Abstract
The biological relevance of long non-coding RNAs (lncRNAs) is emerging. Whether the lncRNAs could form structured precursors for small RNAs (sRNAs) production remains elusive. Here, 172 713 DCL1 (Dicer-like 1)-dependent sRNAs were identified in Arabidopsis. Except for the sRNAs mapped onto the microRNA precursors, the remaining ones led us to investigate their originations. Intriguingly, 65 006 sRNAs found their loci on 5891 lncRNAs. These sRNAs were sent to AGO (Argonaute) enrichment analysis. As a result, 1264 sRNAs were enriched in AGO1, which were then subjected to target prediction. Based on degradome sequencing data, 109 transcripts were validated to be targeted by 96 sRNAs. Besides, 44 lncRNAs were targeted by 23 sRNAs. To further support the origination of the DCL1-dependent sRNAs from lncRNAs, we searched for the degradome-based cleavage signals at either ends of the sRNA loci, which were supposed to be produced during DCL1-mediated processing of the long-stem structures. As a result, 63 612 loci were supported by degradome signatures. Among these loci, 6606 reside within the dsRNA-seq (double-stranded RNA sequencing) read-covered regions of 100 nt or longer. These regions were subjected to secondary structure prediction. And, 43 regions were identified to be capable of forming highly complementary long-stem structures. We proposed that these local long-stem structures could be recognized by DCL1 for cropping, thus serving as the sRNA precursors. We hope that our study could inspire more research efforts to study on the biological roles of the lncRNAs in plants.
Keywords: lncRNA (long non-coding RNA), DCL1 (Dicer-like 1), AGO (Argonaute), degradome, dsRNA-seq (double-stranded RNA sequencing), sRNA (small RNA), Arabidopsis thaliana
Introduction
With the advent of the high-throughput sequencing (HTS) technology,1 an unexpected amount of genomic regions has been shown to be transcribed. Interestingly, a huge portion of these transcripts showed weak protein-coding capacities, which were previously called “dark matters.”2-4 In recent years, great efforts have been taken to uncover the biological functions of these non-coding RNAs.
MicroRNAs (miRNAs), a well-studied species of small non-coding RNAs of 20 to 24 nt in length, are critical players in diverse biological processes in eukaryotic organisms, such as cell proliferation, organ development and stress response.5-7 These small molecules are processed from hairpin structure-containing precursors through two-step cropping. In plants, pri-miRNA (primary microRNA) with an internal hairpin structure is transcribed by RNA polymerase (Pol) II, resulting in poly(A) (polyadenylation) tail. The pri-miRNA is processed into pre-miRNA (precursor microRNA) through DCL1 (Dicer-like 1)-mediated cleavage. The hairpin-structured pre-miRNA is further cropped by DCL1, generating a ~21 bp-long small RNA (sRNA) duplex including miRNA and miRNA*. The mature miRNA strand is selectively incorporated into a specific AGO (Argonaute)-associated silencing complex (in most cases, the plant miRNAs are associated with the AGO1 complexes). Then, the miRNAs could guide the silencing complexes to the target transcripts based on sequence complementarity, thus enabling AGO1-mediated target cleavages.6,7
Long non-coding RNAs (lncRNAs), as another part of the “dark matters,” have also been studied. In plants, the lncRNAs are implicated in chromatin modifications and target mimicry (Certain RNA sequences such as lncRNAs possess highly complementary regions which could be recognized by specific miRNAs. However, the RNAs will not be cleaved by the miRNA-associated silencing complexes owing to the existence of mismatches at the position 9th to 11th nucleotide of the miRNAs. Thus, the RNAs serve as decoys to interfere with the binding of the miRNAs to the other genuine target transcripts. This type of inhibitory mechanism of miRNA activities was termed target mimicry in plants.).8-10 A recent study reported 6,480 long intergenic non-coding RNAs (lincRNAs) in Arabidopsis,11 which have been made available in PLncDB (the plant long non-coding RNA database).12 On the other hand, the functional significance of the RNA molecules has been suggested to be embedded in the well-organized structures.13 Thus, one question was raised whether these lncRNAs could form internal structures for certain biological outputs, such as sRNA production just like pri-miRNAs.
To this end, we did a comprehensive search for DCL1-dependent sRNAs in Arabidopsis by using sRNA HTS data sets prepared from the dcl1 mutant. As a result, 172 713 DCL1-dependent sRNAs were identified, among which only 1079 could be perfectly mapped onto the registered pre-miRNAs of Arabidopsis (miRBase, release 20). This led us to investigate the origination of the remaining large portion of the DCL1-dependent sRNAs. Notably, 65 006 sRNAs could find their loci (a total of 154,106 loci) on 5891 lncRNAs retrieved from PLncDB. AGO enrichment analysis showed that some of these sRNAs, with distinct sequence characteristics, could be selectively recruited by specific AGO protein complexes. Based on degradome sequencing data, we showed that the DCL1-dependent, AGO1-enriched sRNAs had great potential of performing target cleavages. To find further evidences to support the origination of the DCL1-dependent sRNAs from lncRNAs, we searched for the degradome-based cleavage signals at either ends of the sRNA loci, which were supposed to be produced during DCL1-mediated processing of the long-stem structures. As a result, 63 612 loci belonging to 19 012 sRNAs were supported by degradome signatures. Among these loci, 6606 reside within 609 regions of 100 nt or longer. Intriguingly, all of these regions are covered by dsRNA-seq (double-stranded RNA sequencing) reads, indicating their great potential of forming long-stem structures in vivo. Thus, these regions were subjected to secondary structure prediction. After manual screening for the long-stem structures with degradome-supported sRNA loci, 43 structures on 39 lncRNAs were obtained. Taken together, our results present a DCL1-dependent biogenesis pathway for the lncRNA-originated sRNAs with potential regulatory activities. We hope that the proposed model could inspire more research efforts to study on the biological roles of lncRNAs in plants.
Results and Discussion
Identification of DCL1-dependent sRNAs potentially originated from lncRNAs
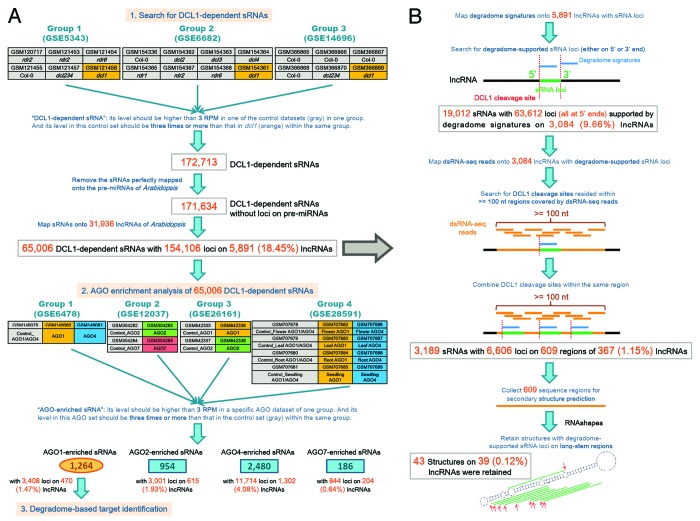
Three groups of sRNA HTS data (GSE5343, GSE6682 and GSE14696) were included, and were analyzed separately (Fig. 1A). For the three groups, in addition to the data sets prepared from the wild type plants of Arabidopsis, the data sets prepared from the mutants rdr1, rdr2, rdr6, dcl2, dcl3, dcl4, and dcl234 (triple mutant) were also recruited as the control sets to do a more comprehensive search for DCL1-dependent sRNAs. It was based on the consideration that the activity of DCL1 is not attenuated in above mutants. For each group, a DCL1-dependent sRNA was defined as follows: its accumulation level should be 3 RPM (reads per million) or higher in at least one of the control sets; and its level in this control set should be three times or more than that in the dcl1 data set within the same group. A rigorous sequence search was performed, and sequence mismatches and length variations were not allowed. In other words, two types of DCL1-dependent sRNAs will be uncovered: (1) the accumulation level of the DCL1-dependent sRNA is 3 RPM or higher in one of the control set, and the exact sequence does not exist in the dcl1 data set within the same group; (2) the exact sequence of DCL1-dependent sRNA exists in both the control set and the dcl1 set within the same group; however, its accumulation level is 3 RPM or higher in the control set, and is three times or more than that in the dcl1 data set. As a result, 172 713 DCL1-dependent sRNAs were identified. Since the research focus of this study is on the lncRNA-originated sRNAs, but not the miRNAs or the byproducts from miRNA precursors, all the DCL1-dependent sRNAs were mapped to all of the registered pre-miRNAs of Arabidopsis (miRBase, release 20), and those have perfect loci on the pre-miRNAs were removed. Interestingly, a dominant portion of the DCL1-dependent sRNAs (a total of 171 634) was retained (Data S1). Then, all of these DCL1-dependent sRNAs were mapped to the Arabidopsis lncRNAs retrieved from PLncDB.12 Notably, 65 006 sRNAs (Data S2) could find their loci (a total of 154 106 loci) on 5891 lncRNAs (Fig. 1A). To date, 3′ modifications (e.g., adenylation and uridylation) of mature miRNAs have been widely observed in both animals and plants.14,15 To exclude the possibility that the DCL1-dependent sRNAs without perfect loci on the Arabidopsis pre-miRNAs was contributed by 3′ end modifications, a systemic search for the 3′ modified candidates of the miRNAs was performed. However, only two DCL1-dependent sRNAs “AGAGGUGACC AUUGGAGAUG G” and “AGGCUUUUAA GAUCUGGUUG CGGU” were identified to contain but be longer than the miRNAs ath-miR5662 and ath-miR5643a/b. Together, our results indicate that lncRNAs are a potentially great contributor for the biogenesis of DCL1-dependent sRNAs in Arabidopsis.
Figure 1. Workflow and result summary of the identification of the DCL1 (Dicer-like 1)-dependent sRNAs (small RNAs) on the long-stem-structured regions of the lncRNAs (long non-coding RNAs) in Arabidopsis. (A) Identification of DCL1-dependent sRNAs. Three groups of sRNA high-throughput sequencing (HTS) data sets were used for this study. The “dcl1” data sets (highlighted in orange background) were prepared from the Arabidopsis mutant dcl1. The data sets highlighted in gray background were treated as control sets. For AGO enrichment analysis, four groups of sRNA HTS data sets were utilized, and the data sets highlighted in gray background were treated as control sets. (B) Identification of potential loci of DCL1-dependent sRNAs on the long-stem-structured lncRNAs.
AGO enrichment analysis
According to the current model,6,7,16,17 incorporation into specific AGO silencing complexes is a prerequisite for the action of miRNAs and other sRNAs. In this regard, AGO enrichment analysis was performed for the 65,006 DCL1-dependent sRNAs identified on the lncRNAs. Four groups of AGO-associated sRNA HTS data sets were included in this analysis, and were treated separately (Fig. 1A). For each group, a sRNA enriched in a specific AGO protein complex was defined as follows: its level in a specific AGO data set should be 3 RPM or higher; and its level in this AGO data set should be three times or more than that in the control set. As a result, 1264 DCL1-dependent sRNAs showed high enrichment in AGO1 complexes (Data S3), and 954, 2480, and 186 sRNAs were enriched in AGO2, AGO4 and AGO7 respectively (Data S4).
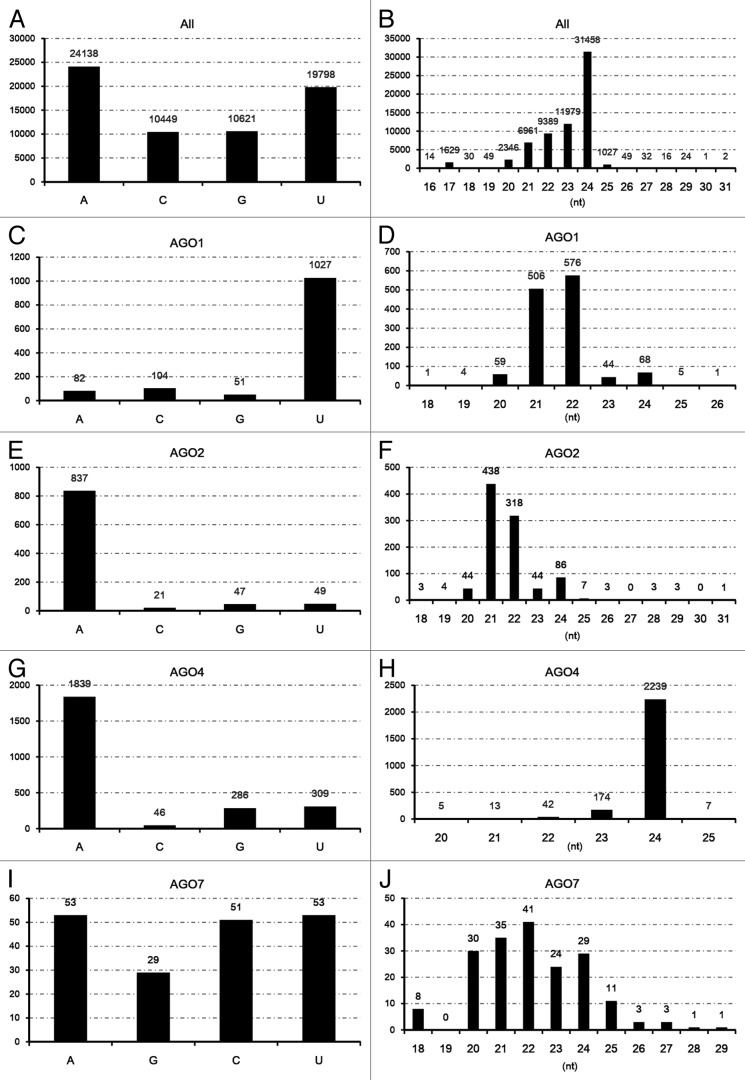
Next, the sequence characteristics of these AGO-enriched sRNAs were analyzed. For all the 65 006 DCL1-dependent sRNAs identified on the lncRNAs, slight enrichment was observed for 5′ A- and 5′ U-started sRNAs (Fig. 2A). However, it is quite different for the AGO-enriched sRNAs. The AGO1-enriched sRNAs dominantly begin with U (Fig. 2C), and the AGO2- and AGO4-enriched ones mainly start with A (Fig. 2E and G). No obvious bias of the 5′ terminal nucleotide composition was observed for the sRNAs enriched in AGO7 (Fig. 2I). For sequence length distribution, a dominant portion of the 65 006 DCL1-dependent sRNAs is enriched in 21 to 24 nt, especially in 24 nt (Fig. 2B). For both the AGO1-enriched and the AGO2-enriched sRNAs, most of the sRNAs are 21 to 22 nt in length (Fig. 2D and F). The AGO4-enriched sRNAs are dominantly 24 nt (Fig. 2H), and the AGO7-enriched ones dominantly range from 20 to 24 nt (Fig. 2J). The above described sequence characteristics of the AGO-enriched sRNAs correlate well with the previously reported sequence features of the AGO1-, AGO2- and AGO4-associated sRNAs in Arabidopsis.18
Figure 2. Sequence characteristics of the DCL1 (Dicer-like 1)-dependent sRNAs (small RNAs) on the long-stem-structured regions of the lncRNAs (long non-coding RNAs) in Arabidopsis. (A) 5′ terminal composition of all the identified DCL1-dependent sRNAs. (B) Sequence length distribution of all the identified DCL1-dependent sRNAs. (C) 5′ terminal composition of DCL1-dependent, AGO1-enriched sRNAs. (D) Sequence length distribution of DCL1-dependent, AGO1-enriched sRNAs. (E) 5′ terminal composition of DCL1-dependent, AGO2-enriched sRNAs. (F) Sequence length distribution of DCL1-dependent, AGO2-enriched sRNAs. (G) 5′ terminal composition of DCL1-dependent, AGO4-enriched sRNAs. (H) Sequence length distribution of DCL1-dependent, AGO4-enriched sRNAs. (I) 5′ terminal composition of DCL1-dependent, AGO7-enriched sRNAs. (J) Sequence length distribution of DCL1-dependent, AGO7-enriched sRNAs.
Target identification for the DCL1-dependent, AGO1-enriched sRNAs
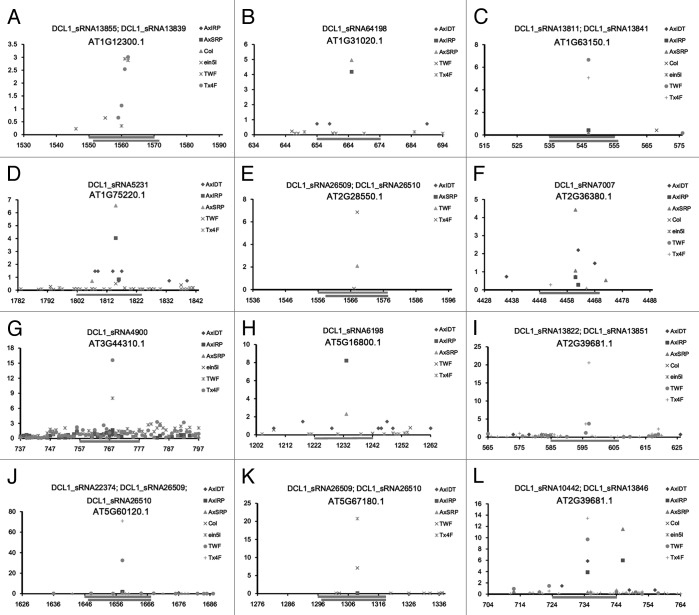
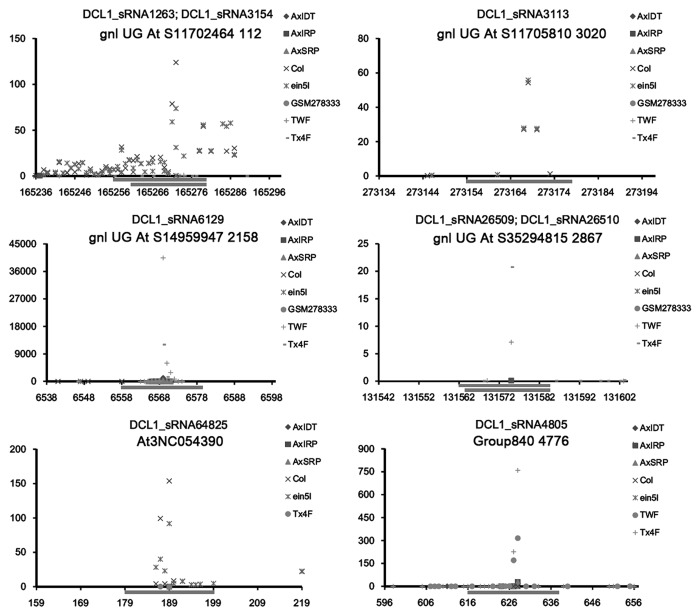
Among the diverse AGO proteins, AGO1 was well characterized to possess RNA slicer activity.17 Thus, the sRNAs associated with AGO1 silencing complexes are likely to guide transcript cleavage-based target gene regulation in plants, which is similar to the action of miRNAs.6,7 In this consideration, a large-scale target identification of the 1,264 DCL1-dependent, AGO1-enriched sRNAs was performed. First, all of the TAIR (The Arabidopsis Information Resource; release 10)-annotated gene transcripts were included as the database for target binding site search by using miRU algorithm19,20 with default parameters. Then, the predicted target transcripts were validated based on degradome sequencing data (see details in “Materials and Methods”). Considering the fact that some evident cleavage signals out of the canonical slicing sites (10th to 11th nt of the regulatory sRNAs) were detected in the previous experiments,21-24 the binding sites with prominent slicing signals resided within 8th to 12th nt of the sRNAs were retained. As a result, 109 transcripts encoded by 78 genes were validated to be targeted by 96 sRNAs, resulting in 248 sRNA—target pairs (Table 1; Fig. S1). Notably, in many cases, the cleavage-based regulation of the targets was supported by evident degradome signatures resided within 9th to 11th nt of the sRNAs (Fig. 3; Fig. S2), indicating the similarity of the action modes between the DCL1-dependent, AGO1-enriched sRNAs and the plant miRNAs. Besides, certain transcripts possess two binding sites which were simultaneously targeted by specific sRNAs. For example, AT2G39681.1 has two binding sites (i.e., 585th to 606th nt and 724th to 744th nt) which were targeted by DCL1_sRNA13822 and DCL1_sRNA13851, and DCL1_sRNA10442 and DCL1_sRNA13846 respectively (Fig. 3). More interestingly, 44 lncRNAs were uncovered to be targeted by 23 sRNAs, resulting in a total of 64 sRNA—target pairs (Fig. S3). Quite prominent cleavage signals were observed within the binding sites on several lncRNAs (Fig. 4). Thus, it raises the possibility that in plants, certain lncRNAs might serve as the precursors for sRNA generation which some other lncRNAs might be treated as the targets of the lncRNA-originated sRNAs.
Table 1. List of genes targeted by Dicer-like 1-dependent, Argonaute 1-enriched small RNAs identified on the long non-coding RNAs of Arabidopsis.
| Small RNA ID | Target transcript | Binding site on target transcript | Cleavage site supported by degradome sequencing data |
Target gene | Target gene annotation |
|---|---|---|---|---|---|
| DCL1_sRNA13855 | AT1G12300.1 | 1550–1570 | 1561 | AT1G12300 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| DCL1_sRNA13839 | AT1G12300.1 | 1551–1571 | 1562 | ||
| DCL1_sRNA13855 | AT1G12620.1 | 1706–1726 | 1717 | AT1G12620 | Pentatricopeptide repeat (PPR) superfamily protein |
| DCL1_sRNA13839 | AT1G12620.1 | 1707–1727 | 1718 | ||
| DCL1_sRNA13855 | AT1G12700.1 | 1503–1523 | 1512 | AT1G12700 | RNA PROCESSING FACTOR 1 (RPF1) |
| DCL1_sRNA13855 | AT1G12775.1 | 1502–1522 | 1514 | AT1G12775 | Pentatricopeptide repeat (PPR) superfamily protein |
| DCL1_sRNA13839 | AT1G12775.1 | 1503–1523 | |||
| DCL1_sRNA5687 | AT1G16890.1 | 618–638 | 627 | AT1G16890 | UBC36/UBC13B encodes a protein that may play a role in DNA damage responses and error-free post-replicative DNA repair |
| DCL1_sRNA5409 | AT1G16890.1 | 617–638 | |||
| DCL1_sRNA5987 | AT1G16890.1 | 615–636 | |||
| DCL1_sRNA5300 | AT1G16890.1 | 616–637 | |||
| DCL1_sRNA5729 | AT1G16890.1 | 617–637 | |||
| DCL1_sRNA5687 | AT1G16890.2 | 637–657 | 646 | ||
| DCL1_sRNA5409 | AT1G16890.2 | 636–657 | |||
| DCL1_sRNA5987 | AT1G16890.2 | 634–655 | |||
| DCL1_sRNA5300 | AT1G16890.2 | 635–656 | |||
| DCL1_sRNA5729 | AT1G16890.2 | 636–656 | |||
| DCL1_sRNA5687 | AT1G16890.3 | 664–684 | 673 | ||
| DCL1_sRNA5409 | AT1G16890.3 | 663–684 | |||
| DCL1_sRNA5987 | AT1G16890.3 | 661–682 | |||
| DCL1_sRNA5300 | AT1G16890.3 | 662–683 | |||
| DCL1_sRNA5729 | AT1G16890.3 | 663–683 | |||
| DCL1_sRNA36672 | AT1G27250.1 | 347–368 | 360 | AT1G27250 | Paired amphipathic helix (PAH2) superfamily protein |
| DCL1_sRNA64198 | AT1G31020.1 | 654–674 | 665 | AT1G31020 | Thioredoxin O2 (TO2) |
| DCL1_sRNA5018 | AT1G45688.1 | 26–47 | 36 | AT1G45688 | Unknown protein |
| DCL1_sRNA5018 | AT1G45688.2 | 12–33 | 22 | ||
| DCL1_sRNA25197 | AT1G56110.1 | 1534–1554 | 1545 | AT1G56110 | NOP56-like protein |
| DCL1_sRNA14038 | AT1G58400.1 | 2646–2667 | 2658 | AT1G58400 | Disease resistance protein (CC-NBS-LRR class) family |
| DCL1_sRNA14056 | AT1G58400.1 | 2645–2666 | |||
| DCL1_sRNA6150 | AT1G59830.1 | 1230–1250 | 1239 | AT1G59830 | Encodes one of the isoforms of the catalytic subunit of protein phosphatase 2A |
| DCL1_sRNA6150 | AT1G59830.2 | 1319–1339 | 1328 | ||
| DCL1_sRNA13855 | AT1G62860.1 | 1492–1512 | 1503 | AT1G62860 | Pseudogene of pentatricopeptide (PPR) repeat-containing protein |
| DCL1_sRNA13839 | AT1G62860.1 | 1493–1513 | 1504 | ||
| DCL1_sRNA13841 | AT1G62910.1 | 535–555 | 547 | AT1G62910 | Pentatricopeptide repeat (PPR) superfamily protein |
| DCL1_sRNA13811 | AT1G62910.1 | 535–556 | |||
| DCL1_sRNA13841 | AT1G62914.1 | 517–537 | 529 | AT1G62914 | Pentatricopeptide (PPR) repeat-containing protein |
| DCL1_sRNA13811 | AT1G62914.1 | 517–538 | |||
| DCL1_sRNA13922 | AT1G62930.1 | 1458–1478 | 1469 | AT1G62930 | RPF3 encodes a pentatricopeptide repeat (PPR) protein involved in 5′ processing of different mitochondrial mRNAs |
| DCL1_sRNA13817 | AT1G62930.1 | 1457–1477 | |||
| DCL1_sRNA13816 | AT1G62930.1 | ||||
| DCL1_sRNA13922 | AT1G63080.1 | 1501–1521 | 1512 | AT1G63080 | Transacting siRNA generating locus |
| DCL1_sRNA13816 | AT1G63080.1 | 1500–1520 | |||
| DCL1_sRNA13841 | AT1G63130.1 | 650–670 | 662 | AT1G63130 | Transacting siRNA generating locus |
| DCL1_sRNA13811 | AT1G63130.1 | 650–671 | |||
| DCL1_sRNA13841 | AT1G63150.1 | 535–555 | 547 | AT1G63150 | Transacting siRNA generating locus |
| DCL1_sRNA13811 | AT1G63150.1 | 535–556 | |||
| DCL1_sRNA13829 | AT1G63230.1 | 1230–1251 | 1242 | AT1G63230 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| DCL1_sRNA13800 | AT1G63230.1 | 1230–1250 | |||
| DCL1_sRNA13829 | AT1G63630.1 | 542–563 | 554 | AT1G63630 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| DCL1_sRNA13800 | AT1G63630.1 | 542–562 | |||
| DCL1_sRNA13829 | AT1G63630.2 | 591–612 | 603 | ||
| DCL1_sRNA13800 | AT1G63630.2 | 591–611 | |||
| DCL1_sRNA5518 | AT1G64720.1 | 1139–1161 | 1151 | AT1G64720 | Membrane related protein CP5 |
| DCL1_sRNA6493 | AT1G64720.1 | 1138–1159 | 1150 | ||
| DCL1_sRNA5389 | AT1G64720.1 | 1140–1161 | 1151 | ||
| DCL1_sRNA4887 | AT1G64720.1 | 1141–1161 | 1151 | ||
| DCL1_sRNA5993 | AT1G72050.1 | 1293–1314 | 1304 | AT1G72050 | Encodes a transcriptional factor TFIIIA required for transcription of 5S rRNA gene |
| DCL1_sRNA5733 | AT1G72050.1 | 1293–1313 | |||
| DCL1_sRNA5031 | AT1G72050.1 | 1292–1313 | |||
| DCL1_sRNA5993 | AT1G72050.2 | 1060–1081 | 1071 | ||
| DCL1_sRNA5733 | AT1G72050.2 | 1060–1080 | |||
| DCL1_sRNA5031 | AT1G72050.2 | 1059–1080 | |||
| DCL1_sRNA46232 | AT1G73500.1 | 127–149 | 140 | AT1G73500 | Member of MAP Kinase Kinase family |
| DCL1_sRNA5231 | AT1G75220.1 | 1802–1823 | 1815 | AT1G75220 | Encodes a vacuolar glucose exporter |
| DCL1_sRNA64537 | AT2G01010.1 | 1649–1669 | 1661 | AT2G01010 | 18SrRNA |
| DCL1_sRNA3154 | AT2G12440.1 | 4625–4648 | 4640 | AT2G12440 | Transposable element gene |
| DCL1_sRNA1263 | AT2G12440.1 | 4629–4648 | |||
| DCL1_sRNA1141 | AT2G17442.1 | 672–695 | 687 | AT2G17442 | Unknown protein |
| DCL1_sRNA1141 | AT2G17442.2 | 696–719 | 711 | ||
| DCL1_sRNA1141 | AT2G17442.3 | 693–716 | 708 | ||
| DCL1_sRNA1141 | AT2G17442.4 | 645–668 | 660 | ||
| DCL1_sRNA1141 | AT2G17442.5 | 644–667 | 659 | ||
| DCL1_sRNA31206 | AT2G27740.1 | 20–43 | 32 | AT2G27740 | Family of unknown function (DUF662) |
| DCL1_sRNA5477 | AT2G28350.1 | 2179–2200 | 2192 | AT2G28350 | AUXIN RESPONSE FACTOR 10 (ARF10), involved in root cap cell differentiation |
| DCL1_sRNA26509 | AT2G28550.1 | 1558–1577 | 1568 | AT2G28550 | Related to AP2.7 (RAP2.7) |
| DCL1_sRNA26510 | AT2G28550.1 | 1556–1577 | |||
| DCL1_sRNA26509 | AT2G28550.2 | 1630–1649 | 1640 | ||
| DCL1_sRNA26510 | AT2G28550.2 | 1628–1649 | |||
| DCL1_sRNA26509 | AT2G28550.3 | 1603–1622 | 1613 | ||
| DCL1_sRNA26510 | AT2G28550.3 | 1601–1622 | |||
| DCL1_sRNA62245 | AT2G33860.1 | 1673–1693/1793–1813 | 1685/1804 | AT2G33860 | ETTIN (ETT) |
| DCL1_sRNA62203 | AT2G33860.1 | 1673–1694/1793–1814 | 1686/1805 | ||
| DCL1_sRNA62205 | AT2G33860.1 | 1674–1694/1794–1814 | 1686/1805 | ||
| DCL1_sRNA62234 | AT2G33860.1 | 1674–1694/1794–1814 | 1686/1805 | ||
| DCL1_sRNA62237 | AT2G33860.1 | 1675–1694/1795–1814 | 1686/1805 | ||
| DCL1_sRNA62218 | AT2G33860.1 | 1675–1695/1795–1815 | 1686/1805 | ||
| DCL1_sRNA62198 | AT2G33860.1 | 1675–1695/1795–1815 | 1686/1805 | ||
| DCL1_sRNA24900 | AT2G33860.1 | 1676–1696/1796–1816 | 1686/1805 | ||
| DCL1_sRNA57442 | AT2G35430.1 | 844–865 | 854 | AT2G35430 | Zinc finger C-x8-C-x5-C-x3-H type family protein |
| DCL1_sRNA7007 | AT2G36380.1 | 4448–4469 | 4461 | AT2G36380 | Pleiotropic drug resistance 6 (PDR6) |
| DCL1_sRNA6129 | AT2G37370.1 | 1365–1386 | 1378 | AT2G37370 | Unknown protein |
| DCL1_sRNA3463 | AT2G37370.1 | 1419–1438 | 1427 | ||
| DCL1_sRNA31206 | AT2G37890.1 | 1447–1470 | 1461 | AT2G37890 | Mitochondrial substrate carrier family protein |
| DCL1_sRNA13846 | AT2G39681.1 | 724–744 | 735 | AT2G39681 | Trans-acting siRNA primary transcript |
| DCL1_sRNA10442 | AT2G39681.1 | 724–744 | |||
| DCL1_sRNA13822 | AT2G39681.1 | 585–606 | 597 | ||
| DCL1_sRNA13851 | AT2G39681.1 | 585–605 | |||
| DCL1_sRNA13822 | AT2G41950.1 | 422–443 | 434 | AT2G41950 | Unknown protein |
| DCL1_sRNA13851 | AT2G41950.1 | 422–442 | 434 | ||
| DCL1_sRNA6760 | AT2G47160.1 | 2252–2273 | 2262 | AT2G47160 | Boron transporter |
| DCL1_sRNA6760 | AT2G47160.2 | 2331–2352 | 2341 | ||
| DCL1_sRNA4906 | AT3G03040.1 | 1099–1120 | 1109 | AT3G03040 | F-box/RNI-like superfamily protein |
| DCL1_sRNA4946 | AT3G03040.1 | 1098–1118 | |||
| DCL1_sRNA9465 | AT3G04730.1 | 752–773 | 762 | AT3G04730 | INDOLEACETIC ACID-INDUCED PROTEIN 16 (IAA16) |
| DCL1_sRNA62220 | AT3G05340.1 | 2296–2318 | 2310 | AT3G05340 | Tetratricopeptide repeat (TPR)-like superfamily protein |
| DCL1_sRNA62220 | AT3G05345.1 | 19–41 | 33 | AT3G05345 | Chaperone DnaJ-domain superfamily protein |
| DCL1_sRNA5844 | AT3G05590.1 | 740–761 | 753 | AT3G05590 | RIBOSOMAL PROTEIN L18 (RPL18) |
| DCL1_sRNA13818 | AT3G15605.1 | 1791–1811 | 1803 | AT3G15605 | Nucleic acid binding |
| DCL1_sRNA13818 | AT3G15605.2 | 1785–1805 | 1797 | ||
| DCL1_sRNA13818 | AT3G15605.3 | 1608–1628 | 1620 | ||
| DCL1_sRNA13818 | AT3G15605.4 | 1540–1560 | 1552 | ||
| DCL1_sRNA4896 | AT3G22010.1 | 715–735 | 726 | AT3G22010 | Receptor-like protein kinase-related family protein |
| DCL1_sRNA5080 | AT3G22990.1 | 877–900 | 890 | AT3G22990 | LEAF AND FLOWER RELATED (LFR) |
| DCL1_sRNA5467 | AT3G23000.1 | 1138–1159 | 1148 | AT3G23000 | CBL-INTERACTING PROTEIN KINASE 7 (CIPK7) |
| DCL1_sRNA30437 | AT3G25470.1 | 1095–1118 | 1110 | AT3G25470 | Bacterial hemolysin-related |
| DCL1_sRNA64537 | AT3G41768.1 | 1649–1669 | 1661 | AT3G41768 | 18SrRNA |
| DCL1_sRNA4900 | AT3G44310.1 | 757–777 | 768 | AT3G44310 | NITRILASE 1 (NIT1) |
| DCL1_sRNA4900 | AT3G44310.2 | 639–659 | 650 | ||
| DCL1_sRNA4900 | AT3G44310.3 | 757–777 | 768 | ||
| DCL1_sRNA26509 | AT3G54990.1 | 965–985 | 976 | AT3G54990 | Encodes a AP2 domain transcription factor that can repress flowering |
| DCL1_sRNA22374 | AT3G54990.1 | ||||
| DCL1_sRNA26510 | AT3G54990.1 | 964–985 | |||
| DCL1_sRNA6738 | AT3G56730.1 | 605–625 | 616 | AT3G56730 | Putative endonuclease or glycosyl hydrolase |
| DCL1_sRNA6738 | AT3G56730.2 | 660–680 | 671 | ||
| DCL1_sRNA31732 | AT4G06708.1 | 15–38 | 28 | AT4G06708 | Transposable element gene |
| DCL1_sRNA6150 | AT4G12730.1 | 1364–1384 | 1376 | AT4G12730 | FASCICLIN-LIKE ARABINOGALACTAN 2 (FLA2) |
| DCL1_sRNA7163 | AT4G15830.1 | 855–876 | 868 | AT4G15830 | ARM repeat superfamily protein |
| DCL1_sRNA47338 | AT4G16830.1 | 119–139 | 130 | AT4G16830 | Hyaluronan/mRNA binding family |
| DCL1_sRNA47338 | AT4G16830.2 | 95–115 | 106 | ||
| DCL1_sRNA47338 | AT4G16830.3 | 119–139 | 130 | ||
| DCL1_sRNA15973 | AT4G18120.1 | 2301–2322 | 2314 | AT4G18120 | MEI2-LIKE 3 (ML3) |
| DCL1_sRNA15973 | AT4G18120.2 | 2152–2173 | 2165 | ||
| DCL1_sRNA7119 | AT4G23450.2 | 180–201 | 192 | AT4G23450 | ABA INSENSITIVE RING PROTEIN 1 (AIRP1) |
| DCL1_sRNA17116 | AT4G29770.1 | 728–748 | 739 | AT4G29770 | Target of trans acting-siR480/255 |
| DCL1_sRNA61901 | AT4G29770.1 | 726–747 | |||
| DCL1_sRNA61963 | AT4G29770.1 | 728–748 | |||
| DCL1_sRNA17095 | AT4G29770.1 | 728–748 | |||
| DCL1_sRNA17297 | AT4G29770.1 | 729–748 | |||
| DCL1_sRNA17118 | AT4G29770.1 | 728–748 | |||
| DCL1_sRNA61866 | AT4G29770.1 | 727–748 | |||
| DCL1_sRNA17098 | AT4G29770.1 | 729–748 | |||
| DCL1_sRNA61962 | AT4G29770.1 | 728–748 | |||
| DCL1_sRNA61865 | AT4G29770.1 | 728–748 | |||
| DCL1_sRNA17097 | AT4G29770.1 | 731–748 | |||
| DCL1_sRNA61867 | AT4G29770.1 | 729–748 | |||
| DCL1_sRNA17116 | AT4G29770.2 | 770–790 | 781 | ||
| DCL1_sRNA61901 | AT4G29770.2 | 768–789 | |||
| DCL1_sRNA61963 | AT4G29770.2 | 770–790 | |||
| DCL1_sRNA17095 | AT4G29770.2 | 770–790 | |||
| DCL1_sRNA17297 | AT4G29770.2 | 771–790 | |||
| DCL1_sRNA17118 | AT4G29770.2 | 770–790 | |||
| DCL1_sRNA61866 | AT4G29770.2 | 769–790 | |||
| DCL1_sRNA17098 | AT4G29770.2 | 771–790 | |||
| DCL1_sRNA61962 | AT4G29770.2 | 770–790 | |||
| DCL1_sRNA61865 | AT4G29770.2 | 770–790 | |||
| DCL1_sRNA17097 | AT4G29770.2 | 773–790 | |||
| DCL1_sRNA61867 | AT4G29770.2 | 771–790 | |||
| DCL1_sRNA7075 | AT4G33280.1 | 808–829 | 818 | AT4G33280 | AP2/B3-like transcriptional factor family protein |
| DCL1_sRNA26509 | AT4G36920.1 | 1329–1349 | 1340 | AT4G36920 | APETALA 2 (AP2) |
| DCL1_sRNA22374 | AT4G36920.1 | ||||
| DCL1_sRNA26510 | AT4G36920.1 | 1328–1349 | |||
| DCL1_sRNA26509 | AT4G36920.2 | 1293–1313 | 1304 | ||
| DCL1_sRNA22374 | AT4G36920.2 | ||||
| DCL1_sRNA26510 | AT4G36920.2 | 1292–1313 | |||
| DCL1_sRNA22374 | AT5G04275.1 | 1–21 | 12 | AT5G04275 | MICRORNA172B (MIR172B), a microRNA that targets several genes containing AP2 domains |
| DCL1_sRNA12080 | AT5G08590.1 | 34–56 | 48 | AT5G08590 | SNF1-RELATED PROTEIN KINASE 2.1 (SNRK2.1) |
| DCL1_sRNA10500 | AT5G11660.1 | 803–824 | 815 | AT5G11660 | Protein of Unknown Function (DUF239) |
| DCL1_sRNA7229 | AT5G13800.1 | 1654–1675 | 1667 | AT5G13800 | PHEOPHYTINASE (PPH) |
| DCL1_sRNA5234 | AT5G14565.1 | 1896–1916 | 1907 | AT5G14565 | MICRORNA398C (MIR398C) |
| DCL1_sRNA13841 | AT5G16640.1 | 581–601 | 593 | AT5G16640 | Pentatricopeptide repeat (PPR) superfamily protein |
| DCL1_sRNA13811 | AT5G16640.1 | 581–602 | 593 | ||
| DCL1_sRNA6198 | AT5G16800.1 | 1222–1242 | 1233 | AT5G16800 | Acyl-CoA N-acyltransferases (NAT) superfamily protein |
| DCL1_sRNA6198 | AT5G16800.2 | 1139–1159 | 1150 | ||
| DCL1_sRNA6198 | AT5G16800.3 | 1148–1168 | 1159 | ||
| DCL1_sRNA17116 | AT5G18040.1 | 675–695 | 686 | AT5G18040 | Unknown protein |
| DCL1_sRNA61901 | AT5G18040.1 | 672–694 | |||
| DCL1_sRNA17095 | AT5G18040.1 | 675–695 | |||
| DCL1_sRNA17297 | AT5G18040.1 | 676–695 | |||
| DCL1_sRNA17118 | AT5G18040.1 | 675–695 | |||
| DCL1_sRNA61866 | AT5G18040.1 | 674–695 | |||
| DCL1_sRNA17098 | AT5G18040.1 | 676–695 | |||
| DCL1_sRNA61865 | AT5G18040.1 | 675–695 | |||
| DCL1_sRNA17097 | AT5G18040.1 | 678–695 | |||
| DCL1_sRNA61867 | AT5G18040.1 | 676–695 | |||
| DCL1_sRNA17116 | AT5G18065.1 | 726–746 | 737 | AT5G18065 | Unknown protein |
| DCL1_sRNA61901 | AT5G18065.1 | 723–745 | |||
| DCL1_sRNA17095 | AT5G18065.1 | 726–746 | |||
| DCL1_sRNA17297 | AT5G18065.1 | 727–746 | |||
| DCL1_sRNA17118 | AT5G18065.1 | 726–746 | |||
| DCL1_sRNA61866 | AT5G18065.1 | 725–746 | |||
| DCL1_sRNA17098 | AT5G18065.1 | 727–746 | |||
| DCL1_sRNA61865 | AT5G18065.1 | 726–746 | |||
| DCL1_sRNA17097 | AT5G18065.1 | 729–746 | |||
| DCL1_sRNA61867 | AT5G18065.1 | 727–746 | |||
| DCL1_sRNA6188 | AT5G33406.1 | 1505–1525 | 1517 | AT5G33406 | hAT dimerization domain-containing protein/transposase-related |
| DCL1_sRNA59503 | AT5G40810.1 | 1015–1038 | 1027 | AT5G40810 | Cytochrome C1 family |
| DCL1_sRNA59503 | AT5G40810.2 | 1344–1367 | 1356 | ||
| DCL1_sRNA7136 | AT5G51300.1 | 2469–2488 | 2477 | AT5G51300 | Splicing factor-related |
| DCL1_sRNA6885 | AT5G51300.1 | 2468–2488 | |||
| DCL1_sRNA5824 | AT5G59732.1 | 1537–1558 | 1548 | AT5G59732 | Potential natural antisense gene, locus overlaps with AT5G59730 |
| DCL1_sRNA26509 | AT5G60120.1 | 1647–1667 | 1658 | AT5G60120 | TARGET OF EARLY ACTIVATION TAGGED 2 (TOE2) |
| DCL1_sRNA22374 | AT5G60120.1 | ||||
| DCL1_sRNA26510 | AT5G60120.1 | 1646–1667 | |||
| DCL1_sRNA26509 | AT5G60120.2 | 1810–1830 | 1821 | ||
| DCL1_sRNA22374 | AT5G60120.2 | ||||
| DCL1_sRNA26510 | AT5G60120.2 | 1809–1830 | |||
| DCL1_sRNA62203 | AT5G60450.1 | 1873–1894/2083–2104 | 1885/2095 | AT5G60450 | AUXIN RESPONSE FACTOR 4 (ARF4) |
| DCL1_sRNA62218 | AT5G60450.1 | 1875–1895/2085–2105 | |||
| DCL1_sRNA24900 | AT5G60450.1 | 1876–1896/2086–2106 | |||
| DCL1_sRNA62205 | AT5G60450.1 | 1874–1894/2084–2104 | |||
| DCL1_sRNA62237 | AT5G60450.1 | 1875–1894/2085–2104 | |||
| DCL1_sRNA62234 | AT5G60450.1 | 1874–1894/2084–2104 | |||
| DCL1_sRNA62245 | AT5G60450.1 | 1873–1893/2083–2103 | |||
| DCL1_sRNA62198 | AT5G60450.1 | 1875–1895/2085–2105 | |||
| DCL1_sRNA62203 | AT5G62000.1 | 1836–1857 | 1848 | AT5G62000 | AUXIN RESPONSE FACTOR 2 (ARF2) |
| DCL1_sRNA62218 | AT5G62000.1 | 1838–1858 | |||
| DCL1_sRNA24900 | AT5G62000.1 | 1839–1859 | |||
| DCL1_sRNA62205 | AT5G62000.1 | 1837–1857 | |||
| DCL1_sRNA62237 | AT5G62000.1 | 1838–1857 | |||
| DCL1_sRNA62234 | AT5G62000.1 | 1837–1857 | |||
| DCL1_sRNA62245 | AT5G62000.1 | 1836–1856 | |||
| DCL1_sRNA62198 | AT5G62000.1 | 1838–1858 | |||
| DCL1_sRNA62203 | AT5G62000.2 | 1734–1755 | 1746 | ||
| DCL1_sRNA62218 | AT5G62000.2 | 1736–1756 | |||
| DCL1_sRNA24900 | AT5G62000.2 | 1737–1757 | |||
| DCL1_sRNA62205 | AT5G62000.2 | 1735–1755 | |||
| DCL1_sRNA62237 | AT5G62000.2 | 1736–1755 | |||
| DCL1_sRNA62234 | AT5G62000.2 | 1735–1755 | |||
| DCL1_sRNA62245 | AT5G62000.2 | 1734–1754 | |||
| DCL1_sRNA62198 | AT5G62000.2 | 1736–1756 | |||
| DCL1_sRNA62203 | AT5G62000.3 | 1734–1755 | 1746 | ||
| DCL1_sRNA62218 | AT5G62000.3 | 1736–1756 | |||
| DCL1_sRNA24900 | AT5G62000.3 | 1737–1757 | |||
| DCL1_sRNA62205 | AT5G62000.3 | 1735–1755 | |||
| DCL1_sRNA62237 | AT5G62000.3 | 1736–1755 | |||
| DCL1_sRNA62234 | AT5G62000.3 | 1735–1755 | |||
| DCL1_sRNA62245 | AT5G62000.3 | 1734–1754 | |||
| DCL1_sRNA62198 | AT5G62000.3 | 1736–1756 | |||
| DCL1_sRNA62203 | AT5G62000.4 | 1734–1755 | 1746 | ||
| DCL1_sRNA62218 | AT5G62000.4 | 1736–1756 | |||
| DCL1_sRNA24900 | AT5G62000.4 | 1737–1757 | |||
| DCL1_sRNA62205 | AT5G62000.4 | 1735–1755 | |||
| DCL1_sRNA62237 | AT5G62000.4 | 1736–1755 | |||
| DCL1_sRNA62234 | AT5G62000.4 | 1735–1755 | |||
| DCL1_sRNA62245 | AT5G62000.4 | 1734–1754 | |||
| DCL1_sRNA62198 | AT5G62000.4 | 1736–1756 | |||
| DCL1_sRNA26509 | AT5G67180.1 | 1297–1318 | 1309 | AT5G67180 | TARGET OF EARLY ACTIVATION TAGGED 3 (TOE3) |
| DCL1_sRNA26510 | AT5G67180.1 | 1296–1318 |
Please refer to Figure S2 for degradome sequencing data-based cleavage evidences.
Figure 3. Degradome sequencing data-based identification of the transcripts targeted by the DCL1 (Dicer-like 1)-dependent, AGO1 (Argonaute 1)-enriched sRNAs (small RNAs) in Arabidopsis. Examples of identified targets are shown. For each figure panel, the target transcript ID and the corresponding sRNA(s) are listed on the top. The y axis measures the intensity (in RPM, reads per million) of the degradome signals, and the x axis indicates the position(s) of the cleavage signal(s) on the target transcript. The binding site of the sRNA regulator on its target transcript was denoted by a gray horizontal line. The figure keys for the signatures from different degradome sequencing libraries are shown on the right. Please note, for AT2G39681.1, two cleavage sites were identified.
Figure 4. Degradome sequencing data-based identification of the lncRNAs (long non-coding RNAs) targeted by the DCL1 (Dicer-like 1)-dependent, AGO1 (Argonaute 1)-enriched sRNAs (small RNAs) in Arabidopsis. Examples of identified targets are shown. For each figure panel, the lncRNA ID and the corresponding sRNA(s) are listed on the top. The y axis measures the intensity (in RPM, reads per million) of the degradome signals, and the x axis indicates the position(s) of the cleavage signal(s) on the lncRNA. The binding site of the sRNA regulator on the lncRNA was denoted by a gray horizontal line. The figure keys for the signatures from different degradome sequencing libraries are shown on the right.
Biological hints inferred from certain sRNA—Target pairs
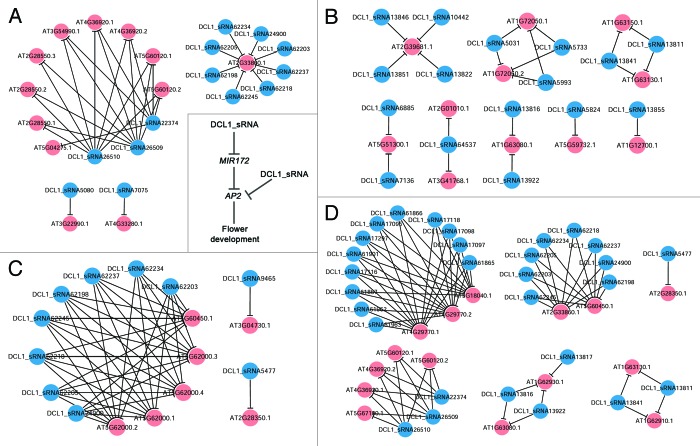
Certain regulatory networks constituted by dozens of degradome-validated sRNA—target pairs were present here, since some biological implications were obtained based on the target gene annotations (TAIR, release 10) and the analytical results from PlantGSEA (The Plant GeneSet Enrichment Analysis Toolkit; http://structuralbiology.cau.edu.cn/PlantGSEA/analysis.php).25 Within the network shown in Figure 5A, AT2G28550, AT3G54990, AT4G33280, AT4G36920 and AT5G60120 were annotated to be the members of APETALA2 (AP2) family, which were potentially involved in flower development.26-28 Two other target genes, AT2G33860 and AT3G22990, were also involved in flower development according to the TAIR annotations. More interestingly, AT5G04275 encoding MIR172B was targeted by DCL1_sRNA22374. miR172 has been demonstrated to play an important role in floral organ development in Arabidopsis.29,30 Based on the above results, a regulatory cascade implicated in flower development, DCL1-dependent sRNA—MIR172—AP2, was proposed (Fig. 5A). For the second network shown in Figure 5B, all of the six target genes are functionally related to RNAs based on TAIR annotations. AT1G72050 encodes a transcription factor TFIIIA required for the transcription of 5S rRNA (rRNA) gene, and AT2G01010 and AT3G41768 encode 18S rRNA. AT2G39681 encodes a primary transcript for trans-acting siRNA (small interfering RNA) production, and AT5G59732 is a potential natural antisense gene overlapping with AT5G59730. Besides, AT5G51300 is involved in RNA splicing. The third network is involved in auxin signaling (Fig. 5C). AT2G28350 encodes ARF10 (Auxin Response Factor 10) involved in root cap formation,31 and AT5G60450 and AT5G62000 encode ARF4 and ARF2 respectively. AT3G04730 encodes IAA16 (indoleacetic acid-induced protein 16) which is also implicated in auxin signal transduction. Based on the analytical results of PlantGSEA,25 certain literature-based evidences were obtained to support some of the sRNA—target pairs whose interactions could be significantly influenced by the activity of DCL1. The 12 target genes (AT1G62910, AT1G62930, AT1G63080, AT1G63130, AT2G28350, AT2G33860, AT4G29770, AT4G36920, AT5G18040, AT5G60120, AT5G60450 and AT5G67180) shown in Figure 5D were significantly upregulated in the dcl1 mutant relative to the wild type plants of Arabidopsis.32,33 On the other hand, the above information partially supports the interactions between the 12 target genes and the corresponding DCL1-dependent sRNAs (Fig. 5D).
Figure 5. Biologically meaningful subnetworks identified from the target lists of the DCL1 (Dicer-like 1)-dependent, AGO1 (Argonaute 1)-enriched sRNAs (small RNAs) in Arabidopsis. (A) Subnetwork containing AP2 (APETALA2)- and flower development-related target genes. (B) Subnetwork containing target genes involved in RNA-level biological processes such as processing, splicing and transcription. (C) Subnetwork containing target genes implicated in auxin signaling. (D) Subnetwork containing target genes whose expression levels were significantly upregulated in the dcl1 mutant of Arabidopsis according to the previous reports.25,32,33 All the subnetworks were generated by using Cytoscape.42
Degradome and dsRNA sequencing data-based evidences supporting the origination of the DCL1-dependent sRNAs from the local long-stem structures of the lncRNAs
Although 65 006 of the 171,634 DCL1-dependent sRNAs could find their loci on the 5891 reported lncRNAs, whether lncRNAs are the genuine precursors for sRNA production through a DCL1-dependent way and how these sRNAs are generated remain unclear. To partially address the above issues, we set out to search for the cleavage signals produced during DCL1-mediated processing of the lncRNA precursors, and to search for the long-stem structures that could be potentially recognized by DCL1 for dicing. It was based on the facts that: the stem region of a hairpin-structured miRNA precursor could be recognized by DCL1 for two-step processing,6,7 and the cropping sites of DCL1 on the miRNA precursors could be mapped by using degradome sequencing data in most cases.23,34
The 154 106 loci of 65 006 DCL1-dependent sRNAs on the lncRNAs were included in this analysis. First, the publicly available degradome signatures were mapped onto the 5891 lncRNAs with sRNA loci. Then, we searched for the sRNA loci with degradome signatures mapped to either 5′ or 3′ ends which were considered to be the evidences for DCL1-mediated cropping (Fig. 1B). As a result, 63 612 loci belonging to 19 012 sRNAs on 3084 lncRNAs were identified to be supported by degradome sequencing data. Strikingly, all of the supportive degradome signatures were found at the 5′ ends of the sRNA loci, indicating the higher stability of the 5′ cleaved remnants relative to the 3′ cleaved ones.
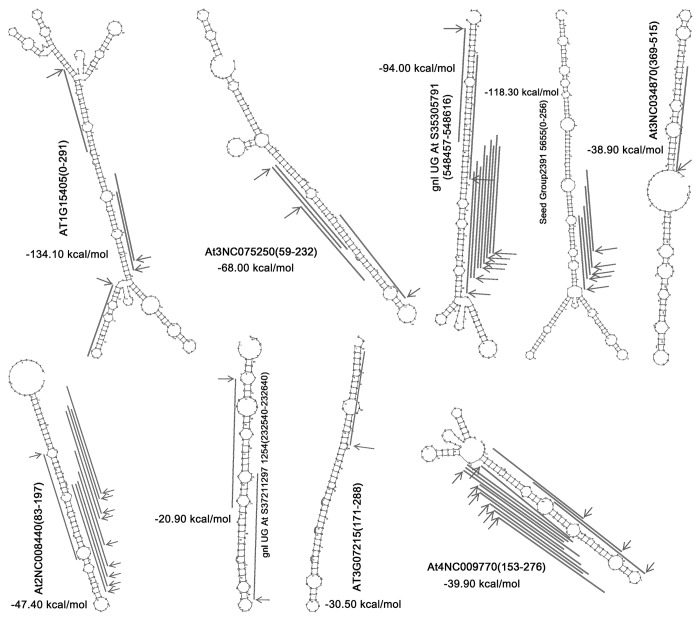
The data generated by dsRNA-seq was quite useful for interrogating the in vivo structures of long transcripts. One dsRNA-seq data set (GSM575244; with two-round rRNA depletion) contributed by a previous study13 was recruited for the following structure-based analysis. First, all the dsRNA-seq reads were mapped onto the 3084 lncRNAs with degradome data-supported sRNA loci. Second, a degradome-supported sRNA locus was retained if it resided within a dsRNA-seq read-covered region of 100 nt or longer. As a result, 6606 loci belonging to 3189 sRNAs were identified. After combining the loci sharing the same regions, a total of 609 dsRNA-seq read-covered regions on 367 lncRNAs were obtained (Fig. 1B; Data S5). All of the 609 sequences were subjected to secondary structure prediction by using RNAshapes,35 since they were likely to form internal long-stem structures. Based on manual screening, the dsRNA-seq read-covered regions with degradome-supported sRNA loci on the predicted long stems were retained. A total of 43 long-stem structures on 39 lncRNAs were identified (Fig. S4). We observed that all of the 43 structures possess highly complementary long-stem regions encoding DCL1-dependent sRNAs (Fig. 6; Fig. S4), which strengthened the possibility of forming stabled internal structures within the lncRNAs. Taken together, based on degradome sequencing data, dsRNA-seq data and structure prediction, we provide further evidences to support the biogenesis model of the DCL1-dependent, lncRNA-originated sRNAs. We deduced that the 39 novel lncRNAs could serve as the sRNA precursors owning to their ability of forming the local long-stem structures (supported by secondary structure prediction and dsRNA-seq data) recognized by DCL1 for cropping (supported by degradome signatures). However, this notion still needs further experimental validations.
Figure 6. Examples of the DCL1 (Dicer-like 1)-dependent sRNA (small RNA) loci identified on the long-stem-structured regions of the lncRNAs (long non-coding RNAs) in Arabidopsis. For all the long-stem structures, the sequence ranges on the lncRNAs were shown in the brackets along with the lncRNA IDs. The DCL1-dependent sRNA loci were denoted by gray lines, and the degradome signature-based evidences for DCL1-mediated cleavages during sRNA generation were marked by gray arrows. All the secondary structures were predicted by using RNAshapes.35 The minimum free energy (kcal/mol) of each optimal secondary structure was provided.
Conclusions
Our results indicate that 43 sequence regions on 39 lncRNAs could form local long-stem structures for sRNA production which relies on the activity of DCL1. The DCL1-dependent sRNAs with different sequence characteristics were associated with different AGO silencing complexes. Specifically, 96 DCL1-dependent, AGO1-enriched sRNAs possess great potential of performing target cleavages on 109 transcripts originated from 78 genes of Arabidopsis. Besides, 44 lncRNAs were discovered to be targeted by 23 DCL1-dependent, AGO1-enriched sRNAs. Summarily, our study could advance the current understanding on the biological roles of lncRNAs in plants.
Materials and Methods
Data sources
The sRNA HTS data sets used for the identification of DCL1-dependent sRNAs and AGO enrichment analysis were retrieved from GEO (Gene Expression Omnibus; http://www.ncbi.nlm.nih.gov/geo/).36 The detailed information of these data sets has been shown in Figure 1A.
The mature miRNAs and the pre-miRNAs of Arabidopsis were obtained from miRBase (release 20; http://www.mirbase.org/).37
The genomic information of lncRNAs (including five groups of lncRNAs, i.e., “lncRNA_EST analysis,” “lncRNA_tiling array analysis 1,” “lncRNA_tiling array analysis 2,” “lncRNA_RepTAS analysis” and “lncRNA_from TAIR”) was retrieved from PLncDB (http://chualab.rockefeller.edu/gbrowse2/homepage.html).12 It was largely contributed by a previous study by Liu et al. (2012).11 According to the above information, the lncRNA sequences were collected from the Arabidopsis Genome available in TAIR (The Arabidopsis Information Resource; release 9; http://www.arabidopsis.org/).38
The dsRNA-seq data set GSM575244 (with two-round rRNA depletion; was retrieved from GEO) is a gift from a previous study by Zheng et al. (2010).13
The transcripts and the annotations of the genes were retrieved from TAIR (release 10; http://www.arabidopsis.org/).
The eight Arabidopsis degradome sequencing data sets (AxIDT, AxIRP, AxSRP, Col, ein5, TWF, Tx4F and GSM278333) were obtained from Next-Gen Sequence Databases (http://mpss.udel.edu/common/web/library_info.php?SITE=at_pare&showAll=true)39 and GEO.
Prediction and validation of the sRNA targets
Target prediction was performed by using miRU algorithm19,20 with default parameters. The degradome sequencing data was utilized to validate the predicted sRNA—target pairs. First, in order to allow cross-library comparison, the normalized read count (in RPM, reads per million) of a short sequence from a specific degradome library was calculated by dividing the raw count of this sequence by the total counts of the library, and then multiplied by 106. Second, all the degradome signatures were mapped onto the predicted target transcripts. Then, the previously proposed criteria40 were applied to extract the potential cleavage sites. Summarily, (1) “Average_Read count_Cleavage site” is the averaged read count (in RPM) of all the degradome signatures (belonging to one library) with their 5′ ends mapped to a potential cleavage site; “Average_Read count_Surrounding” is the averaged read count of all the degradome signatures (also belonging to this library) that mapped to the regions surrounding the cleavage site; “Average_Read count_Cleavage site” should be five times or more than “Average_Read count_Surrounding.” (2) Also for this degradome library, among the degradome signatures mapped to a potential cleavage site, the most abundant tag should be among the top 12-most-abundant degradome signatures that perfectly mapped to the corresponding transcript. (3) The cleavage site should reside within 8—12 nt region of the regulatory sRNA. For any degradome library, if the three rules were fulfilled, the potential slicing sites were retained. Finally, both global and local target plots were drawn to perform manual screening, referring to our previous study.41 Only the transcripts with cleavage signals easy to be recognized were extracted as the potential sRNA—target pairs.
Plant gene set enrichment analysis
The online tool PlantGSEA25 was employed for this analysis. The IDs of all the target genes were submitted for enrichment analysis. “G1” (including “BP,” “CC” and “MF”), “G2,” “G3” (including “PlantCyc,” “KEGG,” “PO” and “Ref”) and “G4” (including “MIR” and “TFT”) were all selected for the analysis. Arabidopsis thaliana was chosen as the species analyzed, and “Suggested background (Whole genome level)” was chosen as the background.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank all the publicly available data sets and the scientists behind them. This work was funded by the National Natural Science Foundation of China [31100937], [31125011] and [31271380], and the Starting Grant funded by Hangzhou Normal University to Yijun Meng [2011QDL60].
Glossary
Abbreviations:
- lncRNA
long non-coding RNA
- sRNA
small RNA
- AGO
Argonaute
- pre-miRNA
precursor microRNA
- pri-miRNA
primary microRNA
- dsRNA-seq
double-stranded RNA sequencing
- DCL1
Dicer-like 1
- GEO
Gene Expression Omnibus
- PLncDB
plant long non-coding RNA database
- TAIR
the Arabidopsis information resource
- RPM
reads per million
- t-plot
target plot
- HTS
high-throughput sequencing
- AP2
APETALA2
- miRNA
microRNA
- Pol
polymerase
- poly(A)
polyadenylation
- lincRNA
long intergenic non-coding RNA
- PlantGSEA
the plant GeneSet enrichment analysis toolkit
- rRNA
ribosomal RNA
- siRNA
small interfering RNA
- ARF
auxin response factor
References
- 1.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iaconetti C, Gareri C, Polimeni A, Indolfi C. Non-coding RNAs: the “dark matter” of cardiovascular pathophysiology. Int J Mol Sci. 2013;14:19987–20018. doi: 10.3390/ijms141019987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins LJ, Penny D. The RNA infrastructure: dark matter of the eukaryotic cell? Trends Genet. 2009;25:120–8. doi: 10.1016/j.tig.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Derrien T, Guigó R, Johnson R. The Long Non-Coding RNAs: A New (P)layer in the “Dark Matter”. Front Genet. 2011;2:107. doi: 10.3389/fgene.2011.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 6.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 7.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–87. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 8.De Lucia F, Dean C. Long non-coding RNAs and chromatin regulation. Curr Opin Plant Biol. 2011;14:168–73. doi: 10.1016/j.pbi.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Wierzbicki AT. The role of long non-coding RNA in transcriptional gene silencing. Curr Opin Plant Biol. 2012;15:517–22. doi: 10.1016/j.pbi.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Wu HJ, Wang ZM, Wang M, Wang XJ. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013;161:1875–84. doi: 10.1104/pp.113.215962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J, Jung C, Xu J, Wang H, Deng S, Bernad L, Arenas-Huertero C, Chua NH. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell. 2012;24:4333–45. doi: 10.1105/tpc.112.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin J, Liu J, Wang H, Wong L, Chua NH. PLncDB: plant long non-coding RNA database. Bioinformatics. 2013;29:1068–71. doi: 10.1093/bioinformatics/btt107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng Q, Ryvkin P, Li F, Dragomir I, Valladares O, Yang J, Cao K, Wang LS, Gregory BD. Genome-wide double-stranded RNA sequencing reveals the functional significance of base-paired RNAs in Arabidopsis. PLoS Genet. 2010;6:e1001141. doi: 10.1371/journal.pgen.1001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyman SK, Knouf EC, Parkin RK, Fritz BR, Lin DW, Dennis LM, Krouse MA, Webster PJ, Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–61. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu S, Sun YH, Chiang VL. Adenylation of plant miRNAs. Nucleic Acids Res. 2009;37:1878–85. doi: 10.1093/nar/gkp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen X. Small RNAs and their roles in plant development. Annu Rev Cell Dev Biol. 2009;25:21–44. doi: 10.1146/annurev.cellbio.042308.113417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaucheret H. Plant ARGONAUTES. Trends Plant Sci. 2008;13:350–8. doi: 10.1016/j.tplants.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–27. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. 2011;39:W155-9. doi: 10.1093/nar/gkr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701-4. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–62. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Addo-Quaye C, Snyder JA, Park YB, Li YF, Sunkar R, Axtell MJ. Sliced microRNA targets and precise loop-first processing of MIR319 hairpins revealed by analysis of the Physcomitrella patens degradome. RNA. 2009;15:2112–21. doi: 10.1261/rna.1774909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010;62:742–59. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 24.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–6. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 25.Yi X, Du Z, Su Z. PlantGSEA: a gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013;41:W98-103. doi: 10.1093/nar/gkt281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–25. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamuro JK, Szeto W, Lotys-Prass C, Jofuku KD. Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell. 1997;9:37–47. doi: 10.1105/tpc.9.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowman JL, Smyth DR, Meyerowitz EM. Genes directing flower development in Arabidopsis. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell. 2003;15:2730–41. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–5. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–16. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–92. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2005;102:12984–9. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng Y, Gou L, Chen D, Wu P, Chen M. High-throughput degradome sequencing can be used to gain insights into microRNA precursor metabolism. J Exp Bot. 2010;61:3833–7. doi: 10.1093/jxb/erq209. [DOI] [PubMed] [Google Scholar]
- 35.Steffen P, Voss B, Rehmsmeier M, Reeder J, Giegerich R. RNAshapes: an integrated RNA analysis package based on abstract shapes. Bioinformatics. 2006;22:500–3. doi: 10.1093/bioinformatics/btk010. [DOI] [PubMed] [Google Scholar]
- 36.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–8. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–5. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–5. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shao C, Chen M, Meng Y. A reversed framework for the identification of microRNA-target pairs in plants. Brief Bioinform. 2013;14:293–301. doi: 10.1093/bib/bbs040. [DOI] [PubMed] [Google Scholar]
- 41.Meng Y, Shao C, Chen M. Toward microRNA-mediated gene regulatory networks in plants. Brief Bioinform. 2011;12:645–59. doi: 10.1093/bib/bbq091. [DOI] [PubMed] [Google Scholar]
- 42.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.