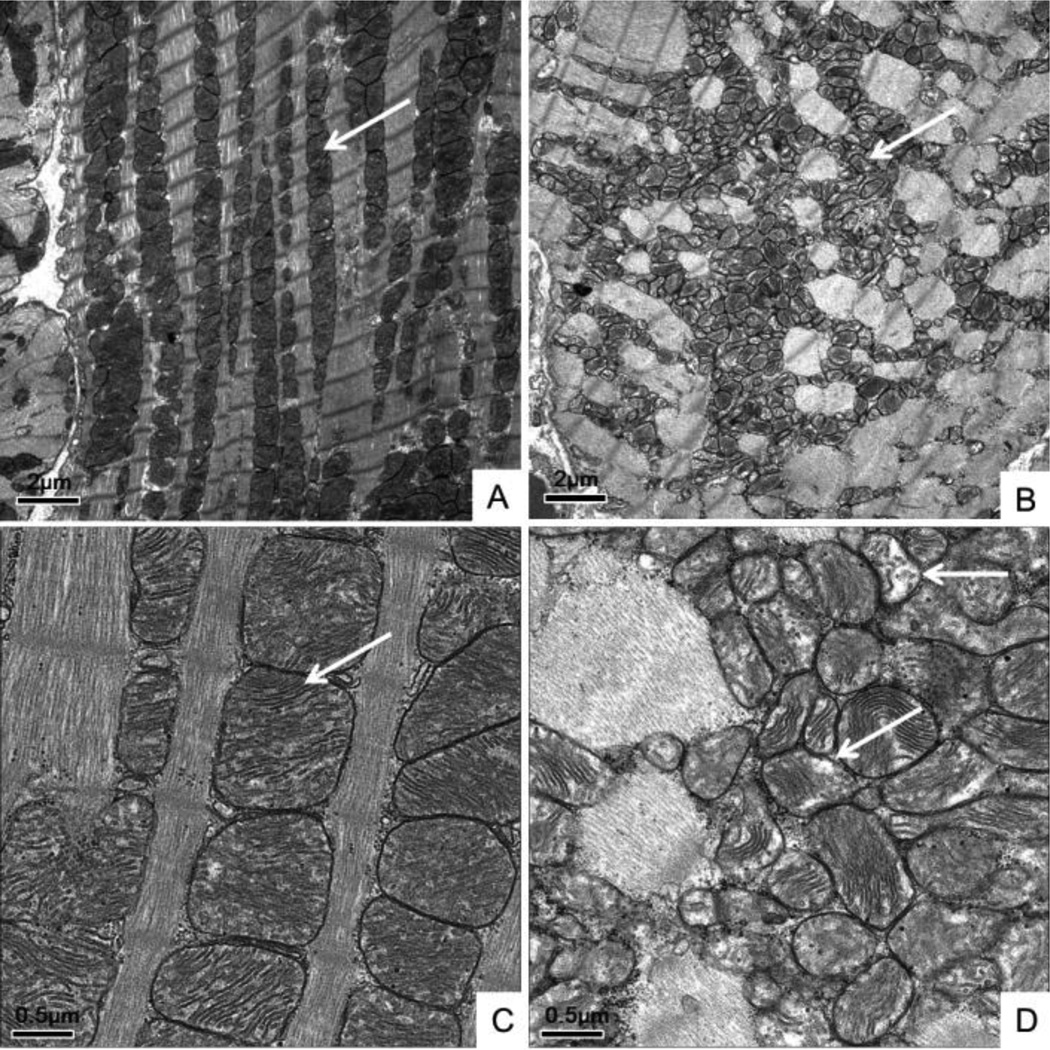
EM demonstrated that mitochondria in the HF hearts (Figure 2B and D) had lost their normal orderly arrangement along the sarcomeres seen in the Shams (Figure 2A and C). In addition, higher magnitude images show that the mitochondria were smaller and fragmented in ischemic HF (Figure 2D), as we and others have previously reported (Chen, Gong, 2009, van Ekeren et al., 1992). Importantly, there was a decrease in mitochondrial density in HF with loss of cristae (Figure 2B and D), consistent with impaired mitochondrial respiratory function. We have previously reported that the population of mitochondria increased from 100/100um2 in the Sham to more than 200/1 00um2 in the HF. The size of mitochondria decreased from 0.6 um2 in the Sham to 0.2 um2 in the HF (Chen, Gong, 2009).
Figure 2.
Electron microscopy (EM) of Sham and HF left ventricles (LV). A) Sham LV. Bar = 2 uM. B) HF LV showing loss of the mitochondrial alignment along the sarcomere (arrows). Bar= 2 uM. C/D) Higher magnification images. C) Sham LV. Bar = 0.5 µM. D) HF LV showing loss of the normal mitochondrial cristae structure in HF (arrows) compared to Sham, consistent with decreased mitochondrial respiratory function. Bar = 0.5 µM.
Mitochondrial Function
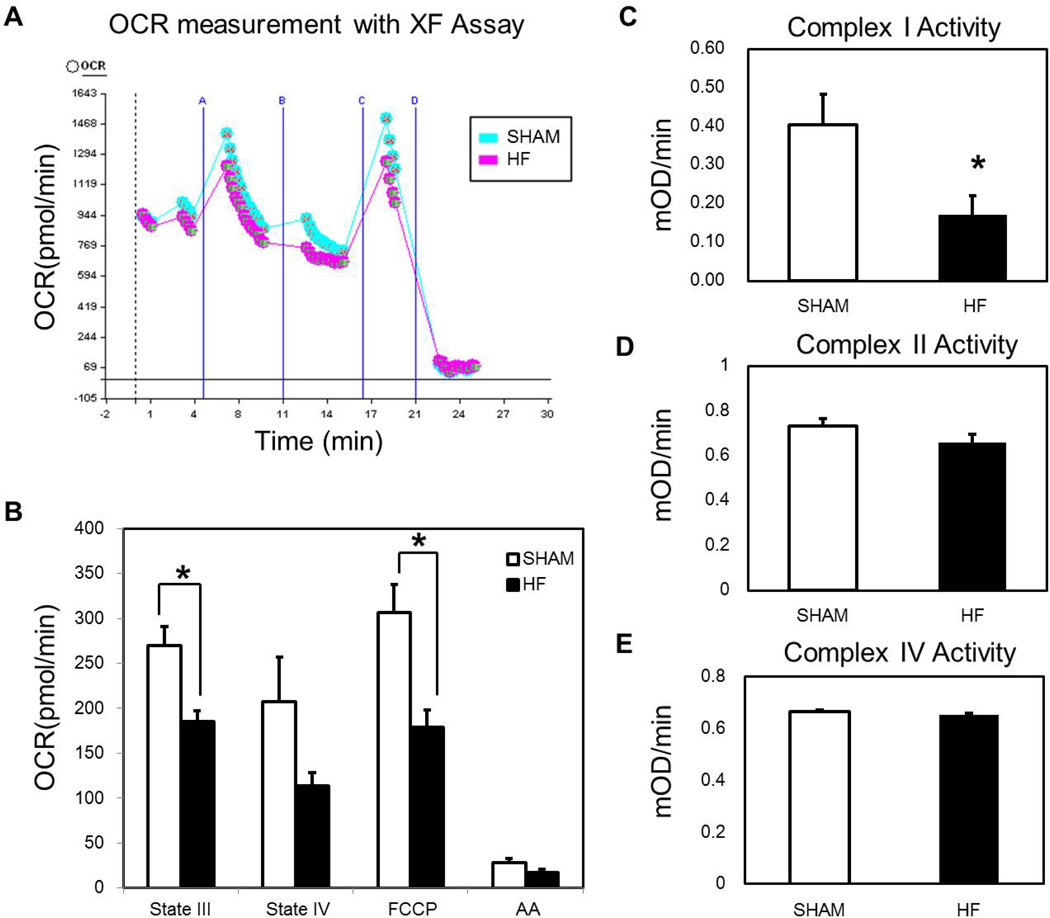
The oxygen consumption rate (OCR) of freshly isolated mitochondria was measured using the Seahorse XF24. Baseline OCR was measured with succinate and rotenone. Baseline studies for each preparation were used to determine the amount of ADP and mitochondria needed to reach peak OCR. In addition, two different quantities of mitochondria (2.5 and 5 µg of protein/well) were added to the plate every time to verify the robustness of the experiment. The final result was normalized to the protein concentration. Respiration was sequentially measured in a coupled state for basal respiration, followed by phosphorylating respiration in the presence of ADP (State III respiration). Following conversion of ADP to ATP, non-phosphorylating respiration was induced by the addition of oligomycin (respiration due to proton leak). Maximal uncoupled respiration was measured after the addition of FCCP. Finally, antimycin A was added to stop all mitochondrial respiration. A representative picture of the Seahorse analysis is shown in Figure 3A, showing that ATP-linked and FCCP-linked respiration is blunted in the HF. Oxygen consumption was significantly decreased in succinate/ ADP-linked (state III) and FCCP-linked maximal respiration capacity (p < 0.05) (Figure 3B). There was no difference seen in the state IV respiration or antimycin A respiration. Thus mitochondrial oxidative phosphorylation and maximal O2 consumption were impaired. Complex I, II, and IV activity were measured using freshly isolated mitochondria to complement the respiration studies (Figure 3C, 3D, 3E). Complex I activity was significantly reduced in HF (Figure 3C). In contrast, complex II and complex IV activity did not differ between HF and Shams (Figure 3D, 3E).
Figure 3.
Mitochondrial Function OCR (oxygen consumption rate) generated from Sham and HF mitochondria. A) Representative OCR curves generated with the Seahorse apparatus. B) Graph summarizes respiration states for HF and Sham mitochondria. Both ADP driven state III respiration and FCCP induced maximal respiration capacity were depressed in HF. N=10/group, *p < 0.05 vs. Sham. C) Complex I Activity - was decreased significantly in HF. D) Complex II activity did not differ between HF and Sham. E) Complex IV activity did not differ between HF and Sham. N=5/group for complex activity assays, *p < 0.05 vs. Sham.
Mitochondrial Proteomics
To gain a better understanding of the critical role of mitochondria in cardiac myocytes function and the changes that occur with ischemic HF, we studied the mitochondrial proteome by LC-MS/MS. More than 500 proteins were identified with 31 having significantly different expression in HF (Table 2). Fifteen proteins were up regulated and sixteen down regulated in the HF model based on analysis and simultaneous comparison of 5 separate Sham and 5 HF mitochondrial samples (Table 2). Changes were seen in key proteins primarily in the following pathways: mitochondrial dysfunction, glutamate degradation, and citrate cycle, based on pathway analysis with IPA software (Table 3). There were 6 proteins with significantly changed expression involved in mitochondrial dysfunction, 2 in glutamate degradation, and 3 in the TCA cycle (Table 3). In the mitochondrial dysfunction category, 4 proteins were up regulated, while 2 proteins were down regulated. Both of the decreased proteins, NDUFA5, and NDUFV1, are both part of complex I, and this is consistent with the observed decrease in complex I activity (Figure 3C).
Table 2.
Thirty-one mitochondrial protein expression changes in ischemic HF summarized from Scaffold analysis with 15 up regulated, and 16 down regulated (p < 0.05). Fold changes in HF is versus respective Sham controls.
| SYMBOL | PROTEIN | Fold Changes | P-value |
|---|---|---|---|
| G5ASL9 | Myosin-6 | 2.811901 | 0.045 |
| NNT | Nicotinamide nucleotide transhydrogenase | 2.259436 | 4E–10 |
| CScs | Citrate synthase | 2.18273 | 0.0043 |
| GW7 | Creatine kinase S-type, mitochondrial | 1.853532 | 3.3E–06 |
| MAOA | Amine oxidase [flavin-containing] A | 1.840772 | 0.000033 |
| MYH7 | Myosin-7 | 1.794734 | 0 |
| SDHC | Succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa |
1.717908 | 0.02 |
| SLC25A11 | Mitochondrial 2-oxoglutarate/malate carrier protein | 1.610646 | 0.0046 |
| ATP5C1 | ATP synthase subunit gamma | 1.592209 | 0.000003 |
| ALDH2 | Aldehyde dehydrogenase, mitochondrial | 1.581248 | 0.0073 |
| IDH2 | Isocitrate dehydrogenase [NADP], mitochondrial | 1.531087 | 0.0001 |
| DLAT | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex |
1.489361 | 0.014 |
| ETFA | Electron transfer flavoprotein subunit alpha, mitochondrial | 1.485936 | 0.007 |
| OGDH | 2-oxoglutarate dehydrogenase, mitochondrial | 1.452112 | 0.00058 |
| GOT2 | glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) |
1.415794 | 0.0046 |
| PGK1 | Phosphoglycerate kinase 1 | 0.699842 | 0.0071 |
| NDUFV1 | NADH dehydrogenase (Ubiquinone) flavoprotein 1 | 0.671429 | 0.016 |
| MCPT1 | Mast cell protease 1 | 0.64121 | 0.0058 |
| SPTAN1 | Spectrin alpha chain | 0.629506 | 1.4E–08 |
| DPP7 | Dipeptidyl peptidase 2 OS=Rattus norvegicus | 0.588844 | 0.00033 |
| ACSL1 | Long-chain-fatty-acid--CoA ligase 1 | 0.579429 | 0.00091 |
| ALDOA | Fructose-bisphosphate aldolase A | 0.535797 | 0.000078 |
| CKM | Creatine kinase M-type | 0.496592 | 0.00078 |
| NDUFA5 | NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | 0.489779 | 0.014 |
| VIM | Vimentin OS=Rattus norvegicus | 0.445656 | 0.00065 |
| VCL | Vinculin | 0.440555 | 0.022 |
| HSPA8 | Heat shock cognate 71 kDa protein | 0.414954 | 0.00075 |
| PYGM | Phosphorylase, glycogen, muscle | 0.401791 | 0.00035 |
| GOT1 | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) |
0.394457 | 0.0011 |
| BDH1 | D-beta-hydroxybutyrate dehydrogenase, mitochondrial | 0.338065 | 0.000098 |
| TRIM72 | Tripartite motif-containing protein 72 | 0.328852 | 1E–10 |
Table 3.
Changes in key proteins involved in mitochondrial dysfunction, glutamate degradation, and TCA cycle based on analysis by Ingenuity Pathway Analysis software.
| Entrez Gene Name | Changes |
|---|---|
| Mitochondrial Dysfunction | |
| ATP synthase subunit gamma | UP |
| Amine oxidase [flavin-containing] A | UP |
| 2-oxoglutarate dehydrogenase, mitochondrial | UP |
| Succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | UP |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 | DOWN |
| NADH dehydrogenase (Ubiquinone) flavoprotein 1 | DOWN |
| Glutamate Degradation | |
| Glutamic-oxaloacetic transaminase 2, mitochondrial (aspartate aminotransferase 2) | UP |
| Glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | DOWN |
| TCA Cycle | |
| Citrate synthase | UP |
| 2-oxoglutarate dehydrogenase, mitochondrial | UP |
| Succinate dehydrogenase complex, subunit C, integral membrane protein, 15kDa | UP |
Western blotting
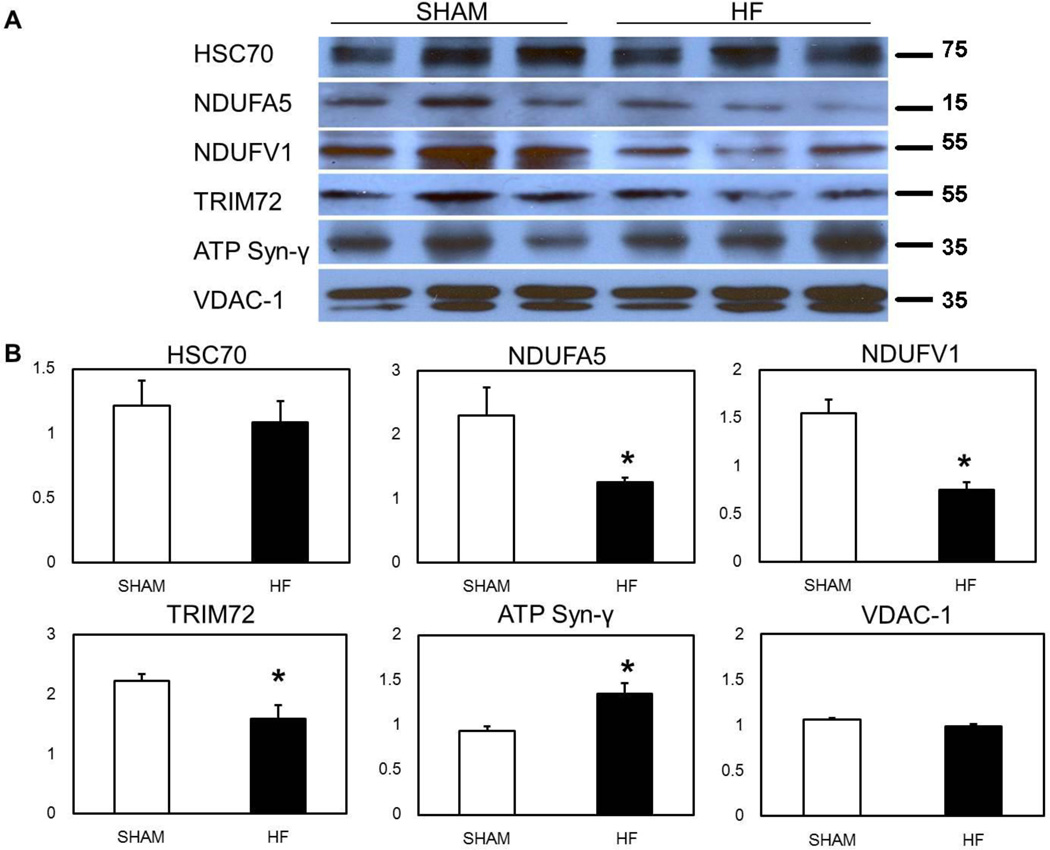
was performed to confirm changes in select proteins of interest. Representative blots are shown in Figure 4A. VDAC-1, which ran as a doublet, was used as a loading control, since it did not differ between Sham and HF groups, by either mass spectrometry or western blot. Densitometry data was normalized to VDAC-1. NDUFV5 and NDUFA1 both decreased about 50% in heart failure mitochondria by mass spec (p < 0.05) and this decrease was confirmed by western blot (Figure 4A and B). HSC70 (HSPA8), which was decreased by mass spec, was decreased approximately 10% by western, but this did not reach significance (Figure 4A and B). TRIM72 was decreased a third by western (Figure 4B), somewhat less than the 70% decrease seen by mass spec, however still a significant decrease (p < 0.05). In addition, ATP synthase gamma (ATP5C1) was increased by mass spectrometry in HF mitochondria, and this was confirmed by western (p < 0.05). We have a longstanding interest in HSP60, and it is a critical mitochondrial protein for protein folding after import. Therefore, we measured mitochondrial HSP60 levels by ELISA. HSP60 in HF was 52.9 ± 12.1 ng/mL, while in Sham it was 65.1 ± 8.3 ng/mL (p=n.s., n=5/group). Thus, HSP60 was not increased in the mitochondria in heart failure mitochondria, consistent with the mass spec findings.
Figure 4.
Western Blot Confirmation of Key Protein Changes Observed by Mass Spectrometry. A) Representative western blots of select proteins. B) Graphs summarize changes in HSC70, NDUFA5, NDUFV1, TRIM72, and ATP synthase gamma. VDAC1 (doublet) was used as a loading control and densities were normalized to VDAC-1. N=5–6/group, *p < 0.05 vs. Sham.
Ingenuity Pathway Analysis of Mitochondrial Proteomics
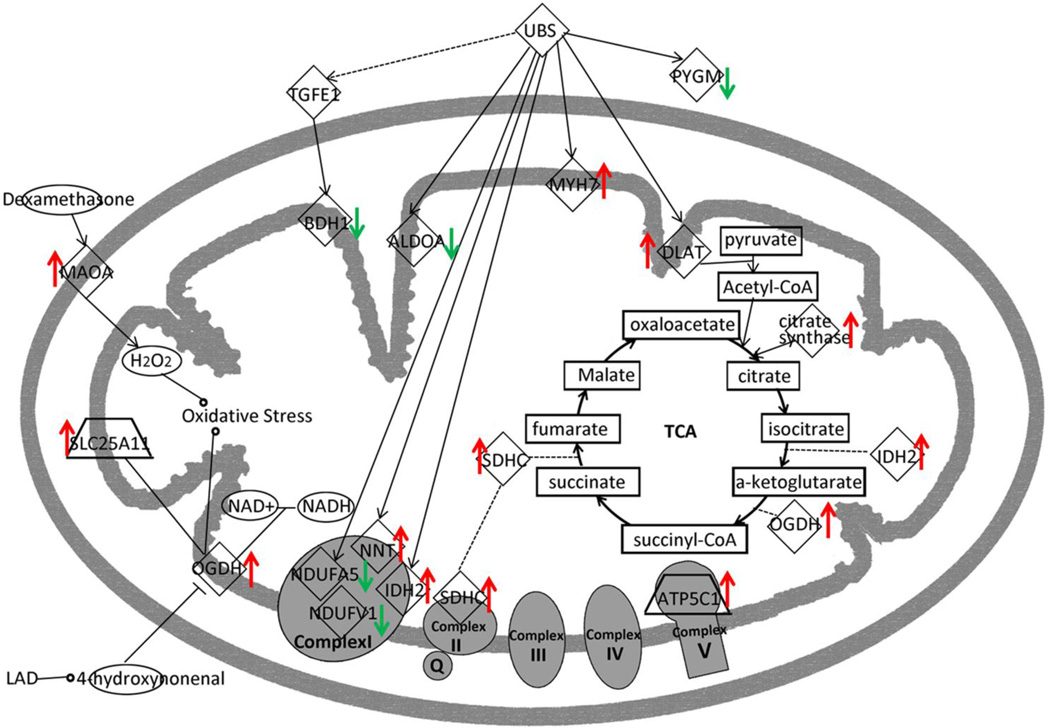
The significantly changed proteins (p < 0.05 vs. Sham) identified by mass spectrometry were analyzed with Ingenuity pathway analysis software (Agnetti et al., 2010, Grant et al., 2009, Zlatkovic et al., 2009). Networks of these proteins were then algorithmically generated based on their connectivity. After analysis, the changed proteins and their interactions can be best summarized by the mitochondrial dysfunction pathway, as shown in Table 3. After re-organization of the proteins based on their location and function in the mitochondria, a graphic signal pathway is generated indicating the most plausible connections among the proteins with significant changes (Figure 5).
Figure 5.
Pathway Analysis. Summary of changes of proteins in the mitochondrial dysfunction pathway by their function and location in the mitochondria. There was decreased expression of complex I proteins and increased expression of TCA cycle proteins.
In the mitochondrial dysfunction pathway, five proteins involved in the TCA cycle all increased significantly. The increased TCA proteins are likely a compensatory response for the impaired mitochondrial respiration. The most important change was the increase in oxoglutarate dehydrogenase (OGDH), a rate limiting step in the citric acid cycle. Six proteins associated with the respiratory chain, had significant, but varied changes in expression. NDUFA5 and NDUFV1 (both complex I) decreased, while SDHC (complex II) and ATP5C1 (complex V) increased. Ubiquitins (UBS)expression is regulated by the balance between regulatory enzyme activities and their regulators, and UBS’s modification plays an essential role in the control of various processes (Kimura and Tanaka, 2010). Although the UBS did not change, it associates with several proteins that changed significantly (Figure 5).
Discussion
Ischemia, hypertension, trans-aortic constriction (TAC) and rapid pacing can all induce heart failure (Bugger, Schwarzer, 2010, Kosar et al., 2006). Although advanced heart failure from many causes looks similar - a dilated, poorly contracting heart with elevated end-diastolic pressures and poor mitochondrial function - the molecular and cellular changes en route to end-stage heart failure may be distinct. Understanding these differences can provide new insights into the underlying mechanisms contributing to the progressive nature of heart failure.
The OCR was abnormal with both significantly reduced ADP stimulated respiration, as well as maximal uncoupled respiration. OCR was measured using succinate/rotenone, a condition where complex I will be inhibited. So the suppressed respiratory function does not reflex complex I function, which was reduced, but other mitochondrial respiration activities.
Mitochondrial Dysfunction Pathway Mitochondrial Respiratory Chain
For the four membrane-bound electron-transporting protein complexes (I–IV) and the ATP synthase (complex V), only a limited number of proteins changed with the most important changes in complex I (Table 2). NDUFV1 and NDUFA5 are subunits of complex I with NDUFV1, having a key role delivering electrons to ubiquinone. NDUFA5, which is an accessory subunit of complex I, localizes to the inner mitochondrial membrane. In addition, two other respiratory complex proteins increased significantly: SDHC (succinate dehydrogenase subunit C, complex II) and ATP5C1 (a mitochondrial ATP synthase subunit, complex V). Both of the complex I related proteins decreased about 50%, which correlated with decreased complex I activity. Mitochondrial complex deficiency has been found in many, disparate diseases including myopathies, heart failure, encephalomyopathies, and neurodegenerative disorders such as Parkinson's disease (Severs et al., 2008) and Leigh syndrome (Hoefs et al., 2011). Complex I dysfunction can lead to increased reactive oxygen species (ROS) production, and ROS affects mitochondrial complex I activity via oxidative damage of cardiolipin, among other targets. Cardiolipin is essential for the function of this multi-subunit enzyme complex (Paradies et al., 2002). Furthermore, oxidative stress generated by H2O2 via MAOA, as well as OGDH (as summarized in figure 5), would lead to more ROS, which would further impair mitochondrial function.
ROS, including superoxide (O2-) and hydrogen peroxide (H2O2), are involved in regulating both growth and death in cardiac myocytes. Mitochondria are considered both a primary source of ROS, and a target for ROS damage (Ide et al., 2000, Kuroda et al., 2010). ROS are markedly increased in failing myocardium (Belch et al., 1991, Ide et al., 1999, Tsutsui et al., 2001, Tsutsui et al., 2006). The primary source of cardiac ROS appears to be uncoupling of the electron transport chain at the level of complexes I and III, but this has been questioned by some (Brown and Borutaite, 2012, Ide et al., 2001). In the failing heart, mitochondrial function was markedly decreased with a significant decrease in complex I activity. NDUFA5 (NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit) and NDUFV1 (NADH dehydrogenase [ubiquinone] flavoprotein 1), both components of complex I were decreased. This decrease in NDUFA5 and NDUFV1 would be expected to decrease complex I function leading to increased ROS. NDUFV1 is also reported to be decreased in patients with dilated cardiomyopathy (Ono et al., 2010). Mutation of NDUFV1 is associated with several diseases that involve impaired mitochondrial energy metabolism (Ortega-Recalde et al., 2013). Interestingly, NDUFA5 expression is reduced in the brain in autism (Anitha et al., 2013). Thus, impairment of expression of these two proteins is seen in several disease states involving high energy organs: the brain and the heart.
To the authors’ knowledge, this is the first report of decreased NDUFA5 in the setting of ischemic HF. After ligation of the coronary, ischemia immediately decreases mitochondrial NADH oxidation, increasing the NADH/NAD+ ratio. It has been shown that reversible inhibition of complex I by S-nitrosation can be cardioprotective by limiting excess ROS formation during reperfusion (Burwell and Brookes, 2008). It is possible, that reduction in NFUFA5 and NDUFV1 is a protective response to excess ROS in heart failure that runs awry over time. Although this idea is intriguing, it remains to be proven.
Interestingly, Nicotinamide nucleotide transhydrogenase (NNT), aka NAD(P) transhydrogenase in mitochondria, increased more than 2-fold. NNT couples transhydrogenation between NAD(H) and NADP(+) and functions as a proton pump across the inner mitochondrial membrane. Under most physiological conditions, the enzyme uses energy from the mitochondrial proton gradient to produce high concentrations of NADPH for biosynthesis and in free radical detoxification (Sheeran et al., 2010). Thus, the increase of NNT could be a response to increased cellular oxidative stress.
Mitochondrial Changes Disparate in Different Types of Systolic Heart Failure
Given the different disease mechanisms leading to heart failure, we hypothesized that the mitochondrial changes would differ to a degree for different etiologies of heart failure (Jeon et al., 2011, Jiang and Wang, 2012, Wong et al., 2010). We compared our findings with those from a comprehensive study on TAC-induced heart failure in male Sprague-Dawley rats (Bugger, Schwarzer, 2010). Although this study and the current one used the same rat strain with similar age and weight and both developed heart failure, there were significant differences in mitochondrial changes between ischemic HF and TAC induced HF. In the current study, mitochondrial respiration was impaired, with significantly lower complex I activity and reduction in some protein components of complex I. This decrease in complex I proteins did not occur in the TAC model. Many of the proteomic changes showed similar trends; however there were select proteins that differed. Table 4 compares key mitochondrial protein changes in our ischemic HF model with those of Bugger et al.’s TAC induced HF model (Bugger, Schwarzer, 2010). For the proteins involved in fatty acid oxidation and glucose oxidation, proteins showed very similar trends. Differences occurred in proteins involved in the citric acid cycle and OXPHOS complex I and IV. Citrate synthase increased more than 50% in ischemic HF vs. no change in TAC induced HF. There were major differences in NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 between ischemic and TAC-induced HF. In the current study, ubiquinone did not change significantly vs. an almost 4 fold increase in the TAC model. Consistent with this, we found markedly decreased complex I function, while the results in the TAC study (Bugger, Schwarzer, 2010), suggest minimal impairment, if any, of complex I.
Table 4.
Comparison of key protein changes in ischemic HF of LAD model vs. pressure over-load HF model from Bugger et al (Bugger, Schwarzer, 2010) (p < 0.05).
| Protein name | Ische mic HF |
Pressure overload HF(Bugger et al., 2010) |
|---|---|---|
| Fatty acid oxidation | ||
|
Acetyl-Coenzyme A acyltransferase 2 (Mitochondrial 3-oxoacyl-Coenzyme A thiolase), isoform CRA_f |
1.27 | 1.14 |
| Acyl-Coenzyme A dehydrogenase, very long chain | 0.83 | 0.75 |
| Trifunctional enzyme subunit alpha, mitochondrial | 0.67 | 0.8 |
| Trifunctional enzyme subunit beta, mitochondrial | 0.77 | 0.8 |
| Glucose oxidation | ||
|
Pyruvate dehydrogenase E1 component subunit beta mitochondrial |
1.58 | 1.25 |
| Citric acid cycle | ||
| Citrate synthase | 2.18 | 0.98 |
| Malate dehydrogenase, mitochondrial | 1.26 | 1.05 |
|
Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha mitochondrial |
1.21 | 0.99 |
| OXPHOS complex I | ||
|
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 4 [Rattus norvegicus] |
1.11 | 3.82 |
|
NADH dehydrogenase (Ubiquinone) Fe-S protein 3 (Predicted), isoform CRA_c |
0.91 | 0.8 |
| NADH dehydrogenase (Ubiquinone) Fe-S protein 7 | 0.70 | 0.76 |
| NADH dehydrogenase (Ubiquinone) flavoprotein 1 | 0.67 | 0.8 |
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial |
1.11 | 1.73 |
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 2 |
0.79 | 0.74 |
|
NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 5 |
0.49 | 0.82 |
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial | 0.79 | 0.79 |
| OXPHOS complex IV | ||
| Cytochrome c oxidase subunit 2 | 1.04 | 0.79 |
| Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | 1.09 | 0.79 |
| Cytochrome c oxidase subunit 5A, mitochondrial | 0.99 | 0.92 |
| Cytochrome c oxidase subunit 5B, mitochondrial | 1.39 | 0.84 |
Another important difference between the two models was expression of cytochrome c oxidase subunit 5B, which is in involved in protein complex binding. In the current study cytochrome c oxidase subunit 5B was slightly increased, while in the pressure overload model of HF this protein was significantly decreased. Thus even in established HF there are clear differences in the mitochondrial proteome as well as differences in mitochondrial dysfunction. Hence, although HF looks the same by echocardiography and physical exam, clearly there are important differences at the molecular and cellular level. These findings suggest that treatment, which now is essentially the same for different types of systolic HF, might one day be targeted to select abnormalities based on the original cause of HF.
Conclusion
Ischemic heart failure mitochondria manifest distinct changes, which differed at key points from the well-established TAC model of pressure overload. Interestingly only complex I activity was depressed in ischemic HF, with complex II and IV unchanged compared to Sham controls. Studies of OCR showed depressed mitochondrial oxidative phosphorylation and depressed maximal O2 consumption in ischemic HF. Analysis of protein pathways provides insight into the changes in function. The mitochondrial dysfunction pathway was the focus of key changes with decreased complex I protein expression and increased TCA cycle proteins. Greater understanding of the specific cellular and molecular changes in different types of HF can provide new insights into differing mechanisms and potentially lead to new, targeted treatments.
Acknowledgements
Supported by the National Institutes of Health (grant numbers HL077281 and HL079071 both to AAK), a VA Merit Award (AAK) and HHMI-MIG (TL)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None
Conflict of Interest Statement
None
References
- Agnetti G, Kaludercic N, Kane LA, Elliott ST, Guo Y, Chakir K, et al. Modulation of Mitochondrial Proteome and Improved Mitochondrial Function by Biventricular Pacing of Dyssynchronous Failing Hearts. Circulation Cardiovascular genetics. 2010;3:78–87. doi: 10.1161/CIRCGENETICS.109.871236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitha A, Nakamura K, Thanseem I, Matsuzaki H, Miyachi T, Tsujii M, et al. Downregulation of the Expression of Mitochondrial Electron Transport Complex Genes in Autism Brains. Brain Pathol. 2013;23:294–302. doi: 10.1111/bpa.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayeva M, Gheorghiade M, Ardehali H. Mitochondria as a Therapeutic Target in Heart Failure. Journal of the American College of Cardiology. 2013;61:599–610. doi: 10.1016/j.jacc.2012.08.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belch JJ, Bridges AB, Scott N, Chopra M. Oxygen Free Radicals and Congestive Heart Failure. British heart journal. 1991;65:245–298. doi: 10.1136/hrt.65.5.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner C, Dietl A, Deutzmann R, Schroder J, Schmid P, Jungbauer C, et al. Proteomic Profiling Implies Mitochondrial Dysfunction in Tachycardia-Induced Heart Failure. Journal of cardiac failure. 2012;18:660–673. doi: 10.1016/j.cardfail.2012.06.418. [DOI] [PubMed] [Google Scholar]
- Brown GC, Borutaite V. There Is No Evidence That Mitochondria Are the Main Source of Reactive Oxygen Species in Mammalian Cells. Mitochondrion. 2012;12:1–4. doi: 10.1016/j.mito.2011.02.001. [DOI] [PubMed] [Google Scholar]
- Bugger H, Schwarzer M, Chen D, Schrepper A, Amorim PA, Schoepe M, et al. Proteomic Remodelling of Mitochondrial Oxidative Pathways in Pressure Overload-Induced Heart Failure. Cardiovascular research. 2010;85:376–484. doi: 10.1093/cvr/cvp344. [DOI] [PubMed] [Google Scholar]
- Burwell LS, Brookes PS. Mitochondria as a Target for the Cardioprotective Effects of Nitric Oxide in Ischemia-Reperfusion Injury. Antioxidants & redox signaling. 2008;10:579–799. doi: 10.1089/ars.2007.1845. [DOI] [PubMed] [Google Scholar]
- Chen L, Gong Q, Stice JP, Knowlton AA. Mitochondrial Opa1, Apoptosis, and Heart Failure. Cardiovascular research. 2009;84:91–109. doi: 10.1093/cvr/cvp181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Knowlton AA. Mitochondrial Dynamics in Heart Failure. Congest Heart Fail. 2011;17:257–361. doi: 10.1111/j.1751-7133.2011.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Liu T, Tran A, Lu X, Tomilov AA, Davies V, et al. Opa1 Mutation and Late-Onset Cardiomyopathy: Mitochondrial Dysfunction and Mtdna Instability. Journal of the American Heart Association. 2012;1:e003012. doi: 10.1161/JAHA.112.003012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai DF, Hsieh EJ, Liu Y, Chen T, Beyer RP, Chin MT, et al. Mitochondrial Proteome Remodelling in Pressure Overload-Induced Heart Failure: The Role of Mitochondrial Oxidative Stress. Cardiovascular research. 2012;93:79–88. doi: 10.1093/cvr/cvr274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Scorrano L. Organelle Isolation: Functional Mitochondria from Mouse Liver, Muscle and Cultured Fibroblasts. Nature protocols. 2007;2:287–395. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- Grant JE, Bradshaw AD, Schwacke JH, Baicu CF, Zile MR, Schey KL. Quantification of Protein Expression Changes in the Aging Left Ventricle of Rattus Norvegicus. Journal of proteome research. 2009;8:4252–5663. doi: 10.1021/pr900297f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefs SJ, van Spronsen FJ, Lenssen EW, Nijtmans LG, Rodenburg RJ, Smeitink JA, et al. Ndufa10 Mutations Cause Complex I Deficiency in a Patient with Leigh Disease. European journal of human genetics : EJHG. 2011;19:270–354. doi: 10.1038/ejhg.2010.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollander JM, Baseler WA, Dabkowski ER. Proteomic Remodeling of Mitochondria in Heart Failure. Congest Heart Fail. 2011;17:262–358. doi: 10.1111/j.1751-7133.2011.00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Tsutsui H, Hayashidani S, Kang D, Suematsu N, Nakamura K, et al. Mitochondrial DNA Damage and Dysfunction Associated with Oxidative Stress in Failing Hearts after Myocardial Infarction. Circulation research. 2001;88:529–635. doi: 10.1161/01.res.88.5.529. [DOI] [PubMed] [Google Scholar]
- Ide T, Tsutsui H, Kinugawa S, Suematsu N, Hayashidani S, Ichikawa K, et al. Direct Evidence for Increased Hydroxyl Radicals Originating from Superoxide in the Failing Myocardium. Circulation research. 2000;86:152–257. doi: 10.1161/01.res.86.2.152. [DOI] [PubMed] [Google Scholar]
- Ide T, Tsutsui H, Kinugawa S, Utsumi H, Takeshita A. Amiodarone Protects Cardiac Myocytes against Oxidative Injury by Its Free Radical Scavenging Action. Circulation. 1999;100:690–782. doi: 10.1161/01.cir.100.7.690. [DOI] [PubMed] [Google Scholar]
- Jeon J, Jeong JH, Baek JH, Koo HJ, Park WH, Yang JS, et al. Network Clustering Revealed the Systemic Alterations of Mitochondrial Protein Expression. PLoS computational biology. 2011;7:e1002093. doi: 10.1371/journal.pcbi.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Wang X. Comparative Mitochondrial Proteomics: Perspective in Human Diseases. Journal of hematology & oncology. 2012;5:11 doi: 10.1186/1756-8722-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model to Estimate the Accuracy of Peptide Identifications Made by Ms/Ms and Database Search. Analytical chemistry. 2002;74:5383–6592. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Tanaka K. Regulatory Mechanisms Involved in the Control of Ubiquitin Homeostasis. Journal of biochemistry. 2010;147:793–988. doi: 10.1093/jb/mvq044. [DOI] [PubMed] [Google Scholar]
- Kosar F, Ermis N, Erdil N, Battaloglu B. Anomalous Lad and Cx Artery Arising Separately from the Proximal Right Coronary Artery--a Case Report of Single Coronary Artery with Coronary Artery Disease. Journal of cardiac surgery. 2006;21:309–512. doi: 10.1111/j.1540-8191.2005.00141.x. [DOI] [PubMed] [Google Scholar]
- Kuroda J, Ago T, Matsushima S, Zhai P, Schneider MD, Sadoshima J. Nadph Oxidase 4 (Nox4) Is a Major Source of Oxidative Stress in the Failing Heart. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:15565–16870. doi: 10.1073/pnas.1002178107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Kim SC, Wang Y, Gupta S, Davis B, Simon SI, et al. Hsp60 in Heart Failure: Abnormal Distribution and Role in Cardiac Myocyte Apoptosis. American journal of physiology Heart and circulatory physiology. 2007;293:H2238–H6547. doi: 10.1152/ajpheart.00740.2007. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Analytical chemistry. 2003;75:4646–5858. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ono H, Nakamura H, Matsuzaki M. A Nadh Dehydrogenase Ubiquinone Flavoprotein Is Decreased in Patients with Dilated Cardiomyopathy. Intern Med. 2010;49:2039–3642. doi: 10.2169/internalmedicine.49.3710. [DOI] [PubMed] [Google Scholar]
- Ortega-Recalde O, Fonseca DJ, Patino LC, Atuesta JJ, Rivera-Nieto C, Restrepo CM, et al. A Novel Familial Case of Diffuse Leukodystrophy Related to Ndufv1 Compound Heterozygous Mutations. Mitochondrion. 2013;13:749–854. doi: 10.1016/j.mito.2013.03.010. [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Pistolese M, Ruggiero FM. Reactive Oxygen Species Affect Mitochondrial Electron Transport Complex I Activity through Oxidative Cardiolipin Damage. Gene. 2002;286:135–241. doi: 10.1016/s0378-1119(01)00814-9. [DOI] [PubMed] [Google Scholar]
- Rosca MG, Hoppel CL. Mitochondrial Dysfunction in Heart Failure. Heart failure reviews. 2012 doi: 10.1007/s10741-012-9340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severs NJ, Bruce AF, Dupont E, Rothery S. Remodelling of Gap Junctions and Connexin Expression in Diseased Myocardium. Cardiovascular research. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeran FL, Rydstrom J, Shakhparonov MI, Pestov NB, Pepe S. Diminished Nadph Transhydrogenase Activity and Mitochondrial Redox Regulation in Human Failing Myocardium. Biochimica et biophysica acta. 2010;1797:1138–1248. doi: 10.1016/j.bbabio.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Chernushevic I, Wilm M, Mann M. "De Novo" Sequencing of Peptides Recovered from in-Gel Digested Proteins by Nanoelectrospray Tandem Mass Spectrometry. Molecular biotechnology. 2002;20:107–188. doi: 10.1385/mb:20:1:107. [DOI] [PubMed] [Google Scholar]
- Sterba M, Popelova O, Lenco J, Fucikova A, Brcakova E, Mazurova Y, et al. Proteomic Insights into Chronic Anthracycline Cardiotoxicity. Journal of molecular and cellular cardiology. 2011;50:849–962. doi: 10.1016/j.yjmcc.2011.01.018. [DOI] [PubMed] [Google Scholar]
- Stice JP, Chen L, Kim SC, Jung JS, Tran AL, Liu TT, et al. 17beta-Estradiol, Aging, Inflammation, and the Stress Response in the Female Heart. Endocrinology. 2011;152:1589–1598. doi: 10.1210/en.2010-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL. What's Driving False Discovery Rates? Journal of proteome research. 2008;7:45–56. doi: 10.1021/pr700728t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H, Ide T, Hayashidani S, Suematsu N, Shiomi T, Wen J, et al. Enhanced Generation of Reactive Oxygen Species in the Limb Skeletal Muscles from a Murine Infarct Model of Heart Failure. Circulation. 2001;104:134–146. doi: 10.1161/01.cir.104.2.134. [DOI] [PubMed] [Google Scholar]
- Tsutsui H, Ide T, Kinugawa S. Mitochondrial Oxidative Stress, DNA Damage, and Heart Failure. Antioxidants & redox signaling. 2006;8:1737–1844. doi: 10.1089/ars.2006.8.1737. [DOI] [PubMed] [Google Scholar]
- van Ekeren GJ, Sengers RC, Stadhouders AM. Changes in Volume Densities and Distribution of Mitochondria in Rat Skeletal Muscle after Chronic Hypoxia. International journal of experimental pathology. 1992;73:51–60. [PMC free article] [PubMed] [Google Scholar]
- Voss MR, Stallone JN, Li M, Cornelussen RN, Knuefermann P, Knowlton AA. Gender Differences in the Expression of Heat Shock Proteins: The Effect of Estrogen. American journal of physiology Heart and circulatory physiology. 2003;285:H687–H792. doi: 10.1152/ajpheart.01000.2002. [DOI] [PubMed] [Google Scholar]
- Wong R, Aponte AM, Steenbergen C, Murphy E. Cardioprotection Leads to Novel Changes in the Mitochondrial Proteome. American journal of physiology Heart and circulatory physiology. 2010;298:H75–H91. doi: 10.1152/ajpheart.00515.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, VerBerkmoes NC, Langston MA, Uberbacher E, Hettich RL, Samatova NF. Detecting Differential and Correlated Protein Expression in Label-Free Shotgun Proteomics. Journal of proteome research. 2006;5:2909–3218. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lin A, Powers J, Lam MP, Lotz C, Liem D, et al. Perspectives On: Sgp Symposium on Mitochondrial Physiology and Medicine: Mitochondrial Proteome Design: From Molecular Identity to Pathophysiological Regulation. The Journal of general physiology. 2012;139:395–406. doi: 10.1085/jgp.201210797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wen Z, Washburn MP, Florens L. Refinements to Label Free Proteome Quantitation: How to Deal with Peptides Shared by Multiple Proteins. Analytical chemistry. 2010;82:2272–3281. doi: 10.1021/ac9023999. [DOI] [PubMed] [Google Scholar]
- Zlatkovic J, Arrell DK, Kane GC, Miki T, Seino S, Terzic A. Proteomic Profiling of Katp Channel-Deficient Hypertensive Heart Maps Risk for Maladaptive Cardiomyopathic Outcome. Proteomics. 2009;9:1314–1425. doi: 10.1002/pmic.200800718. [DOI] [PMC free article] [PubMed] [Google Scholar]