Abstract
MRI connectomics is an emerging approach to study the brain as a network of interconnected brain regions. Understanding and mapping the development of the MRI connectome may offer new insights into the development of brain connectivity and plasticity, ultimately leading to improved understanding of normal development and to more effective diagnosis and treatment of developmental disorders. In this review, we describe the attempts made to date to map the whole-brain structural MRI connectome in the developing brain and pay a special attention to the challenges associated with the rapid changes that the brain is undergoing during maturation. The two main steps in constructing a structural brain network are (i) choosing connectivity measures that will serve as the network “edges” and (ii) finding an appropriate way to divide the brain into regions that will serve as the network “nodes”. We will discuss how these two steps are usually performed in developmental studies and the rationale behind different strategies. Changes in local and global network properties that have been described during maturation in neonates and children will be reviewed, along with differences in network topology between typically and atypically developing subjects, for example, owing to pre-mature birth or hypoxic ischaemic encephalopathy. Finally, future directions of connectomics will be discussed, addressing important steps necessary to advance the study of the structural MRI connectome in development.
It is becoming increasingly common to model the brain as a network, an idealized mathematical construction that refers to a set of interconnected components that together serve a specific function.1 At the cellular level, these components might be neurons and their synaptic connections. At a higher level, the brain can be viewed as a network of interconnected brain regions that together integrate vast amounts of information and perform highly complicated cognitive and regulatory functions. These distributed neural networks are not invariant throughout life and their continuing development requires the co-ordination of an extraordinarily complex set of neurodevelopmental events. Understanding the evolving structure of the brain network through infancy and childhood promises to provide new insights into normal and abnormal brain maturation and plasticity. As a clinical motivation, we consider the question: why do two children with apparently similar injuries have very different outcomes? Ultimately, such knowledge may lead to better diagnosis and treatment of developmental disorders.
An important scientific goal of neuroimaging is to improve our basic understanding of brain function; a strategic way to approach this goal is to characterize the physical connections that mediate information transfer between cortical regions.2 In the past several years, the changes of the macroscopic whole-brain MRI-based structural network—the MRI connectome—during the course of human development have been explored by a number of different research groups. Three reviews to date have examined the development of the connectome, each with a somewhat different focus. Hagmann et al3 discussed concepts in imaging of developing structural connectivity, functional connectivity and structure–function coupling. Dennis and Thompson4 gave a detailed overview of the development of structural and functional connectivity in healthy subjects and in subjects with a range of developmental disorders; they also discussed issues such as hemispheric asymmetry, sex differences and imaging genetics. Collin and van den Heuvel5 reviewed current insights into functional and structural connectome organization throughout development, maturation and ageing, in order to explore the existence of consistent underlying patterns.
This review focuses on methods for constructing and comparing structural MRI connectomes, both across developing subjects and longitudinally, with an emphasis on the challenges that arise in brains that are rapidly changing during development. We limit our review to structural whole-brain connectivity (to which the tools of graph theory can be applied) in neonates and children and will discuss the ways in which this task differs from similar approaches aimed at studying the adult brain.
Biological substrate and trajectory of normal brain development
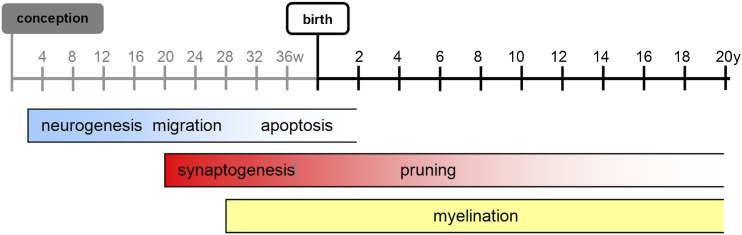
Brain maturation follows an organized, pre-determined pattern that correlates with the functions that a newborn performs at various stages of development.6 While the brain undergoes the most rapid and substantial physical and chemical changes in utero, a number of processes affecting brain structure, including myelination, refinement of white matter pathways, synaptogenesis and dendritic pruning persist into adulthood7 (Figure 1).
Figure 1.
Trajectory of key neurodevelopmental processes in the human brain.
Neurogenesis and apoptosis, synaptogenesis and pruning
After an initial period of patterning, the majority of cortical neurons develop and proliferate within the germinal matrix and subventicular zone between 12 and 20 gestational weeks and migrate to their final destination within the cortex, following a scaffold of glial cells.8 A period of rapid programmed cell death, apoptosis, occurs after this migration, reducing the neuronal number by half from 24 weeks of gestation to 4 weeks after birth.9 Another major developmental process is the proliferation and organization of synapses, which begins slightly later, around the 20th week of gestation.9 Synaptic density increases rapidly after birth; at 1–2 years of age, the number of synapses is approximately 50% greater than that typically seen in adults.10 This is followed by a regionally specific loss of synaptic connections called pruning (Figure 1).
Sulcation
On a macroscopic level, sulcation is commonly cited as an indicator of developmental stage. The first sulci are seen at approximately 15 weeks after conception11 and slowly develop in an orderly sequence, starting from the areas of the auditory, sensorimotor and visual pathways. Most large- and moderate-sized sulci and gyri are present by birth; however, smaller sulci continue to develop after birth, resulting in an increase in sulcal complexity. For reasons that are not yet clear, sulcal development is delayed in pre-term neonates in comparison with a foetus of the same post-conceptual age.6
Myelination
The second important indicator of the developmental stage, from the imaging perspective, is the change in signal intensity of the white matter secondary to the process of myelination. Myelination of the brain begins before term and continues at a much slower rate throughout life (Figure 1). Regionally, it grossly proceeds from caudal to cephalad and from central to peripheral.12–14 Another generalization is that myelination (and brain development in general, including sulcation) progresses from more primitive functions utilized in early life such as sensory and, later, motor, to those that are not utilized until the child is older, such as heteromodal association fibres. Although most major tracts are significantly myelinated by early childhood, axons within the cortex and in some pathways that mediate higher order functions, such as the arcuate fasciculus, continue to myelinate into the second and third decades of life.15
Imaging of brain development before connectomics
MRI is the most commonly used imaging modality for the evaluation of brain maturation.6 MRI is well suited for studies of the developing brain because it uses no ionizing radiation, making it the modality of choice for not only clinical indications but also for research and repeated scans of the same individual. Such longitudinal studies are critical for accurately characterizing normal developmental curves, which are often non-linear. However, the inherently long acquisition times of MRI often result in subject motion and sometimes necessitate sedation. Routine clinical scans include anatomic MRI (standard T1 and T2 weighted images), often along with diffusion tensor imaging (DTI) and proton spectroscopy. Resting-state functional MRI (fMRI), perfusion MRI, susceptibility-weighted imaging and magnetization transfer imaging have been advocated for specific indications6 (Table 1). As already mentioned, sulcation and myelination are the main indicators currently used in deciding whether the brain is developing normally. Table 1 lists other markers and MRI sequences used to assess them.
Table 1.
Most commonly used MRI sequences in studying brain development
| Imaging sequence | Information obtained |
|---|---|
| T1 weighted | Gross morphologic changes, development of sulci, myelination |
| T2 weighted | Gross morphologic changes, development of sulci, myelination |
| Diffusion tensor imaging | Changes in free water diffusion, microstructure, myelination, tracks |
| MR spectroscopy | Biochemical metabolites |
| BOLD functional MRI | Changes in location of specific brain activities |
| Perfusion MRI | Cerebral perfusion (blood delivery to brain tissue per unit time) |
| Susceptibility-weighted imaging | Intracerebral haemorrhages, myelination |
| Magnetization transfer | Myelination |
BOLD, blood oxygenation level dependent.
Because DTI is the principal technique used to calculate connections of a structural connectome, we will briefly discuss how it has been used in paediatric neuroimaging. DTI assesses water molecule displacement at a microstructural level (in the order of microns) and allows for calculating the average rate of water diffusion [apparent diffusion coefficient (ADC)] and its orientational coherence [fractional anisotropy (FA)]. FA is typically high in white matter, as hydrophobic cell membranes and myelin sheaths hinder water diffusion. Therefore, the main direction of the diffusion tensor reflects the underlying orientation of white matter tracts. We extract this information by performing computational fibre tractography. Fibre tracking is able to delineate specific white matter tracts by following local vector orientation from the three-dimensional (3D) vector field. Different tractography algorithms have been developed; the most widely used remains streamline deterministic fibre tracking.16 The 3D fibre track is allowed to continue unless it enters a region of FA less than a pre-defined value, turns at an angle greater than a pre-defined angle between consecutive voxels or exceeds outside pre-determined spatial boundaries. Fibre crossing reduces intravoxel FA and thereby causes one of the major downfalls of fibre tracking. Several approaches, such as the diffusion spectrum imaging (DSI),17 have been proposed to solve this issue.
DTI has been used extensively to explore the structural basis of white matter development (for a review, see Lodygensky et al18). In normally developing white matter, the ADC decreases as it matures, while anisotropy steadily increases.19–21 FA increases with age, partially owing to increasing myelin, but also exists in the “pre-myelinating state” owing to axonal growth and elongation and changes in the axonal membrane.22 DTI tractography has been also performed even in pre-term neonate brains for almost a decade now,23–25 despite the small brain volume and low anisotropy.
MRI connectomics and network analysis
The connectome is a relatively new term, introduced in analogy to the genome, and is used to describe the network of connections of an organism's neural system.26 Depending on the scale at which connections are defined, one can speak about the microscopic or macroscopic connectome. The complete microscopic connectome at the level of the synapse is known only for the worm Caenorhabditis elegans' nervous system, which has as few as 302 neurons, and is unlikely to be mapped in the human nervous system, comprising an estimated 1011 neurons, with 1015 connections between them. On the macroscopic scale, a brain can be viewed as a network of anatomically segregated local brain regions communicating with other local regions via longer inter-regional white matter pathways.
It recently became possible to study the macroscopic connectome non-invasively using MRI.26,27 MRI connectomics treat the brain as a network of connections between brain regions and can provide new information about the topological arrangement of brain connections, how these connections develop and remodel, the efficiency of these connections and, in pathological cases, the mechanisms and impact of disrupted connections. In this approach, the surface of the brain is partitioned, and the partitions serve as nodes of the network, while the white matter fibres, usually reconstructed using diffusion tractography, serve as connections or edges of the network. Alternatively, connections can be derived from the fMRI signal; however, the highly variable and dynamic patterns of functional connectivity are distinct from connectome maps28 and will not be the focus of this review. The suffix “ome” generally describes a permanent set of constituents of a system considered collectively and is assumed to be invariant and context independent, like the base pairs in the genome.
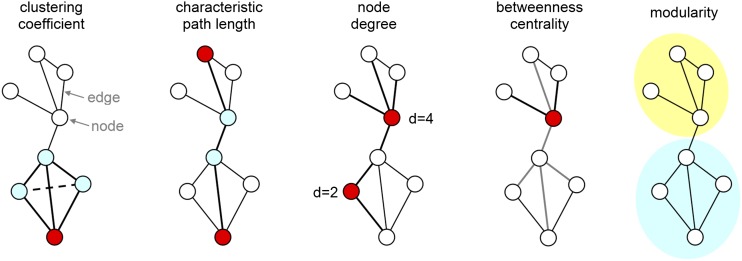
A network can be mathematically represented as a graph or connectivity matrix (Figure 2). A graph is defined as a set of nodes (in our case, brain regions), connected by a set of edges (e.g., white matter tracts). In our system, the edges are often “binarized”, meaning that they are only recognized if there is sufficient evidence of a connection based on MR diffusion tractography. Streamline counts, where each streamline is defined by following the preferred direction of the water diffusion in the brain, are one method for defining connections, for example. Graph theory-based analysis is applied to the connectivity matrices for the extraction of important network characteristics, such as node degree (the number of connections of a node), characteristic path length (the average number of steps between nodes), average clustering coefficient (a measure of regional connectivity) and other quantifiable measures of network connectivity (Figure 2) (for a review, see Rubinov and Sporns29). Studying the human connectome using graph theory offers a unique opportunity to better understand interindividual differences in neural connectivity. In adults, this approach has been applied to studying network disruption in Alzheimer's disease, schizophrenia, multiple sclerosis and other neurological and psychiatric disorders (for a recent review, see Griffa et al30).
Figure 2.
Fundamental graph metrics. Brain networks can be described as a graph consisting of elements (nodes) and their connections (edges). In macroscopic brain networks, the nodes correspond to brain regions and edges to the fibre bundles connecting them. The clustering coefficient of a node is given by the fraction of triangles around an individual node. The average clustering coefficient of the entire graph is used as a measure of segregation, representing the degree to which the network is organized into functionally distinct groups. The characteristic path length of a graph is defined as the average number of steps required to travel between two nodes of the graph; it is commonly used as a measure of integration, describing the ability of a network for distribution of information. The degree of a node is the number of edges attached to that node. The betweenness centrality of a node reflects the (relative) number of shortest paths between all node pairs that pass through it. The modularity of a graph describes the extent to which the graph can be subdivided into weakly linked clusters of densely interconnected nodes.
Whereas the majority of connectome studies examine the adult brain, this review focuses on the developing brain. In general, the major caveat of connectomics is that graph analysis of neuroimaging data is not a straightforward automatic step but a model building exercise, entailing arbitrary assumptions and decisions, which can have influential effects on the results of the analysis.31 This is especially true for the assumptions and decisions made when analysing the developing brain networks, which will be the focus of the following sections.
MAPPING THE STRUCTURAL CONNECTOME IN A DEVELOPING BRAIN
Understanding of how the structural MRI connectome develops in normal children can help to understand the development of brain function, its maturation and plasticity (Figure 3). This may lead to better diagnosis and, ultimately, to the development of targeted therapeutic interventions for paediatric neurological and psychiatric disorders, particularly in the many children with developmental disorders but “normal” anatomic MRIs. A series of studies have recently explored the changes in the human brain network architecture occurring during the course of typical (Table 2) and atypical development (Table 3). Although many studies have investigated microstructural or local connectivity changes in neonates and infants (see review by Dennis and Thompson4), we consider here only structural connectome studies employing the construction and analysis of whole-brain networks and the use of graph theoretic tools. Our specific focus is the challenge of characterizing networks in the changing brain, which is most arduous during early brain development.
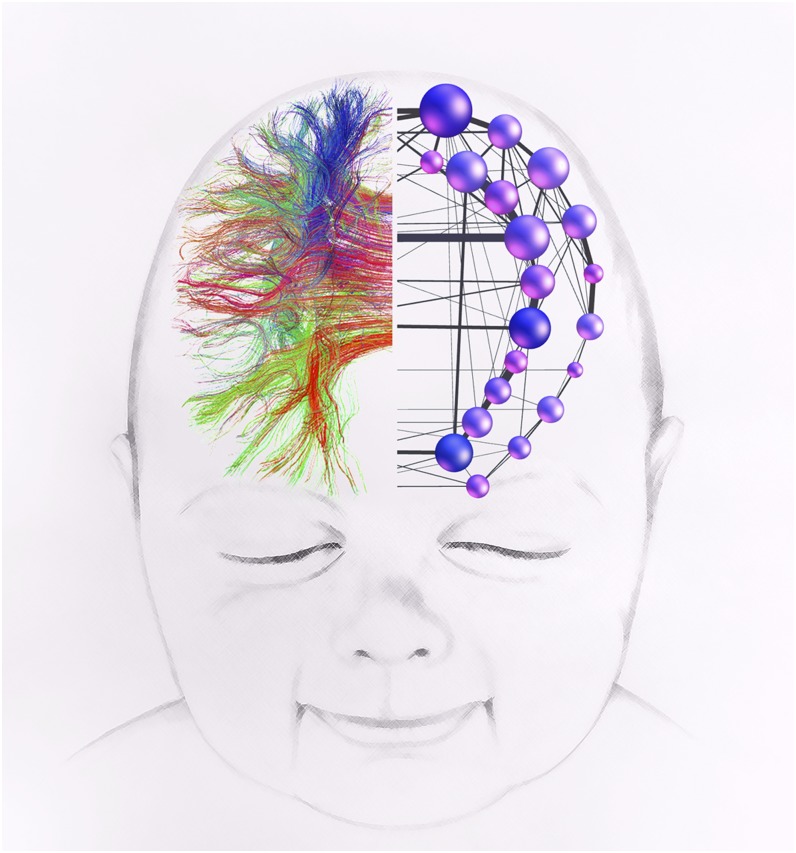
Figure 3.
A schematic image of structural connectivity in a 6-month old infant. A whole-brain tractogram obtained from diffusion tensor imaging is used to construct a network of corticocortical connections.
Table 2.
MRI studies of structural brain networks in typical development
| Study | Subjects | Connectivity measure | Parcellation | Network changes with age |
|---|---|---|---|---|
| Hagmann et al32 | 30 subjects: 18 months to 18 years | DTI and q-ball | 66 or 241 nodes, landmark-based | Global efficiency ↑, clustering ↓, small-worldness ↓ |
| Fan et al33 | 28 subjects: 1 month, 1 year, 2 years longit. + 27 adults | Grey matter volume correlation | 90 nodes, AAL34 | Global efficiency ↑, modularity ↑ from 1 to 2 years |
| Yap et al35 | 39 subjects: 2 weeks, 1 year, 2 years longit. | DTI | 78 nodes, AAL34 | Global efficiency constant, local efficiency ↑ |
| Dennis et al36 | 102 subjects: 12 years, 16 years + 337 adults | HARDI | 70 nodes, gyral-based | Global efficiency ↑, clustering ↓, small-worldness ↓, modularity ↓ |
| Huang et al37 | 25 neonates, 13 toddlers, 25 pre-adolescents + 18 adults | DTI, probabilistic tracking | 80 nodes, AAL,34 adult ICBM-152 template38 | Global efficiency ↑, clustering ↓, small-worldness ↓ |
| Khundrakpam et al39 | 203 subjects: 5–8, 8–11, 11–15 and 15–18 years | Cortical thickness covariance | 78 nodes, AAL34 | Local efficiency ↓, modularity ↓, global efficiency ↑ in late childhood |
| Tymofiyeva et al40 | 26 subjects: pre-term, term 1–14 days, 6 months + 7 adults | DTI | 100 nodes, template-free equal-area sphere partitioning | Global efficiency ↑, clustering ↓, modularity ↓, small-worldness ↓ (except pre-term) |
AAL, anatomical automatic labelling atlas; DTI, diffusion tensor imaging; ICBM, International Consortium for Brain Mapping; HARDI, high angular resolution diffusion imaging; longit., longitudinal.
Table 3.
MRI studies of structural brain networks in atypical development
| Study | Condition/subjects | Connectivity measure | Parcellation | Network changes with condition/outcome |
|---|---|---|---|---|
| Batalle et al41 | IUGR/42 subjects + 41 controls, scanned at 12 ± 2 months | DTI | 93 nodes, AAL adapted to1 year-old population42 | Decreased global and local weighted efficiency; a pattern of altered regional graph theory features; correlation of connectivity measures with BSID-III at 2 years |
| Tymofiyeva et al43 | HIE/17 subjects, scanned at 6 months | DTI | 40 nodes, template-free (1) equal-area sphere partitioning; (2) “gridded” parcellation into regions of equal spatial extent | A trend of declining brain network integration and segregation with increasing neuromotor deficit scores |
| Pandit et al44 | Pre-mature birth/49 subjects, scanned at 11–31 months | DTI, probabilistic tracking | 83 nodes, manual segmentation of adult brains45 | Lower connection strength in tracts involving all cortical lobes and several subcortical structures with increasing pre-maturity at birth |
| Pannek et al46 | Pre-mature birth/9 term neonates, 18 pre-term-born infants | DTI, probabilistic tracking | 48 nodes, John Hopkins University neonate space47 | Affected components of the network identified using NBS48 mainly involved left frontal and temporal cortical areas |
| Shi et al49 | Autism/49 subjects + 51 controls, scanned at 6–15 years | Cortical thickness correlations | 68 nodes, Freesurfer, Desikan–Killiany cortical atlas50 | Reduced network modularity; a larger number of intermodule connections; increased intra- and inter-module connectivity in middle frontal gyrus, inferior parietal gyrus and cingulate |
| Ziv et al51 | HIE/24 subjects, scanned at 6 months | DTI | 100 nodes, template-free equal-area sphere partitioning | Separating normal and abnormal outcome using a machine learning approach |
AAL, anatomical automatic labelling atlas; BSID-III, Bayley scale for infancy and toddler development, 3rd ed.; DTI, diffusion tensor imaging; HIE, hypoxic ischaemic encephalopathy; IUGR, intrauterine growth restriction; NBS, network-based statistics.
Network construction
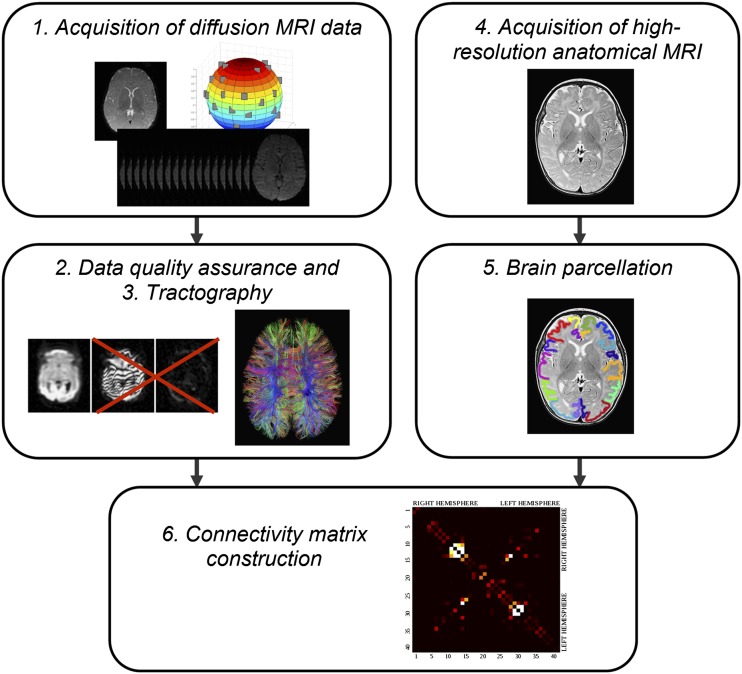
As mentioned above, any network can be represented as a graph or connectivity matrix, also called adjacency matrix, which consists of nodes and edges. All of the reviewed studies performed similar basic steps to construct connectivity matrices and to assess structural networks in development. A typical path from diffusion-weighted images to a connectivity matrix included the following steps (Figure 4):
1. Acquisition of diffusion MRI data. High-resolution diffusion images are acquired of the entire brain. Ideally, these should be acquired at high field strength with multiple diffusion directions (30 or more) and with multiple b-values. Additional b-values employed in multitensor modelling approaches enable a better differentiation of complex white matter anatomy.
2. Data quality assurance. MRI data quality suffers from bulk motion, particularly in unsedated infants. In order to address this issue, different measures may be taken; these include rejection of volumes affected by motion in diffusion MRI, which may distort the matrix, correction for susceptibility distortions, intensity inhomogeneity correction, correction for head movement and eddy current distortions, among others (see review by Pannek et al52).
3. Structural connectivity measures. In this step, whole‐brain diffusion tractography is performed (as in the majority of studies) to characterize connectivity between different brain regions. Alternatively, measurements of cortical thickness derived from a high-resolution anatomical image can be used, as will be discussed in the following section.
4. A high-resolution anatomical MRI is acquired and registered to a standardized brain/atlas. This step is performed in the majority of studies, with the exception of template-free approaches.40,43,51
5. Brain parcellation. To define distinct brain regions used as network nodes, atlases and templates are most commonly used. These are created using sulcal landmarks as the limits of hand-drawn regions of interest (ROIs) on the outer surface of the brain.34 This step will also be discussed in more detail later.
6. Connectivity matrix construction. Whatever measure was derived in Step 3 is combined with Step 5 in order to construct an N × N connectivity matrix, defining all connections among the nodes, where N is the number of nodes.
Figure 4.
A typical pipeline for assembling a structural connectome in the developing brain. After a set of diffusion-weighted images is acquired (1), a quality assurance step is performed in which data affected by motion are rejected and the remaining images are corrected for eddy current distortions and affine head motion (2). Although this step may not be necessary in co-operative adults, it is essential for high-quality tractography in infants. The diffusion tensor is calculated for the resulting data and whole-brain streamline fibre tractography is undertaken (3). Commonly, a high-resolution anatomical MRI is acquired (4), which is registered to a standardized brain/atlas that enables automatic brain parcellation into nodes (5). Steps (3) and (5) are combined and the connectivity matrix is constructed (6).
Network analysis
The connectivity matrix, formed using the steps described above, is used to perform network analyses, for example, using standard toolboxes, such as the brain connectivity toolbox developed for MATLAB® (http://www.brain-connectivity-toolbox.net).29 These analyses yield information concerning the connectivity of the brains studied.
Two of the steps, choosing structural connectivity measures that will serve as the network edges and finding an appropriate way to divide the brain into nodes have a decisive influence on the resulting network measures. There is an ongoing debate regarding the proper definition of network edges and nodes, even for the study of the adult brain. Available choices for the construction of the developing connectome will be discussed next.
Structural connectivity measures
To characterize the structural brain network, an appropriate measure of connectivity is required. This measure can be defined in different ways based on different kinds of MRI data (see review by Bullmore and Bassett31). In diffusion MRI, streamlines from tractographic analysis can serve as a connectivity measure between any pair of grey matter regions. Using anatomical (for example, T1 weighted) MRI data, connectivity can be inferred from inter-regional covariation in cortical thickness or volumes, a process that results in connectivity estimates that resemble those derived from diffusion tensor maps.53 As an example, this approach was used by Fan et al,33 Khundrakpam et al39 and Shi et al49 in their developmental connectome studies.
Because it probes inter-regional connections in a more direct way, diffusion MRI has emerged as the method of preference for inferring structural connectivity in the adult and the developing structural connectome (Tables 2 and 3). The problem of motion in paediatric population and difficulty of performing longer scans make the simpler and faster DTI model the technique of choice. However, it needs to be emphasized that DTI-based connectivity inevitably misrepresents anatomical connectivity to some extent, as it is unable to resolve fibre bundles, or even tracts, with complex configurations. High angular resolution diffusion (HARDI) models provide a more accurate white matter tractography result than the simple tensor model by resolving crossing fibre bundles, for example as used in the study by Dennis et al.36 DSI is probably the most accurate method for probing structural connectivity, but long experimental times preclude its routine use.
Whether DTI, HARDI or DSI is used to acquire and represent intravoxel diffusion, tractography is a fundamental step in transforming these measurements into actual network connections. Different brain regions will be connected by different number of streamlines, mathematical constructs that define a single realization of a potential 3D trajectory linking any two brain areas. These streamlines, or tracks, are approximations and do not represent actual fibres or even an average fibre within a tract, which raises the question as to how to quantify connection strength.
In the language of graph theory, one speaks of weighted vs binary networks. Accordingly, there are two main approaches to defining connectivity:
1. Thresholding connectivity to reduce the complexity of the data set and focus on the pattern of connectivity. Using binarized networks, in which connections consisting of more than a threshold number of streamlines are considered as present and those with a number of streamlines below that threshold are not present, simplifies the calculation and interpretation of many network measures, but also results in a loss of information about the network edges. Information transfer in vivo is not only governed by the connectivity pattern but also by the density of axons in a bundle, axonal diameter, axonal length and, importantly, myelination.3 Since at later stages of development, the strongest changes are most likely to be related to myelin modulation rather than the change of the macroscopic binary pattern, using weighted networks might be more appropriate.
2. While many different approaches to weighting network connection have been proposed (streamline count being the most straightforward), no tractography-derived index proposed so far can quantify “connection strength” in a physiological or anatomical context.54 Changes associated with brain development add an additional difficulty. Owing to methodological biases related to brain volume and diffusion anisotropy changes, tractography studies do not register the higher number of axonal branches present in early development; on the contrary, tractography shows more tracts with increasing age.3 This should not be wrongly interpreted as infants having fewer connections, as would result from simply counting the streamlines. In other words, an equal number of streamlines have different meaning at different maturation stages; therefore, streamline count is not an appropriate weighting factor, especially for longitudinal developmental studies. In probabilistic tractography, the regional connectivity probability can be used to weight cortical regions. For example, Pandit et al44 adapted a probabilistic tracking algorithm to delineate tracts between all ROI pairs, as described by Robinson et al.55 However, it remains unclear whether any of the suggested measures provide a quantitative estimate of “connection strength”. More comprehensive analysis methods such as tractometry, in which different metrics of white matter microstructure are sampled (myelination, axon density, axon diameter), might provide a more meaningful biological quantification of connectivity and a more meaningful weighting scheme.56
Binary and weighted approaches make different assumptions about the analysed networks. In their (weighted) study of late development, Hagmann et al32 assumed no change in axonal number and, therefore, in the pattern of connectivity, and used the inverse of the average ADC (1/ADC) as a measure of “connection efficacy” (a rough estimate of maturation). The number of non-zero entries in the connectivity matrices was fixed and the weights varied, representing maturational changes. This approach has its roots in MRI studies of white matter57,58 showing that all the major white matter tracks are present at birth, but in an immature state. The opposite (binary) approach is to analyse the connectivity pattern after fixing the weight of connections by applying a threshold (or a range of different thresholds), as in the study by Tymofiyeva et al.43 A condition such as hypoxic ischaemic injury of the neonate, for example, would be expected to impact the connectivity pattern and not just the weights. Hagmann et al3 emphasized that it must be clearly hypothesized at the beginning whether, for example, a developmental disease impacts the connectivity pattern or only the physiological weights of the connections, since it is difficult to differentiate these two phenomena.
The choice of the connectivity measure, even the specific variation of the method, can have a strong effect on the resulting network properties. For example, Li et al59 applied three connection reconstruction methods, based on probabilistic tractography, to compile inter-regional connectivity maps of brain networks using the same set of diffusion MRI data, and then compared the resultant connectivity matrices. Although the three methods showed moderate-to-high correlations among the different graph-theoretic measures, significant between-method variability in terms of small-world properties, brain-hub identification and hemispheric asymmetry were demonstrated, suggesting that reconstruction method has a significant impact on derived brain networks.
Brain parcellations
Whether tractography results or cortical thickness correlations are used to define brain network connections, the properties of the resulting connectome depend strongly on the cortical parcellation, the method by which the brain's grey mater is divided into nodes. No single parcellation scheme is universally used. The majority of adult brain studies have employed anatomical parcellation, in which common gyral and sulcal anatomy is used as landmarks to define boundaries of grey matter zones, such as the anatomical automatic labelling (AAL) atlas.34 The main advantage of using an anatomically defined template for nodal parcellation is that it enables a direct comparison of results across studies. One of the limitations of this approach, however, is that the size of different template regions varies considerably, affecting network properties. A more serious limitation stems from the significant intersubject variability in the location and size of these common brain areas, as well as in the non-trivial relationship between macrostructure (sulci and gyri) and microstructure (cytoarchitectonics and cytochemistry). For example, variability of the sulcal anatomy on the cerebral hemispheres has been measured to reach 17–19 mm after an affine stereotaxic normalization.60 The situation becomes even more challenging in the rapidly changing and relatively undeveloped newborn brain, when cortical surface anatomy is dynamic and the signal intensity of the brain changes with age. Even the anatomical co-registration step preceding the parcellation cannot be easily performed by directly applying pipelines developed for the adult brain. For example, widely used methods such as Freesurfer (Laboratory for Computational Neuroimaging, Boston, MA) do not work with incompletely myelinated brain.3
Despite these limitations, most prior work on the developing brain (Tables 2 and 3) has employed adult brain templates in the calculation of network nodes. For example, Fan et al33 noted that anatomical brain regions defined in the AAL atlas might not match very well with function and anatomy during early brain development. Recently, a neonatal brain atlas, referred to as John Hopkins University neonate space, with detailed anatomic information derived from DTI and coregistered anatomical MRI, was developed,47 following a similar concept to the adult group-averaged atlas International Consortium for Brain Mapping (ICBM)-152.38 It has been used by Pannek et al46 to perform network analysis of structural connectivity in the pre-term brain at term equivalent age.
An alternative approach is to use template-free parcellation. Tymofiyeva et al43 suggested a method for anatomically unconstrained parcellation that utilizes equal area partitioning on the sphere. This technique makes intersubject comparison more challenging but enables calculation of single subject network parameters without imposing anatomical bias. An empirical method for finding the optimal number of nodes for equal-area parcellation schemes—for given study group, acquisition and tractography parameters—has also been proposed,51,61 defined as the finest parcellation that covers the entire cortical surface and does not have isolated (lacking connections) components. Although brain networks obtained in this way are not automatically co-registered, it can be done using matrix alignment algorithms.61
Comparing connectomes
The comparison of connectomes can be undertaken either on a global or a local (at the node or edge) level, two complementary ways of analysis.62 At the global level, a set of summary metrics for the entire network is calculated for each subject, and statistical tests are performed to assess the differences between groups while controlling for nuisance covariates. Although several tests are usually performed on the same dataset using different global network metrics, multiplicity correction is, unfortunately, rarely applied.
When global network measures such as characteristic path length or the average clustering coefficient are used, insights regarding plasticity effects, maturational or pathological processes are difficult to infer, as local phenomena are diluted in the global mean.62 Therefore, the analyses of both global and node-wise or edge-wise comparisons are important in order to identify connections associated with a particular effect or outcome, such as a group difference in a case–control comparison or a correlation with clinical measures. A statistical test contrasting two or more groups is computed for each individual connection of the connectivity matrix. This approach is statistically intractable, as the number of comparisons that must be performed grows rapidly with the number of network nodes, virtually ensuring that statistical significance is observed by random chance.
To address this problem, two approaches are suggested: (i) mass-univariate testing of the hypothesis followed by controlling the family-wise error rate (FWE) with a generic procedure such as the false discovery rate;63 or (ii) the network-based statistics (NBS) for group comparison.48 The NBS can provide greater statistical power if the set of connections at which the null hypothesis is rejected constitutes a large component. It should be noted that the NBS is of no use if the contrast does not form an interconnected component without isolated nodes. However, when it does, the NBS has a greater utility than the FWE, as demonstrated in resting-state fMRI networks in patients with schizophrenia.48 An additional advantage of NBS is that it operates directly on raw measures of structural or functional connectivity, rather than on binary connectivity matrices. This approach was used in the study of the pre-mature connectome by Pannek et al.46
Alternatively, comparison can be performed by means of more sophisticated nonparametric statistical testing or classification approaches.62 For example, Ziv et al51 used a machine learning approach to predict neonatal encephalopathy based on structural networks derived from DTI data and demonstrated low testing error (21 ± 4%).
WHAT WE HAVE LEARNT SO FAR: HOW DOES THE CONNECTOME DEVELOP?
Typical connectome development
The essence of brain function is reciprocal communication among many locations, based on two main organizational principles: segregation and integration. High segregation indicates presence of specialized, locally efficient neighbourhoods in the network and is reflected, for example, in a high average clustering coefficient.64 High integration, on the other hand, indicates efficient communication among nodes of the network and is reflected in a short characteristic path length. The interplay of these two organizational principles enables the main brain function: information processing that is simultaneously specialized and distributed across cognitive domains. It has been previously demonstrated that higher integration of brain networks is associated with higher intelligence.65
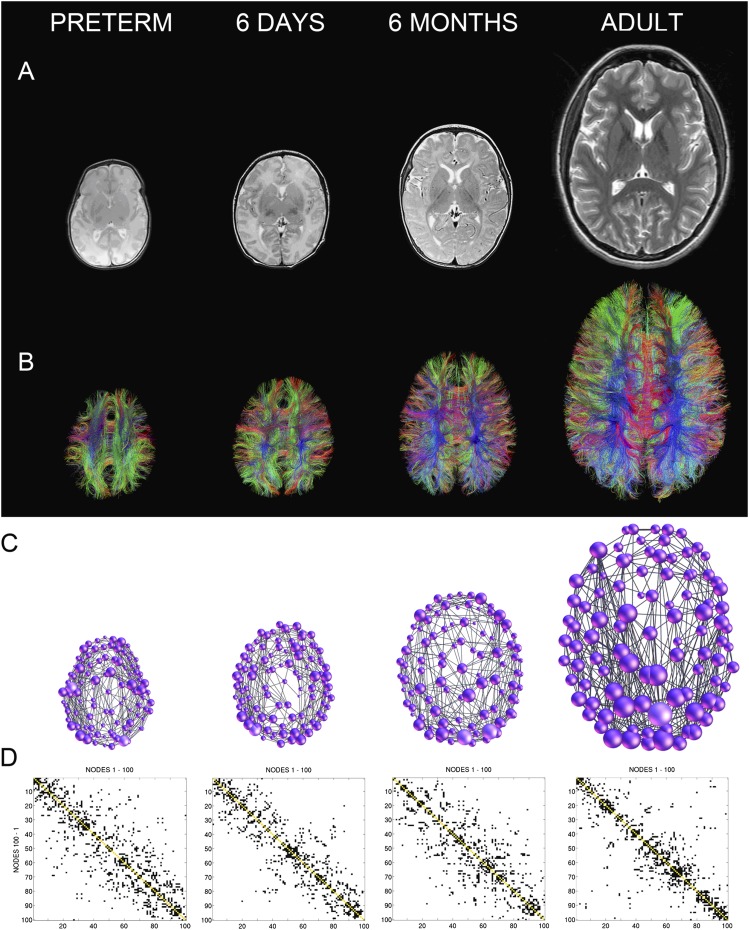
Reported studies (Table 2) have shown changes in measures of integration and segregation (the network's global and local efficiency, respectively) with maturation. Examination of two parcellations with differing numbers of nodes revealed increasing global efficiency and decreasing clustering coefficients (and, hence, local efficiency) in the developing brain throughout childhood and adolescence (2–18 years).32 In a study in 439 adolescents and adults, Dennis et al36 also observed increasing global brain network efficiency and decreasing local connectivity with age: path length, mean clustering coefficient and normalized clustering all decreased with age, suggesting that this period of development is marked by increasing network integration. Our group examined the maturational changes of the cortical connectome across pre-mature neonates, term-born neonates scanned in the first days of life, 6-month-old infants and adults using a template-free analysis.40 Figure 5 illustrates the developmental trajectory of the template-free structural brain network. In line with the observations by Hagmann et al,32 Dennis et al36 and Huang et al,37 this approach also showed that scaled characteristic path length and clustering coefficient both decrease with higher age in term-born subjects.
Figure 5.
Maturation of the structural connectome: examples of brain networks at four different ages. (a) Anatomic T2 weighted MRI images. (b) Tractograms reconstructed based on diffusion tensor imaging data. (c) Brain networks represented as graphs. The size of the nodes is proportional to the node degree. (d) Binary connectivity matrices, reordered in a way that maximizes the number of connections close to the main diagonal. Note: the 6 days and 6 months networks were mapped in the same infant longitudinally. Image modified from Tymofiyeva et al40 with permission from Public Library of Science.
Somewhat different results have been reported by other groups. Constant global efficiency and increased local efficiency were measured in a longitudinal DTI-based study of subjects at the ages of 2 weeks, 1 year and 2 years.35 A separate longitudinal study of subjects at 1 month, 1 year and 2 years, in which the brain networks were derived from correlations between regional grey matter volumes, reported increasing global efficiency, and, from 1 to 2 years, increasing local efficiency.33 Khundrakpam et al39 reported the presence of a critical time window in late childhood (8.5–11.3 years) with increased global efficiency and decreased local efficiency, indicating that structural brain networks may transiently take on a more random configuration during this developmental period.
Increases in global efficiency that were observed in most studies to date can be explained by the strengthening of axonal projections, particularly longer range association projections, during the first years of life. This increasingly integrated topology during development seems to be followed by a plateau lasting through the majority of adulthood before reverting to an increasingly localized topology in late life.5
All studies to date (Table 2) have confirmed that developing structural brain networks exhibit small-worldness.66 This property reflects the fact that the minimum number of steps necessary to travel between any two individual brain regions remains small throughout the brain, even though a direct connection may not always exist between two areas. The small-world index has been observed to decrease with age in a number of studies,32,36,37,40 mediated by a stronger decrease of the clustering coefficient than characteristic path length.
In network science, nodes that are most highly interconnected with other nodes are referred to as network hubs. One can use the node degree or other measures of centrality to identify these nodes. The high centrality of hubs also renders them points of vulnerability, susceptible to disconnection in brain disorders. In the adult brain, network analyses have shown that the praecuneus, anterior and posterior cingulate cortex, insular cortex, superior frontal cortex, temporal cortex and lateral parietal cortex are hubs in the brain network.67 Hagmann et al32 reported that hub regions remain consistent between the ages of 2 and 18 years: the praecuneus, posterior cingulate cortex, superior frontal cortex and superior parietal cortex. In the study of structural brain networks at three landmark cross-sectional ages (neonates, toddlers and pre-adolescents), Huang et al37 found that three regions were the common hubs for all the three groups: the praecuneus, posterior cingulate gyrus and right cuneus.
Recent observations have suggested that structural brain hubs are not only highly connected with the rest of the brain, but are also more highly interconnected among each other than predicted on the basis of their degree alone, hence giving rise to the concept of a “structural core” or “rich club”.67,68 Rich-club organization of neural hubs tends to further boost the robustness of interhub communication, promoting efficient communication across many large regions of the brain. Dennis et al69 charted the developmental trajectory of the rich club in brain networks from 438 subjects aged 12–30 years. The adult and younger cohorts had rich clubs that included different nodes; the rich club effect intensified with age.
Another important way of analysing the organization of brain networks is to detect network communities, or modules—sets of network nodes that are more densely linked among each other than with other nodes in the network. Hagmann et al32 identified modules by optimizing the modularity score and found that no major reorganization of structural modules was observed after 2 years of age. They also observed that, regardless of age, the lengths of fibre pathways linking regions within the same module were significantly shorter than pathways linking regions in different modules. This pattern of connection length distribution reflects the tendency, conserved between 2 and 18 years, of spatially adjacent regions to belong to the same module. The same tendency was observed by Tymofiyeva et al40 who used the maximized modularity to group equal-area nodes into modules. They reported a relatively consistent optimal number of modules in all studied age groups: 5–6 modules (average, 5.38) in the pre-term-born neonates, 5–7 modules (average, 5.75) in the term-born neonates, 5–8 modules (average, 6.6) in the 6-month-old infants and 5–7 modules (average, 5.71) in the adults. Although no prior anatomical information was used to define nodes and modules, the result matched anatomy reasonably well. Similar numbers of modules were detected by Huang et al37 in an atlas-based network analysis of brain connectivity in neonates, toddlers and pre-adolescents. There was also some convergence among studies regarding the decrease of the modularity measure with age (which quantifies decomposability of the network into smaller subnetworks).
Disrupted connectome
According to the World Health Organization (2012),70 more than one in ten babies are born pre-term, which puts them at risk of a lifetime of disability. This population, as well as those infants who sustained perinatal injury, depends on effective therapies and our ability to detect and monitor individual deviations from the normal developmental trajectory. Although visual and motor disabilities often have imaging correlates, it is very difficult to predict other developmental abnormalities in these babies using anatomic imaging. We expect that understanding of the structure, performance and plasticity of the human brain network and its evolution across the lifespan will help in identifying patients who are at risk of developing behavioural or cognitive abnormalities.
Some recent studies have utilized whole-brain structural connectivity analysis in different clinical paediatric populations (Table 3), for example, correlating global network properties with the outcome after hypoxic ischaemic encephalopathy.43,51 Intrauterine growth restriction alters brain network topology at 1 year.41 Pandit et al44 used an optimized processing pipeline, combining anatomical and tissue segmentations with probabilistic diffusion tractography to map mean tract anisotropy in pre-maturely born infants. The “diffusive exchange” between voxels (a measure of anisotropy) integrated across the whole tract was used as a marker of the connection strength. White matter tracts, where diffusive exchange was related to age of delivery or imaging, were identified. Older children had stronger connected tracts predominantly in frontal lobe structures. Increasing pre-maturity at birth was related to widespread reductions in connectivity in all cortical lobes and several subcortical structures.
Unexpected results in pre-mature vs term-born neonates were found by Tymofiyeva et al.40 Pre-mature neonates deviated from the general trends in global network properties. This finding might be due to non-linearity of the developmental curve, differences between developmental curves of pre-maturely and term-born subjects or owing to technical difficulties that accompany MRI scanning of this challenging population.
Shi et al49 studied organizational changes in 49 children with autism and 51 typically developing controls scanned at the ages of 6–15 years, by analysing their brain networks mapped based on inter-regional cortical thickness correlations. In comparison with controls, autistic children showed a significant reduction in network modularity. At the same time, the autistic brain network demonstrated an increase in intra- and intermodule connectivity in brain regions, including middle frontal gyrus, inferior parietal gyrus and cingulate. Since these regions are associated with self-reference and episodic memory, the observed differences might be indicative of an underlying compensatory mechanism in children with autism.
MRI connectomics has potential to become an imaging biomarker of poor neurodevelopmental outcome in infants with pre-natal or perinatal diseases. Additionally, monitoring the brain plasticity changes could provide a basis for developing and optimizing therapies to improve outcomes after acquired brain injuries. A review by Dennis and Thompson4 provides greater detail on changes of structural and functional connectivity in atypical brain development.
FUTURE DIRECTIONS
Although MRI connectomics is still in an early phase of development, it is clear that its application to the study of the developing brain will significantly benefit our understanding of this complex but very important topic. In particular, the field could benefit by addressing the following issues:
1. Longitudinal studies. The majority of paediatric imaging studies to date have made longitudinal or developmental inferences from cross-sectional data, a practice that can be problematic at best for reasons that include biases associated with differences in sampling across age groups, time displacement and unequal error variances.71 To address these difficulties, longitudinal connectome studies are required. Such studies will allow direct examination of the trajectories of post-natal human brain development and, presumably, more valid inferences about their functional correlates.
2. Improved tractography algorithms and biologically meaningful quantification of connectivity (weights). Foreseeable improvements in diffusion data acquisition and analysis tools can be expected to improve the accuracy of structural networks in both paediatric and adult subjects. As highlighted in this review, there is a demand for quantitative estimates of the connection strength that will accurately reflect the complex microstructure of white matter and serve as weights of the edges in the connectome. A number of techniques using diffusion imaging are being developed to provide more accurate estimates of axonal density and diameter and the degree to which individual tracts are myelinated.72–74 Acquisition of multiple b-values is required for complex model fitting.
3. Direct comparison of parcellation schemes. Another advance would be to find robust alternative means of brain parcellation, which are not based on sulcal and gyral landmarks. Direct comparison and standardization of parcellation schemes would benefit the developing field of connectomics.
4. Inclusion of subcortical areas. Despite persistent controversy regarding how subcortical areas should be included and weighted, together with cortical regions, these structures are important for an unbiased brain network study.75 Li et al59 showed a prominent structural role of the subcortical regions: among the top 20% nodes with the highest betweenness centrality values, the subcortical regions constitute 37.5–43.8%, much higher than their representation as a proportion of the total number of brain regions included (17.1%). Therefore, connectome mapping studies that exclude subcortical areas may be missing important connections involving subcortical–cortical coupling and/or corticocortical circuits that loop through subcortical structures. Ball et al76 studied the influence of pre-term birth on the developing thalamocortical connectivity (called by the authors “thalamocortical connectome”), demonstrating that connections between the thalamus and the frontal cortex, supplementary motor areas, occipital lobe and temporal gyri were significantly diminished in the pre-term infants. Indeed, thalamocortical connectivity is known to be a critical component of cerebral cortex development and of cerebrocerebellar communications. Therefore, future models should be constructed to include the deep cerebral nuclei.
5. Understanding the relationship between structural and functional connectivity. Although functional connectivity, strictly speaking, does not provide a “connectome” description,28 it offers a complementary approach, and the full description of both structural and functional connectivity is crucial in understanding normal and abnormal maturation of the brain as a whole. Several reviews have summarized explorations of the functional organization of the developing brain.77–79 The combined use of structural and functional imaging in the same subject would ultimately allow for degenerate (many-to-one, depending on the task) function–structure mapping, which is crucial for understanding the nature of brain networks.
6. Validating models against clinical data. Models of perturbed formation of the connectome need to be further tested and verified in larger clinical samples.
FUNDING
This work is supported by the National Institutes of Health grants R01EB09756, R01HD72074 and R01NS046432.
REFERENCES
- 1.Tau GZ, Peterson BS. Normal development of brain circuits. Neuropsychopharmacology 2010; 35: 147–68. doi: 10.1038/npp.2009.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones DK. Challenges and limitations of quantifying brain connectivity in vivo with diffusion MRI. Imaging Med 2010; 2: 341–55. [Google Scholar]
- 3.Hagmann P, Grant PE, Fair DA. MR connectomics: a conceptual framework for studying the developing brain. Front Syst Neurosci 2012; 6: 43. doi: 10.3389/fnsys.2012.00043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dennis EL, Thompson PM. Mapping connectivity in the developing brain. Int J Dev Neurosci 2013; 31: 525–42. doi: 10.1016/j.ijdevneu.2013.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collin G, van den Heuvel MP. The ontogeny of the human connectome: development and dynamic changes of brain connectivity across the life span. Neuroscientist 2013; 19: 616–28. [DOI] [PubMed] [Google Scholar]
- 6.Barkovich AJ, Raybaud C, eds. Pediatric neuroimaging. 5th edn. Philadelphia, PA: Lippincott Williams & Wilkins; 2012. pp. 1–80. [Google Scholar]
- 7.Giedd JN, Rapoport JL. Structural MRI of pediatric brain development: what have we learned and where are we going? Neuron 2010; 67: 728–34. doi: 10.1016/j.neuron.2010.08.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakic P. The neocortex: ontogeny and phylogeny. In: Finlay BL, Innocenti G, Scheich H, eds. New York, NY: Plenum Press; 1990. pp. 21– 32. [Google Scholar]
- 9.Lenroot RK, Giedd JN. Brain development in children and adolescents: insights from anatomical magnetic resonance imaging. Neurosci Biobehav Rev 2006; 30: 718–29. doi: 10.1016/j.neubiorev.2006.06.001 [DOI] [PubMed] [Google Scholar]
- 10.Huttenlocher PR. Synaptic density in human frontal cortex—developmental changes and effects of aging. Brain Res 1979; 163: 195–205. [DOI] [PubMed] [Google Scholar]
- 11.Levine D, Barnes PD. Cortical maturation in normal and abnormal fetuses as assessed with prenatal MR imaging. Radiology 1999; 210: 751–8. doi: 10.1148/radiology.210.3.r99mr47751 [DOI] [PubMed] [Google Scholar]
- 12.Kinney HC, Brody BA, Kloman AS, Gilles FH. Sequence of central nervous system myelination in human infancy. II. Patterns of myelination in autopsied infants. J Neuropathol Exp Neurol 1988; 47: 217–34. [DOI] [PubMed] [Google Scholar]
- 13.Barkovich AJ, Kjos BO, Jackson DEJr, Norman D. Normal maturation of the neonatal and infant brain: MR imaging at 1.5 T. Radiology 1988; 166: 173–80. doi: 10.1148/radiology.166.1.3336675 [DOI] [PubMed] [Google Scholar]
- 14.Deoni SC, Mercure E, Blasi A, Gasston D, Thomson A, Johnson M, et al. Mapping infant brain myelination with magnetic resonance imaging. J Neurosci 2011; 31: 784–91. doi: 10.1523/JNEUROSCI.2106-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yakovlev PI, Lecours A. The myelogenetic cycles of regional maturation of the brain. In: Minkovski A, ed. Regional development of the brain in early life. Oxford, UK: Blackwell; 1967. pp. 3–65. [Google Scholar]
- 16.Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 1999; 45: 265–9. [DOI] [PubMed] [Google Scholar]
- 17.Wedeen VJ, Wang RP, Schmahmann JD, Benner T, Tseng WY, Dai G, et al. Diffusion spectrum magnetic resonance imaging (DSI) tractography of crossing fibers. Neuroimage 2008; 41: 1267–77. doi: 10.1016/j.neuroimage.2008.03.036 [DOI] [PubMed] [Google Scholar]
- 18.Lodygensky GA, Vasung L, Sizonenko SV, Hüppi PS. Neuroimaging of cortical development and brain connectivity in human newborns and animal models. J Anat 2010; 217: 418–28. doi: 10.1111/j.1469-7580.2010.01280.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huppi PS, Maier SE, Peled S, Zientara GP, Barnes PD, Jolesz FA, et al. Microstructural development of human newborn cerebral white matter assessed in vivo by diffusion tensor magnetic resonance imaging. Pediatr Res 1998; 44: 584–90. [DOI] [PubMed] [Google Scholar]
- 20.Neil JJ, Shiran SI, McKinstry RC, Schefft GL, Snyder AZ, Almli CR, et al. Normal brain in human newborns: apparent diffusion coefficient and diffusion anisotropy measured by using diffusion tensor MR imaging. Radiology 1998; 209: 57–66. [DOI] [PubMed] [Google Scholar]
- 21.Dudink J, Lequin M, van Pul C, Buijs J, Conneman N, van Goudoever J, et al. Fractional anisotropy in white matter tracts of very-low-birth-weight infants. Pediatr Radiol 2007; 37: 1216–23. doi: 10.1007/s00247-007-0626-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huppi P. DTI in brain development. In: Jones DK. ed. Diffusion MRI: theory, methods, and applications. New York, NY: Oxford University Press; 2010. [Google Scholar]
- 23.Partridge SC, Mukherjee P, Henry RG, Miller SP, Berman JI, Jin H, et al. Diffusion tensor imaging: serial quantitation of white matter tract maturity in premature newborns. Neuroimage 2004; 22: 1302–14. doi: 10.1016/j.neuroimage.2004.02.038 [DOI] [PubMed] [Google Scholar]
- 24.Partridge SC, Mukherjee P, Berman JI, Henry RG, Miller SP, Lu Y, et al. Tractography-based quantitation of diffusion tensor imaging parameters in white matter tracts of preterm newborns. J Magn Reson Imaging 2005; 22: 467–74. doi: 10.1002/jmri.20410 [DOI] [PubMed] [Google Scholar]
- 25.Yoo SS, Park HJ, Soul JS, Mamata H, Park H, Westin CF, et al. In vivo visualization of white matter fiber tracts of preterm- and term-infant brains with diffusion tensor magnetic resonance imaging. Invest Radiol 2005; 40: 110–5. [DOI] [PubMed] [Google Scholar]
- 26.Sporns O, Tononi G, Kötter R. The human connectome: a structural description of the human brain. PLoS Comput Biol 2005; 1: e42. doi: 10.1371/journal.pcbi.0010042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hagmann P. From Diffusion MRI to Brain Connectomics. PhD thesis. Lausanne: Ecole Polytechnique Fédérale de Lausanne; 2005.
- 28.Sporns O. Making sense of brain network data. Nat Methods 2013; 10: 491–3. doi: 10.1038/nmeth.2485 [DOI] [PubMed] [Google Scholar]
- 29.Rubinov M, Sporns O. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 2010; 52: 1059–69. doi: 10.1016/j.neuroimage.2009.10.003 [DOI] [PubMed] [Google Scholar]
- 30.Griffa A, Baumann PS, Thiran JP, Hagmann P. Structural connectomics in brain diseases. Neuroimage 2013; 80: 515–26. doi: 10.1016/j.neuroimage.2013.04.056 [DOI] [PubMed] [Google Scholar]
- 31.Bullmore ET, Bassett DS. Brain graphs: graphical models of the human brain connectome. Annu Rev Clin Psychol 2011; 7: 113–40. doi: 10.1146/annurev-clinpsy-040510-143934 [DOI] [PubMed] [Google Scholar]
- 32.Hagmann P, Sporns O, Madan N, Cammoun L, Pienaar R, Wedeen VJ, et al. White matter maturation reshapes structural connectivity in the late developing human brain. Proc Natl Acad Sci USA 2010; 107: 19067–72. doi: 10.1073/pnas.1009073107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fan Y, Shi F, Smith JK, Lin W, Gilmore JH, Shen D. Brain anatomical networks in early human brain development. Neuroimage 2011; 54: 1862–71. doi: 10.1016/j.neuroimage.2010.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage 2002; 15: 273–89. doi: 10.1006/nimg.2001.0978 [DOI] [PubMed] [Google Scholar]
- 35.Yap PT, Fan Y, Chen Y, Gilmore JH, Lin W, Shen D. Development trends of white matter connectivity in the first years of life. PLoS One 2011; 6: e24678. doi: 10.1371/journal.pone.0024678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dennis EL, Jahanshad N, McMahon KL, de Zubicaray GI, Martin NG, Hickie IB, et al. Development of brain structural connectivity between ages 12 and 30: a 4-Tesla diffusion imaging study in 439 adolescents and adults. Neuroimage 2013; 64: 671–84. doi: 10.1016/j.neuroimage.2012.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang H, Shu N, Mishra V, Jeon T, Chalak L, Wang ZJ, et al. Development of human brain structural networks through infancy and childhood. Cereb Cortex 2013; December 2011. Epub ahead of print. doi: 10.1093/cercor/bht335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, et al. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philos Trans R Soc Lond B Biol Sci 2001; 356: 1293–322. doi: 10.1098/rstb.2001.0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khundrakpam BS, Reid A, Brauer J, Carbonell F, Lewis J, Ameis S, et al. Developmental changes in organization of structural brain networks. Cereb Cortex 2013; 23: 2072–85. doi: 10.1093/cercor/bhs187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tymofiyeva O, Hess CP, Ziv E, Lee PN, Glass HC, Ferriero DM, et al. A DTI-based template-free cortical connectome study of brain maturation. PLoS One 2013; 8: e63310. doi: 10.1371/journal.pone.0063310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batalle D, Eixarch E, Figueras F, Muñoz-Moreno E, Bargallo N, Illa M, et al. Altered small-world topology of structural brain networks in infants with intrauterine growth restriction and its association with later neurodevelopmental outcome. Neuroimage 2012; 60: 1352–66. [DOI] [PubMed] [Google Scholar]
- 42.Shi F, Yap PT, Wu G, Jia H, Gilmore JH, Lin W, et al. Infant brain atlases from neonates to 1- and 2-year-olds. PLoS One 2011; 6: e18746. doi: 10.1371/journal.pone.0018746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tymofiyeva O, Hess CP, Ziv E, Tian N, Bonifacio SL, McQuillen PS, et al. Towards the “baby connectome”: mapping the structural connectivity of the newborn brain. PLoS One 2012; 7: e31029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pandit AS, Robinson E, Aljabar P, Ball G, Gousias IS, Wang Z, et al. Whole-brain mapping of structural connectivity in infants reveals altered connection strength associated with growth and preterm birth. Cereb Cortex March 2013. Epub ahead of print. doi: 10.1093/cercor/bht086 [DOI] [PubMed]
- 45.Gousias IS, Rueckert D, Heckemann RA, Dyet LE, Boardman JP, Edwards AD, et al. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. Neuroimage 2008; 40: 672–84. doi: 10.1016/j.neuroimage.2007.11.034 [DOI] [PubMed] [Google Scholar]
- 46.Pannek K, Hatzigeorgiou X, Colditz PB, Rose S. Assessment of structural connectivity in the preterm brain at term equivalent age using diffusion MRI and T2 relaxometry: a network-based analysis. PLoS One 2013; 8: e68593. doi: 10.1371/journal.pone.0068593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oishi K, Mori S, Donohue PK, Ernst T, Anderson L, Buchthal S, et al. Multi-contrast human neonatal brain atlas: application to normal neonate development analysis. Neuroimage 2011; 56: 8–20. doi: 10.1016/j.neuroimage.2011.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zalesky A, Fornito A, Bullmore ET. Network-based statistic: identifying differences in brain networks. Neuroimage 2010; 53: 1197–207. doi: 10.1016/j.neuroimage.2010.06.041 [DOI] [PubMed] [Google Scholar]
- 49.Shi F, Wang L, Peng Z, Wee CY, Shen D. Altered modular organization of structural cortical networks in children with autism. PLoS One 2013; 8: e63131. doi: 10.1371/journal.pone.0063131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006; 31: 968–80. [DOI] [PubMed] [Google Scholar]
- 51.Ziv E, Tymofiyeva O, Ferriero DM, Barkovich AJ, Hess CP, Xu D. A machine learning approach to automated structural network analysis: application to neonatal encephalopathy. PLoS One 2013; 8: e78824. doi: 10.1371/journal.pone.0078824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pannek K, Guzzetta A, Colditz PB, Rose SE. Diffusion MRI of the neonate brain: acquisition, processing and analysis techniques. Pediatr Radiol 2012; 42: 1169–82. doi: 10.1007/s00247-012-2427-x [DOI] [PubMed] [Google Scholar]
- 53.Lerch JP, Worsley K, Shaw WP, Greenstein DK, Lenroot RK, Giedd J, et al. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage 2006; 31: 993–1003. doi: 10.1016/j.neuroimage.2006.01.042 [DOI] [PubMed] [Google Scholar]
- 54.Jones DK, Knösche TR, Turner R. White matter integrity, fiber count, and other fallacies: the do’s and don’ts of diffusion MRI. Neuroimage 2013; 73: 239–54. [DOI] [PubMed] [Google Scholar]
- 55.Robinson EC, Hammers A, Ericsson A, Edwards AD, Rueckert D. Identifying population differences in whole-brain structural networks: a machine learning approach. Neuroimage 2010; 50: 910–9. doi: 10.1016/j.neuroimage.2010.01.019 [DOI] [PubMed] [Google Scholar]
- 56.Bells S, Cercignani M, Deoni S, Assaf Y, Pasternak O, Evans CJ, et al. Tractometry–comprehensive multi-modal quantitative assessment of white matter along specific tracts. Proc Intl Soc Mag Reson Med 2011; 19. [Google Scholar]
- 57.Dubois J, Hertz-Pannier L, Dehaene-Lambertz G, Cointepas Y, Le Bihan D. Assessment of the early organization and maturation of infants’ cerebral white matter fiber bundles: a feasibility study using quantitative diffusion tensor imaging and tractography. Neuroimage 2006; 30: 1121–32. [DOI] [PubMed] [Google Scholar]
- 58.Hermoye L, Saint-Martin C, Cosnard G, Lee SK, Kim J, Nassogne MC, et al. Pediatric diffusion tensor imaging: normal database and observation of the white matter maturation in early childhood. Neuroimage 2006; 29: 493–504. doi: 10.1016/j.neuroimage.2005.08.017 [DOI] [PubMed] [Google Scholar]
- 59.Li L, Rilling JK, Preuss TM, Glasser MF, Hu X. The effects of connection reconstruction method on the interregional connectivity of brain networks via diffusion tractography. Hum Brain Mapp 2012; 33: 1894–913. doi: 10.1002/hbm.21332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson PM, Schwartz C, Lin RT, Khan AA, Toga AW. Three-dimensional statistical analysis of sulcal variability in the human brain. J Neurosci 1996; 16: 4261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tymofiyeva O, Ziv E, Barkovich AJ, Hess CP, Xu D. Brain without anatomy: construction and comparison of fully network-driven structural MRI connectomes. PLoS One 2014; 9: e96196. doi: 10.1371/journal.pone.0096196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meskaldji DE, Fischi-Gomez E, Griffa A, Hagmann P, Morgenthaler S, Thiran JP. Comparing connectomes across subjects and populations at different scales. Neuroimage 2013; 80: 416–25. doi: 10.1016/j.neuroimage.2013.04.084 [DOI] [PubMed] [Google Scholar]
- 63.Genovese CR, Lazar NA, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. Neuroimage 2002; 15: 870–8. doi: 10.1006/nimg.2001.1037 [DOI] [PubMed] [Google Scholar]
- 64.Kaiser M. A tutorial in connectome analysis: topological and spatial features of brain networks. Neuroimage 2011; 57: 892–907. doi: 10.1016/j.neuroimage.2011.05.025 [DOI] [PubMed] [Google Scholar]
- 65.Li Y, Liu Y, Li J, Qin W, Li K, Yu C, et al. Brain anatomical network and intelligence. PLoS Comput Biol 2009; 5: e1000395. doi: 10.1371/journal.pcbi.1000395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Watts DJ, Strogatz SH. Collective dynamics of “small-world” networks. Nature 1998; 393: 440–2. [DOI] [PubMed] [Google Scholar]
- 67.van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci 2013; 17: 683–96. [DOI] [PubMed] [Google Scholar]
- 68.Colizza V, Flammini A, Serrano MA, Vespignani A. Detecting rich-club ordering in complex networks. Nat Phys 2006; 2: 110–15. [Google Scholar]
- 69.Dennis EL, Jahanshad N, Toga AW, McMahon KL, de Zubicaray GI, Hickie I, et al. Development of the “rich club” in brain connectivity networks from 438 adolescents and adults aged 12 to 30. 2013 IEEE 10th International Symposium on Biomedical Imaging (ISBI.); San Francisco, CA; 2013; 624–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.WHO. Born too soon: the global action report on preterm birth. [cited 24 May 2014]. Available from: http://www.who.int/maternal_child_adolescent/documents/born_too_soon/en/
- 71.Kraemer HC, Yesavage JA, Taylor JL, Kupfer D. How can we learn about developmental processes from cross-sectional studies, or can we? Am J Psychiatry 2000; 157: 163–71. [DOI] [PubMed] [Google Scholar]
- 72.Assaf Y, Basser PJ. Composite hindered and restricted model of diffusion (CHARMED) MR imaging of the human brain. Neuroimage 2005; 27: 48–58. doi: 10.1016/j.neuroimage.2005.03.042 [DOI] [PubMed] [Google Scholar]
- 73.Assaf Y, Blumenfeld-Katzir T, Yovel Y, Basser PJ. AxCaliber: a method for measuring axon diameter distribution from diffusion MRI. Magn Reson Med 2008; 59: 1347–54. doi: 10.1002/mrm.21577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang H, Schneider T, Wheeler-Kingshott CA, Alexander DC. NODDI: practical in vivo neurite orientation dispersion and density imaging of the human brain. Neuroimage 2012; 61: 1000–16. doi: 10.1016/j.neuroimage.2012.03.072 [DOI] [PubMed] [Google Scholar]
- 75.Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, et al. Mapping the structural core of human cerebral cortex. PLoS Biol 2008; 6: e159. doi: 10.1371/journal.pbio.0060159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ball G, Boardman JP, Aljabar P, Pandit A, Arichi T, Merchant N, et al. The influence of preterm birth on the developing thalamocortical connectome. Cortex 2013; 49: 1711–21. doi: 10.1016/j.cortex.2012.07.006 [DOI] [PubMed] [Google Scholar]
- 77.Power JD, Fair DA, Schlaggar BL, Petersen SE. The development of human functional brain networks. Neuron 2010; 67: 735–48. doi: 10.1016/j.neuron.2010.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Smyser CD, Snyder AZ, Neil JJ. Functional connectivity MRI in infants: exploration of the functional organization of the developing brain. Neuroimage 2011; 56: 1437–52. doi: 10.1016/j.neuroimage.2011.02.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hoff GE, Van den Heuvel MP, Kersbergen KJ. On development of functional brain connectivity in the young brain. Front Hum Neurosci 2013; 7: 650. doi: 10.3389/fnhum.2013.00650 [DOI] [PMC free article] [PubMed] [Google Scholar]