Abstract
Continuous communication between cells is necessary for development of any multicellular organism and depends on the recognition of secreted signals. A wide range of molecules including proteins, peptides, amino acids, nucleic acids, steroids and polylketides are used as intercellular signals in plants and animals. They are also used for communication in the social amoeba Dictyostelium discoideum when the solitary cells aggregate to form multicellular structures. Many of the signals are recognized by surface receptors that are seven-transmembrane proteins coupled to trimeric G proteins, which pass the signal on to components within the cytoplasm. Dictyostelium cells have to judge when sufficient cell density has been reached to warrant transition from growth to differentiation. They have to recognize when exogenous nutrients become limiting, and then synchronously initiate development. A few hours later they signal each other with pulses of cAMP that regulate gene expression as well as direct chemotactic aggregation. They then have to recognize kinship and only continue developing when they are surrounded by close kin. Thereafter, the cells diverge into two specialized cell types, prespore and prestalk cells, that continue to signal each other in complex ways to form well proportioned fruiting bodies. In this way they can proceed through the stages of a dependent sequence in an orderly manner without cells being left out or directed down the wrong path.
Keywords: Intercellular communication, signal transduction, dependent sequence
A SYSTEMS APPROACH
Embryos develop from fertilized eggs through a series of differentiated states. As a zygote is cleaved into more and more cells, anterior/posterior and dorsal/ventral axes are established by localization of internal signals as well as signals from surrounding cells. Each lineage differentiates further to generate the defining physiological identities that distinguish one tissue from the next. Waddington (1942) used his concept of canalisation to account for the robustness of biological systems in the face of environmental and genotypic variability. He envisioned the steps of embryogenesis as passage through a landscape in which an initial valley splits into several broader valleys which split again to form many smaller canals. In his metaphor, cells would follow the contours from the valleys to the canals in a probabilistic manner, always moving downhill. Some cells might take the left side and others take the right side. The chances of retracing their steps and taking another route would become less and less likely as they descended. However, at the next bifurcation, they could be directed down one or the other canal by relatively gentle signals. Temporal progression of each lineage would have to be integrated with progression in other independent lineages to keep the whole system working. Such integration requires almost continuous conversations between the cells as they follow the epigenetic landscape and could be mediated by diffusion of soluble compounds between cells or subcellular localization of interacting surface components between adjacent cells. A wide variety of signals are used in eukaryotic organisms and many of them date back to the common ancestor of plants and animals.
Embryogenesis has been intensely studied in model organisms for plants, insects, nematodes, echinoderms and vertebrates (Gilbert, 2013). Insights gained from one system often apply to other systems and add to the understanding of development in general. A process which might be technically difficult to study in one organism might be much easier in another because its role had been expanded. Model systems that offer the possibility of harnessing the power of genetics have a considerable advantage in these studies. This reasoning favors inclusion of organisms with advanced genetic techniques such as yeast and amoebae even if their development is limited. Yeast have amazingly good genetics but are essentially unicellular. Amoebae, on the other hand, grow as single cells that permit microbial genetic techniques, and yet develop as multicellular organisms. They present a range of developmental processes that can be studied by classical and molecular genetics.
The amoebozoa branched off shortly after plants and animals separated in evolution and generated a large number of species, most of which have a cooperative stage in their life cycle (Raper, 1984; Eichinger et al., 2002; Schilde and Schaap, 2013). The best characterized amoebozoan species is Dictyostelium discoideum (Bonner, 1959; Loomis, 1975, 1982; Kessin, 2001). Since development of Dictyostelium is far simpler than that of mammals, it can be approached in a systems manner (Figure 1). It uses many of the same signals that are found to function in plants and animals. The signal transduction pathways by which the cells respond to these signals can be studied using the excellent molecular genetics of D. discoideum (Loomis, 1987; Nellen et al., 1987; Newell et al., 1993; Kuspa and Loomis, 2006). A review of the known signaling systems that function at various stages in the 24 hour life cycle gives an idea of what it takes for a group of genetically and physiologically similar cells to form a fruiting body with specialized stalk cells and spores.
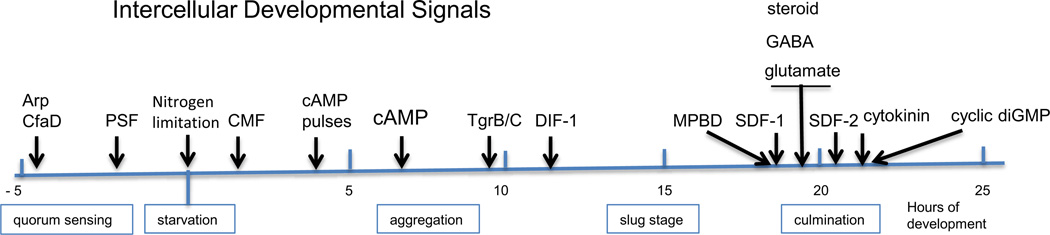
Figure 1.
Signaling during development. The signals used to integrate development of Dictyostelium are indicated at the stages at which they act. In the 5 hours preceeding the initiation of development while the cells are still growing secreted proteins function as quorum sensors. Morphogenesis occurs over 24 hours following the initiation of development by nitrogen limitation; the stages are indicated below the temporal line. The structure of each signal is given in Figure 2 and the mode of action described in the text.
Dictyostelium discoideum was isolated from the forest floor at Little Butt Gap, near Asheville, North Carolina, by Ken Raper about 80 years ago (Raper, 1935). He observed that cells of this new species, like many other soil amoebae, aggregated into mounds when they depleted the local sources of food. He realized that such aggregation requires cells to communicate but did not know how it was done. In a simple but elegant experiment that had aspects of modern day microfluidics, John Bonner showed that starving cells secreted a chemoattractant to which cells downstream responded by moving up the gradient (Bonner, 1947). This was the first convincing evidence for chemotaxis in eukaryotes. It took 20 years to define the chemoattractant chemically but it was finally shown to be cAMP (Konijn et al., 1967). This finding opened up the analysis of cell signaling to biochemical and molecular biological techniques with which it was possible to recognize and characterize the enzymes that synthesize cAMP, the surface receptors for cAMP, and many of the components of the signal transduction pathways (Klein et al., 1987; Pupillo et al., 1988; Insall et al., 1994; Maeda et al., 1996; Swaney, Huang and Devreotes, 2010). These advances solidified the position of Dictyostelium as a model organism to study chemotactic motility and multicellular development.
Raper thought of Dictyostelium as a developmental system because the life cycle was simple and rapid enough that it could be considered as a whole (Raper, 1940; 1984). He described and analyzed a wide variety of processes that occur during development of aggregated cells as they organize into slug-shaped structures that go on to form fruiting bodies. He showed that the two cells types, spores and stalk cells, that are found in fruiting bodies were preceded by prespore and prestalk cells at the slug stage. He found that prestalk cells were at the front of the slugs, where they make up the anterior quarter, and that prespore cells were all in the back. He could distinguish them by grafting red cells from the anterior of slugs generated from populations fed on colored bacteria onto unstained posteriors. The resulting stalks made by these chimeric slugs had red stalks. Moreover, he could show that the proportion of prestalk cells to prespore cells was always constant at 1 : 4 no matter the size of the slug (Raper, 1940).
Since the size of slugs and the total number of cells in each slug can vary by more than 20 fold, there must be an intercellular signal that acts throughout the slug and determines the proportions of prespore and prestalk cells. It has been proposed that prespore cells secrete an inhibitor of prespore differentiation to which prespore cells are resistant and in this way establish the proportion of prespore cells (Loomis, 1993; Soderbom and Loomis, 1998). While the inhibitor is synthesized only by prespore cells, it is removed by all cells. This gives the model its size invariance. The inhibitor diffuses within the slug and keeps any cells that have not differentiated into prespores from doing so after the inhibitor reaches a threshold. The model can also account for regulation of the ratio of the cell types when the proportions are preturbed, but it does not define the nature of the inhibitor. Despite considerable effort no one has come up with an idea on how to isolate the proposed inhibitor or directly test the model. This is one of the many cases in developmental biology where a signal is clearly implicated but still chemically undefined.
The life cycle of Dictyostelium makes it easy to recognize when a mutation affects the release of an intercellular signal rather than altering or inactivating an internal process. Strains carrying mutations affecting intercellular signals are able to synergize with wild type cells while strains carrying mutations affecting signal receptors or internal functions will not benefit from the presence of wild type cells. A large number of non-sporulating mutant strains can be screened for signaling defects by mixing them with an equal number of wild type cells and allowing them to aggregate together. After development is complete, spores can be collected from the chimeric fruiting bodies, diluted appropriately and plated out such that each viable spore forms an independent plaque. If a strain carries a mutation that affects release of a signal, at least half of the plaques will develop with the phenotype of the original mutant strain. The other half will be wild type. In this way a large number of synergizable mutants have been isolated. Cloning and sequencing of the affected genes has given insight on the nature of the signals and in some cases has conclusively proven the critical roles they play in development.
INTERCELLULAR SIGNALS
When they are still growing, Dictyostelium cells ingest bacteria and multiply by binary division. As the population density approaches peak, quorum sensors act to reduce the growth rate and alter the transcriptional profile to prepare for post-mitotic development. Abrupt removal of all nutrients induces the initiation of development and triggers dramatic changes in transcription that adapt the cells to a period of starvation as well as allow them to communicate in novel ways. Within a few hours the cells begin to accumulate and secrete cAMP, which is used both as a chemoattractant and an intercellular signal, such that thousands of cells can aggregate into mounds and change the pattern of gene expression as they go (Gerisch et al., 1975; Klein, 1975; Mann and Firtel, 1987; 1989; Kimmel, 1987; Reymond et al., 1995; Iranfar, Fuller and Loomis 2003). Once the cells have formed aggregates of about 105 cells, they continue to communicate with each other using a series of signals. The resulting transcriptional changes ultimately lead to differentiation into either dormant spores or vacuolized stalk cells (Anjard and Loomis, 2005; 2006; 2008; Iranfar et al., 2006) .
Changes in specific mRNAs and proteins have been well documented at multiple stages in the developmental cycle. In some cases one of the stage specific components has been shown to trigger progression to the next stage, but there is no complete cause-and-effect line connecting the different stages in development. Likewise, the signal transduction pathways for many of the intercellular signals of Dictyostelium do not extend to specific transcription factors that can directly modify the pattern of gene expression. The search for specific DNA binding proteins that regulate stage specific transcription is presently in high gear.
Biochemical Nature of the Signals
Independently of the signal transduction pathways, the properties of the signals can give some insight into development in this organism as well as into parallel processes in other organisms including metazoan animals (Figure 2). It is beginning to become apparent that the signals are as conserved during evolution as their receptors and downstream components. The roles of specific proteins, peptides, polyketides, nucleotides and steroids in intercellular communication in Dictyostelium have come as somewhat of a surprise. Together these studies show that the relatively simple development of Dictyostelium still requires a large number of signals for accurate two-way communication between cells at every stage. These signals may be only the tip of the iceberg and we can expect many more to be found when other aspects of development are investigated.
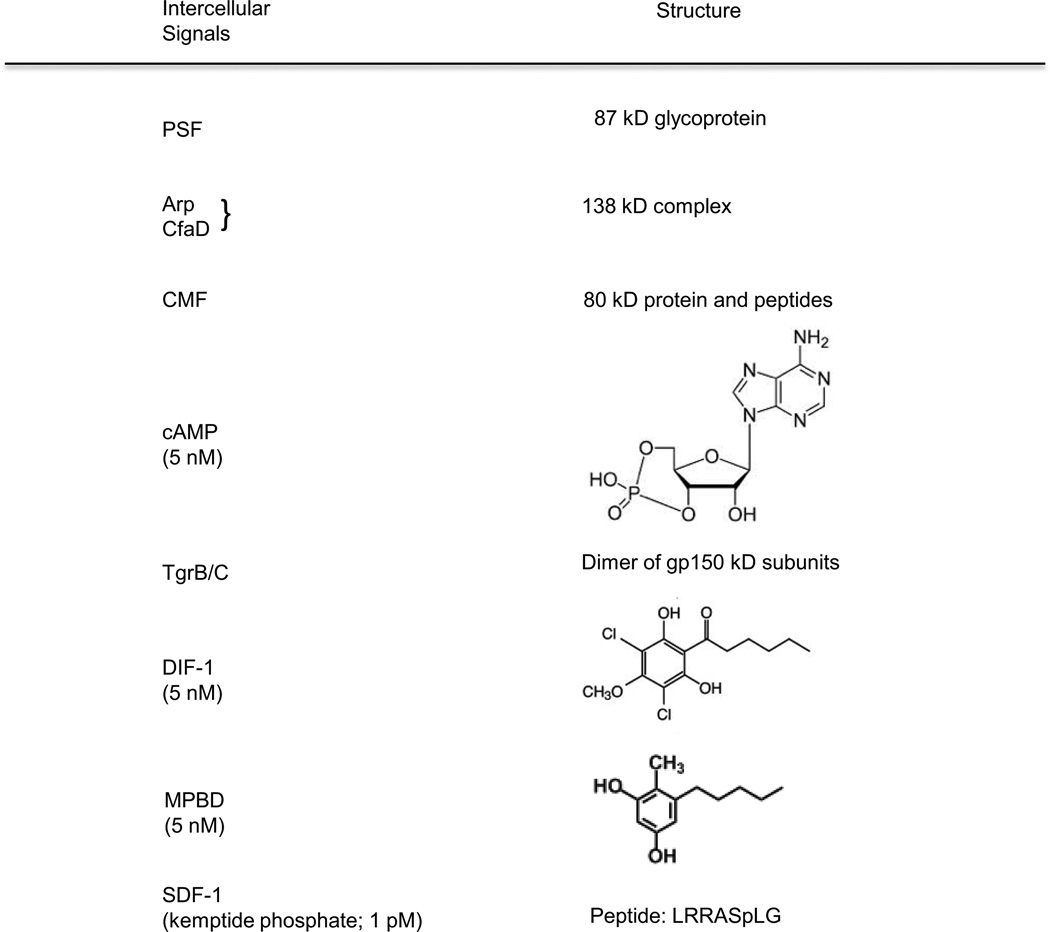
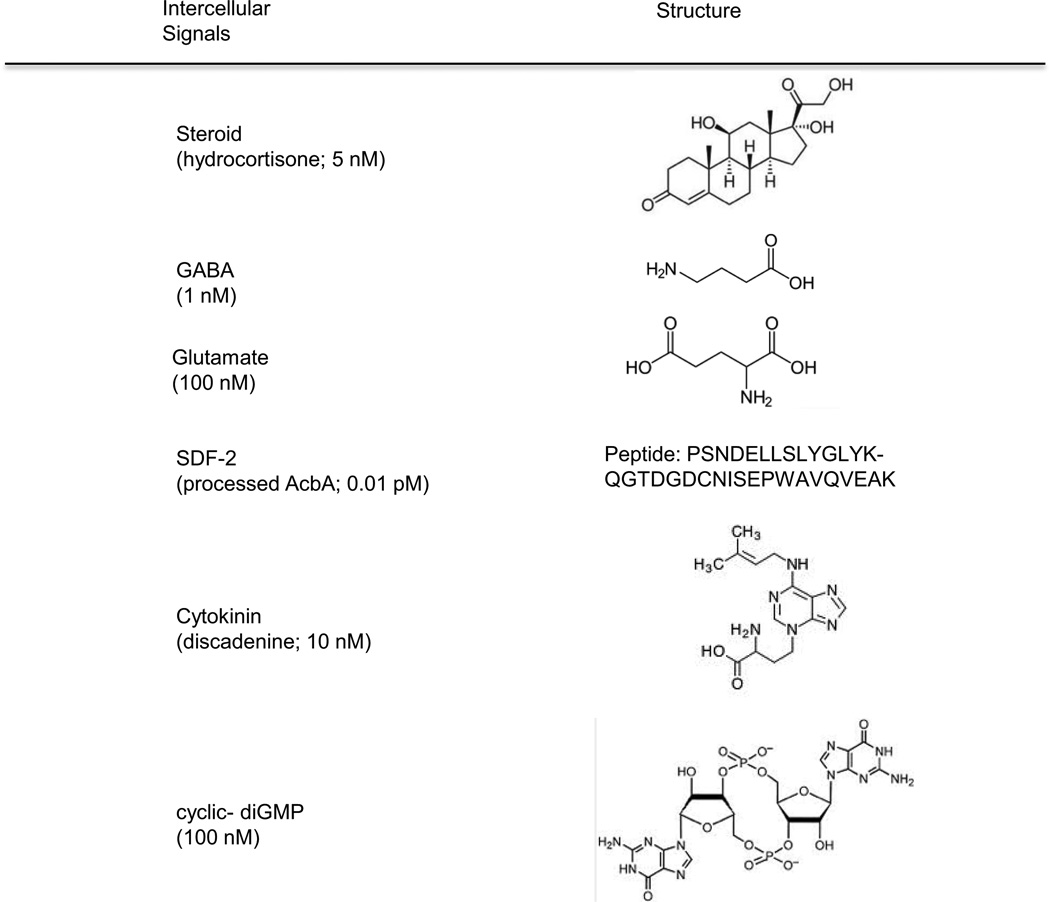
Figure 2.
Structures of the intercellular signals. The signals used by Dictyostelium for communication among developing cells are highly diverse and include proteins, peptides, polyketides, nucleic acids, amino acids and steroids. Each signal is recognized by a unique receptor on the surface that initiates a signal transduction pathway within the cells. Many of the small molecules are similar or identical to signals used by mammalian tissues. Others are similar to signals used by plants, yeast or bacteria.
QUORUM EFFECTORS
The first indication of a quorum effect in growing Dictyostelium cells was the observation that the specific activity of the enzyme N-acetyl glucosaminidase was low as long as the cell density remained below 106/ml and increased dramatically as the density increased (Grabel and Loomis, 1978). The cells could be shown to secrete a small heat stable factor which induced the enzyme when its concentration reached a threshold. Unfortunately, the factor could not be fully purified and characterized because of the difficulty of the bioassay. Nevertheless, it could be shown that the cells continuously secreted the factor which accumulated at a rate proportional to the cell density. As a result its concentration could be used by the cells to determine their cell density and allow them to respond when it was likely they were about to run out of nutrients. Such a quorum sensing mechanism can be used to establish the total number of cells in a tissue (Gomer, Jang and Brazill, 2011).
Prestarvation Factor: PSF
A similar phenomenon was observed while analyzing the accumulation of the lectin, discoidin-I, after cells grew to high concentrations (Clarke, Kayman, and Riley, 1987; Clarke et al., 1992; Clarke and Gomer, 1995; Burdine and Clarke, 1995). The secreted factor in this case could be shown to be a heat-labile 87 kD glycoprotein, called PSF. Growing Dictyostelium cells continuously secrete PSF which reaches a threshold value when the cells are about 106/ml. However, the response to PSF is highly sensitive to the presence of bacteria in the surroundings. Only when the bacterial titer begins to drop, as a result of increased predation by the amoebae, do the cells respond to PSF by inducing the expression of genes such as discoidin-I and the protein kinase YakA (Figure 3).
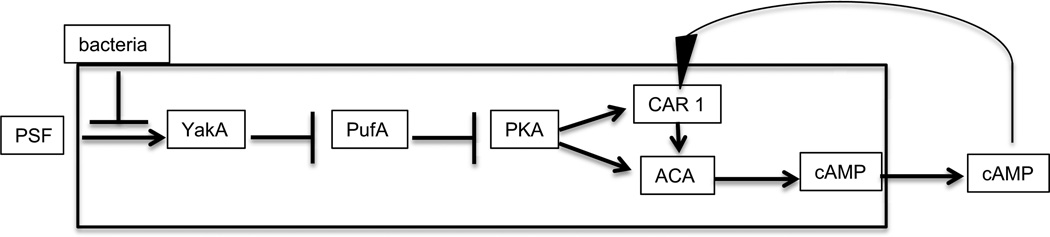
Figure 3.
The growth- differentiation transition pathway. Growing amoebae secrete PSF continuously such that the concentration increases as the cell density increases. When it reaches threshold, it activates the protein kinase YakA. If bacteria are still around, the threshold is higher. YakA activity inhibits PufA which inhibits translation of the catalytic subunit of PKA. Inhibiting an inhibitor results in activation of PKA. This cAMP dependent protein kinase leads to the accumulation of the cAMP receptor CAR1 and the adenylyl cyclase ACA which synthesizes cAMP. Most of the newly made cAMP is secreted into the surrounding fluid where it can diffuses to bind to the receptor on the same cell (autocrine) or other cells (paracrine). Ligand binding to CAR1 stimulates ACA thereby forming a positive feedback loop.
YakA mRNA starts to increase in bacterially grown cells about two generations before all the bacteria are consumed and accumulates until development starts (Souza, Lu and Kuspa, 1998). When grown in liquid media, addition of PSF from media conditioned by high density cell populations induces yakA mRNA at least 10 fold in low density populations. YakA is essential for the initiation of development as shown by the fact that yakA− null strains fail to even begin aggregation. They also continue to express high levels of growth phase genes that normally are turned off within the first hour following the initiation of development. The genes for the cAMP receptor, carA, and adenylyl cyclase, acaA, that are induced within the first 2 hours of development in wild type cells, are not expressed in yakA− null cells. Not surprisingly these mutant cells are unable to aggregate since they cannot synthesize or respond to the chemoattractant, cAMP.
YakA also seems to be responsible for slowing the growth and limiting the maximum cell density during growth (Souza, Lu and Kuspa, 1998). Cells of the yakA− null strains are smaller than wild type cells, grow faster and reach a 50% higher density. Over-expresssion of yakA inhibits growth and accelerates early developmental events. It appears that this protein kinase plays a critical role in the growth to development transition.
The role of YakA in the initiation of development was further elucidated by isolating a suppressor mutation that permitted aggregation in a yakA− null strain (Souza, da Silva and Kuspa, 1999). The mutation turned out to inactivate pufA which is responsible for the synthesis of a translational inhibitor. A potential target for translational control by PufA was the catalytic subunit of the cyclic AMP dependent protein kinase, PKA. Inspection of the sequence of the mRNA for PKA found several putative PufA binding sites at the 3' end and gel mobility shift experiments showed that, indeed, PufA bound to that region. In the absense of PufA the level of PkaC protein and PKA activity was much higher than in wild type cells while the mRNA levels were unchanged as would be expected if PufA were a translational inhibitor of pkaC. Studies in a variety of strains carrying different combinations of mutations in yakA, pufA, and pkaC showed that YakA inhibits PufA, probably by phosphorylation, while PufA inhibits translation of pkaC mRNA by binding to its 3' end (Souza et al., 1998; 1999). When PSF reaches threshold, it induces accumulation of YakA which then inhibits PufA from blocking translation of pkaC and the catalytic subunit accumulates to higher levels than the regulatory subunit. The resulting constitutive activity of PKA is thought to lead to accumulation of the cAMP receptor, CAR1, and adenylyl cyclase, ACA, such that the cells can signal each other with cAMP as well as further activate PKA by binding of cAMP to the regulatory subunit. PKA activity has been found to be involved in developmental timing throughout the life cycle of Dictyostelium and it is not surprising that it is needed for expression of the early genes that lead to sensitivity to cAMP (Mann et al., 1997; Loomis, 1998). YakA also leads to the loss of pufA mRNA in the first hour following the initiation of development, thereby ensuring that PKA activity stays high. Thus, the induction of yakA by PSF accounts for most of the effects of this quorum sensor.
AprA /CfaD Complex
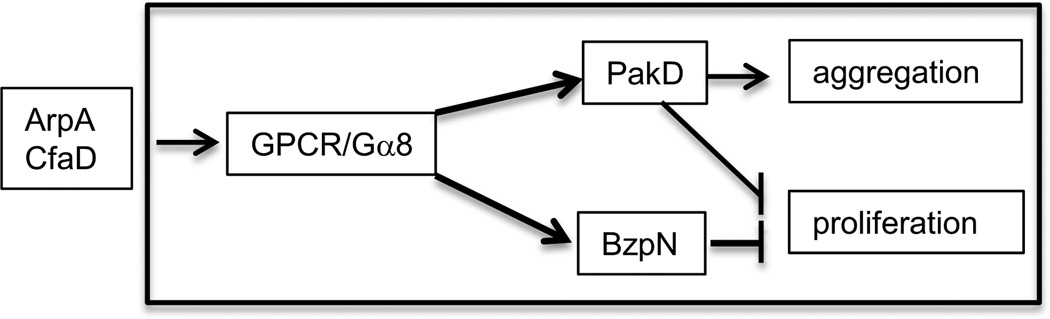
Two other proteins, AprA and CfaD, are secreted during growth and accumulate in the medium in parallel with the increase in cell density. They form a 138 kDa complex that binds to cells and slows their rate of proliferation (Bakthavatsalam et al., 2008, 2009). They also limit the maximum cell density to which the cells can grow. Mutations that knock out either of the genes encoding these proteins result in an increase in proliferation while addition of recombinant AprA or CfaD inhibits cell proliferation. The receptor may be a G Protein Coupled Receptor (GPCR) since the G protein subunit Gα8 is required for cells to respond to AprA (Bakthavatsalam et al., 2009). Inhibition of proliferation during the exponential phase of growth is predominantly controlled by the basic leucine zipper transcription factor BzpN (Phillips et al., 2011). Cells lacking BzpN proliferate more rapidly and do not respond to either AprA or CfaD. The maximum cell density that can be reached during growth is limited by the protein kinase PakD following activation by AprA and CfaD (Garcia et al., 2014; Phillips and Gomer, 2014). Thus, it seems likely that when AprA /CfaD reaches threshold, it affects the transcriptional profile by activating BzpN such that proliferation slows and the cells can prepare for starvation (Figure 4). A parallel pathway activates PakD which inhibits proliferation at high cell densities. This protein kinase also regulates the subcellular positioning of F-actin and is essential for developmental aggregation.
Figure 4.
Quorum sensing. Growing amoebae secrete two proteins, AprA and CfaD, that act as quorum sensors and limit cell proliferation before exogenous nutrients have been fully used up, giving the cells a little extra time. A G protein coupled receptor (GPCR) is implicated by the requirement for the G protein subunit Ga8. The DNA binding protein BzpN inhibits proliferation predominantly at low cell density and the protein kinase PakD inhibits proliferation predominantly at high cell density. PakD is also essential for developmental aggregation.
Conditioned Medium Factor: CMF
There is another cell density sensing system that functions in the first few hours after the removal of nutrients. An 80 kDa protein called CMF is synthesized in growing cells but is only secreted when development is induced by starvation (Gomer, Yuen and Firtel, 1991). Likewise, a gene encoding a receptor for CMF is expressed in vegetative cells but only positioned on the surface following starvation (Jain and Gomer, 1994). As a result the concentration of CMF can be used as a quorum sensor for starved cells at the initiation of development. When vegetative cells are washed free of nutrients and suspended in buffer to which 300 µM cAMP is added after 6 hours, they will express late aggregation stage genes if they are incubated for 18 hours at cell densities greater than 106 cells/ml but will fail to express these genes when incubated at less than 105 cells/ml (Gomer, Yuen and Firtel, 1991). However, if purified CMF is added to cells being shaken at low cell densities, those genes are expressed. In these assays the spore coat protein, CotB, and the prestalk enriched protease, CprB, were measured by staining single cells with antibodies specific to CotB or CprB and counting the number of positive cells (Gomer, Yuen and Firtel, 1991). The more commonly used Western or Northern techniques could not be carried out because each assay of the low cell density cultures would have required cells to be collected from several liters of the suspension which is very tedious.
Blocking expression of the gene encoding CMF, by expressing an antisense copy of the gene, drastically reduced the number of cells in high density cultures that were CotB or CprB positive unless purified CMF was added to the cultures (Jain and Gomer, 1994). Full length recombinant CMF was found to be maximally effective at 10 ng/ml. To delineate the active region of the 80 kDa protein, the gene was subdivided into much smaller fragments and expressed in bacteria. Expression of a short sequence near the N-terminus generated an 88 amino acid peptide that was fully able to rescue CotB and CprD expression in the CMF antisense cells. The small peptide was found to be 100 fold more active than the full length protein. This probably accounts for the initial observation that a number of apparent breakdown products of the 80 kDa protein had equal or greater activity than the full length factor (Gomer, Yuen and Firtel, 1991). The activity of the small (0.5 to 6 kDa) signaling peptides was found to be heat stable, a property shared with the small secreted factor necessary for expression of the marker enzyme N-acetyl glucosaminidase in low density cultures (Grabel and Loomis, 1978).
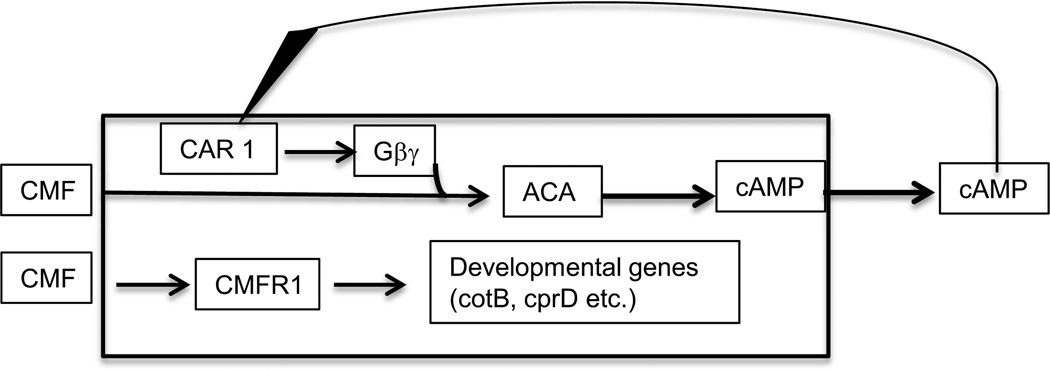
Cells that fail to accumulate CMF because of antisense expression not only do not accumulate CotB or CprB but also fail to efficiently produce the chemoattractant cAMP, and as a result, do not aggregate. It was found that the cells require a period of exposure to CMF before activated G proteins can stimulate the aggregation stage adenylyl cyclase ACA (Yuen et al., 1995). Surprisingly, the cells only had to be exposed to CMF for 10 seconds for the ACA activity to become responsive to CAR1 signaling. The speed of this response rules out any involvement of CMF induced changes in gene expression in relay of the cAMP chemotactic signal but raises the question of how CMF affects ACA so rapidly. It doesn't seem to require that CMF bind to its surface receptor, because CMFR can be knocked out without affecting modification of the ACA response by CMF (Deery and Gomer, 1999). On the other hand, stimulation of expression of cotB and cprD is dependent on the binding of CMF to its receptor, CMFR (Deery and Gomer, 1999). This 50 kDa transmembrane protein binds CMF in a manner that can be competed by addition of the 88 amino acid peptide suggesting that the true quorum effector is the processed peptide. If the density of starving cells is sufficiently high, the CMF peptide will accumulate above threshold and facilitate expression of developmental genes (Figure 5).
Figure 5.
Modulation of cAMP signaling and control of developmental genes. The 80 kDa protein CMF affects early development in at least two independent manners. Shortly after the initiation of development CMF is secreted and signals whether there is a sufficient density of cells to make it worthwhile to aggregate and form fruiting bodies. If CMF is present at 10 ng/ml or higher, G proteins activated by CAR1 are able to stimulate the adenylyl cyclase ACA to synthesize cAMP. Most of the newly made cAMP is secreted so that it can bind to the CAR1 receptor thereby closing a positive feedback loop. CMF and small peptides that are cleaved from it can bind to the receptor CMFR1 which leads to a signal transduction pathway ending in expression of the marker genes cotB and cprD.
EARLY DEVELOPMENTAL SIGNALS
The signal that initiates development in Dictyostelium is the lack of available nutrients. It may seem a bit of a stretch to consider starvation as intercellular signaling but it will only occur when the cells reach a high enough density to clear the majority of nearby bacteria and limit ingestion. So, in a sense, starvation is a measure of cell density just as the concentration of PSF or AprA /CfaD is an indication of cell density. However, the molecular basis for signaling starvation is presently unknown. By adding back components of a defined growth medium to cells incubated in buffer it could be shown that neither glucose nor the vitamins in the medium would delay development. However, adding back the amino acids in the medium was found to delay the initiation of development at least 30 hours (Marin, 1976). These amino acids were further subdivided into "essential" and "conditional" amino acids. The "conditional" group consists of 7 amino acids that are not required for growth: glutamate, glutamine, aspartate, asparagine, alanine, serine, and proline (Marin, 1976; Payne and Loomis, 2006). When the two groups are added together to cells in buffer, inhibition of aggregation is dependent on the concentration of the "conditional" amino acids which must be above 1 mg/ml to be effective (Marin, 1976). This high concentration requirement suggests that the "conditional" amino acids are serving as a food source. The cells are certainly not starving for a carbon source when incubated in buffer alone since they have large glycogen reserves that are metabolized to sugars during development (Garrod and Ashworth, 1972). However, when the medium is replaced by buffer, they are starved for nitrogen since the cells have no known nitrogen reserves. The cells may recognize nitrogen limitation as a change in the ratio of the metabolic flux of carbon to the metabolic flux of nitrogen. As pointed out in a study of catabolite repression of the lac operon in the bacterium Escherichia coli, keto-acids will accumulate when the ratio of carbon to nitrogen flux increases because insufficient nitrogen is available to convert such metabolites as α-ketoglutarate into glutamate or other amino acids (You et al., 2013). High levels of α-ketoglutarate limit transcriptional initiation of various genes including those of the lac operon. It is possible that this signal is also used to recognize nitrogen starvation in Dictyostelium.
During the first few hours after removal of nutrients, genes encoding ribosomal proteins are repressed and genes necessary for cAMP signaling are induced. mRNAs for the cAMP receptor, CAR1, the G protein α2 subunit that is coupled to CAR1 and the extracellular cAMP phosphodiesterase, PdsA, all increase at least 5 fold (Iranfar et al., 2003; 2006). Once the mRNAs are translated the cells can start to signal each other by secreting cAMP that can bind to the G-protein coupled receptor CAR1. Signal transduction from ligand bound receptor to the cytoskeleton directs the formation of pseudopods in the direction of highest cAMP so that the cells can respond by chemotaxis. The cAMP signal transduction pathway also leads to the nucleus, where the expression of a considerable number of developmental genes is affected.
cAMP Signaling
When washed cells are incubated in buffer at 107 cells/ml, they express only a few developmental genes unless treated with pulses of 30 nM cAMP (Firtel, 1996; Iranfar, Fuller and Loomis, 2003). Early experiments relied on biochemical assays or hybridization of developmental cDNAs to Northern blots to monitor the effects of cAMP (Gerisch et al., 1975; Kline, 1975; Kimmel 1987; Firtel, 1991; 1996). More recently, microarrays have made it possible to follow a large number of developmental genes that respond to cAMP (Iranfar, Fuller and Loomis, 2003). By using chips with probes made from cDNA libraries of developing cells, the majority of genes expressed at high levels preferentially during early development could be quantitatively analyzed. It was found that 24 genes started to accumulate in the first 4 hours, 42 genes were first expressed between 4 and 6 hours, and 23 genes were first expressed between 6 and 8 hours. Expression of all but 3 of these genes was dependent on the addition of cAMP pulses to the cells.
During the first 8 hours of development the synthesis and secretion of cAMP is periodic (Gerisch and Wick, 1975; Tomchik and Devreotes, 1981; Loomis, 1979). When the surface receptor CAR1 binds extracellular cAMP, it activates the exchange of GTP for GDP bound to the α subunit of the trimeric G protein associated with CAR1. The trimeric G protein then dissociates into Gα2 and Gβγ subunits which activate GTP-Exchange Factors (GEFs) for Ras proteins that act as molecular switches when they have exchanged GDP for GTP. Ras-GTP leads indirectly to the activation of membrane associated adenylyl cyclase such that it produces about 5 fold more cAMP per minute (Swaney, Huang and Devreotes, 2010). Most of the cAMP is secreted to the extracellular buffer where it can diffuse to adjacent cells. However, extracellular cAMP is rapidly degraded by the extracellular cAMP phosphodiesterase and must be continuously replenished.
The level and shape of the cAMP waves that are relayed outwards from the center of an aggregate are controlled by the activity of the extracellular cAMP phosphodiesterase, PdsA (Orlow et al., 1981; Franke and Kessin, 1992; Sucgang et al., 1997). Since each cell secretes PdsA, the extracellular activity is a gage of the cell density. However, the activity is also controlled by a 26 kDa protein that specifically inhibits PdsA (Kessin et al., 1979; Franke and Kessin, 1981). This inhibitor is secreted when the concentration of extracellular cAMP is low and reduces the affinity of PdsA for cAMP (Kessin et al., 1979; Rossier et al., 1983). When the concentration of extracellular cAMP is high, expression of the inhibitor is repressed (Rossier et al., 1983). Under these conditions most of the PdsA is active and can return the cAMP concentration to near optimum levels.
Ligand bound CAR1 activates GTPase Activating Proteins (GAPs) for Ras that counteract the GEFs (Zhang, Charest, and Firtel, 2008; Takeda et al., 2012). The kinetics of GAP activation appear to be slower than those for GEF such that Ras-GTP generated by GEF can accumulate for 20 seconds or so before the GAP activity prevails and converts Ras-GTP to the inactive Ras-GDP. When Ras-GTP accumulates, it stimulates ACA activity by pathways involving PI3 kinase and TORC2 kinase (Swaney, Huang and Devreotes, 2010). Although most of the newly made cAMP is secreted, some is retained and will activate the protein kinase PKA.
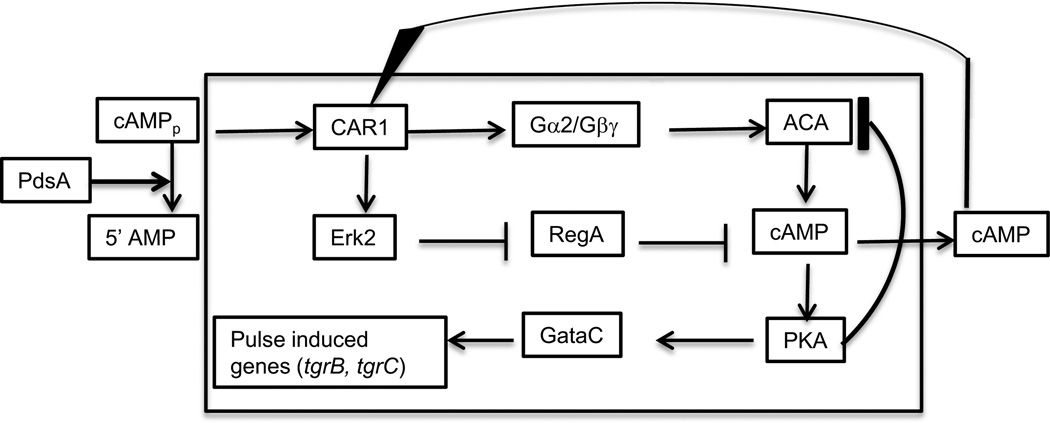
PKA has been found to affect a large number of processes, one of which is inhibition of the activation of ACA itself (Mann et al., 1997; Laub and Loomis, 1998). This cuts off the supply of cAMP and the internal concentration is rapidly reduced by the internal cAMP phosphodiesterase RegA. When cAMP returns to its basal level, PKA is no longer activated. This negative feedback loop limits the time during which both ACA and PKA are active (Figure 6). Kinetic equations can be written for the reactions of this circuit that correctly predict the characteristics of the components in wild type and mutant strains (Laub and Loomis, 1998; Maeda et al., 2004). These analyses have shown that nearby cells will entrain each other such that they all synchronously produce cAMP in waves with a 6 to 8 minute periodicity. These cAMP pulses are used both for chemotactic directionality and induction of the early pulse dependent genes. The communal aspects of extracellular cAMP to which all cells respond resynchronizes the transcriptional profiles of cells after 6 to 8 hours of development.
Figure 6.
The PKA oscilatory circuit. Binding of cAMP to its receptor CAR1 not only activates ACA through its trimeric G protein but also activates the MAP kinase Erk2. This protein kinase inhibits the internal cAMP phosphodiesterase RegA such that it no longer reduces the internal level of cAMP. Increasing the rate of synthesis of cAMP and decreasing its rate of degradation leads to a surge in the concentration of cAMP. Most of the newly synthesized cAMP is secreted where it can further stimulate the circuit. However, internal cAMP activates the protein kinase PKA. Acting indirectly, PKA leads to a block in the activation of ACA and also activates the transcription factor GataC. The reduction in ACA activity lowers the levels of cAMP and the circuit proceeds to reset. External cAMP is reduced by the secreted cAMP phosphodiesterase PdsA which interfers with stimulation of the circuit. Although phosphorylation activates GataC, it also leads to its exit from the nucleus. As a result, GataC is able to stimulate transcription of developmental genes only for a brief period following each pulse of cAMP. The pulse induced genes are still expressed in mutant strains lacking ACA as the result of sufficient cAMP being synthesized by the minor adenylyl cyclase ACR to activate PKA when RegA is inhibited by Erk2.
Cells which are unable to make their own internal cAMP as the result of mutations in the two adenylyl cyclases, ACA and ACR, express only carA,pdsA, and gpaB during development and do so whether or not they are treated with cAMP pulses (Iranfar, Fuller and Loomis, 2003). These genes are considered pulse-independent and their protein products alter the cells so that they can respond to cAMP pulses.
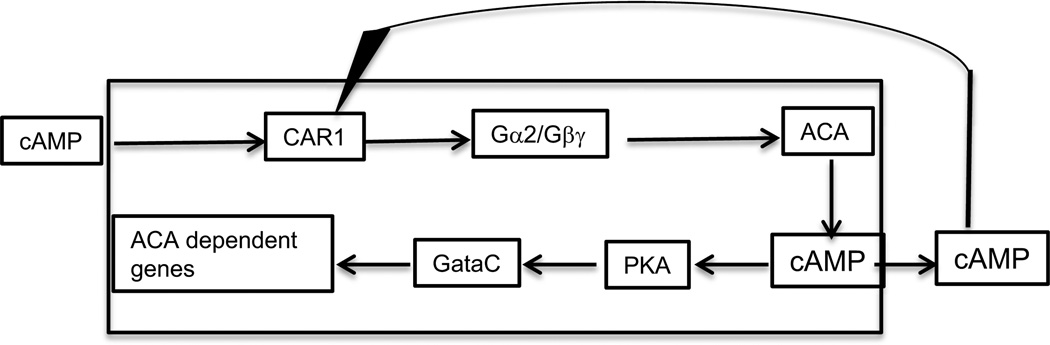
A set of 15 genes was found to be much more strongly expressed when cells were pulsed with cAMP (Iranfar, Fuller and Loomis, 2003). These pulse-dependent genes include those coding for the aggregation stage adenylyl cyclase, ACA, the cell adhesion proteins, CsaA (gp80) and TgrC1, and 5 calcium binding proteins. Another set of 13 genes was found that responded to addition of cAMP but only if the major early adenylyl cyclase ACA was functioning. It appears that expression of this class of ACA dependent genes is mediated by the rise in internal cAMP elicited by addition of external cAMP. The positive feedback loop in which extracellular cAMP induces acaA and ACA synthesizes more cAMP will tend to lock down expression of pulse-induced genes and can explain why the subsequent set of genes is dependent on ACA (Figure 7).
Figure 7.
Control of ACA dependent genes. A set of at least 13 developmental genes are only expressed if the gene for ACA is wild type. The signal transduction pathway from the extracellular cAMP signal to transcriptional control is almost the same as that in the PKA oscilatory circuit (Fig. 6) but requires the robust synthesis of cAMP that ACA can provide following a pulse of extracellular cAMP. It is possible that transcription of these genes depends on higher levels of PKA than the pulse induces genes.
Neither the 15 pulse-dependent nor the 13 ACA-dependent genes are expressed at all in the mutant cells lacking both adenylyl cyclases (acaA- acrA-) whether or not they are given cAMP pulses. However, if these mutant cells are transformed with a vector that leads to over expression of the catalytic subunit of PKA (pkaCOE), they express all the genes of these sets even in the absence of cAMP pulses (Iranfar, Fuller and Loomis, 2003). Over expression of the PKA catalytic subunit results in constitutive activity because there is insufficient regulatory subunits to keep all the catalytic subunits inactive. It appears that transcription of these early genes is regulated in large part by the activity of PKA. As expected, cells lacking PKA activity because of disruption of the pkaC gene or expression of a dominant inhibitory regulatory subunit Rm have almost no adenylyl cyclase (Mann and Firtel, 1991; Mann et al., 1997; Schulkes and Schaap, 1995). As a consequence they fail to aggregate or develop further.
A mechanism that can distinguish periodic cAMP signaling from continuous signaling has recently been characterized (Cai et al., 2014). A pulse of cAMP was found to result in exit of the DNA binding protein GataC from the nucleus. When the levels of cAMP dropped, GataC returned to the nucleus because it has a nuclear localization signal. It turned out that cAMP pulses, acting through the GPCR, CAR1, result in the phosphorylation of GataC which leads to rapid induction of pulse-dependent genes before its exit from the nucleus to the cytoplasm. Phosphorylation not only overrides the nuclear localization signal but probably also activates GataC such that it can induce transcription of its target genes. For a brief period following each pulse of cAMP, GataC is present in the nucleus in an active form where it can regulate the transcriptional profile. However, phosphorylation counteracts the nuclear localization signal and the transcription factor soon exits the nucleus. Such a control circuit is referred to as an incoherent feedforward loop. Differences in the kinetics of the positive and the negative branches set the activation time. If cells are continuously presented with high levels of cAMP, GataC remains phosphorylated in the cytoplasm where is does not have access to its target genes and so fails to activate them. In this manner GataC distinguishes between oscillatory and constant levels of cAMP.
Using RNA-seq to quantitate specific mRNAs, it was shown that the mRNAs of 181 genes that accumulated at least two fold during the first 5 hours of development failed to accumulate in cells lacking GataC (Cai et al., 2014). Clearly, GataC plays an essential role in transcriptional regulation during early development. The GataC dependent genes included acaA, carA, csaA, csbC, dagA (CRAC), dscD, erkA, gbfA, gpaB, pkaR, regA, tgrB1, and tgrC1. Many of these genes had been shown to be members of the pulse-independent group, the pulse-dependent group, or the ACA dependent group (Iranfar, Fuller and Loomis, 2003). The products of several of these genes play critical roles in transcriptional regulation during the subsequent developmental stage.
Some time ago, Kimmel (1987) showed that addition of cAMP pulses induces the accumulation of cAMP receptors on the surface of cells but that addition of constant high levels of cAMP rapidly represses this accumulation. Several other genes such as discoidin have been shown to be induced by pulses and repressed by constant high cAMP (Berger et al., 1985). Now that we know that these genes are controlled by GataC and that this transcription factor is only active for a brief period after extracellular cAMP goes up, it is clear why they are pulse dependent. Throughout the aggregation stage cAMP synthesis is oscillatory with a period of about 6 minutes as the result of control of ACA activity by the PKA circuit (Figure 6). However, when the cells become close-packed in the center of the aggregate and the cAMP generated by each cell is immediately added to the total, the concentration stays at a constant high level. Many of the early genes are repressed by this level of cAMP, while others that accumulate during later stages are induced (Berger et al., 1985; Kimmel, 1987; Iranfar, Fuller and Loomis, 2006).
Cell Contact Signaling
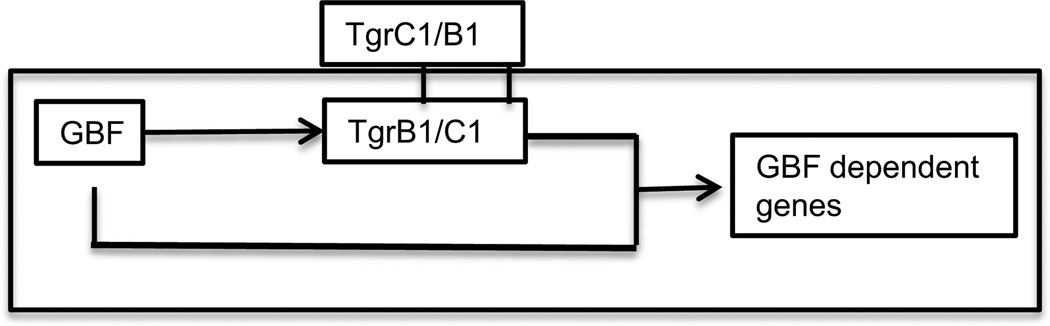
One of the pulse-dependent genes, tgrC, is also dependent on the DNA binding protein GBF (Iranfar, Fuller and Loomis, 2006). None of the other pulse-dependent genes are dependent on GBF. Moreover, transcription of tgrC is initiated almost immediately following the initiation of development which is about 6 hours earlier than the other GBF dependent genes. There is clearly something special about the regulation of expression of tgrC.
TgrC1 (which was originally named LagC) is a membrane protein that functions in both cell-cell adhesion and intercellular signaling (Dynes et al., 1994; Wang et al., 2000). Mutants lacking TgrC1 have arrested development at the loose mound stage, express all of the pulse-independent, pulse-dependent and ACA-dependent genes but none of the GBF-dependent genes. Their transcriptional profile is identical to that of gbfA− cells (Iranfar, Fuller and Loomis, 2006). However, over-expression of TgrC1 in a gbfA− background does not change the transcriptional profile indicating that the TgrC1 dependent signal is essential but not sufficient for expression of these post-aggregative genes. The architecture of this regulatory step is referred to as a feed-forward loop where GBF is necessary for expression of TgrC1 and both GBF and TgrC1 are necessary for expression of post-aggregative genes (Figure 8). Such a loop acts as a filter to avoid the perils of short lived fluctuations in the signal. It also integrates temporal signals with morphological signals to ensure that post-aggregation genes are only expressed when cells have aggregated and formed cellular associations.
Figure 8.
Cell contact signaling. A feedforward loop controls the expression of GBF dependent genes. GBF is a DNA binding protein that regulates expression of many developmental genes including the tgr genes that encode transmembrane proteins for cell-cell adhesion. The Tgr proteins are highly polymorphic in nature and act in intercellular signaling to indicate the level of kinship. If TrgB and TgrC are compatible in adjacent cells, then the GBF dependent developmental genes are expressed as long as GBF is also functional. Such a feedforward loop acts as a low pass filter to avoid reacting to short lived fluctuations in the signals.
TgrC1 is a member of a multi-gene family with 5 paralogous genes and 3 pseudogenes. Its product, gp150, forms a heterodimer with TgrB1 that holds the cells together (Wang et al., 2000; Benabentos et al., 2009; Hirose et al., 2011). Both TgrC1 and TgrB1 are highly polymorphic in independent isolates from the wild. Unless the Tgr genes are compatible, independent strains will sort out after they have co-aggregated. This ensures that most fruiting bodies are homogeneous genetic clones and protects from cheater strains that make more than their fair share of spores (Ho et al., 2013). Thus, the Tgr system functions as a type of kin recognition system that only permits normal post-aggregative gene expression when strict criteria for compatability have been passed.
Group Size
The size of an aggregate is determined in part by the local density of cells; thick streams that bring a large number of cells into an aggregate can form in dense monolayers. However, there is a system that controls the upper limit so as to keep the resulting fruiting bodies from being excessively large (Brock and Gomer, 1999). During early development, cells release a complex of at least 5 proteins called counting factor (CF) that causes aggregation streams to break up and form multiple independent aggregates of about 2 × 104 cells (Tang et al., 2002). High levels of CF decrease cell-cell adhesion and increase random motility such that aggregation streams break up and form small aggregates as predicted by a computer simulation (Jang and Gomer, 2008). CF also decreases the amplitude of cAMP-stimulated cAMP pulses which could limit spread of the signal. The aggregation streams of mutant strains lacking either of two CF subunits, CtnA or CF50, do not break up, resulting in huge fruiting bodies (Brock and Gomer, 1999).
CF appears to regulate cell-cell adhesion and cellular motility by affecting glucose metabolism (Jang, Schwartz and Gomer, 2009). CF decreases the activity of the gluconeogenic enzyme glucose-6-phosphatase which may account for the abnormally low levels of glucose in cells over-secreting CF (Jang et al., 2002; Jang and Gomer, 2005). Adding glucose to cells that over-secrete CF as they aggregate increases the size of the fruiting bodies that are formed such that they resemble those of wild type strains (Jang and Gomer, 2005). There were other changes in metabolic intermediates, such as pyruvate and lactate, in strains with altered CF secretion that might also affect behavior of cells in aggregation streams (Jang, Schwartz and Gomer, 2009).
Size regulation is critical to many tissues and organs in all multicellular organisms. However, it is only in the social amoeba that the size of the terminal structures is determined by the number of cells recruited from the surroundings. In both plants and animals, the size of organs is predominantly established by the relative growth of different tissues. There is no significant growth following the initiation of development in Dictyostelium, so size has to be controlled in another manner. Secretion of a protein complex during aggregation that determines whether the streams break up or not is a solution that couples the number of cells in a stream to the ultimate size of aggregates. Large streams would initially have higher CF levels which could modify gluconeogenesis and the level of metabolites such that cells would be encouraged to leave the stream and take off on their own. The level of CF would then drop leading to an increase in cell-cell adhesion and a decrease in random motility that would stabilize the smaller streams.
PRESTALK SIGNALS
DIF-1 (Differentiation Inducing Factor-1)
The transcriptional profiles of the two main cell types, prespore and prestalk cells, start to diverge during aggregation. In fact, about half of the GBF dependent genes are preferentially expressed in prespore cells and the other half is preferentially expressed in prestalk cells. It is still not clear what determines the choice between one or the other path of differentiation nor how the proportions of the cell types are regulated but it undoubtedly involves cell signaling.
The first step towards answering these questions is to develop an assay where both the cells and the signals can be controlled and quantitatively measured. Shortly after discovering that cAMP was the chemoattractant of D. discoideum, Bonner (1970) showed that adding 1 mM cAMP to the agar on which cells were spread at low density would induce about 10% of the cells to become vacuolized and encased in cellulose much like stalk cells. The assay was further improved by replacing the standard laboratory stock, NC4, with a V12 strain that comes from a seperate isolate from nature (Town, Gross and Kay, 1976). When spread at 105 cells/ cm2 on agar containing 5 mM cAMP, > 90% of V12 M2 cells differentiated into stalk cells in a few days. Many were found in small clumps. The proportion of cells that would differentiate into stalk cells was lower when the cell density was reduced. However, at low cell density (103 cells/ cm2) cells would efficiently form stalk cells if they were incubated on a sheet of cellophane over a layer of cells at high cell density indicating that the cells on top were receiving a signal from the cells below. The signal has been called Differentiation Inducing Factor or DIF (Kay, Dhokia and Jermyn, 1983; Kay and Jermyn, 1983).
The high density conditions provided a way to select for mutant strains that did not all become non-viable stalk cells. After developing for several days at high density with added cAMP, viable cells were isolated, grown up and once again developed under conditions that induce stalk cell differentiation. This protocol was continued until a mutant strain, sci-1, was isolated that formed both spores and stalk cells when incubated at high cell density with 5 mM cAMP (Town, Gross and Kay, 1976). Although the gene affected in strain sci-1 has never been established, the mutant phenotype suggests that the cAMP and density dependence for stalk cell differentiation also holds for spore differentiation.
Using the fact that DIF-1 can cross through the pores of cellophane which exclude molecules larger than 5 kDa, the stalk-inducing factor was partially purified by collecting the buffer below sheets of cellophane on which cells had been developed (Kay, Dhokia and Jermyn, 1983). The active ingredient was isolated from the buffer with non-ionic resin, eluted and further purified by reverse-phase high pressure liquid chromotography (HPLC). A few micrograms of purified DIF-1 were obtained from 1012 cells. The factor was subjected to various biochemical tests and analyses by mass spectroscopy before it was finally shown to be the chlorinated hexanophenone 1-(3,5-dichloro-2,6-dihydroxy-4-methoxyphenyl)-1-hexanone (Morris et al., 1987). This unusual small molecule has been found to be secreted during late aggregation and to affect the transcriptional profile of a subset of prestalk cells (Kay 1998; Thompson and Kay, 2000; Maeda et al., 2003).
The first step specific to the synthesis of this chlorinated alkyl phenone is catalyzed by the polyketide synthetase StlB (Austin et al., 2006). The polyketide product is then modified by a flavin-dependent halogenase encoded by chlA and methylated by the methyltransferase encoded by dmtA (Neumann, Walsh and Kay, 2010; Thompson and Kay, 2000 ). These latter two genes are expressed immediately after the initation of development but the gene for the polyketide synthetase, stlB, is not expressed until after 4 hours of development. DIF-1 can only accumulate after the cells have sufficient polyketide synthetase activity.
Both stlB and dmtA are expressed preferentially in prespore cells. On the other hand, the gene that encodes the dechlorinase enzyme that breaks down DIF-1, drcA, is expressed preferentially in prestalk cells (Velazquez et al., 2011; Parikh et al., 2010). It appears that prespore cells make DIF-1 and prestalk cells break it down resulting in a sharp gradient near the prespore/prestalk boundary.
Molecular characterizations of prestalk cells have been able to subdivide them on the basis of marker gene expression, position within a slug, and ultimate fate (Jermyn et al., 1989; Jermyn and Williams, 1991; Maeda et al., 2003). Prestalk cells are found in the front of slugs and make up about 20% of the total number of cells in a slug. A subtype of prestalk cells, PstA cells, are found at the most anterior of the slug. They are followed by another subtype of prestalk cells, PstO cells, that are found behind the PstA cells and ahead of prespore cells. There is also a group of prestalk cells, PstB cells, found at the ventral side of slugs surrounded by prespore cells.
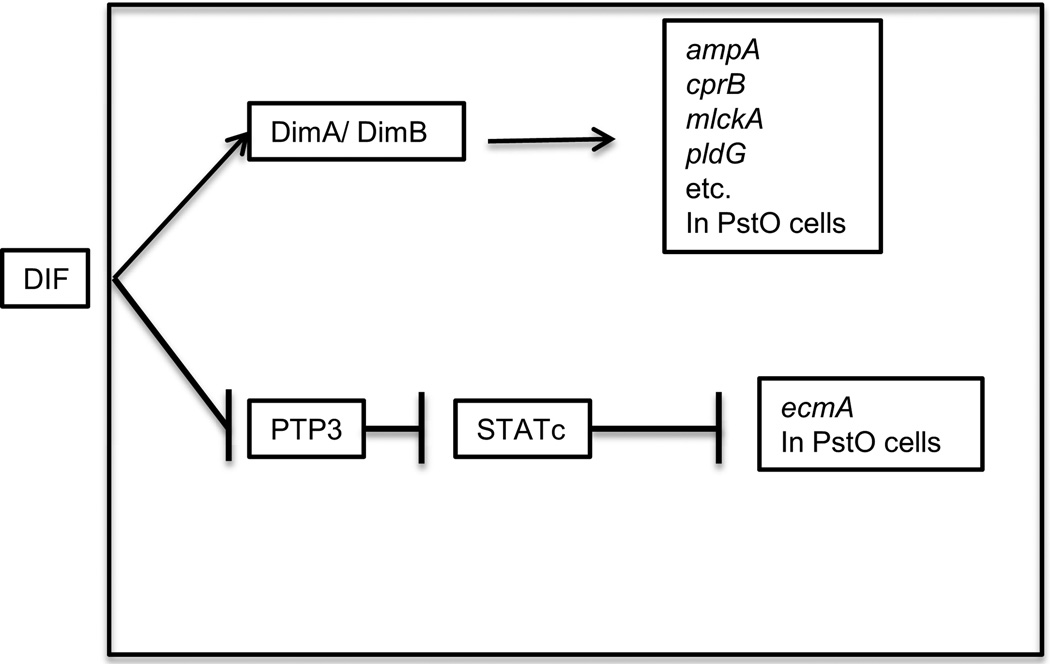
PstA cells are responsible for making the stalk in fruiting bodies while PstO and PstB cells are responsible for the lower cup and the outer basal disc (Fukuzawa, 2011). Mutant strains that fail to make DIF-1 as the result of mutations in stlB or dmtA develop almost normally, making well formed fruiting bodies with tapering stalks but lack PstB cells and do not make either a lower cup or basal disks (Thompson and Kay, 2000; Saito et al., 2008). In situ hybridizations found 30 genes that are uniquely expressed in PstO cells; 18 of these genes had significantly reduced expression in the dmtA- mutant cells, but 12 others were expressed normally in cells lacking DIF-1 (Maeda et al., 2003). It appears that DIF-1 is responsible for some, but not all, of the differentiations of PstO cells (Figure 9).
Figure 9.
Control of prestalk genes by DIF. The chlorinated hexaphenone DIF-1 is synthesized by prespore cells and degraded by prestalk cells. It induces PstO genes via the heterodimer of DNA binding proteins DimA/DimB. Not all genes expressed in PstO cells depend on DIF-1 indicating that it acts in a gene specific manner and not just as a general inducer of PstO differentiation.
Addition of DIF-1 to monolayers of developing cells induces many of the cells to differentiate into vacuolized stalk cells. Since stalk cells are not viable, DIF-1 resistant strains can be enriched by treatment of randomly mutagenized populations with DIF-1 and selection of viable cells. By repeating this treatment several times, DIF-insensitive mutant strains were isolated (Thompson et al., 2004). One of the genes where insertional mutations gave DIF-insensitivity encodes a basic leucine zipper protein, DimA (Figure 9). In wild type cells, addition of DIF-1 results in entry of DimA into the nucleus within 3 minutes. Subsequent work showed that another bZIP protein, DimB, is also rapidly induced to enter the nucleus by DIF-1 (Huang et al., 2006). DimA and DimB form homodimers and heterodimers by interactions of their leucine zipper domains. Moreover, DimA requires DimB for entrance into the nucleus. DimB was found to be a DNA binding protein that recognizes the regulatory region of ecmA critical for its expression in PstO cells. However, rather than inducing expression, it appears to repress it (Zhukovskaya et al., 2006). In response to DIF-1 DimB appears to act as an inducer for some genes and as a repressor for other genes (Yamada et al., 2010). Two other DNA binding proteins, STATc and GataC, respond to addition of DIF-1 by rapid entry into the nucleus where they affect differentiation of PstO and PstB cells (Fukuzawa, Abe and Williams, 2003; Keller and Thompson, 2008; Fukuzawa, 2011). As mentioned before, GataC also plays a role in deciphering cAMP pulses during aggregation (Figures 6 and 7). DIF-1 signaling inhibits the protein tyrosine phosphatase PTP3 which keeps STATc out of the nucleus by dephosphorylating it (Araki et al. 2008). In the presence of DIF-1, STATc is localized in the nuclei of PstO and anterior-like cells where it represses expression from the proximal (PstA specific) enhancer of ecmA (Fukuzawa et al. 2001). GataC plays a critical role in differentiation of PstB cells since a basal disc is not formed in its absence. The exact roles of these regulators are still poorly understood but each one seems to work slightly differently. Clearly, transcriptional regulation by DIF-1 is complex.
SDF-1 (Spore Differentiation Factor-1)
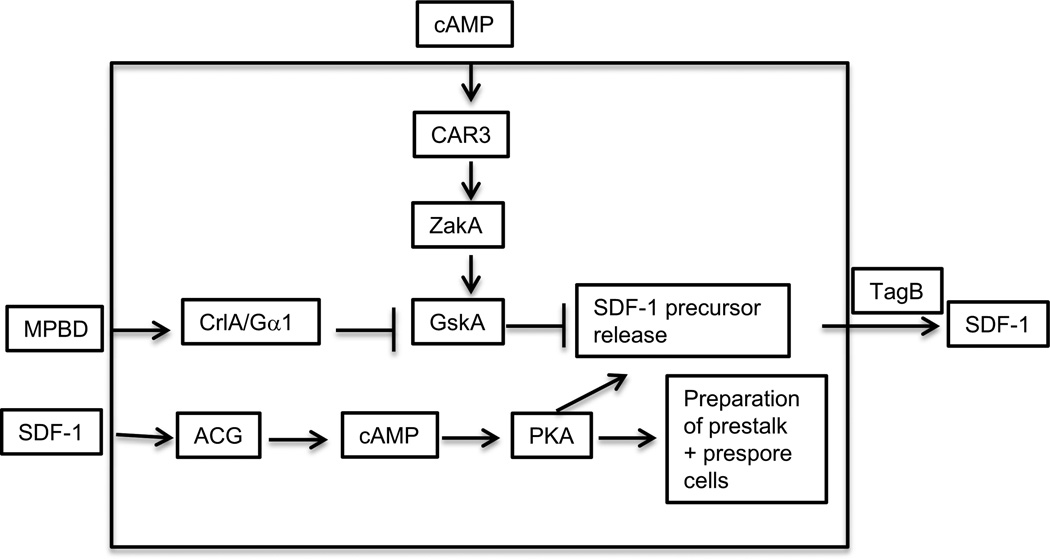
Another polyketide was found to be secreted a little later in development when cells were beginning to construct fruiting bodies (Saito et al., 2006; Anjard, Su and Loomis, 2011). This molecule, MPBD (4-methyl-5-pentylbenzene-1,3 diol), induced terminal differentiation of stalk cells in monolayers of V12M2 cells. It would also induce terminal differentiation of spores in a sporogenous strain (Saito et al., 2006). Using a sporogenous assay, it was shown that MPBD, acting through the surface receptor CrlA and its associated G protein that contains the Gα1 subunit, inhibits the protein kinase GskA that blocks release of the precursor of the signal peptide SDF-1 (Anjard, Su and Loomis, 2011). The activity of GskA is stimulated by exogenous cAMP acting through the surface receptor CAR3 to activate the protein kinase ZakA (Kim et al., 1999). The opposing actions of MPBD and cAMP on GskA activity ensures that the phosphoprotein that is processed extracellularily to give rise to SDF-1 is not released until the enzyme responsible for synthesis of MPBD, StlA, accumulates just before culmination (Austin et al., 2006; Anjard, Su and Loomis, 2011). Once secreted, the precursor is processed by the extracellular protease activity of TagB to give rise to a short phosphopeptide that can be mimicked by the sequence LRRASpLG (Figure 10). This phosphopeptide, SDF-1, does not directly induce either spores or stalk cells but prepares cells in culminants for terminal differentiation when further signals are recieved. There is a 90 minute period following addition of SDF-1 to sporogenous cells before the rate of encapsulation is seen to increase. RNA and protein synthesis are essential during this period for SDF-1 to have its effects (Anjard et al., 1998). New proteins appear to be necessary to prepare both prespore and prestalk cells for terminal differentiation.
Figure 10.
SDF-1 signaling. The small phosphopeptide SDF-1 is secreted after 18 hours of development and signals both prestalk and prespore to prepare themselves for terminal differentiation. This process requires both RNA and protein synthesis. SDF-1 is cleaved from a larger phosphoprotein that is secreted in response to signaling by a polyketide, MPBD, or an increase in the activity of PKA. MPBD is synthesized by the Steely protein, StlA, and binds to the GPCR receptor CrlA. The signal is transduced by the trimeric protein that contains the Gα1 subunit such that the protein kinase GskA is inhibited and no longer blocks release of the SDF-1 precursor protein. The precursor is processed into SDF-1 by the extracellular protease domain of the prestalk specific protein TagB. SDF-1 stimulates PKA activity in a process that depends on the adenylyl cyclase ACG. High PKA activity can also trigger release of the SDF-1 precursor protein such that low levels of SDF-1 are amplified by "priming'.
The ability of SDF-1 to induce encapsulation in sporogenous strains is dependent on the late adenylyl cyclase ACG (Anjard, Su and Loomis, 2011). This membrane associated enzyme may also be the surface receptor for SDF-1 since it has a conserved CHASE domain that can bind peptides. The most likely scenario is that when SDF-1 binds ACG the synthesis of cAMP increases and activates PKA, which then phosphorylates the SDF-1 precursor, thereby facilitating its release and processing to generate more SDF-1 (Figure 10). This positive feedback loop leads to a rapid increase in extracellular SDF-1. Mutational loss of any of the five genes encoding components of the SDF-1 signal transduction pathway (stlA, crlA, gpaA, tagB, acgA) results in very few viable spores being made (Anjard, Su and Loomis, 2011).
When added to cells of the stalk differentiation test strain, V12M2, SDF-1 induces the massive vacuolization characteristic of terminal stalk cell differentiation (Anjard et al., 1998). It can also overcome the ability of cAMP to block stalk cell induction by DIF-1 (Berks and Kay, 1988; Anjard et al., 1998; 2011). It is likely that the increase in internal cAMP that occurs when SDF-1 binds ACG is countering the effects of exogenous cAMP on GskA activity. MPBD is also able to overcome this effect of exogenous cAMP, possibly through its ability to inhibit GskA, or possibly, just as a consequence of stimulating the production of SDF-1.
TERMINAL DIFFERENTIATION SIGNALS
SDF-2 (Spore Differentiation Factor-2)
The first indication that terminal differentiation of spores might involve an extracellular signal during culmination was the observation that expression of the spore specific gene, spiA, started near the top of fruiting bodies in the prespore cells nearest to the prestalk cells and subsequently swept down through underlying prespore cells (Richardson, Loomis, and Kimmel, 1994). It looked like prestalk cells were generating a diffusible signal that induced specific gene expression in prespore cells. Such a circuit would ensure that one cell type did not differentiate without the other. Subsequent microscopic analyses found that encapsulation also started near the top of the ball of prespore cells as they climbed the stalk and then rapidly spread throughout the prespore population.
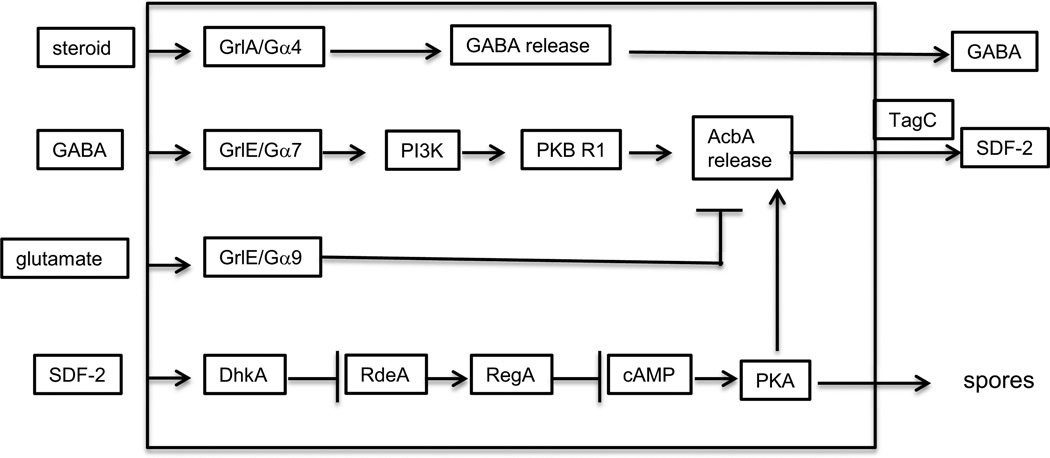
Using an assay in which cells with partially constitutive PKA activity can be induced to form spores even when incubated at low cell density, a sporulation inducing factor was found to be secreted during culmination (Anjard et al., 1998). The factor was shown to be a peptide distinct from SDF-1, but like SDF-1, the peptide is cleaved from a precursor protein in the extracellular space (Anjard and Loomis, 2005). However, release of the precursor into the extracellular space is inhibited by glutamate which accumulates to high levels in the mass of prespore cells as they are rising up the stalk during culmination. Glutamate is also an intercellular signal in mammals where its major role is as an excitatory neurotransmitter in the brain. There are several different kinds of glutamate receptors in neural cells. The so-called metabolic receptors are seven-transmembrane G-protein coupled receptors (GPCRs). Likewise, in Dictyostelium the glutamate receptor is a GPCR. When this receptor, GrlE, binds glutamate, it is coupled to a G protein with the Gα9 subunit and inhibits release of the SDF-2 precursor (Anjard and Loomis, 2006).
When culmination is nearly complete, the cells secrete a steroid similar to hydrocortisone that binds another GPCR, GrlA, that triggers the rapid release of GABA (γ-aminobutyric acid) which overcomes the effects of glutamate (Anjard, Su and Loomis, 2009). It was somewhat surprising to find that a steroid binds to a 7 transmembrane surface receptor since steroid hormones in mammals are well known to cross the plasma membrane and bind to internal nuclear receptors. There are a few exceptions where a steroid binds to a GPCR in vertebrates but they are rare (Norman, Mizwicki and Norman, 2004; Thomas et al., 2006; Prossnitz, Arterburn and Sklar, 2007). Dictyostelium cells lacking the steroid surface receptor or the specific Gα subunit to which it is coupled, due to mutations in grlA or gpaD, do not respond to hydrocortisone by release of GABA or production of SDF-2 (Anjard, Su and Loomis, 2009) (Figure 11). It seems clear that Dictyostelium relies on this GPCR to recognize the steroid.
Figure 11.
SDF-2 signaling. A cascade of intercellular signals amplifies the first signal leading to extracellular production of SDF-2 which is mediated by a steroid that is very similar to hydrocortisone. The steroid receptor is a surface GPCR coupled to a trimeric G protein with the Ga4 subunit. This pathway leads to the release of GABA. The GABA receptor is a surface GPCR coupled to trimeric G protein with the Ga7 subunit. This same receptor binds glutamate when it is coupled to the trimeric G protein with the Ga9 subunit. Glutamate binding inhibits release of the SDF-1 precursor acyl-coA-binding protein, AcbA, but has a lower affinity to GrlE than GABA. When GABA accumulates in the intercellular space the lipid kinase PI3K and the protein kinase PKB R1 are activated and release of AcbA is stimulated. AcbA is cleaved by the extracellular protease domain of the prestalk specific protein TagC to generate SDF-2. This 34 amino acid peptide has to diffuse back to the prespore cells where it triggers rapid encapsulation into spores. The SDF-2 receptor is the membrane embedded histidine kinase DhkA that is converted to a protein phosphatase when SDF-2 is bound. It then removes the phosphate from the small H2 RdeA so that it can no longer phosphorylate and activate the internal cAMP phosphodiesterase RegA. In fact, it actively dephosphorylates RegA thereby inhibiting it from degrading cAMP. As cAMP builds up, PKA is activated and can trigger spore formation as well as release of further AcbA.
GABA is an inhibitory neurotransmitter in mammals that binds to specific GABA receptors on neurons. Most GABA receptors (GABAA) are ionotrophic ligand gated chloride channels. Dictyostelium does not have homologs of this class of receptors. There are other GABA receptors on neurons that are coupled to G proteins and referred to as GABAB metabolic receptors. Dictyostelium has multiple genes encoding receptors of the GABAB family, one of which, GrlE, is the glutamate receptor mentioned above. It appears that in Dictyostelium, unlike in neurons, this GPCR binds both glutamate and GABA. In neurons, the metabolic glutamate receptors show no afffinity for GABA and the GABAB receptors show no affinity to glutamate. But in Dictyostelium, GABA competes with glutamate for binding to GrlE. When GABA is bound, GrlE is coupled to a G protein with a different α subunit, Gα7, rather than Gα9 to which it is coupled when it has bound glutamate (Anjard and Loomis, 2006). Such a ligand induced switch in coupled G protein seems like an effective way to switch between signal transduction pathways in response to different environmental signals, but has not been found in other systems.
When GABA is bound to GrlE, the SDF-2 precursor, acyl-CoA- binding protein (AcbA) is rapidly secreted by an unconventional pathway (Kinseth et al., 2007). The signal transduction pathway leading to AcbA release involves the enzymes PI3K and PKB R1 based on mutant and pharmacological analyses (Figure 11). PKA activity is also essential for AcbA release.
Although cells isolated from early culminants have a high affinity to steroids and can respond by secreting GABA when as little as 5 nM hydrocortisone is added to the buffer, it takes the output of almost every cell in a culminant to reach this threshold (Anjard, Su and Loomis, 2009). However, they respond by secreting at least 10 times more GABA. This triggers the cells to produce several thousand fold excess SDF-2 that can rapidly induce encapsulation of prespore cells into spores. This dramatic amplification of the original steroid signal ensures an all-or-none response and can compensate for variations in shape or water retention among fruiting bodies.
AcbA is a 84 amino acid protein that binds acyl-CoA and is found exclusively in prespore cells. It is processed to the 34 amino acid peptide, SDF-2, by the protease TagC on the external surface of the cells (Anjard et al., 1998; Anjard and Loomis, 2006). AcbA is highly conserved in Dictyostelium, yeast and animals. In mammals it is procesed into a neuropeptide, DBI, that gets its name (Diazepam Binding Inhibitor) because it competes with diazepam (Valium) for binding to ionotropic glutamate receptors in the central nervous system. Human DBI is so similar to SDF-2 that it can substitute for SDF-2 in Dictyostelium (Anjard and Loomis, 2005).
TagC is a membrane protein found exclusively in prestalk cells. Its protease domain is not exposed to the intercellular space until after GABA induction thereby coordinating the presentation of extracellular AcbA with the ability to cleave it to the active peptide. The prestalk response to GABA involves many of the same components that are used in the prespore response, such as GrlE and Gα7, but in prestalk cells the signal transduction pathway triggers presentation of the TagC protease on the surface rather than secretion of AcbA as it does in prespore cells.
The back-and-forth of cell signaling is particularily clear in the case of SDF-2. GABA induces prespore cells to secrete AcbA and induces prestalk cells to expose the protease domain of TagC on the surface. AcbA has to diffuse from prespore cells to prestalk cells to be processed into SDF-2. There is a positive feedback loop in prestalk cells such that the SDF-2 signal is amplified. SDF-2 then has to diffuse back to prespore cells where it can induce rapid encapsulation.
Addition of SDF-2 to cells dissociated from early culminants or monolayer cells of a strain that has partially constitutive PKA results in an increase in encapsulation within 10 minutes (Anjard, Su and Loomis, 2009). SDF-2 binds to its surface receptor DhkA and converts it from a protein kinase into a protein phosphatase (Wang et al. 1996, 1999; Anjard and Loomis, 2005). DhkA is a member of the family of two component protein kinases that autophosphorylate on a histidine and transfer the phosphate to a response regulator region via a small H2 protein. RdeA is the sole H2 protein of Dictyostelium. It transfers a phosphate to RegA to activate this cAMP phosphodiesterase. When DhkA binds SDF-2, it no longer passes phosphate to RdeA but instead it hydrolyzes the phosphate off RdeA. Unphosphorylated RdeA is then able to reversibly retrieve the phosphate from RegA. Dephosphorylated RegA is no longer active and so cAMP can build up and activate PKA. Ligand binding to DhkA results in taking off the accelerator and applying the brakes to RegA. This dual relationship doubles the sharpness of the response.
High levels of PKA trigger the release of more AcbA which can be processed to SDF-2. In this way low levels of SDF-2 "prime" cells to release their full complement of AcbA thereby producing high levels of SDF-2. This amplification sets a sharp threshold for the response to SDF-2 and ensures that PKA activity rapidly increases to levels that induce sporulation.
Cytokinin
Derivatized adenine compounds that stimulate cell division in plants are referred to as cytokinins. They occur in a wide variety of organisms including bacteria, fungi and plants (Mok and Mok, 2001). In Dictyostelium discoideum a cytokinin, discadenine, is secreted from fruiting bodies and inhibits spore germination (Abe et al., 1976; Tanaka et al., 1978). Discadenine also induces rapid sporulation in the sporogenous strain that expresses partially constitutive PKA (Anjard and Loomis, 2008). Addition of the cytokinins 10 nM isopentyladenine, 1 µM zeatin or 10 nM discadenine to a monolayer of the test cells resulted in an increase in the rate of encapsulation within 10 minutes. Isopentyl transferase catalyzes the de novo synthesis of isopentyladenine which is further modified to form discadenine. The gene encoding this enzyme, iptA, is only expressed after 20 hours of development which will restrict the formation of cytokinins to late culmination. When iptA was inactivated by mutation, the rate of accumulation of the cytokinins in fruiting bodies was reduced about 10 fold and sporulation reduced to half of that in wild type strains. It appears that cytokinins play significant roles in triggering efficient sporulation (Anjard and Loomis, 2008).
The response to discadenine, isopentyladenine and zeatin is not seen in the test strain when the genes encoding the late adenylyl cyclase ACR or the prespore specific histidine kinase DhkB are inactivated by insertions (Anjard and Loomis, 2008). Likewise, fruiting bodies of mutant strains carring the acrA- and dhkB- mutations are defective in sporulation, forming only 1% and 15% of the number of viable spores found in wild type fruiting bodies (Wang et al., 1999; Soderbom et al., 1999). Both ACR and DhkB are membrane bound, but neither is essential for binding isopentyladenine to the cell surface (Anjard and Loomis, 2008). Although the surface receptor for isopentyladenine is developmentally regulated, it appears to be distinct from ACR and DhkB.
Besides the domains characteristic of adenyl cyclases, ACR carries a degenerate histidine kinase domain that has lost its ability to autophosphorylate but may still participate in dimerization with other histidine kinases (Wang et al., 1999). Assuming that DhkB acts as a histidine kinase in this signal transduction pathway, it may activate ACR by phosphorelay to the response regulatory region. However, it does not use the cannonical relay pathway that relies on an H2 protein intermediate because the sole H2 in Dictyostelium, RdeA, can be mutated without affecting the response to cytokinins (Anjard and Loomis, 2008). It is likely that ACR and DhkB form a heterodimer in which the phosphate on the histidine of DhkB is transfered directly to the aspartate in the response regulator of ACR which would activate it and result in a rise in cAMP and PKA activity (Figure 12). This pathway works in parallel but independently of the SDF-2 signal transduction pathway. The additive effects of these pathways on PKA activity makes them a coincidence detector for the induction of sporulation.
Figure 12.
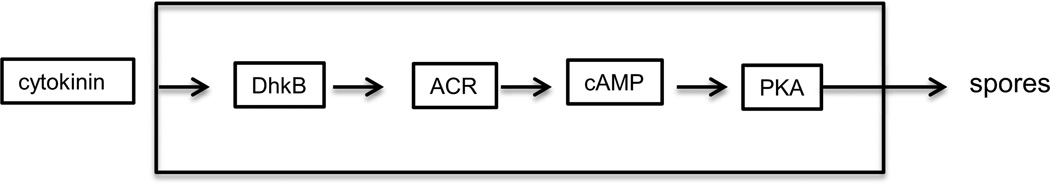
Cytokinin signaling. Cytokinins are thought of as plant hormones but their use as intercellular signals in Dictyostelium results from inheritance from a common ancestor of amoebozoa and plants. The isopentyladenine derivative discadenine is a cytokinin produced by Dictyostelium that leads to an increase in PKA activity in a manner dependent on the histidine kinase, DhkB and the late adenylyl cyclase ACR. As is the case in SDF-2 signaling, the increase in PKA activity leads to rapid encapsulation of prespore cells. DhkB and ACR are preferentially found in prespore cells.
Cyclic di-GMP
By scanning the genome of Dictyostelium discoideum, Chen and Schaap (2012) found a motif characteristic of the bacterial enzyme responsible for the synthesis of the signaling molecule cyclic-di-GMP. This signal is widely used in prokaryotes to regulate cell division and differentiation but had never been found in eukaryotes. The Dictyostelium gene, dgcA, was sequenced and knocked out to define its role. To their delight, they found that dgcA− null mutants went through the early stages of development normally but stopped at the slug stage and failed to make spores or stalks (Chen and Schaap, 2012). Addition of 1 µM cyclic-di-GMP to the mutant cells at the slug stage restored their ability to form stalks, spores, and complete fruiting body formation. Addition of cAMP, cGMP, or cyclic-di-AMP had no effect on the mutant cells. Moreover, they found that mixing 10% wild type cells to the dgcA− resulted in the mutant cells forming normally proportioned fruiting bodies, thereby clearly showing that the wild type cells were signaling the dgcA- cells. Finally, they showed that addition of 10 µM cyclic-di-GMP to monolayers of the V12M2 test strain induced the differentiation of stalk cells. It seems clear that cells release cyclic-di-GMP that serves as an essential signal for stalk formation. But could cyclic-di-GMP also signal for spore formation? Probably not directly, since the gene is only expressed in prestalk cells and addition of 10 µM cyclic-di-GMP to wild type cells developing at low density does not induce sporulation (Chen and Schaap, 2012). It appears that the lack of sporulation in dgcA− cells in the absense of added cyclic-di-GMP may result from the lack of stalk cell differentiation rather than the lack of cyclic-di-GMP signaling. Signals essential for encapsulation may depend on the function of prestalk specific genes expressed during terminal differentiation.
DISCUSSION
A dependent sequence
The order and tempo of events is critical to multicellular development and is carefully monitored by regular checks and balances as development progresses. Skipping or inverting stages is usually disastrous and has to be avoided at all costs. Later stages depend on earlier stages in a dependent sequence that is rigidly maintained by natural selection (Loomis, White and Dimond, 1976). Likewise, speeding up or slowing down progression from one stage to the next can disrupt the coordination between cells that is essential for successful development. Differentiating cells have to frequently broadcast their physiological state in a manner that their interacting partners can recognize and interpret. In most cases this means stage specific synthesis and release of signaling molecules that diffuse to their interacting partners where they are recognized by surface receptors, but in some cases involves direct cell-cell contact.
Embryogenesis rapidly generates a large number of different cell types which have to integrate differentiation amongst themselves as well as coordinate subsequent steps with other cell types. The number and diversity of signaling systems increases rapidly during the early cleavage stages of most embryos such that the number of cells making each specific signal is often quite small. As a consequence, biochemical characterization of the signals and their signal transduction pathways can be problematic. The development of Dictyostelium provides an opportunity for high resolution analyses of cell signaling by capitalizing on the ability to synchronize morphogenesis and apply the highly developed molecular genetics available in this system. While it is not meaningful to extrapolate from Dictyostelium to specific embryonic processes such as dorsal/ventral polarity, gastrulation or neurulation, studies on this model system can lead to a basic understanding of general cell biological processes essential for multicellular development. Just as an embryo must time events in separate tissues, Dictyostelium has to coordinate cells at either ends of slugs. The relative simplicity of Dictyostelium development in which only two major cell types arise encourages a holistic approach.
Universal signals
Half of the intercellular signals now recognized in Dictyostelium development have been previously shown to signal during vertebrate embryogenesis, synaptic transmission, or cellular interactions in plants or bacteria. The other half are novel but go to show that almost any secreted component can act as an intercellular signal.
Nitrogen limitation initiates differentiation in Dictyostelium and does so as well in yeast and bacteria. Do they all monitor the internal concentration of keto-acids to respond appropriately? Another commonly used signal is cAMP. Almost all cells, including Dictyostelium, use cAMP as a second message in regulating gene expression and PKA activity. But Dictyostelium also acquired the ability to secrete cAMP and use it to guide cells into aggregates. However, there is nothing special about using cAMP as the chemoattractant as shown by the use of a peptide and a pterin as chemoattractants in related social amoebae. It appears that using cAMP as an intercellular signal is a fairly recent evolutionary invention (Schaap, 2011).
Steroids are common hormones in a wide variety of organisms. Dictyostelium cells synthesize and secrete a steroid that is very similar to hydrocortisone. Somewhat surprisingly, it is recognized by a surface receptor that is a member of the GPCR class of 7-transmembrane proteins rather than a cytoplasmic receptor as we have come to expect. In a few cases, mammalian cells have also been shown to bind steroids to GPCR surface receptors. This may have been the ancestral mechanism.
Glutamate and GABA control the release of AcbA in Dictyostelium. Steroids control the release of AcbA from the glial cells of mammalian brains (Loomis et al., 2010). Both Dictyostelium and glial cells proteolytically cleave secreted AcbA to generate signalling peptides. In mammals the peptide is referred to as DBI which is so similar to SDF-2 that it can replace it in Dictyostelium mutants. Cytokinins are well known plant growth hormones but also function in Dictyostelium where they trigger sporulation and maintain dormancy. Cyclic di-GMP is known to regulate the formation of biofilms in some bacteria and is now known to induce stalk cell differentiation in Dictyostelium.
If we look at the range of molecules that are known to act as intercellular signals in Dictyostelium and other organisms, it becomes apparent that molecules with very different biophysical properties are quite acceptable as signals. The fact that only a limited subset of molecules is used for biological signaling indicates that there was a strong advantage to using proven signaling molecules and adapting them to the regulation of new processes.
Signal transduction pathways
Surface receptors are known for 8 of the 13 intercellular signals. Of these, 5 are G-protein coupled receptors (GPCRs) that are highly similar to such receptors in all the animal phylla. While GABA is recognized by both ionotropic GABAA-type receptors and a GABAB-type GPCR in mammals, GABA is recognized only by a GABAB-type GPCR in Dictyostelium. DBI binds to ionotrophic glutamate receptors on neurons while its close homolog, SDF-2, binds to a histidine kinase in Dictyostelium. In this case, the signal has been more conserved than the receptor.
Downstream of the receptors, positive feedback loops, mostly involving cAMP, are prevalent in Dictyostelium. They are likely to stabilize progression from one developmental stage to the next and reduce the chances of rescinding. Negative feedback is used to ensure oscillation of PKA activity during the aggregation phase. Double-negative control occurs 4 times in activating pathways and seems to work just as well as simple positive activation. A feedforward loop was found which can dampen signaling noise. Other types of control circuits have been observed in Dictyostelium such as an incoherent feedforward loop and ultrasensitive balancing that provide adaptation and amplification of chemotactic responses (Takeda et al., 2012; Skoge et al., 2014).
About half of the signal transduction pathways that coordinate development in Dictyostelium do not affect transcriptional patterns and are not expected to end with a transcriptional regulator. However, 3 pathways control transcriptional regulators that carry basic leucine zipper motifs (bZIP proteins) and 2 others end with transcriptional regulators that carry multiple zinc-fingers. One of these (GataC) has GATA-type zinc-fingers which recognize the DNA sequence GATA in both Dictyostelium and mammalian cells. An incoherent feedforward loop regulates GataC activity in the nucleus (Cai et al., 2014). Such a mechanism is only activated in response to pulsatile signaling and can act as a counting mechanism for the number of pulses that a cell has experienced. Such a mechanism may be advantageous whenever oscillatory signals are used.
Some of the first molecular genetic studies of embryogenesis were carried out on fertilized Drosophila eggs. A story unfolded that early zygotic transcriptional regulators controled later transcriptional regulators that controlled subsequent transcriptional regulators until most segments were specified (Nusslein-Volhard and Wieschaus, 1980; Lawrence, 1992). Some thought that this was a powerful paradigm for early embryogenesis and went looking for zygotic transcriptional regulators in all sorts of organisms. However, it turned out that such a simple cascade of transcription factors is mostly restricted to Drosophila and related insects and occurs hardly anywhere else. It appears to have been selected to facilitate rapid development of larvae rather than adaptability. Most metazoan embryos take a more measured approach that involves specific cell-cell communication before the transcriptional machinery is changed. Dictyostelium also enters into development cautiously and requires detailed cellular communications before proceeding along the dependent sequence. Only when cells have signaled that they have completed a stage will they move to the next stage and modify transcriptional patterns appropriately.
Regenerative medicine is based on the hope that we can learn to program pluripotent stem cells through the appropriate stages to reproducibly differentiate into specific cell types that can replace missing or diseased cells. The approach may use the patient's own stem cells either in situ or after in vitro selection. The ability to isolate and culture totipotent embryonic stem cells (ESCs) has come along in leaps and bounds (Martin, 1981; Evans and Kaufman, 1981; Thomson et al., 1998; Takahashi and Yamanaka, 2006). It is now possible to convert mature human cells into induced pluripotent stem cells (iPSCs) by providing a cocktail of 4 stem cell associated transcription factors (Yu et al., 2007; Takahashi et al., 2007). After culturing for a month, about 0.1% of the cells grow as colonies resembling those of embryonic stem cells that can be subcultured. By adding specific hormones and cell signals to the culture medium some of the iPSCs could be coaxed into differentiating as specific cell types such as cardiomyocytes that spontaneously start beating. However, the efficiency of terminal differentiation is low and the continued presence of growing cells that may cause cancer cannot be ruled out.
Learning how to program ES cells rapidly and efficiently may benefit from studies on the regulation of differentiation in Dictyostelium. There is a lot in common between directin programming of ES cells down specific pathways of differentiation and controlling the progress of cell type specific cell differentiation in Dictyostelium. The challenge is to understand the full effects of various intercellular signals on the physiology and potential of the cells. In some cases, the differentiations may best be considered as probabilistic changes (Waddington, 1942). In other cases, it might be productive to consider the interplay of kinetic equilibria. There is no simple, linear way to uncover new ways of thinking about multicellular development, but it certainly helps when the test subject is small, simple, rapidly developing and genetically tractable. Dictyostelium may provide surprises and insights pertinent to multicellular differentiation for many years to come.
While considerable progress has been made in the last 10 years to define some of the signaling modes used by Dictyostelium, we have probably only recognized the ones that stand out the most clearly - the tips of the icebergs. Further check points can be expected when more aspects of development are explored. Since there is little or no selection against adding complexity to existing pathways, even the known signaling systems are likely to turn out to be more complicated than presently appreciated. Such subtleties may only be characterized when the pathways are studied in single cells or reconstructed in naive cells.
Highlights.
Cells communicate at almost every stage in development
Signals include peptides, amino acids, nucleic acids, steroids and polyketides
G protein coupled receptors (GPCRs) are prevalent in cell signaling
Positive and negative feedback loops and incoherent feedforward loops play roles
Progress through a dependent sequence is regulated by extracellular signals
ACKNOWLEDGEMENTS
I thank Margarita Behrens for helpful comments on this review. Work in my laboratory is supported by the U.S. National Institutes of Health (PO GM078586).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain
REFERENCES
- Abe H, Uchiyama M, Tanaka Y, Saito H. Structure of discadenine, a spore germination inhibitor from the cellular slime mold Dictyostelium discoideum. Tetrahedron Lett. 1976;42:3807–3810. [Google Scholar]
- Anjard C, Loomis WF. Peptide signaling during terminal differentiation of Dictyostelium. Proc. Natl. Acad. Sci. USA. 2005;102:7607–7611. doi: 10.1073/pnas.0501820102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C, Loomis WF. GABA induces terminal differentiation of Dictyostelium through a GABA(B) receptor. Development. 2006;133:2253–2261. doi: 10.1242/dev.02399. [DOI] [PubMed] [Google Scholar]
- Anjard C, Loomis WF. Cytokinins induce sporulation in Dictyostelium. Development. 2008;135:819–827. doi: 10.1242/dev.018051. [DOI] [PubMed] [Google Scholar]
- Anjard C, Su Y, Loomis WF. Steroids initiate a signaling cascade that triggers rapid sporulation in Dictyostelium. Development. 2009;136:803–812. doi: 10.1242/dev.032607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C, Su Y, Loomis WF. The polyketide MPBD initiates the SDF-1 signaling cascade that coordinates terminal differentiation in Dictyostelium. Eukaryot Cell. 2011;10:956–963. doi: 10.1128/EC.05053-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C, Zeng C, Loomis WF, Nellen W. Signal transduction pathways leading to spore differentiation in Dictyostelium discoideum. Dev. Biol. 1998;193:146–155. doi: 10.1006/dbio.1997.8804. [DOI] [PubMed] [Google Scholar]
- Araki T, Langenick J, Gamper M, Firtel RA, Williams JG. Evidence that DIF-1 and hyper-osmotic stress activate a Dictyostelium STAT by inhibiting a specific protein tyrosine phosphatase. Development. 2008;135:1347–1353. doi: 10.1242/dev.009936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin MB, Saito T, Bowman ME, Haydock S, Kato A, Moore BS, Kay RR, Noel JP. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat. Chem Biol. 2006;2:494–502. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatsalam D, Brock DA, Nikravan NN, Houston KD, Hatton RD, Gomer RH. The secreted Dictyostelium protein CfaD is a chalone. J Cell Sci. 2008;121:2473–2480. doi: 10.1242/jcs.026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthavatsalam D, Choe JM, Hanson NE, Gomer RH. A Dictyostelium chalone uses G proteins to regulate proliferation. BMC Biol. 2009;7:44. doi: 10.1186/1741-7007-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabentos R, Hirose S, Sucgang R, Curk T, Katoh M, Ostrowski EA, Strassmann JE, Queller DC, Zupan B, Shaulsky G, Kuspa A. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr Biol. 2009;19:567–572. doi: 10.1016/j.cub.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger EA, Bozzone DM, Berman MB, Morgenthaler JA, Clark JM. Regulation of discoidin I gene expression in Dictyostelium discoideum by cell-cell contact and cAMP. J. Cell Biochem. 1985;27:391–400. doi: 10.1002/jcb.240270408. [DOI] [PubMed] [Google Scholar]
- Berks M, Kay RR. Cyclic AMP is an inhibitor of stalk cell differentiation in Dictyostelium discoideum. Dev Biol. 1988;126:108–114. doi: 10.1016/0012-1606(88)90244-8. [DOI] [PubMed] [Google Scholar]
- Bonner JT. Evidence for the formation of cell aggregates by chemotaxis in the development of the slime mold Dictyostelium discoideum. J. Exp. Zool. 1947;106:1–26. doi: 10.1002/jez.1401060102. [DOI] [PubMed] [Google Scholar]
- Bonner JT. The cellular slime molds. Princeton, NJ: Princeton Univ. Press; 1959. [Google Scholar]
- Bonner JT. Induction of stalk cell differentiation by cyclic AMP in the cellular slime mold Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 1970;65:110–113. doi: 10.1073/pnas.65.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock DA, Gomer RH. A cell-counting factor regulating structure size in Dictyostelium. Genes Devel. 1999;13:1960–1969. doi: 10.1101/gad.13.15.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdine V, Clarke M. Genetic and physiologic modulation of the prestarvation response in Dictyostelium discoideum. Mol. Biol. Cell. 1995;6:311–325. doi: 10.1091/mbc.6.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai H, Katoh-Kurasawa M, Muramoto T, Santhanam B, Long Y, Li L, Ueda M, Iglesias P, Shaulsky G, Devreotes P. Nucleocytoplasmic shuttling of a GATA transcription factor functions as a development timer. Science. 2014;343(1329):1249531. doi: 10.1126/science.1249531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZH, Schaap P. The prokaryote messenger c-di-GMP triggers stalk cell differentiation in Dictyostelium. Nature. 2012;488:680–683. doi: 10.1038/nature11313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke M, Dominguez N, Yuen IS, Gomer RH. Growing and starving Dictyostelium cells produce distinct density-sensing factors. Dev. Biol. 1992;152:403–406. doi: 10.1016/0012-1606(92)90147-9. [DOI] [PubMed] [Google Scholar]
- Clarke M, Gomer RH. PSF and CMF, autocrine factors that regulate gene expression during growth and early development of Dictyostelium. Experientia. 1995;51:1124–1134. doi: 10.1007/BF01944730. [DOI] [PubMed] [Google Scholar]
- Clarke M, Kayman SC, Riley K. Density-dependent induction of discoidin-I synthesis in exponentially growing cells of Dictyostelium discoideum. Differentiation. 1987;34:79–87. doi: 10.1111/j.1432-0436.1987.tb00053.x. [DOI] [PubMed] [Google Scholar]
- Deery WJ, Gomer RH. A putative receptor mediating cell-density sensing in Dictyostelium. J. Biol Chem. 1999;274:34476–34482. doi: 10.1074/jbc.274.48.34476. [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, Tunggal B, Kummerfeld S, Madera M, Konfortov BA, Rivero F, Bankier AT, Lehmann R, Hamlin N, Davies R, Gaudet P, Fey P, Pilcher K, Chen G, Saunders D, Sodergren E, Davis P, Kerhornou A, Nie X, Hall N, Anjard C, Hemphill L, Bason N, Farbrother P, Desany B, Just E, Morio T, Rost R, Churcher C, Cooper J, Haydock S, vanDriessche N, Cronin A, Goodhead I, Muzny D, Mourier T, Pain A, Lu M, Harper D, Lindsay R, al e. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M, Kaufman M. Establishment in culture of pluripotent cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Firtel RA. Signal transduction pathways controlling multicellular development in Dictyostelium. Trends Genet. (TIG) 1991;7:381–388. doi: 10.1016/0168-9525(91)90260-w. [DOI] [PubMed] [Google Scholar]
- Firtel RA. Interacting signaling pathways controlling multicellular development in Dictyostelium. Curr. Opin. Genet. Devel. 1996;6:545–554. doi: 10.1016/s0959-437x(96)80082-7. [DOI] [PubMed] [Google Scholar]
- Franke J, Kessin RH. The cyclic nucleotide phosphodiesterase inhibitory protein of Dictyostelium discoideum. Purification and characterization. J. Biol. Chem. 1981;256:7628–7637. [PubMed] [Google Scholar]
- Franke J, Kessin RH. The cyclic nucleotide phosphodiesterases of Dictyostelium discoideum: Molecular genetics and biochemistry. Cell. Signal. 1992;4:471–478. doi: 10.1016/0898-6568(92)90016-2. [DOI] [PubMed] [Google Scholar]
- Fukuzawa M. Control of prestalk-cell differentiation by transcription factors. Dev Growth Differ. 2011;53:538–47. doi: 10.1111/j.1440-169X.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Abe T, Williams JG. The Dictyostelium prestalk cell inducer DIF regulates nuclear accumulation of a STAT protein by controlling its rate of export from the nucleus. Development. 2003;130:797–804. doi: 10.1242/dev.00303. [DOI] [PubMed] [Google Scholar]
- Fukuzawa M, Araki T, Adrian I, Williams JG. Tyrosine phosphorylation-independent nuclear translocation of a Dictyostelium STAT in response to DIF signaling. Mol Cell. 2001;7:779–788. doi: 10.1016/s1097-2765(01)00222-2. [DOI] [PubMed] [Google Scholar]
- Garcia R, Ray S, Brown I, Irom J, Brazill D. PakD, a Putative p21-Activated Protein Kinase in Dictyostelium discoideum Regulates Actin. Eukaryot. Cell. 2014;13:119–126. doi: 10.1128/EC.00216-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod DR, Ashworth JM. Effect of growth conditions on development of the cellular slime mould, Dictyostelium discoideum. J. Embryol. Exp. Morphol. 1972;28:463–479. [PubMed] [Google Scholar]
- Gerisch G, Fromm H, Huesgen A, Wick U. Control of cell-contact sites by cyclic AMP pulses in differentiating Dictyostelium cells. Nature. 1975;255:547–549. doi: 10.1038/255547a0. [DOI] [PubMed] [Google Scholar]
- Gerisch G, Wick U. Intracellular oscillations and release of cyclic AMP from Dictyostelium cells. Biochem. Biophys. Res. Commun. 1975;65:364–370. doi: 10.1016/s0006-291x(75)80102-1. [DOI] [PubMed] [Google Scholar]
- Gilbert S. Developmental Biology. Sunderland, Massachusetts: Sinauer Assoc; 2013. [Google Scholar]
- Gomer RH, Jang W, Brazill D. Cell density sensing and size determination. Dev Growth Differ. 2011;53:482–494. doi: 10.1111/j.1440-169X.2010.01248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomer RH, Yuen IS, Firtel RA. A secreted 80×10(3)Mr protein mediates sensing of cell density and the onset of development in Dictyostelium. Development. 1991;112:269–278. doi: 10.1242/dev.112.1.269. [DOI] [PubMed] [Google Scholar]
- Grabel L, Loomis WF. Effector controlling accumulation of N-acteylglucosaminidase during development of Dictyostelium discoideum. Dev. Biol. 1978;64:203–209. doi: 10.1016/0012-1606(78)90072-6. [DOI] [PubMed] [Google Scholar]
- Hirose S, Benabentos R, Ho HI, Kuspa A, Shaulsky G. Self-recognition in social amoebae is mediated by allelic pairs of tiger genes. Science. 2011;333:467–470. doi: 10.1126/science.1203903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho HI, Hirose S, Kuspa A, Shaulsky G. Kin recognition protects cooperators against cheaters. Curr. Biol. 2013;23:1590–1595. doi: 10.1016/j.cub.2013.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Blagg SL, Keller T, Katoh M, Shaulsky G, Thompson CRL. bZlP transcription factor interactions regulate DIF responses in Dictyostelium. Development. 2006;133:449–458. doi: 10.1242/dev.02240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R, Kuspa A, Lilly PJ, Shaulsky G, Levin LR, Loomis WF, Devreotes P. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J. Cell Biol. 1994;126:1537–1545. doi: 10.1083/jcb.126.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranfar N, Fuller D, Loomis WF. Genome-wide expression analyses of gene regulation during early development of Dictyostelium discoideum. Euk. Cell. 2003;2:664–670. doi: 10.1128/EC.2.4.664-670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranfar N, Fuller D, Loomis WF. Transcriptional regulation of post-aggregation genes in Dictyostelium by a feed-forward loop involving GBF and LagC. Dev. Biol. 2006;290:460–469. doi: 10.1016/j.ydbio.2005.11.035. [DOI] [PubMed] [Google Scholar]
- Jang W, Chiem B, Gomer RH. A secreted cell number counting factor represses intracellular glucose levels to regulate group size in Dictyostelium. J. Biol. Chem. 2002;277:39202–39208. doi: 10.1074/jbc.M205635200. [DOI] [PubMed] [Google Scholar]
- Jang W, Gomer RH. Exposure of cells to a cell number-counting factor decreases the activity of glucose-6-phosphatase to decrease intracellular glucose levels in Dictyostelium discoideum. Euk. Cell. 2005;4:72–81. doi: 10.1128/EC.4.1.72-81.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Gomer RH. Combining experiments and modelling to understand size regulation in Dictyostelium discoideum. J R Soc Interface. 2008;5(Suppl 1):S49–s58. doi: 10.1098/rsif.2008.0067.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang W, Schwartz OG, Gomer RH. A cell number counting factor alters cell metabolism. Commun Integr Biol. 2009;2:293–297. doi: 10.4161/cib.2.4.8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Gomer RH. A developmentally regulated cell surface receptor for a density-sensing factor in Dictyostelium. J. Biol. Chem. 1994;269:9128–9136. [PubMed] [Google Scholar]
- Jermyn KA, Duffy KT, Williams JG. A new anatomy of the prestalk zone in Dictyostelium. Nature. 1989;340:144–146. doi: 10.1038/340144a0. [DOI] [PubMed] [Google Scholar]
- Jermyn KA, Williams JG. An analysis of culmination in Dictyostelium using prestalk and stalk-specific cell autonomous markers. Development. 1991;111:779–787. doi: 10.1242/dev.111.3.779. [DOI] [PubMed] [Google Scholar]
- Kay RR. The biosynthesis of differentiation-inducing factor, a chlorinated signal molecule regulating Dictyostelium development. J. Biol. Chem. 1998;273:2669–2675. doi: 10.1074/jbc.273.5.2669. [DOI] [PubMed] [Google Scholar]
- Kay RR, Dhokia B, Jermyn KA. Purification of stalk-cell-inducing morphogens from Dictyostelium discoideum. Eur. J. Biochem. 1983;136:51–56. doi: 10.1111/j.1432-1033.1983.tb07703.x. [DOI] [PubMed] [Google Scholar]
- Kay RR, Jermyn KA. A possible morphogen controlling differentiation in Dictyostelium. Nature. 1983;303:242–244. doi: 10.1038/303242a0. [DOI] [PubMed] [Google Scholar]
- Keller T, Thompson CR. Cell type specificity of a diffusible inducer is determined by a GATA family transcription factor. Development. 2008;135:1635–1645. doi: 10.1242/dev.020883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessin RH, Orlow SJ, Shapiro RI, Franke J. Binding of inhibitor alters kinetic and physical properties of extracellular cyclic AMP phosphodiesterase from Dictyostelium discoideum. Proc. Natl. Acad. Sci. USA. 1979;76:5450–5454. doi: 10.1073/pnas.76.11.5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessin RH. Dictyostelium - Evolution, cell biology, and the development of multicellularity. Cambridge, UK: Cambridge Univ. Press; 2001. [Google Scholar]
- Kim L, Liu JC, Kimmel AR. The novel tyrosine kinase ZAK1 activates GSK3 to direct cell fate specification. Cell. 1999;99:399–408. doi: 10.1016/s0092-8674(00)81526-3. [DOI] [PubMed] [Google Scholar]
- Kimmel AR. Different molecular mechanisms for cAMP regulation of gene expression during Dictyostelium development. Dev. Biol. 1987;122:163–171. doi: 10.1016/0012-1606(87)90342-3. [DOI] [PubMed] [Google Scholar]
- Kinseth MA, Anjard C, Fuller D, Guizzunti G, Loomis WF, Malhotra V. The golgi-associated protein Grasp is required for unconventional protein secretion during development. Cell. 2007;130:524–534. doi: 10.1016/j.cell.2007.06.029. [DOI] [PubMed] [Google Scholar]
- Klein C. Induction of phosphodiesterase by cyclic adenosine 3',5'-monophosphate in differentiating Dictyostelium discoideum. J. Biol. Chem. 1975;250:7134–7138. [PubMed] [Google Scholar]
- Klein P, Vaughan R, Borleis J, Devreotes P. The surface cAMP receptor in Dictyostelium - Levels of ligand-induced phosphorylation, solubilization, identification of primary transcript, and developmental regulation of expression. J. Biol. Chem. 1987;262:358–364. [PubMed] [Google Scholar]
- 1154.Konijn TM, van de Meene JGC, Bonner JT, Barkley DS. The acrasin activity of adenosine-3',5'-cyclic phosphate. Proc. Natl. Acad. Sci. USA. 1967;58:1152. doi: 10.1073/pnas.58.3.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. The genome of Dictyostelium discoideum. Meth. Mol. Biol. 2006;346:15–30. doi: 10.1385/1-59745-144-4:15. [DOI] [PubMed] [Google Scholar]
- Laub MT, Loomis WF. A molecular network that produces spontaneous oscillations in excitable cells of Dictyostelium. Mol. Biol. Cell. 1998;9:3521–3532. doi: 10.1091/mbc.9.12.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence P. The Making of a Fly: The Genetics of Animal Design. Oxford, UK: Blackwell Sci. Pub; 1992. [Google Scholar]
- Loomis WF. A developmental system. New York: Ac. Press; 1975. Dictyostelium discoideum. [Google Scholar]
- Loomis WF. Biochemistry of aggregation in Dictyostelium. Dev. Biol. 1979;70:1–12. doi: 10.1016/0012-1606(79)90002-2. [DOI] [PubMed] [Google Scholar]
- Loomis WF. The development of Dictyostelium discoideum. New York: Ac. ress; 1982. [Google Scholar]
- Loomis WF. Genetic tools for Dictyostelium discoideum. Meth. Cell Biol. 1987;28:31–65. doi: 10.1016/s0091-679x(08)61636-2. [DOI] [PubMed] [Google Scholar]
- Loomis WF. Lateral inhibition and pattern formation in Dictyostelium. Curr. Topics Dev. Biol. 1993;28:1–46. doi: 10.1016/s0070-2153(08)60208-2. [DOI] [PubMed] [Google Scholar]
- Loomis WF. Role of PKA in the timing of developmental events in Dictyostelium cells. Microbiol. Mol. Biol. Rev. 1998;62:684. doi: 10.1128/mmbr.62.3.684-694.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF, Behrens MM, Williams Ma, Anjard C. Pregnenolone sulfate and cortisol induce secretion of acyl CoA binding protein and its conversion into endozepines from astrocytes. J. Biol. Chem. 2010;285:21359–21365. doi: 10.1074/jbc.M110.105858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis WF, White SRD. A sequence of dependent stages in the development of Dictyostelium discoideum. Devel. Biol. 1976;53:171–177. doi: 10.1016/0012-1606(76)90221-9. [DOI] [PubMed] [Google Scholar]
- Maeda M, Aubry L, Insall R, Gaskins C, Devreotes PN, Firtel RA. Seven helix chemoattractant receptors transiently stimulate mitogen-activated protein kinase in Dictyostelium - Role of heterotrimeric G proteins. J. Biol. Chem. 1996;271:3351–3354. doi: 10.1074/jbc.271.7.3351. [DOI] [PubMed] [Google Scholar]
- Maeda M, Lu SJ, Shaulsky G, Miyazaki Y, Kuwayama H, Tanaka Y, Kuspa A, Loomis WF. Periodic signaling controlled by an oscillatory circuit that includes protein kinases ERK2 and PKA. Science. 2004;304:875–878. doi: 10.1126/science.1094647. [DOI] [PubMed] [Google Scholar]
- Maeda M, Sakamoto H, Iranfar N, Fuller D, Maruo T, Ogihara S, Morio T, Urushihara H, Tanaka Y, Loomis WF. Changing patterns of gene expression in Dictyostelium prestalk cell subtypes recognized by in situ hybridization with genes from microarray analyses. Euk. Cell. 2003;2:627–637. doi: 10.1128/EC.2.3.627-637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SK, Firtel RA. Two-phase regulatory pathway controls cAMP receptor-mediated expression of early genes in Dictyostelium. Proc. Natl. Acad. Sci. USA. 1989;86:1924–1928. doi: 10.1073/pnas.86.6.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SKO, Brown JM, Briscoe C, Parent C, Pitt G, Devreotes PN, Firtel RA. Role of cAMP-dependent protein kinase in controlling aggregation and postaggregative development in Dictyostelium. Dev. Biol. 1997;183:208–221. doi: 10.1006/dbio.1996.8499. [DOI] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. Cyclic AMP regulation of early gene expression in Dictyostelium discoideum: Mediation via the cell surface cyclic AMP receptor. Mol. Cell. Biol. 1987;7:458–469. doi: 10.1128/mcb.7.1.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann SKO, Firtel RA. A developmentally regulated, putative serine/threonine protein kinase is essential for development in Dictyostelium. Mech. Devel. 1991;35:89–101. doi: 10.1016/0925-4773(91)90060-j. [DOI] [PubMed] [Google Scholar]
- Marin FT. Regulation of development in Dictyostelium discoideum: I. Initiation of the growth to developmental transition by amino acid starvation. Dev. Biol. 1976;48:110–117. doi: 10.1016/0012-1606(76)90050-6. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok DW, Mok MC. Cytokinin metabolism and action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:89–118. doi: 10.1146/annurev.arplant.52.1.89. [DOI] [PubMed] [Google Scholar]
- Morris HR, Taylor GW, Masento MS, Jermyn KA, Kay RR. Chemical structure of the morphogen differentiation inducing factor from Dictyostelium discoideum. Nature. 1987;328:811–814. doi: 10.1038/328811a0. [DOI] [PubMed] [Google Scholar]
- Morrissey JH, Devine KM, Loomis WF. The timing of cell-type specific differentiation in Dictyostelium discoideum. Dev. Biol. 1984;103:414–424. doi: 10.1016/0012-1606(84)90329-4. [DOI] [PubMed] [Google Scholar]
- Neumann CS, Walsh CT, Kay RR. A flavin-dependent halogenase catalyzes the chlorination step in the biosynthesis of Dictyostelium differentiation-inducing factor 1. Proc Natl Acad Sci U S A. 2010;107:5798–5803. doi: 10.1073/pnas.1001681107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell PC, Williams KL, Kuspa A, Loomis WF. Genetic map of Dictyostelium. In: O'Brien SJ, editor. Genetic maps: locus maps of complex genomes. 6th edition. CSH Lab. Press, Cold Spring Harbor; 1993. pp. 3.1–3.10. [Google Scholar]
- Norman AW, Mizwicki MT, Norman DP. Steroid-hormone rapid actions, membrane receptors and a conformational ensemble model. Nat Rev Drug Discov. 2004;3:27–41. doi: 10.1038/nrd1283. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- Orlow SJ, Shapiro I, Franke J, Kessin RH. The extracellular cyclic nucleotide phosphodiesterase of Dictyostelium discoideum. Purification and characterization. J. Biol. Chem. 1981;256:7620–7627. [PubMed] [Google Scholar]
- Parikh A, Miranda ER, Katoh-Kurasawa M, Fuller D, Rot G, Zagar L, Curk T, Sucgang R, Chen R, Zupan B, Loomis WF, Kuspa A, Shaulsky G. Conserved developmental transcriptomes in evolutionarily divergent species. Genome Biol. 2010;11:R35. doi: 10.1186/gb-2010-11-3-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne SH, Loomis WF. Retention and loss of amino acid biosynthetic pathways based on analysis of whole-genome sequences. Euk. Cell. 2006;5:272–276. doi: 10.1128/EC.5.2.272-276.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J, Gomer R. The p21-activated kinase (PAK) family member PakD is required for chemorepulsion and proliferation inhibition by autocrine signals in Dictyostelium discoideum. PLoS One. 2014 doi: 10.1371/journal.pone.0096633. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Huang E, Shaulsky G, Gomer RH. The putative bZIP transcription factor BzpN slows proliferation and functions in the regulation of cell density by autocrine signals in Dictyostelium. PLoS One. 2011;6:e21765. doi: 10.1371/journal.pone.0021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prossnitz ER, Arterburn JB, Sklar LA. GPR30: A G protein-coupled receptor for estrogen. Mol. Cell Endocrinol. 2007;265:138–142. doi: 10.1016/j.mce.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raper KB. Dictyostelium discoideum, a new species of slime mold from decaying forest leaves. J. Agr. Res. 1935;50:135–147. [Google Scholar]
- Raper KB. Pseudoplasmodium formation and organization in Dictyostelium discoideum. J. Elisha Mitchell Sci. Soc. 1940;56:241–282. [Google Scholar]
- Raper KB. The Dictyostelids. Princeton, NJ: Princeton Univ. Press; 1984. [Google Scholar]
- Reymond CD, Schaap P, Veron M, Williams JG. Dual role of cAMP during Dictyostelium development. Experientia. 1995;51:1166–1174. doi: 10.1007/BF01944734. [DOI] [PubMed] [Google Scholar]
- Richardson DL, Loomis WF, Kimmel AR. Progression of an inductive signal activates sporulation in Dictyostelium discoideum. Development. 1994;120:2891–2900. doi: 10.1242/dev.120.10.2891. [DOI] [PubMed] [Google Scholar]
- Rossier C, Franke J, Mullens IA, Kessin RH. Regulation of the mRNA for the inhibitor of extracellular cyclic AMP phosphodiesterase of Dictyostelium discoideum. Experientia. 1983;39:672. doi: 10.1111/j.1432-1033.1983.tb07474.x. [DOI] [PubMed] [Google Scholar]
- Saito T, Kato A, Kay RR. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev Biol. 2008;317:444–453. doi: 10.1016/j.ydbio.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Taylor GW, Yang JC, Neuhaus D, Stetsenko D, Kato A, Kay RR. Identification of new differentiation inducing factors from Dictyostelium discoideum. Biochim. Biophys. Acta. 2006;1760:754–761. doi: 10.1016/j.bbagen.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Schaap P. Evolution of developmental cyclic adenosine monophosphate signaling in the Dictyostelia from an amoebozoan stress response. Dev Growth Differ. 2011;53:452–462. doi: 10.1111/j.1440-169X.2011.01263.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilde C, Sschaap P. The amoebozoa. Methods Mol Biol. 2013;983:1–15. doi: 10.1007/978-1-62703-302-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulkes C, Schaap P. cAMP-dependent protein kinase activity is essential for preaggregative gene expression in Dictyostelium. FEBS Lett. 1995;368:381–384. doi: 10.1016/0014-5793(95)00676-z. [DOI] [PubMed] [Google Scholar]
- Skoge M, Yue H, Erickstad M, Bae A, Levine H, Groisman A, Loomis WF, Rappel W-J. Cellular memory in eukaryotic chemotaxis. 2014 doi: 10.1073/pnas.1412197111. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderbom F, Anjard C, Iranfar N, Fuller D, Loomis WF. An adenylyl cyclase that functions during late development of Dictyostelium. Development. 1999;126:5463–5471. doi: 10.1242/dev.126.23.5463. [DOI] [PubMed] [Google Scholar]
- Soderbom F, Loomis WF. Cell-cell signaling during Dictyostelium development. Trends Microbiol. 1998;6:402–406. doi: 10.1016/s0966-842x(98)01348-1. [DOI] [PubMed] [Google Scholar]
- Souza GM, da Silva AM, Kuspa A. Starvation promotes Dictyostelium development by relieving PufA inhibition of PKA translation through the YakA kinase pathway. Development. 1999;126:3263–3274. doi: 10.1242/dev.126.14.3263. [DOI] [PubMed] [Google Scholar]
- Souza GM, Lu S, Kuspa A. YakA, a protein kinase required for the transition from growth to development in Dictyostelium. Development. 1998;125:2291–2302. doi: 10.1242/dev.125.12.2291. [DOI] [PubMed] [Google Scholar]
- Sucgang R, Weijer CJ, Siegert F, Franke J, Kessin RH. Null mutations of the Dictyostelium cyclic nucleotide phosphodiesterase gene block chemotactic cell movement in developing aggregates. Dev. Biol. 1997;192:181–192. doi: 10.1006/dbio.1997.8720. [DOI] [PubMed] [Google Scholar]
- Swaney KF, Huang CH, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. doi: 10.1146/annurev.biophys.093008.131228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takeda K, Shao D, Adler M, Charest P, Loomis WF, Levine H, Groisman A, Rappel W-J, Firtel RA. Incoherent feedforward control governs adaptation of activated Ras in a eukaryotic chemotaxis pathway. Science Signalling. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Shao D, Adler M, Charest P, Loomis WF, Levine H, Groisman A, Rappel W-J, Firtel RA. Incoherent feedforward control governs adaptation of activated Ras in a eukaryotic chemotaxis pathway. Science Signaling. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K, Shao D, Adler M, Charest P, Loomis WF, Levine H, Groisman A, Rappel W-J, Firtel RA. Incoherent feedforward control governs adaptation of activated Ras in a eukaryotic chemotaxis pathway. SSci. Signaling. 2012;5:ra2. doi: 10.1126/scisignal.2002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Abe H, Uchiyama M, Taya Y, Nishimura S. Isopentenyladenine from Dictyostelium discoideum. Phytochemistry. 1978;17:543–544. [Google Scholar]
- Tang L, Gao T, McCollum C, Jang W, Vicker MG, Ammann RR, Gomer RH. A cell number-counting factor regulates the cytoskeleton and cell motility in Dictyostelium. Proc. Natl. Acad. Sci. USA. 2002;99:1371–1376. doi: 10.1073/pnas.022516099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CRL, Fu Q, Buhay C, Kay RR, Shaulsky G. A bZIP/bRLZ transcription factor required for DIF signaling in Dictyostelium. Development. 2004;131:513–523. doi: 10.1242/dev.00939. [DOI] [PubMed] [Google Scholar]
- Thompson CRL, Kay RR. The role of DIF-1 signaling in Dictyostelium development. Mol. Cell. 2000;6:1509–1514. doi: 10.1016/s1097-2765(00)00147-7. [DOI] [PubMed] [Google Scholar]
- Thomson J, Itskovitz-Eldor J, Shapiro S, Waknitz M, Swiergiel J, Marshall V, Jones J. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Tomchik KJ, Devreotes PN. Adenosine 3',5'-monophosphate waves in Dictyostelium discoideum: A demonstration by isotope dilution-fluorography technique. Science. 1981;212:443–446. doi: 10.1126/science.6259734. [DOI] [PubMed] [Google Scholar]
- Town CD, Gross JD, Kay RR. Cell differentiation without morphogenesis in Dictyostelium discoideum. Nature. 1976;262:717–719. doi: 10.1038/262717a0. [DOI] [PubMed] [Google Scholar]
- Velazquez F, Peak-Chew SY, Fernandez IS, Neumann CS, Kay RR. Identification of a eukaryotic reductive dechlorinase and characterization of its mechanism of action on its natural substrate. Chem Biol. 2011;18:1252–1260. doi: 10.1016/j.chembiol.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH. Canalization of development and the inheritance of acquired characters. Nature. 1942;150:563–565. doi: 10.1038/1831654a0. [DOI] [PubMed] [Google Scholar]
- Wang J, Hou LS, Awrey D, Loomis WF, Firtel RA, Siu CH. The membrane glycoprotein gp150 is encoded by the lagC gene and mediates cell-cell adhesion by heterophilic binding during Dictyostelium development. Dev. Biol. 2000;227:734–745. doi: 10.1006/dbio.2000.9881. [DOI] [PubMed] [Google Scholar]
- Wang N, Shaulsky G, Escalante R, Loomis WF. A two-component histidine kinase gene that functions in Dictyostelium development. EMBO J. 1996;15:3890–3898. [PMC free article] [PubMed] [Google Scholar]
- Wang N, Soderbom F, Anjard C, Shaulsky G, Loomis WF. SDF-2 induction of terminal differentiation in Dictyostelium discoideum is mediated by the membrane-spanning sensor kinase DhkA. Mol. Cell. Biol. 1999;19:4750–4756. doi: 10.1128/mcb.19.7.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y, Kay RR, Bloomfield G, Ross S, Ivens A, Williams JG. A new Dictyostelium prestalk cell sub-type. Dev Biol. 2010;339:390–397. doi: 10.1016/j.ydbio.2009.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C, Okano H, Hui S, Zhang Z, Gunderson CW, Wang YP, Lenz P, Yan D, Hwa T. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature. 2013;500:301–306. doi: 10.1038/nature12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik M, Smuga-Otto K, Antosiewicz-Bourget J, Frane J, Tian S, Nie J, Jonsdottir G, Ruotti V, Stewart R, Slukvin I, Thomson J. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Yuen IS, Jain R, Bishop JD, Lindsey DF, Deery WJ, van Haastert PJM, Gomer RH. A density-sensing factor regulates signal transduction in Dictyostelium. J. Cell Biol. 1995;129:1251–1262. doi: 10.1083/jcb.129.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Charest PG, Firtel RA. Spatiotemporal regulation of Ras activity provides directional sensing. Curr Biol. 2008;18:1587–1593. doi: 10.1016/j.cub.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhukovskaya NV, Fukuzawa M, Yamada Y, Araki T, Williams JG. The Dictyostelium bZIP transcription factor DimB regulates prestalk-specific gene expression. Development. 2006;133:439–448. doi: 10.1242/dev.02190. [DOI] [PubMed] [Google Scholar]