Abstract
Aim:
To investigate the roles of P21-activated kinase 5 (PAK5) in proliferation and tumorigenicity of human hepatocellular carcinoma (HCC).
Methods:
HCC and matched paraneoplastictis tissue samples were obtained from 30 patients. Human HCC cell lines SMMC7721, HepG2, Hep3B, SK-HEP-1, Huh-7, and liver cell line HL-7702 were examined. The expression of PAK5 gene was studied using real-time qPCR and Western blotting. Cell proliferation was quantified with the MTT assay. Cell cycle was analyzed with flow cytometry. The tumorigenicity of Lv-shRNA-transfected HepG2 cells was evaluated in BALB/cA nude mice.
Results:
The mRNA level of PAK5 was significantly higher in 25 out of 30 HCC samples compared to the matched paraneoplastic tissues. The HCC cell lines showed varying expression of PAK5 protein, and the highest level was found in the HepG2 cells. PAK5 gene silencing in HepG2 cells markedly reduced the cell proliferation and colony formation, and induced cell cycle arrest in the G1 phase. Furthermore, PAK5 gene silencing suppressed the tumor formation in nude mice, and significantly decreased the expression of HCC-related genes Cyclin D1 and beta-catenin.
Conclusion:
PAK5 may play essential roles in the initiation and progression of human HCC. Thus, it may be an effective therapeutic target or perhaps serve as a clinical diagnostic or prognostic marker in human HCC.
Keywords: P21-activated kinase 5 (PAK5), human hepatocellular carcinoma, HepG2 cell, Cyclin D1, beta-catenin, cell cycle arrest, cell proliferation, tumorigenesis, gene silencing
Introduction
P21-activated kinases (PAKs) are a family of serine/threonine kinases that play an important role in regulating cell shape, movement, proliferation and survival1,2. PAKs are characterized by a highly conserved amino-terminal Cdc42/Rac interactive binding (CRIB) domain and a carboxyl terminal kinase domain3. PAKs are categorized into group I (PAK1, 2, and 3) and II (PAK4, 5, and 6) based on their amino acid sequences and functions4. PAK5 is the latest PAK family member to be identified, in 2002, and is considered a group II PAK based on its sequence5. Previous work by several groups has found that PAK5 has different properties from those of other members of the PAK family; for example, the Cdc42/Rac GTPase binds to PAK5 but does not regulate PAK5 kinase activity6. Unlike the group I PAKs, binding of Cdc42 is not necessary for the kinase activity of PAK55. Additionally, the CRIB domain of PAK5 is critical for appropriate subcellular localization of the protein7, and this localization is independent of kinase activity or Cdc42 binding6. PAK5 phosphorylates BAD on Ser-112, which prevents the localization of BAD to the mitochondria and inhibits the apoptotic cascade6. Finally, the multiple locations of the protein indicate that PAK5 most likely plays an important role in distinct signaling pathways. Research suggests that both, the mitochondrial localization and kinase activity of PAK5, are vital to its effects on apoptosis6,8.
PAK5 is primarily expressed in adult neuronal tissue9, with higher levels of expression detected in the cerebellum, cerebral cortex and olfactory bulb5. PAK5 has been found to regulate neurite development by operating downstream of Cdc42 and Rac and antagonizing Rho in this pathway9. The other members of the PAK family have been identified as regulators of tumor formation in the breast10, colon11, ovary12, pancreas13, and prostate14. Although the role of PAK5 in cancer progression has not been fully investigated, two recent reports suggest that PAK5 is overexpressed in colorectal cancer and can promote cancer-cell invasion15,16. Furthermore, the combined inhibition of PAK7, MAP3K7, and CK2a kinases was shown to inhibit the growth of MiaPaCa2 pancreatic cancer cell xenografts17.
Hepatocellular carcinoma (HCC) is the most common human cancer, with approximately 750 000 new cases occurring worldwide each year18. Despite its high prevalence, the molecular mechanisms underlying HCC initiation, maintenance, and progression are poorly understood. As noted above, the significance of PAK5 in cancer cell biology is being studied although its function in HCC remains unknown. In our current study, we found that PAK5 gene expression is highly elevated in HCC cell lines and tumor samples. Moreover, silencing PAK5 gene expression in the HCC cell line HepG2 significantly reduced cell proliferation and colony formation in vitro, tumor formation in nude mice, and HCC-related gene expression. Thus, these studies provide new insight into the biological effects of PAK5 in HCC and may be a useful reference for clinical diagnosis of HCC.
Materials and methods
Tissue preparation
The tissue samples utilized in this study were collected retrospectively from 30 HCC patients; the disease was confirmed by histopathological analyses after surgical resection. Paraneoplastic tissues were also obtained from the same group of 30 patients. All paraffin-embedded human tissue samples were archival remnants of tissues resected for clinical purposes and were obtained from the Department of Hepatobiliary Surgery of Taizhou Hospital (Linhai, China). Tissue samples were collected and used for qPCR and immunohistochemical assays to measure PAK5 gene expression. The study was performed in accordance with the ethical standards of the Helsinki Declaration.
Cell culture
The HCC cell lines SMMC7721, HepG2, Hep3B, SK-HEP-1, and Huh-7 and the liver cell line HL-7702 (L-02) were purchased from the American Type Culture Collection. These cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovin serum (FBS, Invitrogen, Carlsbad, CA, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin (Invitrogen, Carlsbad, CA, USA). All cells were maintained in 5% CO2.
Lentiviral vector construction, lentivirus packaging, and transfection
Oligodeoxyribonucleotides encoding two different short hairpin RNAs (shRNA) targeting PAK5 (GenBank Accession: NM 020341.3) were designed and cloned into the pGCSIL-GFP plasmid vector (GeneChem, Shanghai, China). This plasmid contains a CMV-driven eGFP reporter and a U6 promoter upstream of the restriction enzyme sites for Age I and EcoR I used for cloning. Restriction enzyme mapping and DNA sequencing helped confirm the appropriate insertion of the shRNA cassettes. The scrambled sequence 5′-TTCTCCGAACGTGTCACGT-3′ was cloned and used as a negative control (scr-shRNA). The top-strand sequences of the deoxyribonucleotides used for cloning were as follows: PAK5-shRNA1, 5′-CCGGGCCTCCATAAATATGATCTATCTCGAGATAGATCATATTTATGGAGGCTTTTT-3′ and PAK5-shRNA2, 5′-CCGGCGGGATTACCACCATGACAATCTCGAGATTGTCATGGTGGTAATCCCGTTTTT-3′. Lentiviruses were packaged and purified to yield Lv-PAK5-shRNA1, Lv-PAK5-shRNA2, and Lv-scr-shRNA, which were used to transfect cells using previously reported methods19. Silencing by PAK5-targeted shRNAs was confirmed by qPCR and Western blotting assays.
RNA extraction and real-time quantitative PCR (qPCR)
RNA was extracted from cell lines, hepatocellular carcinomas and paraneoplastic tissues with TRIzol Reagent (Gibco BRL) according to the manufacturer's instructions. Two micrograms of RNA per sample was reverse transcribed into cDNA with reverse transcriptase (MMLV-RT, Promega) using a standard protocol with random hexamer primers. MMLV was omitted in the negative control. The qPCR reactions were performed by the two-step method according to the manufacturer's instructions. Amplification was performed under the following conditions: an initial denaturation cycle at 95 °C for 15 s, 45 cycles of denaturation at 95 °C for 15 s and annealing at 60 °C for 15 s. The resulting data were analyzed with the Gene Amp Sequence Detection System (Applied Biosystems, Foster City, CA, USA), and the Ct values of the samples were normalized using GAPDH as the internal control. The expression levels of PAK5 were plotted as the log2 of the ratio between the tumor and the paraneoplastic tissue samples.
Immunohistochemical analysis
Immunohistochemistry was performed using the avidin-biotin complex protocol for paraffin-embedded tissues as previously described20. Antigen retrieval was performed in 10 mmol/L citrate buffer, pH 6, using a pressure cooker. The antibodies used, namely anti-PAK5 antibody (ab110069, 1:100) and goat anti-rabbit IgG-H&L (HRP) (ab6721, 1:100), were acquired from Abcam (Cambridge, MA, USA). For the negative controls, the primary antibody was substituted with rabbit IgG.
Western blotting
The cells were washed twice with PBS, resuspended in 2×Lysis Buffer [100 mmol/L Tris-HCl (pH 6.8), 2% mercaptoethanol, 20% glycerol and 4% SDS], and incubated on ice for 10–15 min. The cells were lysed by ultrasonication and centrifuged at 12 000×g for 15 min at 4 °C. The protein concentrations of the supernatants were measured by the Bradford assay and adjusted to 2 μg/μL protein. An equal volume of 2×loading buffer was added, and the samples were boiled for 5–10 min prior to fractionation by SDS-polyacrylamide gel electrophoresis. Western blots were performed according to standard procedures. Proteins were detected with the ECL Plus Western Blotting System (Amersham), and tubulin was used as the internal control. The antibodies used, namely anti-PAK5 (ab110069, 1:100), anti-Cyclin D1 (ab6152, 1:100), anti-beta catenin (ab32572, 1:100), anti-beta tubulin (ab15568, 1:100) and goat anti-rabbit IgG-H&L (HRP) (ab6721, 1:100), were from Abcam (Cambridge, MA, USA).
Cell proliferation assay
Cellular proliferation was quantified over a course of five days post-transfection by the MTT assay (Trevigen, Gaithersburg, MD, USA) according to the manufacturer's instructions. Briefly, cells were seeded in 96-well culture plates with culture medium at a density of 5×103 cells per well in triplicate wells for the Lv-PAK5-shRNA1, Lv-PAK5-shRNA2, and Lv-scr-shRNA groups. Absorbance was measured at 490 nm with a microtiter plate reader. The values were averaged from triplicate wells at each time point.
Cell cycle assay
The cells were harvested, washed twice with D-Hanks' medium, and fixed in 70% ethanol at 4 °C overnight. Subsequently, the cells were collected by centrifugation and stained with propidium iodide (PI) (50 μg/mL). RNase A (20 μg/mL) was then added, and the samples were incubated for 30 min at 37 °C. Cell cycle analysis was performed with the BD FACS Calibur (Becton Dickinson, San Jose, CA, USA), and the cell cycle phases were analyzed with FlowJo and FCS3.0 software.
Colony formation assay
To determine substrate-independent cell growth, soft agar assays were performed in 6-well plates with a 1.5-mL bottom layer and a 0.5-mL top layer. Cells (1×104/well) were distributed as a cell suspension in 3.4 mg/mL agar (Difco Laboratories, Detroit, MI, USA) in the assay medium and overlaid onto a bottom layer consisting of 5.1 mg/mL agar in the assay medium. The cells were incubated for 2 to 3 h at room temperature and then transferred to 37 °C, 5% CO2. The cell layer was covered with 400 μL of fresh assay medium every two days, and the colonies were Giemsa stained and counted after 10 d.
Tumorigenicity assay with nude mice
The tumorigenicity of Lv-shRNA transfected HepG2 cells was evaluated in BALB/cA nude mice (Taconic). The concentration of the cell suspensions was adjusted to 5×107/mL. Mice were subcutaneously injected with 0.2 mL of the cell suspension from Lv-PAK5-shRNA1, Lv-PAK5-shRNA2, or Lv-scr-shRNA transfected cells (n=5 mice per group), in the right lateral aspect of the thoracic wall. The mice were inspected for tumor growth three times a week. Animal care practices and all experiments were reviewed and approved by the Animal Committee of Tongji University School of Medicine, Shanghai, China (TJmed-010-10).
Statistical analysis
The data were analyzed by one-way ANOVA with SPSS 12.0. P<0.05 was considered statistically significant. Error bars represent the standard deviations.
Results
Elevated expression of PAK5 in HCC tissues and HCC cell lines
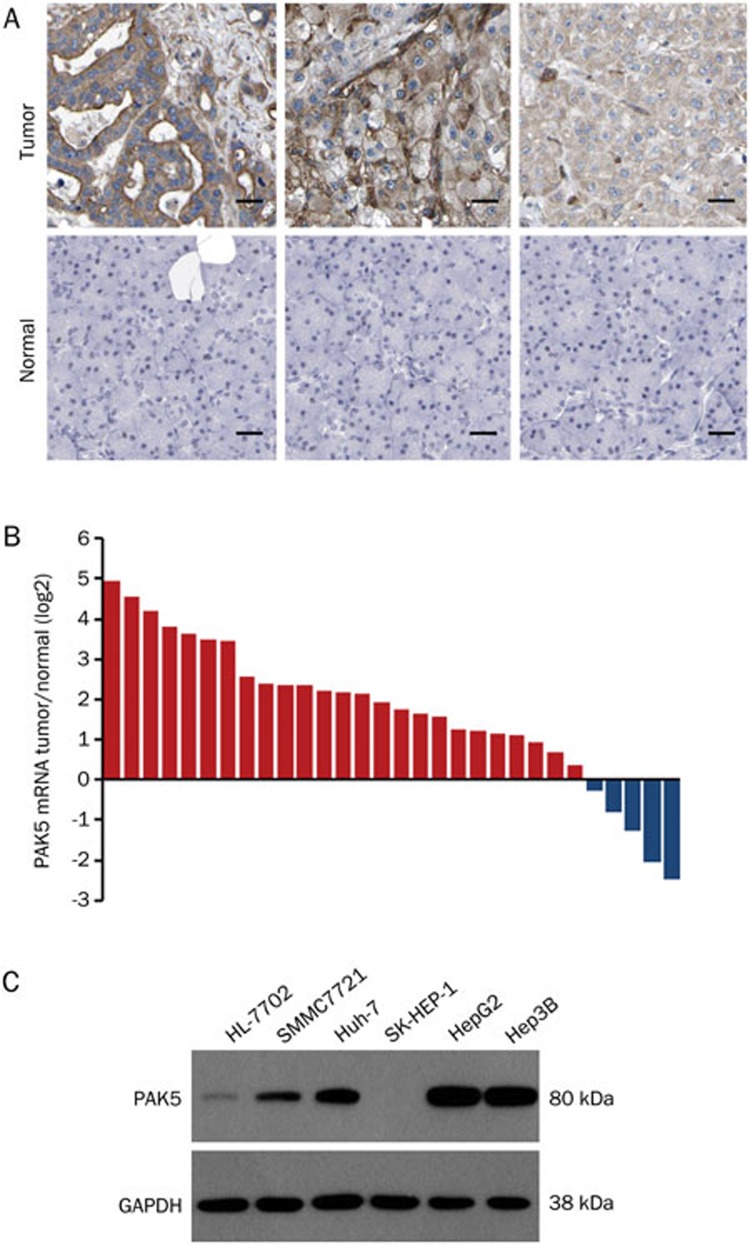
PAK5 gene expression in HCC tissues was compared to that in matched paraneoplastic tissues by qPCR and immunohistochemical analysis. PAK5 mRNA was significantly higher in 25 of 30 HCC samples (83.33%, P<0.05) than that in the corresponding paraneoplastic tissues (Figure 1B). Likewise, immunohistochemical analysis showed higher levels of PAK5 protein in the tumor samples (Figure 1A). The expression of PAK5 protein was also examined in a number of human tumor cell lines, including HCC (SMMC7721, HepG2, Hep3B, SK-HEP-1, and Huh-7), and liver cells (HL-7702). The tumor cell lines presented varying levels of PAK5 protein, and the highest expression in the tumor cell lines was observed in the HepG2 cells (P<0.05) (Figure 1C). Based on these results, we decided to examine the effects of PAK5 on cellular proliferation and tumorigenicity, employing HepG2 cells as a model.
Figure 1.
Expression analyses of PAK5 in HCC tissues and cancer cell lines. (A) The expression of PAK5 was evaluated in HCC tissue samples and normal tissue samples by immunohistochemical analysis. Three representative examples are shown. Scale bar=20 μm. (B) The relative abundance of PAK5 mRNA was evaluated in HCC tissues and matched paraneoplastic tissues by qPCR (n=30). The data are plotted as the log2 of the ratio of PAK5 mRNA. (C) PAK5 protein expression was evaluated in human tumor cell lines, including HCC cell lines (SMMC7721, HepG2, Hep3B, SK-HEP-1, and Huh-7), and a liver cell line (HL-7702) by Western blotting.
Effects of PAK5 gene silencing on cell function
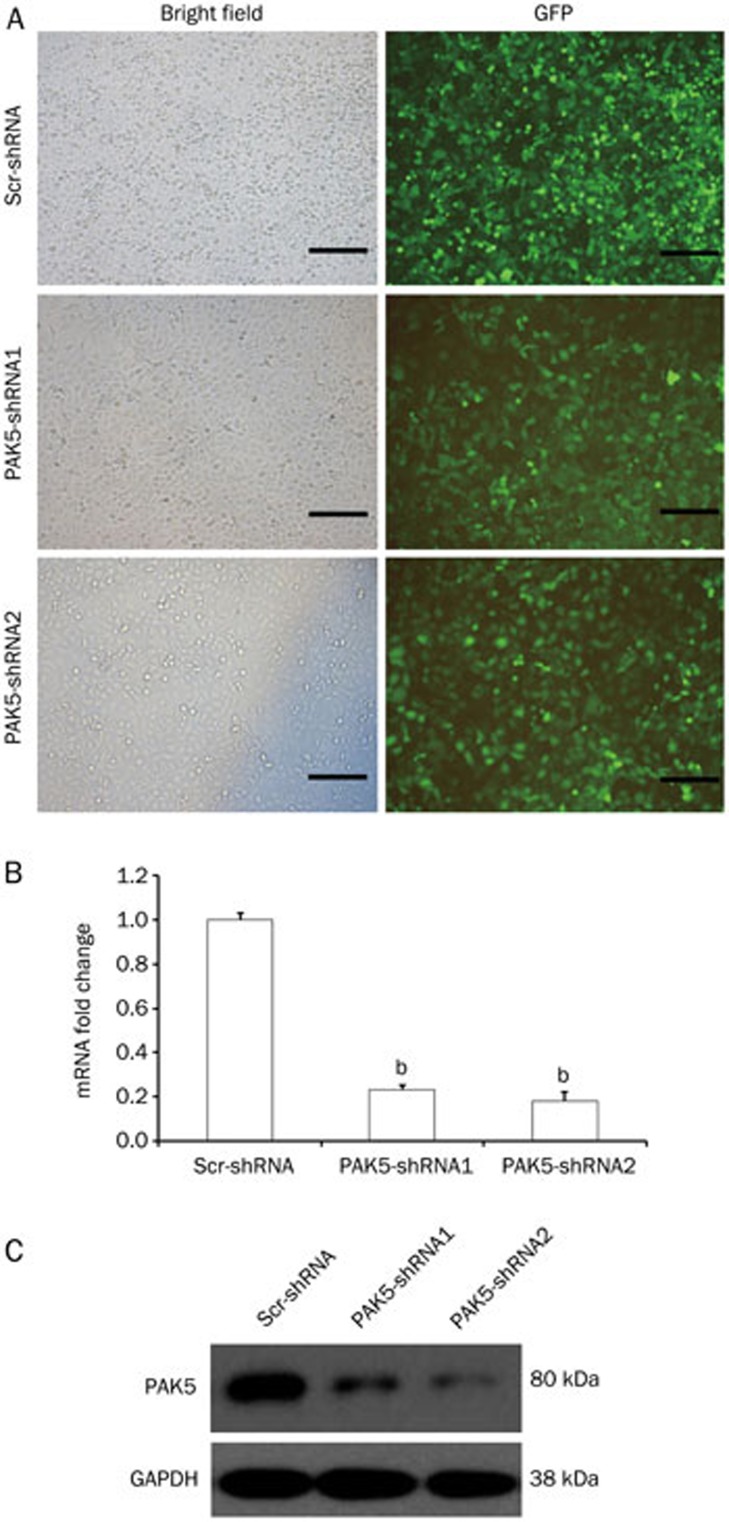
To examine the biological effects of PAK5, we designed two shRNA expression constructs for lentivirus-mediated gene silencing and used scr-shRNA as the control. The constructs also expressed GFP to assess transfection efficiency by fluorescence microscopy. At an MOI of 40, all three lentiviruses transfected HepG2 cells at a high efficiency as observed at 72 h (Figure 2A). Compared to Lv-scr-shRNA transfected cells, both Lv-PAK5-shRNA1 and Lv-PAK5-shRNA2 reduced the PAK5 mRNA levels by ∼75% (Figure 2B; P<0.05), and at the protein level, Lv-PAK5-shRNA2 appeared slightly more effective than Lv-PAK5-shRNA1 (Figure 2C). These results demonstrate the efficacy of the lentivirus constructs for PAK5 silencing.
Figure 2.
Evaluation of PAK5 gene silencing by transfection of cells with lentiviral shRNAs. Two lentiviruses, Lv-PAK5-shRNA1 and Lv-PAK5-shRNA2, each expressing a different shRNA against PAK5, were generated. A control lentivirus expressing a scrambled shRNA, Lv-scr-shRNA, was also prepared. HepG2 cells were transfected with the lentiviruses and PAK5 gene expression was evaluated after 72 h. (A) GFP expression was examined in transfected HepG2 cells by immunofluorescence (right column) (n=5. Scale bar=20 μm). The phase contrast images are also shown (left column). (B and C) PAK5 expression was evaluated in transfected HepG2 cells by qPCR (B) and Western blotting (C) (n=5). In panel (B), qPCR data are plotted as the mRNA fold change compared to Lv-scr-transfected cells, which was set at 1. bP<0.05.
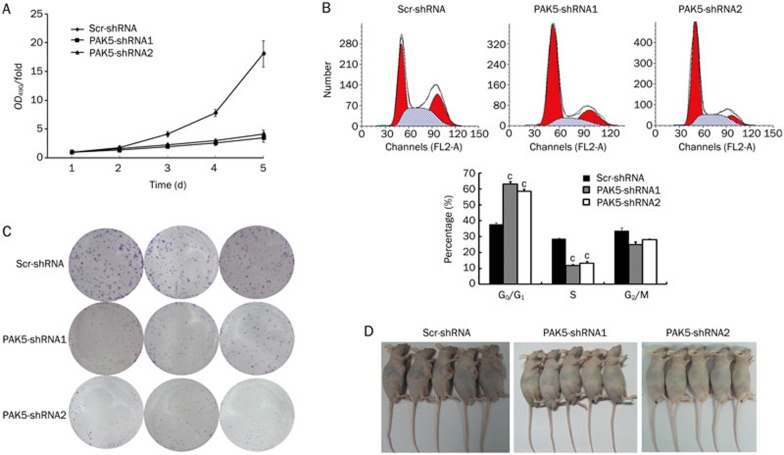
The biological effects of PAK5 silencing in HepG2 cells were further assessed by examining cell proliferation, colony formation in soft agar, and cell cycle distribution. Three days following the transfection of HepG2 cells with Lv-scr-shRNA, Lv-PAK5-shRNA1, or Lv-PAK5-shRNA2, cell proliferation was measured by the MTT assay over the ensuing five days. Transfection with either Lv-PAK5-shRNA construct markedly reduced cell proliferation compared with Lv-scr-shRNA transfected cells (Figure 3A, P<0.05). Likewise, PAK5 gene silencing strongly inhibited the ability of HepG2 cells to form colonies in soft agar (Figure 3B). Finally, flow cytometry was used to measure the DNA content of transfected cells, and the distribution of cells in the G1, S, and G2/M phases was measured. The fraction of Lv-PAK5-shRNA1- and Lv-PAK5-shRNA2-transfected cells in the G1 phase increased to 64% and 58%, respectively, with PAK5 gene silencing compared with 37% of Lv-scr-shRNA–transfected control cells in the G1 phase (Figure 3C, lower panel, P<0.05). In addition, the fraction of cells in the S phase decreased upon PAK5 gene silencing. All of these observations show that PAK5 promotes cell proliferation.
Figure 3.
Effects of PAK5 gene silencing on cell proliferation and tumorigenicity. HepG2 cells were transfected with Lv-PAK5-shRNA1, Lv-PAK5-shRNA2, or the control Lv-scr-shRNA, and biological assays were performed after 72 h. (A) Cell proliferation was evaluated by the MTT assay. The data are plotted as the OD490-fold versus time over the ensuing five days, where the values at d 1 are set to 1. The data points are the mean±SD (n=5). Cell proliferation was significantly reduced upon PAK5 gene silencing. (B) Transfected cells were analyzed by flow cytometry to determine the distribution of cell cycle phases. The data from the flow cytometry analyses (top panel) were quantified (bottom panel). Reduced PAK5 expression induced cell cycle arrest in the G1 phase (n=5). cP<0.01 vs control. (C) Tumorigenicity was evaluated in vitro by colony-formation assays and the plates were stained with Giemsa. Reduced PAK5 expression severely impaired colony formation (n=3). (D) Tumorigenicity was evaluated in vivo in nude mouse assays. Transfected cells were used to grow xenografts in nude mice. The mice were evaluated for tumor development after three weeks, and photographed as shown.The mice that underwent PAK5 gene silencing showed no visible tumors (n=5).
PAK5 gene silencing inhibits tumor formation in nude mice
Based on these observations, we next examined the effect of PAK5 gene silencing on tumor formation by HepG2 cells. The cells were transfected with the lentiviruses expressing control shRNA or shRNAs against PAK5, and tumorigenicity assays were performed with BALB/cA nude mice. The mice injected with control Lv-scr-shRNA-transfected cells developed sizable tumors at the site of the injection (Figure 3D, left), whereas mice injected with cells expressing the shRNA against PAK5 had no visible tumors (Figure 3D, middle and right; P<0.05). Taken together, these data indicate that PAK5 significantly contributes to cell proliferation and tumor formation.
PAK5 gene silencing suppresses hepatocellular cancer-related gene expression
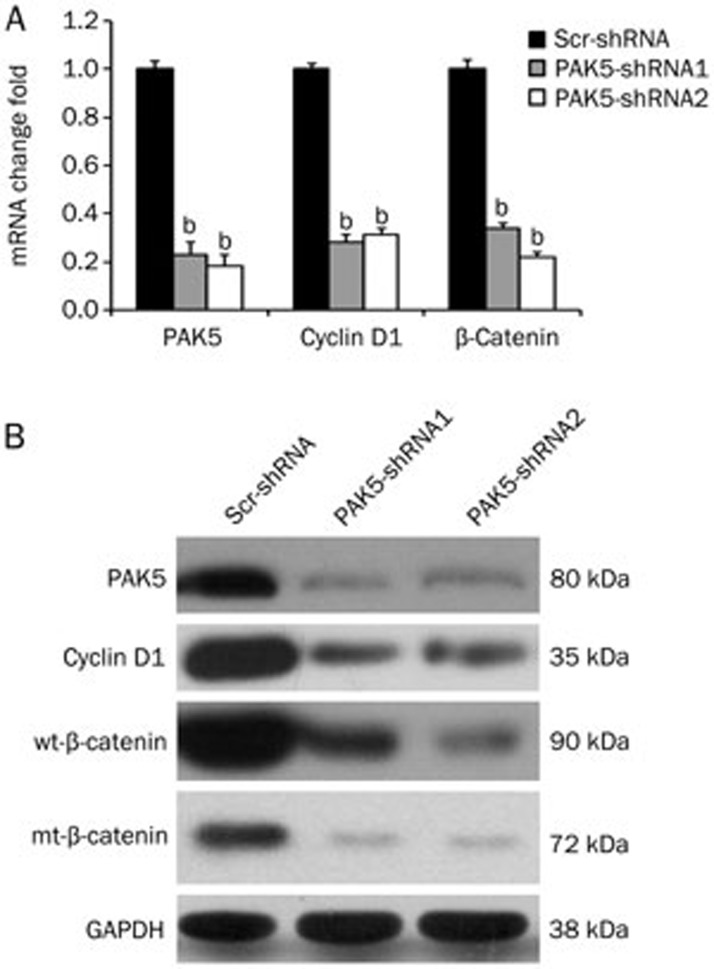
Genes encoding Cyclin D1 and beta-catenin have been implicated in hepatocellular carcinoma21,22,23,24,25,26. To determine whether PAK5 gene silencing affects the expression of these genes, HepG2 cells were transfected with the Lv-PAK5-shRNAs or control Lv-scr-shRNA as before, and the expression of Cyclin D1 and beta-catenin were analyzed by qPCR and Western blotting. We found that silencing of PAK5 reduced the mRNA expression of both Cyclin D1 and beta-catenin by 70%–80% compared to the control shRNA (Figure 4A, P<0.05). Similarly, knockdown of PAK5 protein reduced the expression of Cyclin D1 and beta-catenin proteins (Figure 4B). Whether PAK5 gene silencing has a direct effect on the expression of these genes awaits future evaluation.
Figure 4.
Effects of PAK5 gene silencing on the expression of HCC-related genes. HepG2 cells were transfected with lentiviruses expressing the shRNAs. PAK5, Cyclin D1 and beta-catenin mRNA and protein were assayed by qPCR and Western blots. (A) For the qPCR assays, the data are plotted as the mRNA fold changes relative to the control scr-shRNA for each gene. PAK5 silencing decreased Cyclin D1 and beta-catenin mRNA levels by 70%–80% (bP<0.05 vs control). (B) The Western blot analyses indicated that PAK5 silencing also decreased the Cyclin D1 and beta-catenin protein levels. One representative result is shown.
Discussion
Tumorigenesis is generally driven by the altered expression and/or function of critical genes, particularly those involved in the cell cycle27,28. Deregulation of Rho GTPases and Rho GEFs plays a significant role in oncogenic transformation and metastasis29. PAKs that interact with Rho GTPases and Rho GEFs are therefore closely associated with the signaling events that propagate cell transformation and tumor formation30. Integrins and the small GTPases Cdc42 and Rac, whose targets are the PAK family, also play an important role in tumorigenesis. Previous studies have demonstrated that group II PAK activity can influence cancer-cell behavior31. PAK4 is closely associated with the progression and metastasis of breast cancer32, the adhesion of prostate cancer cells33 and the migration of human gastric cancer cells34, and PAK6 is overexpressed in prostate cancer and breast cancer cell lines29,35.
Several studies have provided detailed insights into the role of the PAK5 protein in colorectal cancer15,16 and pancreatic cancer17. However, the role of PAK5 in HCC tumorigenesis remains poorly understood. In the present study, we first compared PAK5 gene expression in HCC versus matched paraneoplastic tissues and tumor cell lines of different origins. The PAK5 mRNA was significantly elevated in 25 of 30 (83.33%, P<0.05) patient samples, suggesting that PAK5 might play a role in tumor formation, maintenance, and/or progression. Of the six tumor cell lines investigated, four expressed significant levels of PAK5 protein, and the highest expression was observed in the HepG2 HCC cells; in contrast, the HL-7702 liver cells and the SK-HEP-1 HCC cells expressed PAK5 at very low levels.
Subsequently, we employed a strategy to silence PAK5 using a lentiviral vector-based RNAi in HepG2 cells to assess the biological consequences of PAK5 knockdown. Consistent with PAK5's role as a cell cycle regulatory molecule, PAK5 gene silencing slowed cell proliferation, most likely by arresting the cells in the G1 phase (Figure 3). Our future research will study the crosstalk between the DNA replication function of PAK5 and the G1 phase arrest observed in the current work. Furthermore, we found that PAK5 silencing in HepG2 cells reduced colony formation in soft agar, and most importantly, significantly reduced the tumor size in nude mouse xenografts. These results together demonstrated that PAK5 plays an important role in tumorigenesis. The tumor data are quite striking as mice injected with cells expressing the shRNA against PAK5 had no visible tumors in contrast to the control mice (Figure 3D). Genetic evidence supports a role for Cyclin D1 and beta-catenin activation in HCC21,22,23,24,25,26, and our results indicated that silencing of PAK5 gene expression significantly decreased Cyclin D1 and beta-catenin mRNA and protein, although the exact mechanism requires further study. This result is consistent with the reduced tumorigenicity of HepG2 cells upon PAK5 gene silencing.
In summary, our study utilized PAK5 gene silencing to elucidate the role of PAK5 in the proliferation and tumorigenicity of HCC cells. Thus, PAK5 may prove to be an effective therapeutic target or perhaps serve as a clinical diagnostic or prognostic marker in human HCC.
Author contribution
Fa-biao ZHANG, Rui-liang GE, Zhe-ping FANG, Xue-feng GU, Bin ZHAO, and Bei-ge JIANG designed the research; Zhe-ping FANG, Xue-feng GU, and Bei-ge JIANG performed the research; Fa-biao ZHANG, Rui-liang GE, Zhe-ping FANG, and Bei-ge JIANG analyzed the data; and Fa-biao ZHANG, Rui-liang GE, and Xue-feng GU wrote the paper.
Acknowledgments
This study was supported by the Science Technology Program of Zhejiang Province on the Scientific Research Project (2009C33SA800006), the National Natural Science Foundation of China (30872989), and the Scientific Research Foundation for the Junior Teachers of Medicine in the Second Military Medical University (2011QN20).
References
- Bagrodia S, Cerione RA. Pak to the future. Trends Cell Biol. 1999;9:350–5. doi: 10.1016/s0962-8924(99)01618-9. [DOI] [PubMed] [Google Scholar]
- Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–81. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Sells MA, Chernoff J. Emerging from the Pak: the p21-activated protein kinase family. Trends Cell Biol. 1997;7:162–7. doi: 10.1016/S0962-8924(97)01003-9. [DOI] [PubMed] [Google Scholar]
- Jaffer ZM, Chernoff J. p21-activated kinases: three more join the Pak. Int J Biochem Cell B. 2002;34:713–7. doi: 10.1016/s1357-2725(01)00158-3. [DOI] [PubMed] [Google Scholar]
- Pandey A, Dan I, Kristiansen TZ, Watanabe NM, Voldby J, Kajikawa E, et al. Cloning and characterization of PAK5, a novel member of mammalian p21-activated kinase-II subfamily that is predominantly expressed in brain. Oncogene. 2002;21:3939–48. doi: 10.1038/sj.onc.1205478. [DOI] [PubMed] [Google Scholar]
- Cotteret S, Jaffer ZM, Beeser A, Chernoff J. p21-Activated kinase 5 (Pak5) localizes to mitochondria and inhibits apoptosis by phosphorylating BAD. Mol Cell Biol. 2003;23:5526–39. doi: 10.1128/MCB.23.16.5526-5539.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Frost JA. Multiple Rho proteins regulate the subcellular targeting of PAK5. Biochem Bioph Res Commun. 2006;351:328–35. doi: 10.1016/j.bbrc.2006.09.172. [DOI] [PubMed] [Google Scholar]
- Cotteret S, Chernoff J. Nucleocytoplasmic shuttling of Pak5 regulates its antiapoptotic properties. Mol Cell Biol. 2006;26:3215–30. doi: 10.1128/MCB.26.8.3215-3230.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan C, Nath N, Liberto M, Minden A. PAK5, a new brain-specific kinase, promotes neurite outgrowth in N1E-115 cells. Mol Cell Biol. 2002;22:567–77. doi: 10.1128/MCB.22.2.567-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm C, Rayala S, Jirstrom K, Stal O, Kumar R, Landberg G. Association between Pak1 expression and subcellular localization and tamoxifen resistance in breast cancer patients. J Natl Cancer Inst. 2006;98:671–80. doi: 10.1093/jnci/djj185. [DOI] [PubMed] [Google Scholar]
- Carter JH, Douglass LE, Deddens JA, Colligan BM, Bhatt TR, Pemberton JO, et al. Pak-1 expression increases with progression of colorectal carcinomas to metastasis. Clin Cancer Res. 2004;10:3448–56. doi: 10.1158/1078-0432.CCR-03-0210. [DOI] [PubMed] [Google Scholar]
- Schraml P, Schwerdtfeger G, Burkhalter F, Raggi A, Schmidt D, Ruffalo T, et al. Combined array comparative genomic hybridization and tissue microarray analysis suggest PAK1 at 11q13.5-q14 as a critical oncogene target in ovarian carcinoma. Am J Pathol. 2003;163:985–92. doi: 10.1016/S0002-9440(10)63458-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlamaki EH, Kauraniemi P, Monni O, Wolf M, Hautaniemi S, Kallioniemi A. High-resolution genomic and expression profiling reveals 105 putative amplification target genes in pancreatic cancer. Neoplasia. 2004;6:432–9. doi: 10.1593/neo.04130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila M, Frost AR, Grizzle WE, Chakrabarti R. LIM kinase 1 is essential for the invasive growth of prostate epithelial cells: implications in prostate cancer. J Biol Chem. 2003;278:36868–75. doi: 10.1074/jbc.M306196200. [DOI] [PubMed] [Google Scholar]
- Gong W, An Z, Wang Y, Pan X, Fang W, Jiang B, et al. P21-activated kinase 5 is overexpressed during colorectal cancer progression and regulates colorectal carcinoma cell adhesion and migration. Int J Can. 2009;125:548–55. doi: 10.1002/ijc.24428. [DOI] [PubMed] [Google Scholar]
- Wang X, Gong W, Qing H, Geng Y, Wang X, Zhang Y, et al. p21-activated kinase 5 inhibits camptothecin-induced apoptosis in colorectal carcinoma cells. Tumour Biol. 2010;31:575–82. doi: 10.1007/s13277-010-0071-3. [DOI] [PubMed] [Google Scholar]
- Giroux V, Iovanna JL, Garcia S, Dagorn JC. Combined inhibition of PAK7, MAP3K7 and CK2alpha kinases inhibits the growth of MiaPaCa2 pancreatic cancer cell xenografts. Cancer Gene Ther. 2009;16:731–40. doi: 10.1038/cgt.2009.22. [DOI] [PubMed] [Google Scholar]
- Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Can. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- Coleman JE, Huentelman MJ, Kasparov S, Metcalfe BL, Paton JF, Katovich MJ, et al. Efficient large-scale production and concentration of HIV-1-based lentiviral vectors for use in vivo. Physiol Genomics. 2003;12:221–8. doi: 10.1152/physiolgenomics.00135.2002. [DOI] [PubMed] [Google Scholar]
- Perez-Martinez FC, Alonso V, Sarasa JL, Nam-Cha SG, Vela-Navarrete R, Manzarbeitia F, et al. Immunohistochemical analysis of low-grade and high-grade prostate carcinoma: relative changes of parathyroid hormone-related protein and its parathyroid hormone 1 receptor, osteoprotegerin and receptor activator of nuclear factor-κB ligand. J Clin Pathol. 2007;60:290–4. doi: 10.1136/jcp.2006.037853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joo M, Kang YK, Kim MR, Lee HK, Jang JJ. Cyclin D1 overexpression in hepatocellular carcinoma. Liver. 2001;21:89–95. doi: 10.1034/j.1600-0676.2001.021002089.x. [DOI] [PubMed] [Google Scholar]
- Wolfe A, Thomas A, Edwards G, Jaseja R, Guo GL, Apte U. Increased activation of the Wnt/beta-catenin pathway in spontaneous hepatocellular carcinoma observed in farnesoid X receptor knockout mice. J Pharmacol Exp Ther. 2011;338:12–21. doi: 10.1124/jpet.111.179390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmani R, Just PA, Perret C. The Wnt/beta-catenin pathway as a therapeutic target in human hepatocellular carcinoma. Clin Res Hepatol Gas. 2011;35:709–13. doi: 10.1016/j.clinre.2011.05.010. [DOI] [PubMed] [Google Scholar]
- Zekri AR, Bahnassy AA, Alam El-Din HM, Morsy HM, Shaarawy S, Moharram NZ, et al. Serum levels of beta-catenin as a potential marker for genotype 4/hepatitis C-associated hepatocellular carcinoma. Oncol Rep. 2011;26:825–31. doi: 10.3892/or.2011.1355. [DOI] [PubMed] [Google Scholar]
- Xu JM, Wen JM, Zhang M, Lu GL, Wu LZ, Wang WS. A study of gene amplification and expression of cyclin D1 in hepatocellular carcinoma. Zhonghua Bing Li Xue Za Zhi. 2004;33:26–30. [PubMed] [Google Scholar]
- Deane NG, Parker MA, Aramandla R, Diehl L, Lee WJ, Washington MK, et al. Hepatocellular carcinoma results from chronic cyclin D1 overexpression in transgenic mice. Cancer Res. 2001;61:5389–95. [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends Genet. 1993;9:138–41. doi: 10.1016/0168-9525(93)90209-z. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Eswaran J, Soundararajan M, Knapp S. Targeting group II PAKs in cancer and metastasis. Cancer Metast Rev. 2009;28:209–17. doi: 10.1007/s10555-008-9181-4. [DOI] [PubMed] [Google Scholar]
- Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–71. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- Wells CM, Jones GE. The emerging importance of group II PAKs. Biochem J. 2010;425:465–73. doi: 10.1042/BJ20091173. [DOI] [PubMed] [Google Scholar]
- Yang JX, Han YJ, Zheng H, Luo RC. Expression of PAK4 in breast cancer and benign breast pathological changes. Nan Fang Yi Ke Da Xue Xue Bao. 2010;30:981–3. [PubMed] [Google Scholar]
- Wells CM, Whale AD, Parsons M, Masters JR, Jones GE. PAK4: a pluripotent kinase that regulates prostate cancer cell adhesion. J Cell Sci. 2010;123:1663–73. doi: 10.1242/jcs.055707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ke Q, Li Y, Liu F, Zhu G, Li F. DGCR6L, a novel PAK4 interaction protein, regulates PAK4-mediated migration of human gastric cancer cell via LIMK1. Int J Biochem Cell Biol. 2010;42:70–9. doi: 10.1016/j.biocel.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Kaur R, Yuan X, Lu ML, Balk SP. Increased PAK6 expression in prostate cancer and identification of PAK6 associated proteins. Prostate. 2008;68:1510–6. doi: 10.1002/pros.20787. [DOI] [PubMed] [Google Scholar]