Abstract
Control of pre-mRNA splicing is a critical part of the eukaryotic gene expression process. Extensive evidence indicates that transcription and splicing are spatiotemporally coordinated and that most splicing events occur co-transcriptionally. A kinetic coupling model has been proposed in metazoans to describe how changing RNA Polymerase II (RNAPII) elongation rate can impact which alternative splice sites are used. In Saccharomyces cerevisiae, in which most spliced genes have only a single intron and splice sites adhere to a strong consensus sequence, we recently observed that splicing efficiency was sensitive to mutations in RNAPII that increase or decrease its elongation rate. Our data revealed that RNAPII speed and splicing efficiency are generally anti-correlated: at many genes, increased elongation rate caused decreased splicing efficiency, while decreased elongation rate increased splicing efficiency. An improved splicing phenotype was also observed upon deletion of SUB1, a condition in which elongation rate is slowed. We discuss these data in the context of a growing field and expand the kinetic coupling model to apply to both alternative splicing and splicing efficiency.
Keywords: RNA polymerase II, elongation rate, kinetic coupling, splicing, splicing efficiency
Splicing Control Begins Co-transcriptionally
Splicing is the process by which an intron is removed from a pre-mRNA transcript, and the flanking exons are ligated together. Removal of each intron proceeds via numerous consecutive steps, including commitment of a pre-mRNA to splicing by binding of splicing factors to the 5′ splice site and the branch site/3′ splice site, multiple conformational rearrangements, and two catalytic steps.1,2 Splicing in metazoans is particularly complex, as most genes contain multiple introns flanked by weak splice sites, which can give rise to alternatively spliced transcripts. In S. cerevisiae, also known as budding yeast, most spliced genes have only a single intron defined by strong, constitutive (as opposed to alternative) splice sites.
While splicing can occur in a minimal in vitro system, independent of transcription, a major outstanding question in the field is how splicing proceeds in vivo. In both metazoans and budding yeast, splicing factors bind many transcripts while they are still associated with elongating RNA polymerase II (RNAPII), i.e. co-transcriptionally. When assayed by chromatin immunoprecipitation (ChIP), a splicing factor co-immunoprecipitates DNA several hundred base pairs downstream of its encoded binding site, presumably via a nascent transcript and RNAPII.3-7 In budding yeast, while many intron-containing genes (ICGs) are associated with early splicing factors,7 it has proven difficult to measure association of later splicing factors, presumably due to short second (downstream) exons.7,8 However, splicing products have been observed in association with RNAPII,9 and a genome-wide analysis of chromatin-associated transcripts showed that most transcripts are fully spliced before they are released from the chromatin template,10 as is also generally the case in metazoans.11
Several pieces of evidence have pointed to the functional importance of all or part of the splicing process occurring co-transcriptionally. Experiments in budding yeast showed that intron-containing reporter genes with high co-transcriptional association of early-acting splicing factors also exhibit efficient splicing, as measured by the fraction of spliced vs. unspliced mRNA.8 Computational modeling of splicing and transcription using kinetic parameters determined from splicing reporters also predicts that the full splicing reaction is more efficient when occurring co-transcriptionally, as opposed to post-transcriptionally, i.e. after transcription termination.12 Because the retention of an intron promotes transcript degradation via nonsense-mediated decay, thus abrogating its translation into protein, the efficiency of splicing that occurs co-transcriptionally can have profound phenotypic consequences on the cell.
A specific functional connection between transcription by RNAPII and splicing has been referred to as “kinetic coupling,” as the timing of one reaction influences the outcome of a second reaction. A kinetic coupling model was originally proposed in metazoa to describe how perturbing the RNAPII elongation rate can influence alternative splicing. Because the transcribed splice sites emerge from RNAPII in the order defined by transcription (5′ to 3′) and with the timing defined by elongation rate, slowing RNAPII elongation in the vicinity of a weak upstream splice site affords more time for it to be recognized by splicing factors before a competing downstream splice site is transcribed.13 In eukaryotes, transcription occurs in the context of nucleosome-bound DNA, or chromatin. The chromatin environment at any given locus is complex and is defined by variables such as post-translational modifications of histones, nucleosome positioning, and nucleosome remodeling, that together affect RNAPII elongation rate in gene- and intron-specific ways. It is likely because of the chromatin–RNAPII elongation connection that factors such as histone acetylation and nucleosome density are correlated with alternative splicing patterns.14 Earlier experiments in budding yeast using a two-intron gene in which the splice sites have been made artificially weak showed that kinetic coupling is not a phenomenon restricted to metazoans. These experiments revealed that slowing RNAPII elongation favored the use of a weaker upstream splice site,15 as was also observed in metazoans.
More generally, while the number of ICGs in budding yeast is only several hundred of ~6000, the gene ontology categories enriched within this list a priori suggest that splicing may have been retained in this species as a regulatory node within the broader gene expression process. Indeed, the efficiency of the splicing reaction has been shown to be regulated in some cases, in which resulting protein levels are affected. For example, several meiotic transcripts are spliced only during progression through meiosis.16,17 In addition, splicing efficiency of ribosomal protein gene transcripts rapidly decreases upon amino acid starvation, leading to the intron being retained in these transcripts.18,19 Whether this regulated splicing involves coordination with RNAPII elongation rate is unknown; however, more generally, these and other data argue that splicing efficiency in budding yeast makes for a useful system for investigating the nature of kinetic coupling.
Budding yeast as a system offers an unrivalled opportunity to combine genetics and whole-genome splicing analyses to ask whether, and if so, how the efficiency of the splicing reaction is kinetically coupled to transcription speed. We were able to investigate multiple spatiotemporal facets of kinetic coupling—not only by directly modulating transcription elongation rate, but also using high-throughput genetic interaction mapping to begin to interrogate aspects of the co-transcriptional environment that impact kinetic coupling.
Splicing Efficiency is Sensitive to RNAPII Elongation Rate
We recently examined splicing in the context of an allelic series of point mutants in the active site of RNAPII20 known to transcribe in vitro21 at an average rate of ~1.5 nt/s (Rpb1-H1085Q), ~5 nt/s (Rpb1-F1086S), ~30 nt/s (Rpb1-E1103G), and > 50 nt/s (Rpb1-G1097D), compared with ~12 nt/s for wild-type RNAPII.
There are several ways to measure splicing efficiency; here we employed splicing-specific microarrays19 to measure the effect of these RNAPII mutants on the abundance of the intron, exon, and exon–exon junction from intron-containing genes. Mutations in splicing factors tend to exhibit large increases in the abundance of intron, with concomitant decreases in the junction.22 While we acknowledge that additional factors such as pre-mRNA-specific degradation could impact this ratio (see below), we here interpret this readout to mean that the pre-mRNA is spliced less efficiently.
In a strain with the E1103G mutation, which causes a moderately fast elongation rate in vitro,21,23 we observed a phenotype consistent with a decrease in splicing efficiency: many genes exhibited an increase in the intron feature (reflecting an accumulation of pre-mRNA), intron/junction ratio (indicating increased pre-mRNA relative to mature mRNA), and intron/exon (indicating accumulation in pre-mRNA relative to overall expression changes).20 A strain harboring the G1097D point mutation,20 which causes an even faster elongation rate in vitro,21 exhibited an even more extreme splicing defect.
These results were consistent with a model in which splice sites are less efficiently utilized in strains with a fast RNAPII elongation rate, and accordingly, we suspected that strains with a slower elongation rate would exhibit improved splicing. Indeed, when we analyzed splicing in a strain with the F1086S mutation, which caused RNAPII elongation to be moderately slow in vitro,24 we observed decreases in the intron feature, intron/junction ratio, and intron/exon ratio at a large number of genes, all of which are consistent with increased splicing efficiency. We observed an even higher efficiency of splicing in cells harboring the H1085Q mutation in RNAPII,20 which elongates at only ~1.5 nt/s in vitro.21
To confirm that these effects were the result of changing RNAPII speed rather than being an indirect consequence of the specific point mutations used, we analyzed splicing in two double mutant strains, in which mutations that individually cause RNAPII to elongate quickly or slowly in vitro together return RNAPII elongation rate to very near wild-type rate.21 Splicing in these strains was very similar to that in a wild-type strain,20 further arguing for a direct relationship between elongation rate and splicing efficiency.
Interestingly, Khodor et al. showed in Drosophila melanogaster that splicing efficiency of some introns improves upon expression of a mutant RNAPII with lower processivity,25 a result that is consistent with ours. To the best of our knowledge, our study represents the first instance where an RNAPII genetically engineered to elongate at a range of speeds was assayed with respect to its genome-wide effects on splicing in vivo.
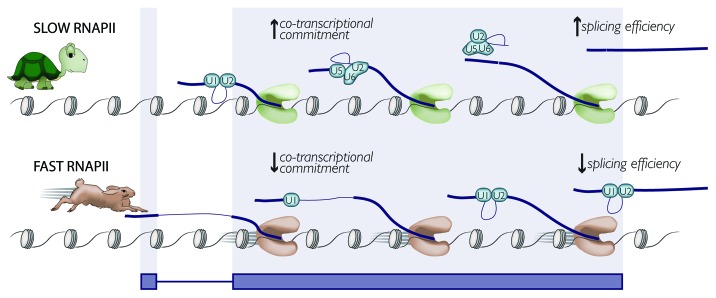
In a general sense, the opposite splicing phenotypes that occur in strains with fast vs. slow RNAPII elongation suggest that, although transcripts do not undergo alternative splicing in budding yeast, splicing efficiency is nonetheless kinetically coupled to transcription (Fig. 1). Importantly, the effects we observed on splicing were most extreme in the RNAPII mutants with the fastest and slowest elongation rates, arguing that kinetic coupling occurs in budding yeast over a ~40-fold range of elongation rates. Our results support the notion that splicing efficiency in yeast (i.e. degree of intron retention) is a bona fide readout of kinetic coupling.
Figure 1. Hypothetical model of how RNAPII elongation rate impacts splicing efficiency. See text for details.
How Might Kinetic Coupling Occur in S. cerevisiae?
Is splicing in competition with termination?
As RNAPII approaches the 3′ end of each gene, nascent transcripts are cleaved from RNAPII and polyadenylated, in a process that is tightly coupled with transcription termination.26 In systems where alternative splicing takes place, altered RNAPII elongation rate impacts the temporal window of opportunity for splicing using upstream vs. downstream splice sites; in the case of budding yeast, it logically follows that co-transcriptional splicing is instead in competition with transcription termination/3′ end cleavage. Consistent with this notion, it was concluded based on sequencing of chromatin-associated transcripts and RNAPII ChIP that genes with high levels of co-transcriptional splicing are those at which RNAPII slows near the 3′ end.10 This was interpreted as evidence for a mechanism to promote co-transcriptional splicing catalysis by delaying transcription termination.
These same experiments revealed that in a wild-type cell, half of intron-containing genes are > 74% spliced while chromatin-associated.10 Despite this abundance of co-transcriptional splicing, this number implies that completion of splicing is not a prerequisite for release from the 3′ end. While we observed changes in splicing efficiency using whole cell RNA as input,20 we might predict that changes in RNAPII elongation rate may alter the fraction of transcripts that are fully spliced before release from the chromatin (Fig. 1). The change in this fraction upon alteration of RNAPII elongation rate cannot be determined on a genome-wide scale without an initial enrichment for nascent transcripts, and thus, poses an important question for the future.
Experiments on splicing of terminal introns in metazoa have revealed that splicing and 3′ end processing may be coordinated.27-30 This could explain why mutants in the 3′ end processing and termination machinery tend to cause splicing defects.31-33 However, if splicing and termination are in direct competition, it should be possible to identify mutants in the transcription termination machinery that would allow more time for splicing completion, in the same way a slow RNAPII mutant does, but without interfering with the coupling between those processes. Furthermore, experiments could be designed to test whether modulating the length of the last exon in yeast would alter the proportion of transcripts that are spliced prior to transcription termination.
An additional possibility is that alterations in RNAPII elongation rate will change the location within the gene at which splicing factor association occurs: nascent transcripts from the faster RNAPII mutants would exhibit lower splicing factor occupancy by ChIP, or a 3′ shift in association, whereas splicing factors in a slow RNAPII mutant would associate with nascent transcripts with a more 5′ distribution (Fig. 1).
Why are the effects on splicing gene-specific?
Our conclusion that transcription rate and splicing efficiency are generally anti-correlated was based on a sizeable subset of genes we term “kinetically coupled,” whose splicing was hindered with an RNAPII mutant with increased elongation rate and enhanced with an RNAPII mutant with decreased elongation rate. Notably, however, not all genes conformed to this trend, provoking the question of what the difference is between genes that responded reciprocally to RNAPII speed and those that did not.
The metazoan model of kinetic coupling would suggest that this gene specificity could come from differences in splice site strength. However, unlike in metazoans, ICGs in budding yeast generally have very strong splice sites that do not diverge far from the consensus; in any case, the introns whose splicing were most kinetically coupled to RNAPII elongation were not enriched for those bearing weak splice sites (data not shown). Munding, et al. have discovered that, in certain settings, spliceosome abundance can be a limiting factor for the splicing of some transcripts.34 Whole transcriptome analyses of RNA from the RNAPII mutant strains showed that the total abundance of RNAPII-transcribed RNA was not altered in these strains;20 thus, we do not think it likely that a limitation in spliceosome availability per se accounts for the connection we observed between polymerase speed and splicing.
In trying to understand the nature of gene-specific effects of RNAPII elongation rate on splicing efficiency, it is important to note that wild-type RNAPII rate likely varies from gene to gene to begin with. At present, assays for measuring genome-wide RNAPII elongation rate in live budding yeast are lacking. The E1103G mutation confers a fast elongation rate in vivo on the GAL1 gene,35 thus strengthening the prediction that in vivo rates should generally mirror those measured in vitro. If we could directly measure gene-specific elongation rates in vivo, perhaps the kinetically coupled genes would be the only genes at which RNAPII elongation rate is altered in the direction and to the degree expected based on in vitro elongation measurements. Alternatively, perhaps the genes that did not respond reciprocally to changes in RNAPII rate have an epistatic mechanism for maintaining normal RNAPII elongation rate even in the context of the RNAPII mutations. Furthermore, identifying pause sites genome-wide9 in these strains could reveal locus-specific effects on RNAPII elongation, and would contribute to a better understanding of the effect of polymerase pauses on co-transcriptional splicing.36
An important aspect of the integration between transcription and splicing is doubtless the chromatin environment in which both processes occur. In fact, it has long been understood that the chromatin environment—nucleosome distribution, histone modifications, and transcription factors more generally—confers gene-specific regulation of transcription initiation, elongation, and termination. Thus, an intriguing possibility is that the kinetically coupled genes we identified exist within chromatin environments that promote co-transcriptional spliceosome assembly and/or catalysis. Direct physical recruitment of splicing factors to histones bearing specific modifications has been reported in metazoans.37,38 Factors in the chromatin environment may directly or indirectly promote spliceosomal rearrangements that lead to a productive splicing reaction, as proposed in budding yeast for the cycle of histone acetylation and deacetylation.39,40
Additional histone post-translational modifiers in yeast39-42 and metazoa43-45 affect splicing via mechanisms that remain poorly understood. These modifiers could act directly, as in the above examples, or indirectly, via altering RNAPII elongation in the vicinity of encoded splice sites. This latter example has been observed in a variety of scenarios, including histone acetylation level, positioning of nucleosomes, and slowing of RNAPII due to localized DNA damage.46
Identifying transcription rate-sensitive factors
An important experimental goal is to identify additional factors that regulate RNAPII elongation rate. In our study, we relied on the power of yeast genetics and performed high-throughput genetic interaction mapping, in which double-mutant strains were generated by crossing a given RNAPII elongation rate mutant to an additional strain carrying a mutation in one of 1200 genes. The growth of the resulting double-mutant strains was then measured as a proxy for fitness. Two gene subsets of interest were identified by this analysis: (1) those whose mutation suppressed the growth defect of the fast RNAPII mutants and exacerbated the growth defect of the slow mutants; (2) vice versa (mutation exacerbated growth in fast mutants and suppressed the growth defect of slow mutants).20 We proposed that these factors may either be sensitive to elongation rate, or might themselves directly influence RNAPII rate. We reasoned that this genetic data set could provide a list of candidate factors that influence RNAPII dynamics, and therefore, splicing, in site- or gene-specific ways. We discuss some of these candidates below.
Transcription factors
We identified two members of the PAF-complex (Rtf1 and Cdc73) whose deletion adversely impacted growth in strains harboring a slow RNAPII mutant, but suppressed the growth defect of the fast RNAPII mutants.20 These genetic data are consistent with the known role of the PAF-C as a positively acting transcription factor that coordinates many events during elongation,47 and led us to suspect that strains lacking this complex would have slow elongation and improved splicing. As it turns out, however, rtf1∆ and cdc73∆ strains do not exhibit slow RNAPII elongation.48 And interestingly, two separate reports have shown that the PAF-C actually promotes splicing, as both observe data consistent with a splicing defect in strains lacking a functional PAF-C.33,49
An additional factor, like members of the PAF-C, whose deletion caused synthetic lethality with the slow RNAPII alleles is Sub1, a gene so named due to its role in transcription initiation as a suppressor of TFIIB.50 These genetic data prompted us to hypothesize that the sub1∆ strain might exhibit a slow elongation rate, which was confirmed by Garcia, et al. while the work by Braberg, et al. was under review: RNAPII elongates slowly on the GAL1 gene in the absence of SUB1.51 Consistent with our predictions, the sub1∆ strain exhibited improved splicing at a large number of genes, similar to what is observed in slow RNAPII mutant strains. It was previously reported that Sub1 localizes to ribosomal protein genes,52 and consistent with this, the genes whose splicing is improved by deletion of SUB1 are enriched for genes encoding the cytosolic ribosome (data not shown). Therefore, the gene-specificity conferred by Sub1 localization may account for its intron specificity.
Histone modifiers
Histone acetylation has been shown to promote transcription through nucleosomal templates,53 a function opposed by histone deacetylation; therefore, complexes harboring these histone acetyltransferases and deacetylases are attractive candidates for gene-specific regulators of RNAPII elongation rate and splicing efficiency. In fact, connections between histone acetylation and splicing have previously been observed.46,39,40 Our genetic interaction mapping identified SAS3, SAS4, and ESA1 (components of NuA3, SAS, and NuA4 histone acetyltransferase complexes, respectively) as causing synthetic lethality in combination with the fast RNAPII point mutant alleles. These data are tantalizing, but to the best of our knowledge, these complexes have not been examined with respect to their gene-specific effects on splicing.
The existence of gene-specific histone modifiers and RNAPII elongation regulators raises the possibility that co-transcriptional splicing is regulated in a gene-specific fashion, depending on the nature of the co-transcriptional environment at each gene. In metazoans, this specificity has been proposed to occur at specific introns within genes.14
Current Questions and Future Directions
Many questions remain about how splicing is regulated co-transcriptionally, and additional factors, not discussed above, are clearly also at play. For instance, not all splicing factors promote splicing upon binding a pre-mRNA—auxiliary splicing regulatory factors can also inhibit the use of specific splice sites. Thus, while the co-transcriptional nature of splicing has been generally thought to improve the recognition of transcribed splice sites,8,12,54 perhaps it is the case that co-transcriptional splicing allows for broader regulation, both positive and negative, from the chromatin environment.
Because completion of splicing often occurs co-transcriptionally, the quality control mechanisms required for splicing fidelity, i.e. the accurate recognition of exon/intron junctions in pre-mRNAs,1 must perforce operate co-transcriptionally as well. This raises the question of whether there is input from RNAPII or other co-transcriptional elements that promotes splicing fidelity. A transcription–splicing checkpoint has been proposed, during which RNAPII pauses briefly at the 3′ splice site, and which may promote splicing fidelity.36 Furthermore, the cycle of histone acetylation and deacetylation has been associated with specific spliceosomal rearrangements,40 and leaves open the possibility that each step in splicing is associated with a chromatin-specific quality control mechanism. In strains where RNAPII elongation is too fast or too slow, do these quality control steps occur faithfully? We are currently testing whether splicing fidelity is improved or hampered in strains harboring fast or slow RNAPII mutant alleles.
Early experiments showed that mRNA secondary structure can impact alternative splicing via occlusion of alternative splice sites.55 RNA secondary structure may be stabilized by kinetic trapping during transcription,56 and therefore, may be sensitive to RNAPII elongation rate. The degree to which RNA secondary structure changes in the RNAPII point mutant strains would be of great interest to determine, and is now possible given recent advances in mapping secondary structure in vivo.57,58
It also remains unknown whether the intron accumulation we observed by microarray in strains with fast RNAPII elongation20 is attributable to pre-mRNAs on which splicing factors have not acted, or to stalled splicing events that might have been released as splicing intermediate products, destined to be discarded from the spliceosome and degraded. Given recent results supporting extensive quality control by way of tight coupling between splicing and RNA degradation,59-61 it is possible that the altered abundances of pre-mRNA and mature mRNA we observed are also due to changes in decay rates of those species. It would be of much interest to inquire about kinetic coupling in the context of strains with defective RNA decay, and this represents an interesting future direction.
In the future, the development of genome-wide assays to analyze the multiple steps in splicing—not only commitment to splicing, but both steps of splicing catalysis, will be critical for studying co-transcriptional splicing regulation. Furthermore, splicing has been called a co-transcriptional process, with the implication that it takes place prior to transcription termination.11 In some cases, transcripts are retained at the 3′ end of a chromatin locus after transcription termination,62-64 and therefore, are still potentially subject to the same regulatory processes as during transcription elongation. With recent advances in chromatin-associated and RNAPII-associated transcript sequencing,9,10,65,66 the way co-transcriptionality is operationally defined may change.
That splicing and transcription occur in the same space and time has allowed for functional coupling to occur. This has been revealed in experiments by us and others in which RNAPII elongation is directly or indirectly altered and leads to the idea that the cell utilizes this coupling to maintain an efficient gene expression program or rapidly adapt this program in response to environmental variables. An exciting example of this from human neuronal culture shows a direct link between chromatin modification and alternative splicing, via an effect on RNAPII elongation rate.67 As additional factors that influence splicing efficiency are defined, and assays to better characterize the timing and nature of the co-transcriptional splicing reaction are developed, we anticipate that a more integrated picture of how individual steps in gene expression function together will emerge.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of the Guthrie and Krogan labs, in particular, Anne de Bruyn Kops, Jaclyn Greimann, Kelly Nissen, Kristin Patrick, and Argenta Price, as well as the anonymous reviewer, for excellent comments and suggestions for improving the manuscript.
References
- 1.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–26. doi: 10.1016/S0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 2.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–18. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Listerman I, Sapra AK, Neugebauer KM. Cotranscriptional coupling of splicing factor recruitment and precursor messenger RNA splicing in mammalian cells. Nat Struct Mol Biol. 2006;13:815–22. doi: 10.1038/nsmb1135. [DOI] [PubMed] [Google Scholar]
- 4.Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23:5768–79. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacadie SA, Rosbash M. Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5’ss base pairing in yeast. Mol Cell. 2005;19:65–75. doi: 10.1016/j.molcel.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Görnemann J, Kotovic KM, Hujer K, Neugebauer KM. Cotranscriptional spliceosome assembly occurs in a stepwise fashion and requires the cap binding complex. Mol Cell. 2005;19:53–63. doi: 10.1016/j.molcel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 7.Moore MJ, Schwartzfarb EM, Silver PA, Yu MC. Differential recruitment of the splicing machinery during transcription predicts genome-wide patterns of mRNA splicing. Mol Cell. 2006;24:903–15. doi: 10.1016/j.molcel.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 8.Tardiff DF, Lacadie SA, Rosbash M. A genome-wide analysis indicates that yeast pre-mRNA splicing is predominantly posttranscriptional. Mol Cell. 2006;24:917–29. doi: 10.1016/j.molcel.2006.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churchman LS, Weissman JS. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 2011;469:368–73. doi: 10.1038/nature09652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrillo Oesterreich F, Preibisch S, Neugebauer KM. Global analysis of nascent RNA reveals transcriptional pausing in terminal exons. Mol Cell. 2010;40:571–81. doi: 10.1016/j.molcel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Brugiolo M, Herzel L, Neugebauer KM. Counting on co-transcriptional splicing. F1000Prime Rep. 2013;5:9. doi: 10.12703/P5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aitken S, Alexander RD, Beggs JD. Modelling reveals kinetic advantages of co-transcriptional splicing. PLoS Comput Biol. 2011;7:e1002215. doi: 10.1371/journal.pcbi.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR, la Mata de M A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–32. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Dujardin G, Lafaille C, Petrillo E, Buggiano V, Gómez Acuña LI, Fiszbein A, Godoy Herz MA, Nieto Moreno N, Muñoz MJ, Alló M, et al. Transcriptional elongation and alternative splicing. Biochim Biophys Acta. 2013;1829:134–40. doi: 10.1016/j.bbagrm.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 15.Howe KJ, Kane CM, Ares M., Jr. Perturbation of transcription elongation influences the fidelity of internal exon inclusion in Saccharomyces cerevisiae. RNA. 2003;9:993–1006. doi: 10.1261/rna.5390803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engebrecht JA, Voelkel-Meiman K, Roeder GS. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–68. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 17.Munding EM, Igel AH, Shiue L, Dorighi KM, Treviño LR, Ares M., Jr. Integration of a splicing regulatory network within the meiotic gene expression program of Saccharomyces cerevisiae. Genes Dev. 2010;24:2693–704. doi: 10.1101/gad.1977410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergkessel M, Whitworth GB, Guthrie C. Diverse environmental stresses elicit distinct responses at the level of pre-mRNA processing in yeast. RNA. 2011;17:1461–78. doi: 10.1261/rna.2754011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–37. doi: 10.1016/j.molcel.2007.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013;154:775–88. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaplan CD, Jin H, Zhang IL, Belyanin A. Dissection of Pol II trigger loop function and Pol II activity-dependent control of start site selection in vivo. PLoS Genet. 2012;8:e1002627. doi: 10.1371/journal.pgen.1002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pleiss JA, Whitworth GB, Bergkessel M, Guthrie C. Transcript specificity in yeast pre-mRNA splicing revealed by mutations in core spliceosomal components. PLoS Biol. 2007;5:e90. doi: 10.1371/journal.pbio.0050090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Malagon F, Kireeva ML, Shafer BK, Lubkowska L, Kashlev M, Strathern JN. Mutations in the Saccharomyces cerevisiae RPB1 gene conferring hypersensitivity to 6-azauracil. Genetics. 2006;172:2201–9. doi: 10.1534/genetics.105.052415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan CD, Larsson K-M, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by α-amanitin. Mol Cell. 2008;30:547–56. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khodor YL, Rodriguez J, Abruzzi KC, Tang C-HA, Marr MT, 2nd, Rosbash M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in Drosophila. Genes Dev. 2011;25:2502–12. doi: 10.1101/gad.178962.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mischo HE, Proudfoot NJ. Disengaging polymerase: terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta. 2013;1829:174–85. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dye MJ, Proudfoot NJ. Terminal exon definition occurs cotranscriptionally and promotes termination of RNA polymerase II. Mol Cell. 1999;3:371–8. doi: 10.1016/S1097-2765(00)80464-5. [DOI] [PubMed] [Google Scholar]
- 28.Vagner S, Rüegsegger U, Gunderson SI, Keller W, Mattaj IW. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1 snRNP. RNA. 2000;6:178–88. doi: 10.1017/S1355838200991854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rigo F, Martinson HG. Functional coupling of last-intron splicing and 3′-end processing to transcription in vitro: the poly(A) signal couples to splicing before committing to cleavage. Mol Cell Biol. 2008;28:849–62. doi: 10.1128/MCB.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martins SB, Rino J, Carvalho T, Carvalho C, Yoshida M, Klose JM, de Almeida SF, Carmo-Fonseca M. Spliceosome assembly is coupled to RNA polymerase II dynamics at the 3′ end of human genes. Nat Struct Mol Biol. 2011;18:1115–23. doi: 10.1038/nsmb.2124. [DOI] [PubMed] [Google Scholar]
- 31.Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chanfreau G, Noble SM, Guthrie C. Essential yeast protein with unexpected similarity to subunits of mammalian cleavage and polyadenylation specificity factor (CPSF) Science. 1996;274:1511–4. doi: 10.1126/science.274.5292.1511. [DOI] [PubMed] [Google Scholar]
- 33.Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong J-L, Chang T-H, Squazzo S, Hartzog G, Ares M., Jr. Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol. 2005;12:175–82. doi: 10.1038/nsmb891. [DOI] [PubMed] [Google Scholar]
- 34.Munding EM, Shiue L, Katzman S, Donohue JP, Ares M., Jr. Competition between pre-mRNAs for the splicing machinery drives global regulation of splicing. Mol Cell. 2013;51:338–48. doi: 10.1016/j.molcel.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hazelbaker DZ, Marquardt S, Wlotzka W, Buratowski S. Kinetic competition between RNA Polymerase II and Sen1-dependent transcription termination. Mol Cell. 2013;49:55–66. doi: 10.1016/j.molcel.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alexander RD, Innocente SA, Barrass JD, Beggs JD. Splicing-dependent RNA polymerase pausing in yeast. Mol Cell. 2010;40:582–93. doi: 10.1016/j.molcel.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sims RJ, 3rd, Millhouse S, Chen C-F, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunderson FQ, Johnson TL. Acetylation by the transcriptional coactivator Gcn5 plays a novel role in co-transcriptional spliceosome assembly. PLoS Genet. 2009;5:e1000682. doi: 10.1371/journal.pgen.1000682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunderson FQ, Merkhofer EC, Johnson TL. Dynamic histone acetylation is critical for cotranscriptional spliceosome assembly and spliceosomal rearrangements. Proc Natl Acad Sci U S A. 2011;108:2004–9. doi: 10.1073/pnas.1011982108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Albulescu L-O, Sabet N, Gudipati M, Stepankiw N, Bergman ZJ, Huffaker TC, Pleiss JAA. A quantitative, high-throughput reverse genetic screen reveals novel connections between Pre-mRNA splicing and 5′ and 3′ end transcript determinants. PLoS Genet. 2012;8:e1002530. doi: 10.1371/journal.pgen.1002530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moehle EA, Ryan CJ, Krogan NJ, Kress TL, Guthrie C. The yeast SR-like protein Npl3 links chromatin modification to mRNA processing. PLoS Genet. 2012;8:e1003101. doi: 10.1371/journal.pgen.1003101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hnilicová J, Hozeifi S, Dušková E, Icha J, Tománková T, Staněk D. Histone deacetylase activity modulates alternative splicing. PLoS One. 2011;6:e16727. doi: 10.1371/journal.pone.0016727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Z, Jones A, Joo HY, Zhou D, Cao Y, Chen S, Erdjument-Bromage H, Renfrow M, He H, Tempst P, et al. USP49 deubiquitinates histone H2B and regulates cotranscriptional pre-mRNA splicing. Genes Dev. 2013;27:1581–95. doi: 10.1101/gad.211037.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F, Yu X. WAC, a functional partner of RNF20/40, regulates histone H2B ubiquitination and gene transcription. Mol Cell. 2011;41:384–97. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luco RF, Alló M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jaehning JA. The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta. 2010;1799:379–88. doi: 10.1016/j.bbagrm.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mason PB, Struhl K. Distinction and relationship between elongation rate and processivity of RNA polymerase II in vivo. Mol Cell. 2005;17:831–40. doi: 10.1016/j.molcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 49.Lacadie SA, Tardiff DF, Kadener S, Rosbash M. In vivo commitment to yeast cotranscriptional splicing is sensitive to transcription elongation mutants. Genes Dev. 2006;20:2055–66. doi: 10.1101/gad.1434706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knaus R, Pollock R, Guarente L. Yeast SUB1 is a suppressor of TFIIB mutations and has homology to the human co-activator PC4. EMBO J. 1996;15:1933–40. [PMC free article] [PubMed] [Google Scholar]
- 51.García A, Collin A, Calvo O. Sub1 associates with Spt5 and influences RNA polymerase II transcription elongation rate. Mol Biol Cell. 2012;23:4297–312. doi: 10.1091/mbc.E12-04-0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tavenet A, Suleau A, Dubreuil G, Ferrari R, Ducrot C, Michaut M, Aude J-C, Dieci G, Lefebvre O, Conesa C, et al. Genome-wide location analysis reveals a role for Sub1 in RNA polymerase III transcription. Proc Natl Acad Sci U S A. 2009;106:14265–70. doi: 10.1073/pnas.0900162106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de la Mata M, Muñoz MJ, Alló M, Fededa JP, Schor IE, Kornblihtt AR. RNA Polymerase II Elongation at the Crossroads of Transcription and Alternative Splicing. Genet Res Int. 2011;2011:309865. doi: 10.4061/2011/309865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Eperon LP, Graham IR, Griffiths AD, Eperon IC. Effects of RNA secondary structure on alternative splicing of pre-mRNA: is folding limited to a region behind the transcribing RNA polymerase? Cell. 1988;54:393–401. doi: 10.1016/0092-8674(88)90202-4. [DOI] [PubMed] [Google Scholar]
- 56.Lewicki BT, Margus T, Remme J, Nierhaus KH. Coupling of rRNA transcription and ribosomal assembly in vivo. Formation of active ribosomal subunits in Escherichia coli requires transcription of rRNA genes by host RNA polymerase which cannot be replaced by bacteriophage T7 RNA polymerase. J Mol Biol. 1993;231:581–93. doi: 10.1006/jmbi.1993.1311. [DOI] [PubMed] [Google Scholar]
- 57.Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505:701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 59.Volanakis A, Passoni M, Hector RD, Shah S, Kilchert C, Granneman S, Vasiljeva L. Spliceosome-mediated decay (SMD) regulates expression of nonintronic genes in budding yeast. Genes Dev. 2013;27:2025–38. doi: 10.1101/gad.221960.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sayani S, Chanfreau GF. Sequential RNA degradation pathways provide a fail-safe mechanism to limit the accumulation of unspliced transcripts in Saccharomyces cerevisiae. RNA. 2012;18:1563–72. doi: 10.1261/rna.033779.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell. 2012;48:409–21. doi: 10.1016/j.molcel.2012.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA. 2009;15:1896–908. doi: 10.1261/rna.1714509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brody Y, Neufeld N, Bieberstein N, Causse SZ, Böhnlein E-M, Neugebauer KM, Darzacq X, Shav-Tal Y. The in vivo kinetics of RNA polymerase II elongation during co-transcriptional splicing. PLoS Biol. 2011;9:e1000573. doi: 10.1371/journal.pbio.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rhee HS, Pugh BF. Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature. 2012;483:295–301. doi: 10.1038/nature10799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhatt DM, Pandya-Jones A, Tong A-J, Barozzi I, Lissner MM, Natoli G, Black DL, Smale ST. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–90. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–8. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schor IE, Fiszbein A, Petrillo E, Kornblihtt AR. Intragenic epigenetic changes modulate NCAM alternative splicing in neuronal differentiation. EMBO J. 2013;32:2264–74. doi: 10.1038/emboj.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]