Abstract
Aim:
To examine whether CYP3A4 overexpression influences the metabolism of anticancer agent imidazoacridinone C-1311 in CHO cells and the responses of the cells to C-1311.
Methods:
Wild type CHO cells (CHO-WT), CHO cells overexpressing cytochrome P450 reductase (CPR) [CHO-HR] and CHO cells coexpressing CPR and CYP3A4 (CHO-HR-3A4) were used. Metabolic transformation of C-1311 and CYP3A4 activity were measured using RP-HPLC. Flow cytometry analyses were used to examine cell cycle, caspase-3 activity and cell apoptosis. The expression of pH 6.0-dependent β-galactosidase (SA-β-gal) was studied to evaluate accelerated senescence. ROS generation was analyzed with CM-H2 DCFDA staining.
Results:
CYP3A4 overexpression did not change the metabolism of C-1311 in CHO cells: the levels of all metabolites of C-1311 increased with the exposure time to a similar extent, and the differences in the peak level of the main metabolite M3 were statistically insignificant among the three CHO cell lines. In CHO-HR-3A4 cells, C-1311 effectively inhibited CYP3A4 activity without affecting CYP3A4 protein level. In the presence of C-1311, CHO-WT cells underwent rather stable G2/M arrest, while the two types of transfected cells only transiently accumulated at this phase. C-1311-induced apoptosis and necrosis in the two types of transfected cells occurred with a significantly faster speed and to a greater extent than in CHO-WT cells. Additionally, C-1311 induced ROS generation in the two types of transfected cells, but not in CHO-WT cells. Moreover, CHO-HR-3A4 cells that did not die underwent accelerated senescence.
Conclusion:
CYP3A4 overexpression in CHO cells enhances apoptosis induced by C-1311, whereas the metabolism of C-1311 is minimal and does not depend on CYP3A4 expression.
Keywords: anticancer drug, C-1311, CHO cell, CYP3A4, cytochrome P450 reductase, drug metabolism, cell cycle, apoptosis, senescence
Introduction
The vital limitation of a drug's effectiveness in a living organism is the individual's level of enzymes, which catalyze drug metabolism. Therefore, a unique response to drug treatment can be expected for each patient. Enzymes are also the molecular targets for drugs, which can lead to the modulation of their enzymatic activity. In addition, the expression level of metabolic enzymes in tumor tissues (in vivo) or tumor cells (in vitro) might affect the final effect of the drug treatment1. Therefore, the results of studies on drug metabolic pathways and drug-enzyme interactions are useful to investigate the molecular mechanism responsible for the biological effects of drugs. In the present study, we examined whether the overexpression of selected metabolic enzymes influenced drug metabolism and the impact of enzyme overexpression on the cellular response of tumor cells treated with an antitumor agent.
The major group of enzymes responsible for drug metabolism is the cytochrome P450 system. Five specific cytochrome P450 isozymes, CYP1A2 (9%), CYP2C9 (17%), CYP2C19 (10%), CYP2D6 (15%), and CYP3A4/3A5 (37%), are most frequently involved in the metabolism of antitumor agents (percentages in the brackets show the involvement of each CYP in drug transformation)2. Cytochrome P450 reductase (CPR) is necessary for efficient catalysis3.
The antitumor agent 5-diethylaminoethylamino-8-hydroxyimidazoacridinone (C-1311) was synthesized in our department4,5,6. It is the leading compound in a novel class of imidazoacridinones that are inhibitors of both topoisomerases and certain receptor kinases, including the FMS-like tyrosine kinase FLT37,8. C-1311 exhibited activity against advanced solid tumors in phase I and II clinical trials9,10, and it has also been evaluated for the treatment of autoimmune diseases11.
Limited mutagenic potential12 and low potency to generate oxygen free radicals, suggesting a lack of cardiotoxic properties, are the outstanding features of C-1311 as an antitumor agent. Drug accumulation in the nucleus is believed to enable its myeloperoxidase-mediated metabolism and rapid interaction with DNA13,14. It was postulated that intercalation and covalent binding to DNA, preceded by metabolic activation of C-1311, are significant steps in its molecular mechanism of action.
Studies on the mode of enzymatic oxidative activation of this drug with peroxidases15 found products of N-dealkylation on the side chain and a metabolite with a dimer-like structure. The formation of dimer-like metabolites under in vitro conditions reflects the potent reactivity of this molecule under cellular conditions in vivo. This reactivity is likely to be responsible for the observed high potency of C-1311 in covalent binding with intracellular nucleophiles such as proteins and nucleic acids.
Further studies of C-1311 showed that it was metabolized by rat and human liver metabolic enzymes but not by any tested human recombinant cytochrome P450. Moreover, two of these, CYP1A2 and, to a lesser extent, CYP3A4, were inhibited by C-1311. In contrast, C-1311 was shown to be a good substrate for selected isoforms of the human recombinant flavin-containing monooxygenase (FMO), resulting in an Nω-oxide metabolite that was also found after microsomal metabolism16. As a result of phase II metabolism, C-1311 was effectively transformed by UDP-glucuronosyltransferase (UGT) as a highly selective substrate of the UGT1A10 isoform, but not UGT2B7; the latter was inhibited by C-1311. UGT-mediated metabolism to 8-O-glucuronides occurred in the liver and intestinal microsomes with comparable efficiency17.
Previous studies on the biological action of C-1311 showed that this drug induced the arrest of cell cycle progression in G2 and subsequent apoptosis of murine leukemia L1210 cells and human cervix carcinoma HeLa S3 cells18,19. In ovarian and osteogenic sarcoma cells, G2 arrest resulted in a low level of apoptosis20, while in the human colon carcinoma HT-29 cell line, drug-treated cells progressed into mitosis after initial arrest in G2 but were unable to undergo cytokinesis and died in a process resembling mitotic catastrophe21. Likewise, the treatment of human leukemia MOLT4 cells with C-1311 resulted in mitotic catastrophe, leading to a massive apoptotic response22.
Taking these aspects of the mode of action of C-1311 into account, namely, metabolism and cell death, we examined the metabolism of this drug in an in vitro CHO cell model (previously, the metabolism of C-1311 was only investigated in cell-free systems), and we focused on the role of cytochrome P450 in the cellular response following drug treatment. In more detail, we investigated the following: (i) whether CYP3A4 overexpression influences the rate and pattern of drug metabolism, (ii) whether the drug modulates CYP3A4 activity in a cellular system and (iii) what the impact of CYP3A4 overexpression on cell cycle progression and the mode of cell death are.
Materials and methods
Chemicals
Imidazoacridinone C-1311 (NSC 645809)4,5 was synthesized by Barbara HOROWSKA, PhD in our department. C-1311 was prepared as a 10 mmol/L stock solution in 50% ethanol and kept at −20 °C until use. Methanol (gradient grade for liquid chromatography) was obtained from Merck (Darmstadt, Germany). The antibody to the cytochrome P450 3A4 isoenzyme was obtained from Sigma-Aldrich (St Louis, MO, USA). The secondary antibody to the goat primary antibody was from Cell Signaling Technology (Beverly, MA, USA). An Annexin-V-FLUOS Staining Kit was purchased from Roche (Mannheim, Germany). The Active Caspase-3 Staining Kit was ordered from BD Pharmingen (San Diego, CA, USA). CM-H2DCFDA (General Oxidative Stress Indicator) was obtained from Molecular Probes, Life Technologies (Carlsbad, CA, USA). Unless otherwise stated, all other chemicals were obtained from Sigma-Aldrich (St Louis, MO, USA).
Cell culture
Chinese hamster ovary cells (CHO)—wild type (CHO-WT), stably transfected CHO-HR and CHO-HR-3A4 cell lines—were kindly provided by Thomas FRIEDBERG and C Roland WOLF from the Biomedical Research Centre at the University of Dundee, Scotland, UK23.
The CHO-WT and CHO-HR cell lines were maintained in monolayer culture at 37 °C in a humidified 5% CO2 atmosphere in high-glucose Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin, 100 μg/mL streptomycin and HAT Supplement (100 μmol/L hypoxanthine, 0.4 μmol/L aminopterin and 16 μmol/L thymidine). The CHO-HR-3A4 cell line was maintained in monolayer culture at 37 °C in a humidified 5% CO2 atmosphere in Minimum Essential Medium (MEM) Alpha modifications supplemented with 10% fetal bovine serum (FBS), 100 units/mL penicillin and 100 μg/mL streptomycin. To maintain the stable overexpression of cytochrome P450 reductase and the CYP3A4 isoenzyme, geneticin (G418) and methotrexate, respectively, were added to the media one day after each passage. All media, supplements and antibiotics were obtained from Gibco Life Technologies (Paisley, Scotland).
Growth inhibition assay
Cell growth inhibition was assessed through cell counting using a Coulter Counter, model ZBI (Beckman, Fullerton, CA, USA). Briefly, cells were seeded in 24-well plates (4×104/well for 48 h, 2×104/well for 72 h, 1×104/well for 96 h) and treated with C-1311 (concentrations ranging from 0.0001 to 10 μmol/L). A dose-response curve was plotted and used to calculate the drug concentration that yielded 50% and 80% inhibition of cell growth (IC50 and IC80). The growth inhibition assay was performed at least three times.
Metabolic transformation of C-1311 in CHO cells
To determine the metabolic transformation of C-1311, CHO cells (2×106) were plated in 60-mm dishes and treated the next day with 25 μmol/L C-1311 for 0 (1 min), 24, 48, or 72 h. Cells were then collected with the media by gentle scraping, washed with ice-cold PBS, suspended in ice-cold 60% methanol and lysed in an ultrasound bath for 15 min. The cell extract was incubated on ice for 1 h. Insoluble material was removed by centrifugation at 16 000×g at 4 °C for 15 min, and the supernatant (200 μL) was directly analyzed by RP-HPLC with UV/VIS detection at 420 nm. The metabolic transformation experiment was performed four times.
Preparation of cell extracts and immunoblotting for CYP3A4
To prepare cell extracts for CYP3A4 immunoblotting, CHO-WT, CHO-HR (as controls) and CHO-HR-3A4 cells were plated in 100-mm dishes and, on the next day, treated with C-1311 for up to 120 h. After the treatment, cells were harvested by trypsin and resuspended in 0.2 mL of 10 mmol/L sodium phosphate buffer (pH 8.0) containing 2 mmol/L MgCl2, 2 mmol/L dithiothreitol and 1 mmol/L EDTA23. Cells were lysed by homogenization on ice for 3 min. The protein concentration was determined using the Bio-Rad protein assay (Bio-Rad Laboratories, Hercules, CA, USA). Samples were mixed with Laemmli buffer and denatured at 100 °C for 5 min, and then 40 μg of total protein was subjected to SDS-PAGE, transferred onto a nitrocellulose membrane and blocked with 5% non-fat milk in TBS-T buffer [150 mmol/L NaCl, 10 mmol/L Tris (pH 7.4) and 0.05% Tween-20] at room temperature for 2 h. The membrane was incubated with rabbit polyclonal anti-CYP3A4 antibody (Sigma-Aldrich, St Louis, MO, USA) diluted at 1:400 in TBS-T buffer containing 0.5% bovine albumin (BSA) at 4 °C overnight with rocking. After washing with TBS-T three times for 15 min each, the membrane was incubated with anti-rabbit horseradish peroxidase-conjugated secondary antibody (Sigma-Aldrich, St Louis, MO, USA) diluted at 1:5000 in TBS-T buffer containing 1% non-fat milk for 1 h at room temperature. The antigen was detected using the Enhanced Chemiluminescence Western blotting detection system (Pierce Biotechnology, Rockford, IL, USA) according to the manufacturer's protocol. Equal protein loading was confirmed by rehybridization of membrane and reprobing with the mouse anti-β-actin primary antibody diluted at 1:1000 (Sigma-Aldrich, St Louis, MO, USA) in TBS-T buffer containing 1% non-fat milk and anti-mouse horseradish peroxidase-conjugated secondary antibody diluted at 1:5000 (Cell Signaling, Danvers, MA, USA) in TBS-T buffer containing 1% non-fat milk. The CYP3A4 protein level was analyzed three times.
Measurement of CYP3A4 activity
The activity of CYP3A4 was evaluated by measuring the α-hydroxylation of testosterone to 6-β-hydroxytestosterone. CHO-HR-3A4 cells (1×106) were plated in 60-mm dishes and, on the next day, treated for up to 120 h with a single concentration of C-1311 (equal to IC80 value or 1 μmol/L) or with increasing concentrations of C-1311 (from 0.01 to 50 μmol/L). After the treatment, the culture medium was replaced, and the testosterone solution was added at a 50 μmol/L final concentration for 24 h. Media from the dishes was taken, mixed with acetonitrile at a 1:1 ratio and shaken for 5 min. After extraction, samples were incubated on ice for 30 min, and the upper phase was directly analyzed by RP-HPLC with UV/VIS detection at 254 nm. The rates of substrate metabolism were calculated as a ratio of the surface area under the substrate HPLC peak compared to the untreated control. The number of cells from each dish was counted to determine the relative ratio of testosterone conversion. CYP3A4 activity assays were carried out three times.
HPLC analysis
Samples obtained according to the procedures described above were analyzed by HPLC with a reversed-phase 5 μm Suplex pKb-100 analytical column (0.46 cm×25 cm, C18, Supelco, Bellefonte, PA, USA) with a Waters Breeze System (Milford, MA, USA). The HPLC analyses were carried out at a flow rate of 1 mL×min−1 with the following eluent system: a linear gradient from 15% to 80% methanol in ammonium formate (0.05 mol/L, pH 3.4) for 25 min, followed by a linear gradient from 80% to 100% methanol in ammonium formate for 3 min.
Cell cycle analysis
Analysis of the DNA content was performed according to the method described by Augustin et al24. Briefly, CHO-WT, CHO-HR, and CHO-HR-3A4 cells (1×106) were plated in 100-mm dishes and, on the next day, treated with C-1311 at a concentration corresponding to the IC80 value for up to 120 h. Cells were collected by trypsinization, washed twice with PBS and fixed in ice-cold 70% ethanol overnight at −20 °C. On the day of the analysis, cells were washed twice in PBS, resuspended in 1 mL buffer of DNA staining solution containing 20 μg/mL propidium iodide (PI) and 100 μg/mL RNase A in PBS, incubated for 30 min in the dark at room temperature and analyzed by flow cytometry using a FACScan (Becton Dickinson, San Jose, CA, USA). The data were analyzed using WinMDI software (The Scripps Research Institute, La Jolla, CA, USA). Cell cycle analysis was performed at least three times.
Phosphatidylserine externalization
An annexin V-FITC binding assay (using Annexin-V-Fluos Staining Kit, Roche, Mannheim, Germany) was carried out according to the manufacturer's instructions using counterstaining with PI and bivariate flow cytometry analysis. Briefly, following treatment with C-1311 for up to 120 h, cells (1.5×106) were harvested by low speed centrifugation, washed twice with ice-cold PBS, pelleted and resuspended in 50 μL of Annexin V–FITC diluted in binding buffer containing propidium iodide. FITC-conjugated Annexin V and PI were added at the manufacturer's recommended concentrations. Cells were incubated for 15 min at room temperature in the dark. After incubation, cell suspensions were diluted with 0.35 mL of binding buffer and analyzed by flow cytometry within 1 h. The phosphatidylserine externalization assay was performed twice and three times for CHO-HR-3A4 cells.
Caspase-3 activity measurement
Caspase-3 activity was determined using the Active Caspase-3 Apoptosis kit (BD Pharmingen, San Diego, CA, USA) according to the manufacturer's instructions. Briefly, untreated and C-1311-treated cells (1.5×106) were washed twice with cold PBS, then fixed and permeabilized using Cytofix/CytopermTM for 20 min on ice, pelleted and washed with Perm/WashTM Buffer. Cells were then stained with anti-active caspase-3 mAb using 20 μL per sample for 30 min at room temperature in the dark. Following incubation with the antibody, cells were washed with Perm/WashTM buffer, resuspended in Perm/WashTM Buffer and analyzed by flow cytometry. Caspase-3 activity measurement was conducted twice.
Morphological examination
The nuclear morphology of cells was examined under a fluorescence microscope (Olympus BX60, Tokyo, Japan) after staining with 4′,6-diamidino-2-phenylindole (DAPI). Following treatment with C-1311 for up to 120 h, cells (approximately 2×105) were spun onto microscopic slides, fixed in methanol:acetic acid (3:1) for 15 min and stained with 1 μg/mL DAPI for 5 min. Twenty random fields containing at least 50 cells were examined at 200×magnification for the presence of apoptotic and multinucleated cells. For each experiment, at least 500 cells were counted. Cells were scored as apoptosis based on the presence of condensed, fragmented chromatin. Enlarged cells containing multiple (≥3), small, evenly stained interphase nuclei were considered to have undergone multinucleation typical for mitotic catastrophe25,26. Photos of stained cells for presentation were made at 400×magnification. Morphological examination was performed twice.
Membrane integrity assay
The number of dead and live cells was evaluated by double staining with fluorescein diacetate (FDA) and propidium iodide (PI). Cells were exposed to C-1311 for up to 120 h. After the treatment, cells were harvested by trypsinization, washed twice with PBS, suspended in 100 μL of PBS containing FDA (1 μg/mL) and incubated for 10 min at 37 °C in the dark. Then, cells were stained with PI by adding 1 μL of 2 mg/mL PI solution (final concentration 10 μg/mL) and immediately analyzed under a fluorescence microscope (magnification, ×200). For each experiment, at least 500 cells were counted. Green cells stained with hydrolyzed FDA were scored as alive, and red cells stained with PI were scored as dead. The membrane integrity assay was performed twice and produced similar results.
ROS generation
Reactive oxygen species (ROS) generation in CHO cells was examined under a fluorescence microscope (Olympus BX60, Tokyo, Japan) after staining with 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA, Molecular Probes) and Hoechst 33342. Following treatment with C-1311 for up to 120 h, cells (approximately 106) were washed twice with PBS and stained with 10 μmol/L CM-H2DCFDA and 2 μg/mL Hoechst 33342 for 30 min in 37 °C in the dark. Then, cells were washed with PBS and spun onto microscopic slides. Photos of stained cells for presentation were made at 400×magnification using light and fluorescence microscopy with a UV spectrum for Hoechst 33342 staining and a blue spectrum for CM-H2DCFDA staining. Analysis of ROS generation was performed twice.
Accelerated senescence examination
The accelerated senescence examination was based on the analysis of pH 6.0-dependent β-galactosidase (SA-β-gal) expression together with senescence-related morphology such as enlarged and flattened cells. Cells were plated in 60-mm plates containing microscope cover slides and treated with C-1311 for up to 120 h. After the treatment, cover slides with attached cells were moved to 35-mm dishes, washed twice with PBS and fixed with 2% glutaraldehyde and 0.2% formaldehyde for 5 min. The cells were then washed twice with PBS and incubated at pH 6.0 with X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactosidase, substrate for SA-β-gal) staining solution (1 mg/mL X-gal, 40 mmol/L citric acid/sodium phosphate, pH 6.0, 5 mmol/L potassium ferrocyanide, 5 mmol/L potassium ferricyanide, 150 mmol/L NaCl, 2 mmol/L MgCl2), which produced blue coloration in SA-β-gal-positive cells. Following overnight incubation at 37 °C, the cells were washed twice with PBS and inspected under light microscopy (magnification, ×200). The accelerated senescence examination was conducted twice and produced similar results.
Colony forming assay
The ability of cells to return to proliferation after C-1311 treatment was measured by a colony forming assay. After exposure time, 300 cells were harvested and seeded in a new 60-mm dish containing fresh media without the drug and incubated for two weeks. Non-treated cells were used as controls. After two weeks, colonies were stained with crystal violet and counted, and photos were taken. The colony assay was performed twice and produced similar results.
Statistical analysis
All of the data reported represented as mean±SD. Pairs of values were compared with Student's unpaired t test, and the difference was considered significant if P<0.05. Analysis of variance (one-way ANOVA) with a post hoc Dunnett's test was used for CYP3A4 activity measurement and colony forming analysis and carried out using GraphPad Prism (GraphPad Software Inc, La Jolla, CA, USA).
Results
Effects of C-1311 on cell proliferation
Cell growth inhibition was monitored using a Coulter Counter. CHO-WT, CHO-HR, and CHO-HR-3A4 cells were treated with C-1311 for 48, 72, and 96 h. A significant concentration-dependent inhibition of cell proliferation was observed in all three CHO cell lines. The estimated drug concentrations required to inhibit cell growth by 50% and 80% are shown in Table 1. CHO-HR cells overexpressing cytochrome P450 reductase (CPR) were the most sensitive to 72-h C-1311 treatment, whereas the wild-type CHO cells were the most resistant. However, after 96 h of incubation, CHO-HR-3A4 cells were the most sensitive to C-1311. In further experiments pertaining to the cell cycle and cellular response, the C-1311 concentration corresponding to the estimated IC80 value after 72 h of incubation was used.
Table 1. Cytotoxic activity of C-1311 in wild type (CHO-WT) and transfected CHO cells overexpressing cytochrome P450 reductase (CHO-HR) or cytochrome P450 reductase and CYP3A4 (CHO-HR-3A4).
| Incubation time | CHO-WT (μmol/L) ± SD | CHO-HR (μmol/L) ± SD | CHO-HR-3A4 (μmol/L) ± SD | ||||
|---|---|---|---|---|---|---|---|
| 48 h | IC50 | 0.024 | 0.003 | 0.009 | 0.002c | 0.012 | 0.004c |
| IC80 | 0.085 | 0.003 | 0.060 | 0.007c | 0.060 | 0.011b | |
| 72 h | IC50 | 0.019 | 0.005 | 0.009 | 0.003 | 0.015 | 0.005 |
| IC80 | 0.078 | 0.006 | 0.032 | 0.002c | 0.057 | 0.012b,e | |
| 96 h | IC50 | 0.008 | 0.001 | 0.004 | 0.001c | 0.003 | 0.001c |
| IC80 | 0.031 | 0.003 | 0.017 | 0.002c | 0.011 | 0.002c,f | |
Each value represented the mean±SD of at least three independent experiments.
bP<0.05,
cP<0.01 vs CHO-WT.
eP<0.05,
fP<0.01 vs CHO-HR.
Metabolic transformation of C-1311 in CHO model cells
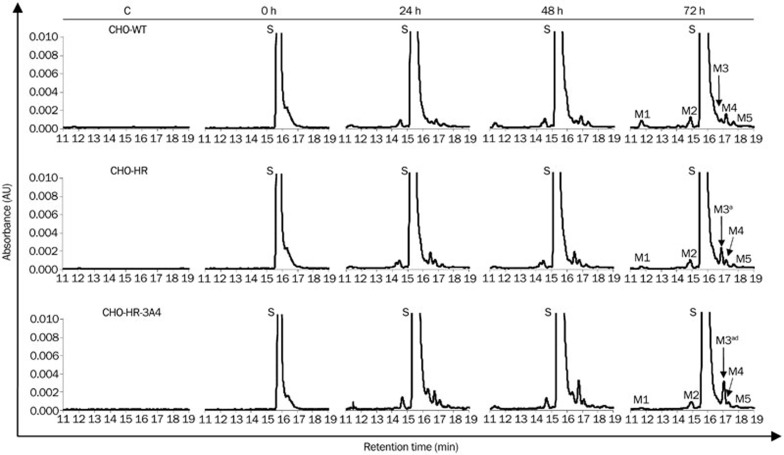
Intracellular biotransformation of C-1311 was monitored by HPLC analysis of cell extracts. CHO cells were treated with the drug at 25 μmol/L for various time intervals (0, 24, 48, and 72 h). Figure 1 shows that a peak of the native compound C-1311 was observed on the chromatograms at a retention time of approximately 16 min, and its metabolism in the three CHO cell lines proceeded in a very limited range. The levels of all metabolites increased with the exposure time to a similar extent in all three CHO cell lines. The differences in the peak height of the main M3 metabolite among CHO-WT, CHO-HR, and CHO-HR-3A4 were statistically insignificant. In addition, the general conversion of C-1311 to M3 was very low, less than 0.2%.
Figure 1.
Metabolism of imidazoacridinone C-1311 in CHO cells. Representative RP-HPLC profiles of methanol extracts from untreated (control) CHO cells and cells treated with 25 μmol/L C-1311 for 0 (1 min), 24, 48, and 72 h. Peaks: S=substrate (C-1311), M1, M2, M3, M4, M5=metabolite No 1, 2, 3, 4, 5, respectively. Chromatographic conditions are described in the Materials and methods section. The statistical analysis was performed on the basis of the M3 peak area. aP>0.05 vs CHO-WT cells. dP>0.05 vs CHO-HR, indicating that the amount of M3 metabolite was not significantly different, n=3, Student's t-test.
Effects of C-1311 on CYP3A4 level and activity
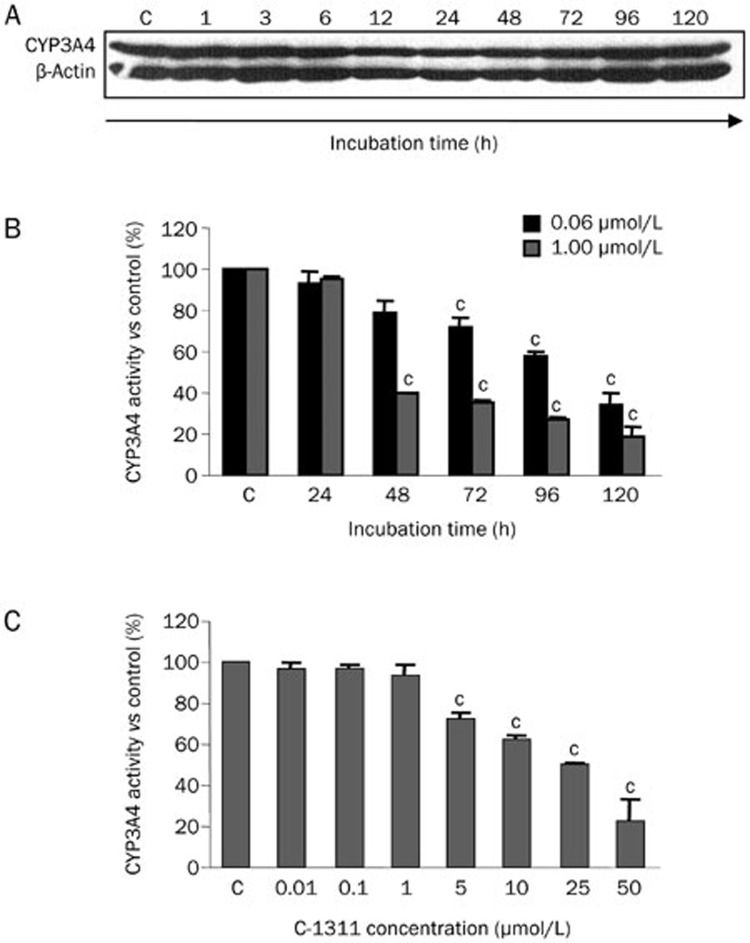
Preliminary results demonstrated that CYP3A4 was expressed in untreated CHO-HR-3A4 cells, and the CYP3A4 protein level and enzymatic activity were very high in these cells (data not shown). Next, the time-dependent influence of C-1311 on the level of CYP3A4 was investigated. CHO-HR-3A4 cells were treated with C-1311 from 1 to 120 h at the concentration corresponding to the IC80 value. CYP3A4 immunostaining results (Figure 2A) showed that the protein level did not change with the duration of exposure to C-1311. To evaluate whether C-1311 modifies CYP3A4 enzymatic activity, the rate of the metabolic transformation of testosterone (a standard CYP3A4 substrate)27 was determined in CHO-HR-3A4 cells and calculated in relation to live cells. The dependence on the incubation time, which was tested from 24 to 120 h at 0.06 μmol/L (IC80) and 1 μmol/L (50×IC50) C-1311, and on the drug concentration, which was tested from 0.01, 0.1, 1, 5, 10, 25, and 50 μmol/L following 24 h of drug treatment, is presented in Figures 2B and 2C. A C-1311-mediated decrease in CYP3A4 activity occurred in a time-dependent manner, reaching an inhibition of 35% after a 120 h incubation at 0.06 μmol/L (IC80). At a higher concentration, 1.0 μmol/L, CYP3A4 activity dropped to 40% after a 48 h incubation and to 19% after 120 h incubation (Figure 2B). The dependence of enzymatic activity on the drug concentration indicated that CYP3A4 activity started to decrease at 5 μmol/L and reached the lowest level at 50 μmol/L at 22% activity. Thus, CYP3A4 activity was inhibited in CHO-HR-3A4 cells in a time- and concentration-dependent manner.
Figure 2.
The effect of C-1311 on the CYP3A4 level and enzymatic activity in CHO-HR-3A4 cells. (A) Representative immunoblots of the level of cytochrome P450 3A4 in CHO-HR-3A4 cells exposed to C-1311 for the indicated time. CHO-HR-3A4 cells were exposed to C-1311 at the concentration corresponding to the IC80 (0.06 μmol/L). After the indicated time, cells were lysed, and total protein was extracted, separated by SDS-PAGE, electrotransferred to a nitrocellulose membrane, and subjected to immunoblotting with anti-CYP3A4 antibody. n=3. (B) The time course of C-1311 influence on CYP3A4 activity in CHO-HR-3A4 cells. Cells were treated with two different concentrations of C-1311, 0.06 μmol/L (corresponding to the IC80) and 1.0 μmol/L (corresponding to 50×IC50), for the times indicated. (C) The concentration-dependent influence of C-1311 on CYP3A4 enzymatic activity in CHO-HR-3A4 cells. Cells were treated with 0.01, 0.1, 1.0, 5, 10, 25, and 50 μmol/L for 24 h. After the treatment, the media was changed, and 50 μmol/L testosterone solution was added for 24 h. The conversion of testosterone to 6-β-hydroxytestosterone was measured using RT-HPLC and calculated as the activity of CYP3A4 versus control (untreated cells). Values are the mean±SD. cP<0.01 vs control non-treated cells, n=3, one-way ANOVA test with Dunnett's post hoc test.
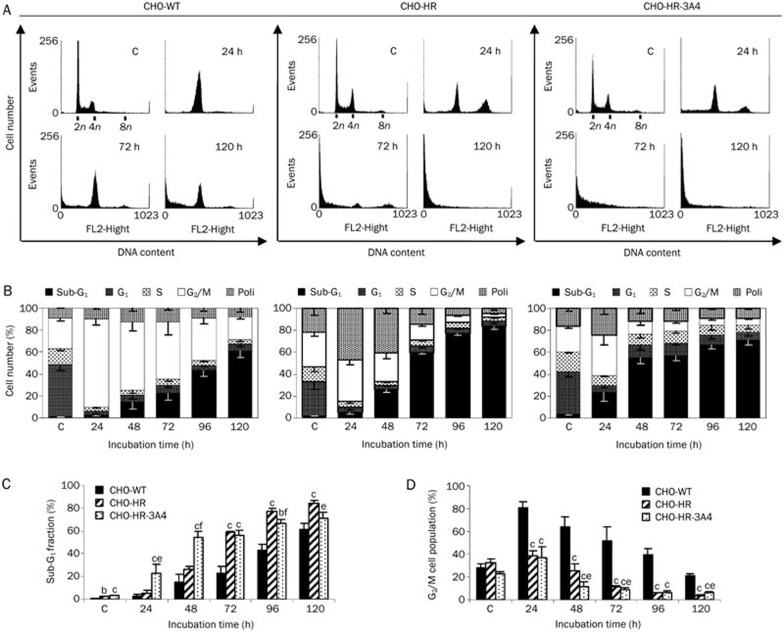
Cell cycle distribution following C-1311 treatment
CHO cells were cultured for various periods of time (6, 12, 24, 48, 72, 96, and 120 h) in the presence of C-1311 at the IC80 concentration, and DNA content was analyzed by flow cytometry. As shown in Figure 3, C-1311 caused different changes in cell cycle progression in the three investigated CHO cell lines. Starting at 6 h of C-1311 treatment, wild-type CHO cells underwent transient arrest in G2/M (not shown), and at 24 h, the number of cells in G2/M reached the highest level, 80% (Figure 3D). Prolonged drug incubation resulted in a decreasing population of cells in this phase concomitant with significant accumulation of the cell population in the sub-G1 compartment (<2n DNA content), which is indicative of apoptosis. After 120 h of C-1311 exposure, 61% of CHO-WT cells were in sub-G1 phase (Figure 3C). C-1311 treatment of CHO-HR cells overexpressing P450 reductase resulted in a higher level of apoptotic population than that observed in CHO wild-type cells. After 120 h of C-1311 exposure, the sub-G1 fraction reached 84% (Figure 3C). Moreover, starting at 12 h, the population of cells containing 4n DNA (G2/M phase) and more than 4n DNA (polyploid cells) began to increase and reached maximum levels of 38% and 47%, respectively, after 24 h. Both populations started to decrease in number following 48 h of C-1311 incubation in association with the increase of the sub-G1 fraction (Figure 3B). In CHO-HR-3A4 cells, transient accumulation of cells in G2/M was also observed at 24 h (35%). After that time, the number of cells in this compartment decreased to 9% (120 h treatment) (Figure 3D). Compared to the control (untreated cells), the population of polyploid cells slightly increased to 26% after 24 h of C-1311 treatment and then remained essentially unchanged for up to 120 h (Figure 3B). Additionally, the population of CHO-HR-3A4 cells with <2N DNA content (sub-G1) started to increase from 12 h of drug treatment and reached 71% after 120 h (Figure 3C). In comparison to the other two CHO cell lines, C-1311 induced DNA damage earlier and to a greater extent in CHO-HR-3A4 cells.
Figure 3.
Analysis of DNA content in CHO cells following treatment with C-1311 at the IC80 concentration for the time indicated. CHO-WT, CHO-HR, CHO-HR-3A4 cells were fixed in ethanol and stained with PI, and their DNA content was analyzed by FACScan. (A) Representative histograms of the selected incubation time. (B) Quantification of cells in specific cell cycle compartments. n=3. (C) Cell population in the sub-G1 fraction and G2/M compartment (D). Values are the mean±SD. bP<0.05, cP<0.01 vs CHO-WT cells. eP<0.05, fP<0.01 vs CHO-HR cells, n=3, Student's t-test.
Induction of apoptosis by C-1311
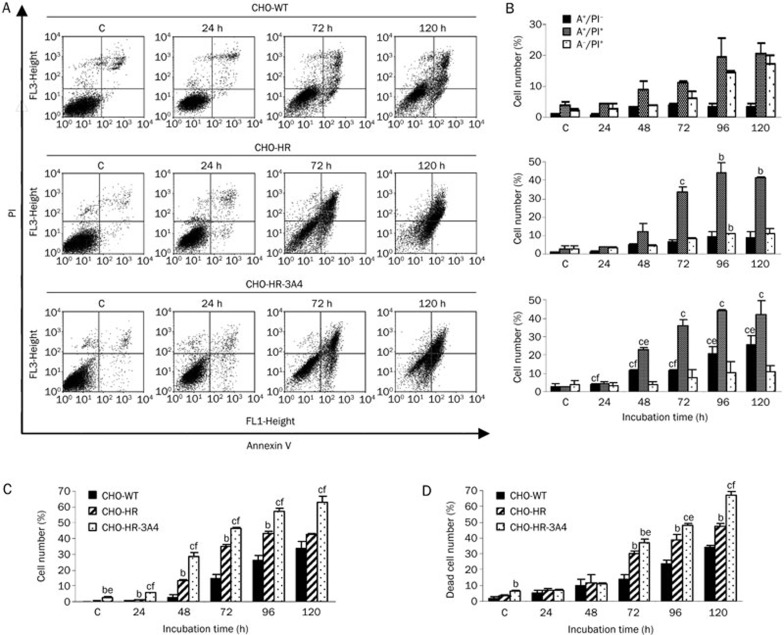
Following C-1311 treatment, analysis of the cell cycle distribution of CHO cells showed a time-dependent increase in the number of hypodiploid cells (sub-G1 cells), which represent the apoptotic cell population. The apoptotic mode of cell death induced by C-1311 was further confirmed by Annexin V/PI dual staining and active caspase-3 analysis. In the early stage of apoptosis, phosphatidylserine is translocated from the inner leaflet of the plasma membrane to the cell surface, and the polarity and asymmetry of the plasma membrane are disturbed. Positive staining with fluorescence-labeled Annexin V correlates with a loss of plasma membrane polarity and precedes the complete loss of membrane integrity that accompanies later stages of cell death resulting from either apoptosis or necrosis. In contrast, PI can only enter cells after the loss of membrane integrity. Thus, dual staining with Annexin V and PI allows the clear discrimination among unaffected cells (Annexin V−/PI−), early apoptotic cells (Annexin V+/PI−), late apoptotic and secondary necrotic cells (Annexin V+/PI+) and primary necrotic cells (Annexin V−/PI+). In the following experiment, flow cytometry analyses were performed with the three CHO cell lines exposed to C-1311 at the IC80 concentration from 24 to 120 h.
As shown in Figure 4, CHO-WT cells remained largely unaffected for the first 24 h of C-1311 treatment. After 48 h, the population of early (Annexin V+/PI−) and late (Annexin V+/PI+) apoptotic cells together amounted to 9% and increased to 26% following prolonged drug incubation up to 120 h (Figure 4B). Induction of apoptosis was also confirmed by measuring the activity of caspase-3, which is crucial to the execution stage of apoptosis28. After 72 h of treatment with C-1311, a small amount of cells with active caspase-3 were observed (Figure 4C). Prolonged incubation with C-1311 resulted in a significant increase in the number of cells with active caspase-3 (32% after 120 h).
Figure 4.
Flow cytometry analysis of the membrane alteration and caspase-3 activity and quantification of dead cells in CHO cells treated with C-1311 at the IC80 concentration for the time indicated. (A) Representative bivariate flow cytometry histograms of the Annexin V signal versus the PI signal. The bottom left quadrant represents live unaffected cells (Annexin V−, PI−); the bottom right quadrant represents early apoptotic cells (Annexin V+, PI−); the top right quadrant represents late apoptotic and secondary necrotic cells (Annexin V+, PI+); and the top left quadrant represents primary necrotic cells (Annexin V−, PI+). (B) Percentages of early and late apoptotic and primary necrotic cells based on flow cytometry analysis. (C) Number of cells with active caspase-3 based on flow cytometry analysis. Values are the mean±SD. n=2 for CHO-WT and CHO-HR cells, n=3 for CHO-HR-3A4 cells. (D) Population of dead cells quantified under fluorescence microscope after double staining with fluorescein diacetate (FDA) and propidium iodide (PI). Cells that hydrolyzed FDA to its green fluorescent derivative were counted as live cells, whereas red cells stained only with PI were considered dead. Values are the mean±SD. n=2. bP<0.05, cP<0.01 vs CHO-WT cells. eP<0.05, fP<0.01 vs CHO-HR cells, Student's t-test.
CHO-HR cells with cytochrome P450 reductase overexpression were more prone to undergo apoptosis than CHO wild-type cells. The proportion of early and late apoptotic cells progressively increased with the duration of C-1311 exposure, and after 120 h, these populations together reached 50% (Figure 4B). The percentage of CHO-HR cells expressing caspase-3 after C-1311 treatment reached 46% after 120 h of drug treatment (Figure 4C).
Treatment of CHO-HR-3A4 cells overexpressing both CPR and CYP3A4 with C-1311 resulted in a significantly higher level of apoptosis compared to wild-type CHO and CHO-HR cells. The number of early and late apoptotic (Annexin V+/PI− and Annexin V+/PI+) cells increased to 68% after 120 h (Figure 4B). Activation of caspase-3 was observed simultaneously with phosphatidylserine translocation from the inner leaflet to the plasma surface. After 120 h of C-1311 exposure, 63% of the cells expressed active caspase-3 (Figure 4C).
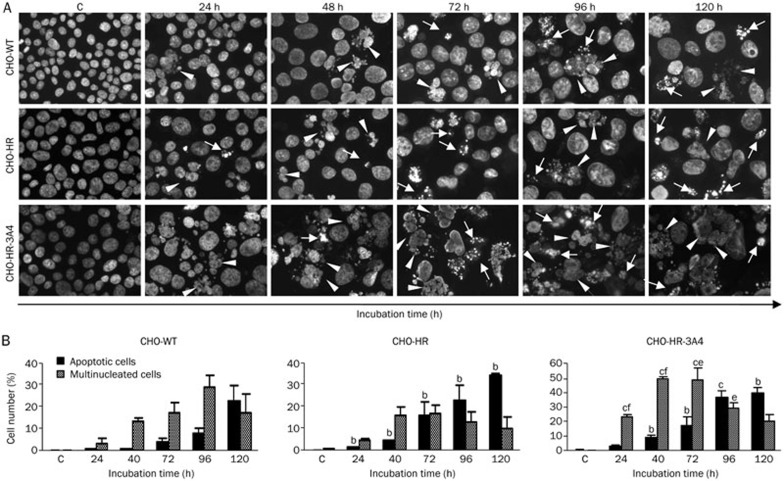
Morphological changes in nuclei triggered by C-1311
The induction of apoptosis was confirmed by morphological observation of cell nuclei stained with DAPI. Compared to the control, untreated CHO cells, C-1311 exposure resulted in the time-dependent appearance of cells exhibiting nuclear morphological changes typical of apoptosis, such as the condensation of chromatin and fragmentation into discrete bodies (Figure 5A). Quantitative analysis revealed that approximately 40% of CHO-HR-3A4 cells showed features of apoptosis after 120 h of C-1311 exposure (Figure 5B), whereas the number of apoptotic CHO-WT and CHO-HR cells was 22% and 34%, respectively. Additionally, C-1311 treatment of wild-type and transfected CHO cells resulted in the appearance of multinucleated cells, which is the hallmark of mitotic catastrophe25,26. Interestingly, after prolonged drug incubation (120 h), the multinucleated cell population decreased, accompanied by an increase in the fraction of apoptotic cells (Figure 5B). It is worth noting that the population of cells with unchanged nuclei was 56% for CHO-WT cells, 50% for CHO-HR cells and only 23% for CHO-HR-3A4 cells after 120 h of C-1311 exposure.
Figure 5.
Changes in the nuclear morphology of CHO cells following exposure to imidazoacridinone C-1311 at the IC80 concentration. (A) Representative pictures of cells stained with DAPI and examined under a fluorescence microscope (magnification 400×). Cells with condensed, intensely stained fragmented chromatin are typical of apoptosis (arrows); enlarged cells containing multiple evenly stained nuclei (multinucleated cells) are characteristic of mitotic catastrophe (arrow heads). (B) Quantification of the percentage of apoptotic and multinucleated cells based on fluorescence microscopy analysis. Values are the mean±SD. n=2. bP<0.05, cP<0.01 vs CHO-WT cells. eP<0.05, fP<0.01 vs CHO-HR cells, Student's t-test.
Necrosis and overall death caused by C-1311
Dual staining with Annexin V and PI (Figure 4A and 4B) showed not only cells that underwent apoptosis (early and late) but also those that underwent secondary (Annexin V+/PI+) and primary necrosis (Annexin V−/PI+), which is defined as necrosis that is not the consequence of apoptosis-triggered cellular damage. To estimate the overall number of cells in which the integrity of the plasma membrane was lost, dual staining with fluorescein diacetate and propidium iodide was applied. FDA is a non-polar agent that can enter all cells, but it is only hydrolyzed to its green fluorescent derivative in live, metabolically active cells. PI can only be taken up through the damaged cell membrane of dead and dying cells, and thus, it intercalates into the DNA and forms red fluorescence only in dead cells29. As shown in Figure 4D, compared to the control (untreated cells), a time-dependent increase of dead cells (stained with PI) was observed in all three CHO cell lines following C-1311 treatment. After 120 h of drug treatment, the fraction of cells that produced red fluorescence reached 35% in wild-type CHO cells, 47% in CHO-HR cells and 67% in CHO-HR-3A4 cells. These data are consistent with the Annexin V/PI analysis (Figure 4B), which showed that after a 120 h incubation with C-1311, the number of CHO-WT necrotic cells (primary and secondary) was 37%. In CHO-HR cells, this population amounted to 50%. In CHO-HR-3A4 cells, the population of necrotic cells was 53%.
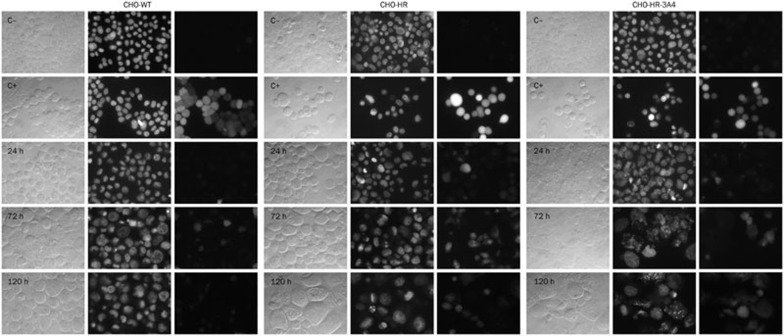
Generation of ROS by C-1311
There is some evidence indicating that overexpression of cytochrome P450 reductase itself induces a higher level of reactive oxygen species (ROS)30. Stimulation of the formation of ROS can lead to oxidative stress, which is involved in the induction of apoptosis31. Therefore, we determined the ability of C-1311 to induce ROS to establish the relevance of this effect to the induction of apoptosis and cytotoxic activity of this drug. CHO cells were incubated with C-1311 and with H2O2 as a positive control for up to 120 h and then immediately analyzed under a fluorescence microscope after staining with 5-(and-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate, acetyl ester (CM-H2DCFDA) and Hoechst 33342. As shown in Figure 6, C-1311 did not induce ROS generation in wild-type CHO cells. Few cells exhibited CM-H2DCFDA fluorescence. For comparison, in the positive control (cells treated with H2O2), high CM-H2DCFDA fluorescence, which demonstrated ROS generation, was observed in all studied CHO cell lines. In CHO-HR cells, a time-dependent and very strong increase in CM-H2DCFDA fluorescence was detected following C-1311 treatment. In contrast, a lower level of CM-H2DCFDA fluorescence was observed in CHO-HR-3A4 cells from 72 h to 120 h of incubation with C-1311. Interestingly, in CHO-HR-3A4 cells, CM-H2DCFDA fluorescence was observed only in cells that presented typical features of apoptosis, while in CHO-HR cells, this fluorescence was visible in every type of cell.
Figure 6.
Generation of reactive oxygen species (ROS) in CHO cells following treatment with C-1311 at the IC80 concentration for the time indicated. After exposure to the drug, CHO-WT, CHO-HR, and CHO-HR-3A4 cells were double stained with CM-H2DCFDA and Hoechst 33342 and immediately analyzed with a fluorescence microscope. C−, negative control (untreated cells); C+, positive control (cells treated with H2O2). Left panel, cells viewed under a light microscope; middle panel, cells stained with Hoechst 33342 (UV spectrum); and right panel, cells stained with CM-H2DCFDA (blue spectrum). Panels were taken from the same microscopic field. The positive control is CHO cells treated with H2O2. n=2.
Induction of accelerated senescence by C-1311
The above results revealed that C-1311 did not induce cell death in the entire population of the investigated cell lines. A fraction of the cells remained alive even after 120 h of drug treatment. Therefore, we investigated whether these cells underwent accelerated senescence following C-1311 exposure. Expression of SA-β-galactosidase (SA-β-gal) and morphological features, such as an enlarged and flattened cell shape, were used as markers of senescence. The characteristic blue color resulting from the metabolism of the SA-β-gal substrate was observed in CHO-HR and CHO-HR-3A4 cells starting at 72 h of incubation (Figure 7A). The number of senescent cells increased with the duration of exposure to C-1311 in both cell lines. In addition to increased SA-β-gal staining, these cells had an enlarged and flattened morphology typical of senescent cells. Moreover, the population of senescent cells was larger in CHO-HR-3A4 cells than in CHO-HR cells. Interestingly, C-1311 did not induce accelerated senescence in wild-type CHO cells.
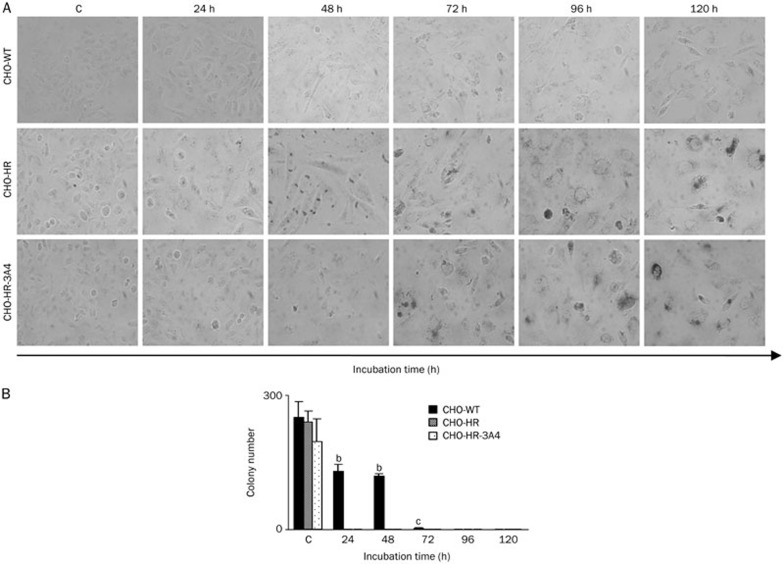
Figure 7.
Accelerated senescence induction and colony-forming ability of live CHO cells treated with C-1311 (IC80). (A) Representative pictures of the time course of senescence associated with β-galactosidase (SA-β-gal) expression in C-1311-treated CHO cells. The expression of SA-β-gal and morphological alterations such as enlarged and flattened cell shape were used as a marker of a senescence-like phenotype (magnification ×200). n=2. (B) The ability of CHO cells to return to proliferation after C-1311 exposure at the IC80 concentration. After the indicated times of drug treatment, approximately 300 CHO-WT, CHO-HR, and CHO-HR-3A4 cells were cultured for two weeks in fresh media, and the number of colonies was calculated. Values are the mean±SD. bP<0.05, cP<0.01 vs control non-treated cells, n=3, one-way ANOVA with Dunnett's post hoc test.
The ability of CHO cells to return to proliferation after C-1311 treatment
A colony-forming assay was performed to investigate whether CHO cells were able to return to proliferation following C-1311 exposure. After the treatment (from 24 to 120 h), approximately 300 cells were incubated for 2 weeks in fresh media, and their ability to form colonies was observed (Figure 7B). The study revealed that only half of wild-type CHO cells treated with C-1311 for 48 h were able to proliferate. After 72 h of drug exposure, only 2 cells were able to undergo mitosis and divide. At 96 h, the proliferation of CHO-WT cells was completely inhibited. In CHO-HR and CHO-HR-3A4 cells, cell division was fully blocked, and colonies failed to form after only 24 h of treatment.
Discussion
The present work investigated a current topic in the field of drug metabolism related to the role of cytochrome P450 isoenzymes in the final cellular response induced by drugs in tumor cells. We raised the question of whether metabolites of C-1311 are responsible for its action in CHO cells. We hypothesized that overexpression of CYP3A4 might mediate the cellular response independent of drug metabolite formation.
The results presented here indicated that the concentrations of all metabolites were very low (up to 0.3% of the transformation rate) and similar in the three CHO cell lines. Furthermore, the metabolic profiles of the C-1311 antitumor agent did not differ significantly between wild-type CHO cells and CPR-overexpressing or CPR- and CYP3A4-overexpressing transductants. These results clearly indicated that CYP3A4 supported by CPR did not participate in C-1311 metabolism in CHO cells. This finding confirms previous results testing C-1311 under non-cellular conditions16. Namely, this compound was not metabolized by CYP3A4, which is the P450 isoenzyme with the highest abundance in the human liver and which contributes to the oxidative metabolism of more than 60% of all clinically used drugs, including antitumor agents such as cyclophosphamide32, ifosfamide33, paclitaxel34, tamoxifen35,36 and doxorubicin37. The resistance of C-1311 to the action of other cytochrome P450 enzymes was shown previously with human recombinant enzymes from the CYP1A, CYP2C and CYP2D families16.
The lack of transformation products does not exclude drug-enzyme interactions between C-1311 and enzyme proteins in tumor cells. Therefore, in this study, the influence of C-1311 on the CYP3A4 level and enzymatic activity was investigated. We demonstrated that the level of P450 protein was independent of the presence of this agent. However, the enzymatic activity of this isoenzyme dropped significantly in CHO cells overexpressing CYP3A4 in a dose- and time-dependent manner, demonstrating the potency of C-1311 in inhibiting the activity of the CYP3A4 isoform of cytochrome P450. It should also be mentioned that C-1311 was identified by molecular modeling to be an inhibitor of the enzyme NQO2, NAD(P)H: quinone oxidoreductase 1 (DT-diaphorase)38.
The inhibition of CYP3A4, an enzyme responsible for the transformation of many clinically used drugs, might have crucial consequences on the final effects of multidrug therapy, which is usually applied in tumor-based diseases. C-1311-mediated inhibition of CYP3A4 might modulate the metabolism of other therapeutics that are sensitive to the action of this enzyme. Many reports have demonstrated the modulation of CYP3A4 activity by drugs such as carbamazepine39,40 or 17α-ethynylestradiol41. The latter strongly inhibited CYP3A4 while also being metabolized by it. However, there are few reports about the inhibition of specific P450 isoenzymes by antitumor agents, eg, by thiotepa42,43, tamoxifen44,45 and the less well-known sorafenib46.
Our results showed that cytochrome P450 CYP3A4 did not metabolize C-1311 in cells, confirming our hypothesis that overexpression of this enzyme might modulate the cellular response of CHO cells following C-1311 treatment, even though CYP3A4 does not participate in metabolic transformations. Cell cycle analysis revealed that wild-type CHO cells underwent G2/M arrest, whereas the populations of two transfected cell lines, CHO-HR and CHO-HR-3A4, only transiently accumulated in this compartment following C-1311 treatment (24 h). Prolonged incubation with C-1311 caused the degradation of DNA in all three CHO cell lines, but it was most effective in the two transfected ones (Figure 3C). Sub-G1 DNA content, representing the apoptotic fraction of cells, reached 71% in CHO-HR-3A4 cells and was slightly higher in CHO-HR cells (84% after 120 h of treatment). However, in the case of CHO-HR-3A4 cells, it was very difficult to determine the cell cycle compartment starting at 72 h because of the massive disruption of nuclei compared to CHO-HR cells (Figure 3A). The sub-G1 population was smaller in wild-type CHO cells and amounted to 61% after 120 h of C-1311 treatment.
Further studies, including morphological examination of cell nuclei, phosphatidylserine externalization, caspase-3 activation, cell membrane integrity and apoptotic DNA breaks (data for the later not shown), confirmed the induction of apoptosis and necrosis by C-1311 in CHO cells. Moreover, cell death was induced to the greatest extent in CHO-HR-3A4 cells coexpressing CPR and the CYP3A4 isoenzyme compared to CHO-HR cells overexpressing P450 reductase or wild-type CHO cells.
The higher sensitivity to C-1311 and increased susceptibility to apoptosis observed in CHO-HR cells compared to CHO-WT cells following drug treatment could be explained by the fact that overexpression of cytochrome P450 reductase itself increased the level of free radicals (ROS) in the cell30. We showed here that C-1311 induced ROS in CHO-HR cells but not in wild-type CHO cells. Generation of ROS was also observed in C-1311-treated CHO-HR-3A4 cells, but the level of ROS was lower than that in CHO-HR cells and was associated with apoptosis induced by C-1311 in the majority of CHO-HR-3A4 cells (Figure 6). A similar explanation of the higher cytotoxicity of 5-fluorouracil in CPR-overexpressing cells compared to wild-type cells has also been reported47.
In further studies of the C-1311-induced cellular response, we demonstrated that transfected CHO cells that did not die through apoptosis or necrosis following C-1311 exposure were in a state of accelerated senescence based on their altered cell morphology and the appearance of senescence-associated β-galactosidase (SA-β-gal) activity compared to untreated cells (Figure 7A). Interestingly, senescence was not induced by C-1311 in wild-type CHO cells. Additionally, the colony-forming assay revealed that after a short period of drug treatment (48 h), half of the population of wild-type CHO cells was able to return to proliferation, but by 96 h of drug treatment, proliferation, ie, colony formation of CHO-WT cells, was completely inhibited. In transfected CHO-HR and CHO-HR-3A4 cells, cell division was completely blocked after only 24 h of C-1311 treatment.
There are some data in the literature suggesting that the CYP3A4 expression level plays an important role in cellular response, including tumor progression, and that this role is independent of the activation of carcinogens or the metabolism of anticancer drugs. For instance, CYP3A4 promoted the progression of the cell cycle from G1 to S phase and induced a hypoxic response in Hep3B cells48. Other reports indicated that CYP3A4 promotes Stat3-mediated breast cancer cell growth49 and that its strong expression may be a key feature in an unfavorable prognosis in breast cancer50. There are also findings that the pregnane X receptor (PXR), which controls the expression of CYP3A451, plays an anti-apoptotic role in cancer cells, and this role is independent of the PXR-regulated expression of other cytochrome P450 enzymes52,53. Our previous studies also demonstrated that cytotoxic activity and cellular effects such as apoptosis, necrosis and senescence induced by the anticancer triazoloacridinone derivative C-1305 in CHO cells were affected by CYP3A4 expression54.
Conclusion
Collectively, our findings showed that C-1311-induced apoptosis, necrosis and senescence in CHO cells were modulated by CYP3A4 overexpression, whereas the metabolic transformation of this drug was independent of CYP3A4 expression. In conclusion, the alteration of the cellular response induced by C-1311 in CYP3A4-overexpressing cells was not dependent on the concentration of CYP3A4-mediated metabolites of this compound. Further studies should elucidate the mechanism of action and the role of CYP3A4 in C-1311-induced pathology.
Author contribution
Monika PAWŁOWSKA performed the research, analyzed the data and wrote the first draft. Ewa AUGUSTIN designed the research, performed experiments, analyzed the data and wrote the manuscript. Zofia MAZERSKA designed the research, analyzed the data and wrote the manuscript.
Acknowledgments
This work was supported by R&D grant No 014668/009 from the Chemical Faculty of Gdańsk University of Technology. We thank Dr Thomas FRIEDBERG and Dr Roland C WOLF from the Biomedical Research Centre, Dundee, Scotland, UK, for providing the CHO cell lines for this study. We also thank Dr Joanna POLEWSKA from our department for help in the flow cytometry experiments.
References
- Zhou SF. Drugs behave as substrates, inhibitors and inducers of human cytochrome P450 3A4. Curr Drug Metab. 2008;9:310–22. doi: 10.2174/138920008784220664. [DOI] [PubMed] [Google Scholar]
- Guengerich FP. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 2006;8:101–11. doi: 10.1208/aapsj080112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CJ, Otto DM, Carrie D, Magnuson MA, McLaren AW, Rosewell I, et al. Inactivation of the hepatic cytochrome P450 system by conditional deletion of hepatic cytochrome P450 reductase. J Biol Chem. 2003;278:13480–6. doi: 10.1074/jbc.M212087200. [DOI] [PubMed] [Google Scholar]
- Cholody WM, Martelli S, Paradziej-Lukowicz J, Konopa J. 5-[(Aminoalkyl)amino]imidazo[4,5,1-de]acridin-6-ones as a novel class of antineoplastic agents. Synthesis and biological activity. J Med Chem. 1990;33:49–52. doi: 10.1021/jm00163a009. [DOI] [PubMed] [Google Scholar]
- Cholody WM, Martelli S, Konopa J. Chromophore-modified antineoplastic imidazoacridinones. Synthesis and activity against murine leukemias. J Med Chem. 1992;35:378–82. doi: 10.1021/jm00080a026. [DOI] [PubMed] [Google Scholar]
- Cholody WM, Horowska B, Paradziej-Lukowicz J, Martelli S, Konopa J. Structure-activity relationship for antineoplastic imidazoacridinones: synthesis and antileukemic activity in vivo. J Med Chem. 1996;39:1028–32. doi: 10.1021/jm950564r. [DOI] [PubMed] [Google Scholar]
- Chau M, Otake Y, Christensen JL, Fernandes DJ, Ajami AM. The imidazoacridinone, C-1311 (Symadex™): The first of a potent new class of FLT3 inhibitors (Abstract) AACR Meeting Abstracts. 2006. p. B35.
- Skwarska A, Augustin E, Sobiesiak S, Koprowska J, Konopa J. Down-regulation of FLT3 kinase phosphorylation and survivin expression contributes to the activity of imidazoacridinone C-1311 against leukemia cells with FLT3 IDT mutation (Abstract) Eur J Cancer. 2010;8:185. [Google Scholar]
- Isambert N, Campone M, Bourbouloux E, Drouin M, Major A, Yin W, et al. Evaluation of the safety of C-1311 (Symadex) administered in a phase I dose-escalation trials as a weekly infusion for 3 consecutive weeks in patients with advanced solid tumors. Eur J Cancer. 2010;46:729–34. doi: 10.1016/j.ejca.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Capizzi RL, Roman LA, Tjulandin S, Smirnova I, Manikhas A, Paterson S, et al. Phase II trial of C-1311, a novel inhibitor of topoisomerase II in advanced breast cancer (Abstract) J Clin Oncol. 2008;26:1055. [Google Scholar]
- Thomas AL, Anthoney A, Scott E, Ahmed S, Lundberg AS, Major A, et al. C-1311, a novel inhibitor of FLT3 and topoisomerase II: A phase 1 trial of once every three week schedule in patients with advanced solid tumours (Abstract) J Clin Oncol. 2008;26:2576. [Google Scholar]
- Berger B, Marquardt H, Westendorf J. Pharmacological and toxicological aspects of new imidazoacridinone antitumor agents. Cancer Res. 1996;56:2094–104. [PubMed] [Google Scholar]
- Mazerska Z, Dziegielewski J, Konopa J. Enzymatic activation of a new antitumor drug, 5-diethylaminoethylamino-8-hydroxyimidazoacridinone, C-1311, observed after its intercalation into DNA. Biochem Pharmacol. 2001;61:685–94. doi: 10.1016/s0006-2952(01)00527-5. [DOI] [PubMed] [Google Scholar]
- Dziegielewski J, Slusarski B, Konitz A, Skladanowski A, Konopa J. Intercalation of imidazoacridinones to DNA and its relevance to cytotoxic and antitumor activity. Biochem Pharmacol. 2002;63:1653–62. doi: 10.1016/s0006-2952(02)00916-4. [DOI] [PubMed] [Google Scholar]
- Mazerska Z, Sowinski P, Konopa J. Molecular mechanism of the enzymatic oxidation investigated for imidazoacridinone antitumor drug, C-1311. Biochem Pharmacol. 2003;66:1727–36. doi: 10.1016/s0006-2952(03)00477-5. [DOI] [PubMed] [Google Scholar]
- Potega A, Dabrowska E, Niemira M, Kot–Wasik A, Ronseaux S, Henderson CJ, et al. The imidazoacridinone antitumor drug, C-1311, is metabolized by flavin monooxygenases but not by cytochrome P450s. Drug Metab Dispos. 2011;39:1423–32. doi: 10.1124/dmd.111.038984. [DOI] [PubMed] [Google Scholar]
- Fedejko-Kap B, Bratton SM, Finel M, Radominska-Pandya A, Mazerska Z. Role of human UDP — glucuronosyltransferases in the biotransformation of the imidazo- and triazoloacridinone antitumor agents C-1305 and C-1311: Highly selective substrates for UGT1A10. Drug Metab Dispos. 2012;40:1736–43. doi: 10.1124/dmd.112.045401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin E, Wheatley DN, Lamb J, Konopa J. Imidazoacridinones arrest cell cycle progression in the G2 phase of L1210 cells. Cancer Chemother Pharmacol. 1996;38:39–44. doi: 10.1007/s002800050445. [DOI] [PubMed] [Google Scholar]
- Lamb J, Wheatley DN. Cell killing by the novel imidazoacridinone antineoplastic agent, C-1311, is inhibited at high concentrations coincident with dose-differentiated cell cycle perturbation. Br J Cancer. 1996;74:1359–68. doi: 10.1038/bjc.1996.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffaroni Z, De Marco C, Villa R, Riboldi S, Daidone MG, Double JA. Cell growth inhibition, G2/M cell cycle arrest and apoptosis induced by the imidazoacridinone C1311 in human tumour cell lines. Eur J Cancer. 2001;37:1953–62. doi: 10.1016/s0959-8049(01)00227-1. [DOI] [PubMed] [Google Scholar]
- Hyzy M, Bozko P, Konopa J, Skladanowski A. Antitumour imidazoacridinone C-1311 induces cell death by mitotic catastrophe in human colon carcinoma cells. Biochem Pharmacol. 2005;69:801–9. doi: 10.1016/j.bcp.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Skwarska A, Augustin E, Konopa J. Sequential induction of mitotic catastrophe followed by apoptosis in human leukaemia MOLT4 cells by imidazoacridinone C-1311. Apoptosis. 2007;12:2245–57. doi: 10.1007/s10495-007-0144-y. [DOI] [PubMed] [Google Scholar]
- Ding S, Yao D, Burchell B, Wolf CR, Friedberg T. High levels of recombinant CYP3A4 expression in Chinese hamster ovary cells are modulated by coexpressed human P450 reductase and hemin supplementation. Arch Biochem Biophys. 1997;348:403–10. doi: 10.1006/abbi.1997.0405. [DOI] [PubMed] [Google Scholar]
- Augustin E, Mos-Rompa A, Skwarska A, Witkowski J, Konopa J. Induction of G2/M phase arrest and apoptosis of human leukemia cells by potent antitumor triazoloacridinone C-1305. Biochem Pharmacol. 2006;72:1668–79. doi: 10.1016/j.bcp.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updat. 2006;4:303–13. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- Mansila S, Bataller M, Portugal J. Mitotic catastrophe as a consequence of chemotherapy. Anticancer Agents Med Chem. 2006;6:589–602. doi: 10.2174/187152006778699086. [DOI] [PubMed] [Google Scholar]
- Nallani SC, Goodwin B, Buckley AR, Buckley DJ, Desai PB. Differences in the induction of cytochrome P450 3A4 by taxane anticancer drugs, docetaxel and paclitaxel, assessed employing primary human hepatocytes. Cancer Chemother Pharmacol. 2004;54:219–29. doi: 10.1007/s00280-004-0799-9. [DOI] [PubMed] [Google Scholar]
- Cohen GM. Caspases: the executioners of apoptosis. Biochem J. 1997;326:1–16. doi: 10.1042/bj3260001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KH, Senft JA. An improved method to determine cell viability by simultaneous staining with fluorescein diacetate-propidium iodide. J Histochem Cytochem. 1985;33:77–9. doi: 10.1177/33.1.2578146. [DOI] [PubMed] [Google Scholar]
- Fussell KC, Udasin RG, Gray JP, Mishin V, Smith PJS, Heck DE, et al. Redox cycling and increased oxygen utilization contribute to diquat-induced oxidative stress and cytotoxicity in Chinese hamster ovary cells overexpressing NADPH-cytochrome P450 reductase. Free Radic Biol Med. 2011;50:874–82. doi: 10.1016/j.freeradbiomed.2010.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Circu ML, Aw TY. Reactive oxygen species, cellular redox systems, and apoptosis. Free Rad Biol Med. 2010;48:749–62. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emadi A, Jones JR, Brodsky RA. Cyclophosphamide and cancer: golden anniversary. Nat Rev Clin Oncol. 2009;6:638–47. doi: 10.1038/nrclinonc.2009.146. [DOI] [PubMed] [Google Scholar]
- Chugh R, Wagner T, Griffith KA, Taylor JMG, Thomas DG, Worden FP, et al. Assessment of ifosfamide pharmacokinetics, toxicity, and relation to CYP3A4 activity as measured by the erythromycin breath test in patients with sarcoma. Cancer. 2007;109:2315–22. doi: 10.1002/cncr.22669. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y, Ando A, Takamura Y, Taguchi T, Tamaki Y, Noguchi S. Prediction of response of docetaxel by CYP3A4 mRNA expression in breast cancer tissues. Int J Cancer. 2002;97:129–32. doi: 10.1002/ijc.1568. [DOI] [PubMed] [Google Scholar]
- Gjerde J, Kisanga ER, Hauglid M, Holm PI, Mellgren G, Lien EA. Identification and quantification of tamoxifen and four metabolites in serum by liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2005;1082:6–14. doi: 10.1016/j.chroma.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Yang SI, Suh JH, Lee MG. Pharmacokinetic interaction between tamoxifen and ondasetron in rats: non competitive (hepatic) and competitive (intestinal) inhibition of tamoxifen metabolism by ondansetron via CYP2D and 3A1/2. Cancer Chemother Pharmacol. 2010;65:407–18. doi: 10.1007/s00280-009-1043-4. [DOI] [PubMed] [Google Scholar]
- Quintieri L, Rosato A, Napoli E, Sola F, Geroni C, Floreani M, et al. In vivo antitumor activity and host toxicity of methoxymorpholinyl doxorubicin: role of cytochrome P450 3A. Cancer Res. 2000;60:3232–8. [PubMed] [Google Scholar]
- Nolan KA, Humphries RA, Stratford JJ. Imidazoacridin-6-ones as a novel inhibitors of the quinone oxidoreductase NQO2. Bioorg Med Chem Lett. 2010;20:2832–6. doi: 10.1016/j.bmcl.2010.03.051. [DOI] [PubMed] [Google Scholar]
- Pearce RE, Uetrecht JP, Leeder JS. Pathways of carbamazepine bioactivation in vitro: II. The role of human cytochrome P450 enzymes in the formation of 2-hydroxyiminostilbene. Drug Metab Dispos. 2005;33:1819–26. doi: 10.1124/dmd.105.004861. [DOI] [PubMed] [Google Scholar]
- Pearce RE, Lu W, Wang Y, Uetrecht JP, Correia MA, Leeder JS. Pathways of carbamazepine bioactivation in vitro: III. The role of human cytochrome P450 enzymes in the formation of 2,3-dihydroxycarbamazepine. Drug Metab Dispos. 2008;36:1637–49. doi: 10.1124/dmd.107.019562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HL, Kent UM, Hollenberg PF. Mechanism-based inactivation of cytochrome P450 3A4 by 17 alpha-ethynylestradiol: Evidence for heme destruction and covalent binding to protein. J Pharmacol Exp Ther. 2002;301:160–7. doi: 10.1124/jpet.301.1.160. [DOI] [PubMed] [Google Scholar]
- Jacobson PA, Green K, Birnbaum A, Remmel RP. Cytochrome P450 isozymes 3A4 and 2B6 are involved in the in vitro metabolism of thiotepa to TEPA. Cancer Chemother Pharmacol. 2010;49:461–7. doi: 10.1007/s00280-002-0453-3. [DOI] [PubMed] [Google Scholar]
- Richter T, Schwab M, Eichelbaum M, Zanger UM. Inhibition of human CYP2B6 by N,N′,N″-triethylenethiophosphoramide is irreversible and mechanism-based. Biochem Pharmacol. 2005;69:517–24. doi: 10.1016/j.bcp.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Sridar C, Kent UM, Notley LM, Gillam EM, Hollenberg PF. Effect of tamoxifen on the enzymatic activity of human cytochrome CYP2B6. J Pharmacol Exp Ther. 2002;301:945–52. doi: 10.1124/jpet.301.3.945. [DOI] [PubMed] [Google Scholar]
- Notley LM, Crewe KH, Taylor PJ, Lennard MS, Gilliam EM. Characterization of the human cytochrome P450 forms involved in metabolism of tamoxifen to its alpha-hydroxy and alpha, 4-dihydroxy derivatives. Chem Res Toxicol. 2005;18:1611–8. doi: 10.1021/tx050140s. [DOI] [PubMed] [Google Scholar]
- Sugiyama M, Fujita K, Marayama N, Akiyama Y, Yamazaki H, Sasaki Y. Sorafenib and sunitinib, two anticancer drugs, inhibit CYP3A4-mediated and activate CYP3A5-mediated midazolam 1′-hydroxylation. Drug Metab Dispos. 2011;39:757–62. doi: 10.1124/dmd.110.037853. [DOI] [PubMed] [Google Scholar]
- Martinez VG, Williams KJ, Stratford IJ, Clynes M, O'Connor R. Overexpression of cytochrome P450 NADPH reductase sensitises MDA 231 breast carcinoma cells to 5-fluorouracil: possibile mechanisms involved. Toxicol in Vitro. 2008;22:582–8. doi: 10.1016/j.tiv.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Oguro A, Sakamoto K, Funae Y, Imaka S. Overexpression of CYP3A4, but not of CYP2D6, promotes hypoxic response and cell growth of Hep3B cells. Drug Metab Pharmacokinet. 2011;26:407–15. doi: 10.2133/dmpk.dmpk-11-rg-017. [DOI] [PubMed] [Google Scholar]
- Mitra R, Guo Z, Milani M, Mesaros C, Rodriguez M, Nguyen J, et al. CYP3A4 mediates growth of estrogen receptor-positive breast cancer cells in part by inducing nuclear translocation of phospho–Stat3 through biosynthesis of (±)-14,15-epoxyeicosatrienoic acid (EET) J Biol Chem. 2011;286:17543–59. doi: 10.1074/jbc.M110.198515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas S, Pierl C, Harth V, Pesch B, Rabstein S, Brüning T, et al. Expression of xenobiotic and steroid hormone metabolizing enzymes in human breast carcinomas. Int J Cancer. 2006;119:1785–91. doi: 10.1002/ijc.21915. [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest. 1998;102:1016–23. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama H, Nakatsukasa H, Takamoto N, Hiramatsu Y. Down-regulation of pregnane X receptor contributes to cell growth inhibition and apoptosis by anticancer agents in endometrial cancer cells. Mol Pharmacol. 2007;72:1045–53. doi: 10.1124/mol.107.037937. [DOI] [PubMed] [Google Scholar]
- Zhou J, Liu M, Zhai Y, Xie W. The antiapoptotic role of pregnane X receptor in human colon cancer cells. Mol Endocrinol. 2008;22:868–80. doi: 10.1210/me.2007-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustin E, Borowa-Mazgaj B, Kikulska A, Kordalewska M, Pawlowska M. CYP3A4 overexpression enhances the cytotoxicity of the antitumor triazoloacridinone derivative C-1305 in CHO cells. Acta Pharmacol Sin. 2013;34:146–56. doi: 10.1038/aps.2012.132. [DOI] [PMC free article] [PubMed] [Google Scholar]