Abstract
Dynamin-related proteins (DRPs) are multi-domain GTPases that function via oligomerization and GTP-dependent conformational changes to play central roles in regulating membrane structure across phylogenetic kingdoms. How DRPs harness self-assembly and GTP-dependent conformational changes to remodel membranes is not understood. Here we present the crystal structure of an assembly-deficient mammalian endocytic DRP, dynamin 1, lacking the proline-rich domain, in its nucleotide-free state. The dynamin 1 monomer is an extended structure with the GTPase domain and bundle signalling element positioned on top of a long helical stalk with the pleckstrin homology domain flexibly attached on its opposing end. Dynamin 1 dimer and higher order dimer multimers form via interfaces located in the stalk. Analysis of these interfaces provides insight into DRP family member specificity and regulation and provides a framework for understanding the biogenesis of higher order DRP structures and the mechanism of DRP-mediated membrane scission events.
Dynamin-related proteins (DRPs) belong to a highly conserved (Supplementary Fig. 1) GTPase superfamily that catalyses diverse membrane remodelling events1,2.Membrane scission DRPs include dynamin 1, which catalyses clathrin-coated vesicle scission at the plasma membrane and Drp1/Dnm1, which divide mitochondria. Despite their functional diversity, all DRPs undergo GTP cycle-dependent conformational changes to regulate self-assembly and disassembly3–8. DRP architecture includes an amino-terminal GTPase domain, a bundle signalling element (BSE), a middle domain (MD) and a GTPase effector domain (GED)9,10. Many DRPs also have a variable region between the MD and GED; in dynamin 1, this is a pleckstrin homology domain (PH) that binds to phosphatidylinositol-4,5-bisphosphate (PtdIns-4,5-P2)-containing membranes to facilitate targeting and possibly membrane remodelling via membrane insertion11–14.
Assembled DRPs can form helical structures in vitro15,16.Within these structures, GTP cycle-driven conformational changes result in membrane remodelling4,5,7,17. The structural basis for DRP self-assembly and GTP cycle-dependent conformational changes are not fully understood. The mechanism of assembly has been informed by the structure of the ‘stalk’ of MxA, a distantly related DRP, in which the MD and part of the GED form an extended helical bundle that mediates self-assembly via conserved interfaces18. Several structures have also provided insight into DRP GTP cycle conformational changes (Supplementary Fig. 2)19–24. Cryo-electron microscopic structures of assembled dynamin in guanosine-5′-[(β,γ)-methyleno]triphosphate (GMPPCP)-bound and nucleotide- free states have provided models for the assembled oligomers and the location of the GTPase and PH domains within the helical structures17,25. However, to understand the basis of DRP self-assembly and mechanism, the architecture of DRP domains within a single molecule must be elucidated. Here, we report the crystal structure of an assembly-deficient dynamin 1 in the nucleotide-free state that lacks only the unstructured carboxy-terminal proline-rich domain (PRD).
G397D, an assembly-deficient dynamin 1 mutant
The propensity of DRPs to assemble has been an obstacle to obtaining crystals suitable for diffraction experiments. We identified the mutation G436D, an invariant MD residue, in the Saccharomyces cerevisiae mitochondrial division DRP Dnm1, by screening for mutations that possessed the same phenotype as assembly-deficient Dnm1 G385D, specifically mutations that shifted Dnm1–green fluorescent protein (GFP) fusion from a punctate to a more diffuse localization pattern in yeast cells16. We expressed and purified (Supplementary Fig. 3a) the orthologous rat dynamin 1 mutant protein lacking the PRD (Dyn1 G397D ΔPRD) and examined its ability to self assemble using light scattering (Fig. 1a). Addition of GMPPCP caused an increase in scattering in Dyn1 ΔPRD samples, but caused no change in scattering inDyn1 G397D ΔPRD samples. Using a combination of size-exclusion chromatography and sucrose gradient centrifugation26, Dyn1 ΔPRD and Dyn1 G397D ΔPRD were estimated to be dimeric (Supplementary Fig. 3b and Supplementary Table 1) under non-assembly conditions, similar to other assembly-deficient DRP mutants16,27. Under assembly conditions, we observed an increase in the sedimentation coefficient of Dyn1ΔPRD, but not forDyn1 G397D ΔPRD (Supplementary Table 1). Dyn1G397D ΔPRD was defective for assembly-stimulated GTP hydrolysis (Fig. 1b and Supplementary Table 2) and failed to assemble into helical structures on phosphatidylinositol 4-phosphate (PtdIns-4P)- containing lipid nanotubes, in contrast to Dyn1 ΔPRD (Fig. 1c). Dyn1 G397D ΔPRD was also soluble at higher concentrations than Dyn1 ΔPRD. Together, these data indicate that the G397D mutation severely hampers dynamin self assembly and substantiates the critical role of the MD in intermolecular interactions16,18,27–29. Given these characteristics, it presented an attractive target for crystallization.
Figure 1. The G397D mutation in Dyn1 ΔPRD blocks self-assembly.
. a, 90° light scattering. Dyn1 ΔPRD (blue, 1 µM) and Dyn1 G397D ΔPRD (red, 1µM) were monitored after addition of 0.5mM GMPPCP and, in the case of Dyn1 ΔPRD, 1mMGTP (blue arrows). Red arrow indicates opening of sample port. AU, arbitrary units. b, Steady-state GTP hydrolysis kinetics of Dyn1 ΔPRD and Dyn1 G397DΔPRD in the absence (light blue and red) and presence (dark blue and orange) of 0.1mgml−1 liposomes containing 10% PtdIns-4P, monitored by a NADH-dependent coupled assay as described43. A representative trace is shown with 1mM GTP. c, Transmission electron microscopy of negative-stained 0.25 mgml−1 10% PtdIns-4P lipid nanotubes with Dyn1 ΔPRD (middle) and Dyn1 G397D ΔPRD (right) and no added protein (left) and 0.5mM GMPPCP. Scale bars, 200 nm.
Crystallization and structure of Dyn1 G397D ΔPRD
We obtained orthorhombic crystals of Dyn1 G397D ΔPRD that diffracted to a minimum Bragg spacing of 3.1Å (Methods and Supplementary Table 3). The structure was solved by molecular replacement, using the nucleotide-free rat dynamin 1 GTPase domain, the human dynamin 1 PH and a portion of the human MxA stalk, as sequential search models (Methods)11,13,18,30. We traced the complete model, except for some disordered loops, and assigned amino acids after refinement of the molecular replacement solution. The model was refined to R/Rfree values of 0.21/0.27. A representative example of the B-sharpened likelihood-weighted 2mFobs − DFcalc electron density map is shown in Fig. 2c.
Figure 2. The crystal structure of Dyn1 G397D ΔPRD.
a, Schematic of the Dyn1 domain structure. Numbers indicate domain-ending amino acid. Colour scheme used here is retained. b, Crystal structure of Dyn1 G397D ΔPRD. Linkers between the NGTPase and GTPase domain and the CGTPase and MD are shown in grey. Loops with no density are represented with dashed lines. Stalk nomenclature is based on that of the MxA stalk structure18. VL, variable loop. c, An example of the refined to B-sharpened 2mFobs – DFcalc map, contoured at 1s. The region shown is part of the stalk boxed by purple dotted lines in (b). d, Three symmetry-related PH domains in the lattice.
Dyn1 G397D ΔPRD forms an extended structure with the GTPase and PH domains separated by a stalk consisting of an anti-parallel helical bundle of the MD and a helix from the GED (Fig. 2a, b). By contrast, the distantly related bacterial dynamin-like protein (BDLP) is compactly folded in its nucleotide-free and GDP-bound states (Supplementary Fig. 2a), with its ‘paddle’, in an analogous region to the PH domain of dynamin, in close proximity to the GTPase domain19,20. Thus, unlike BDLP, GTP binding in dynamin is not harnessed to form an extended structure, consistent with nucleotide-independent assembly of dynamin on liposomes4,8,31,32. Linkers connecting the MD to the PH and the PH to the GED are disordered, indicating a flexibly tethered PH. Thus, any of three crystallographic symmetry-related PH domains could connect to the remainder of the structure (Fig. 2d). The probable PH partner, based on the fit with the envelope of an assembly-deficient dynamin dimer determined by small-angle X-ray scattering33 (SAXS), is shown in Fig. 2b. The structure of the PH is similar to those previously determined11,13, with expected differences concentrated in the variable loops.
The structure of the GTPase domain is similar to that of the previously determined nucleotide-free dynamin GTPase domain structure30. There are minor expected changes in the poorly resolved switch 2 region, the dynamin-specific loop and in the loop connecting the NGTPase to the GTPase domain. The CGTPase helix is kinked at the conserved proline 294 and, together with the NGTPase helix and a helix at the C terminus of the GED (CGED), forms the three-helix BSE9. CGED covers a groove of 937Å 2 between the NGTPase and CGTPase helices. In the nucleotide-free dynamin 1 GTPase and the GDP-bound Dictyostelium discoideum dynamin A GTPase structures, a myosin peptide substitutes for CGED, indicating the importance of this interface for GTPase domain stability30,34.
A linker with elevated crystallographic temperature factors connects the CGTPase to the dynamin stalk (Fig. 2a, b) and contains two prolines (319 and 322), whose equivalents in the distantly related DRP atlastin 1 connect the GTPase and MD (Supplementary Fig. 2b). Although the dynamin stalk sequence shares limited identity to MxA, its overall structure is similar and the MxA nomenclature is retained18 (Supplementary Fig. 4a). As for MxA, helix α1 is split by a disordered loop, L1. The remainder of α1 diverges from MxA and is split into two helices, termed α1C1 and α1C2. Helix α1C2 connects to helix α2 via a short disordered loop, L2. Helices α2 and α3 run the length of the stalk and are joined by a short loop, L3. The stalk is completed by GED-derived helix α4 that spans the stalk and connects to CGED via a linker. Following helix α3 is a coil that folds across α4 that is strongly conserved in dynamins, Drp1s and Dnm1, but absent in MxA.
The dynamin–dimer interface
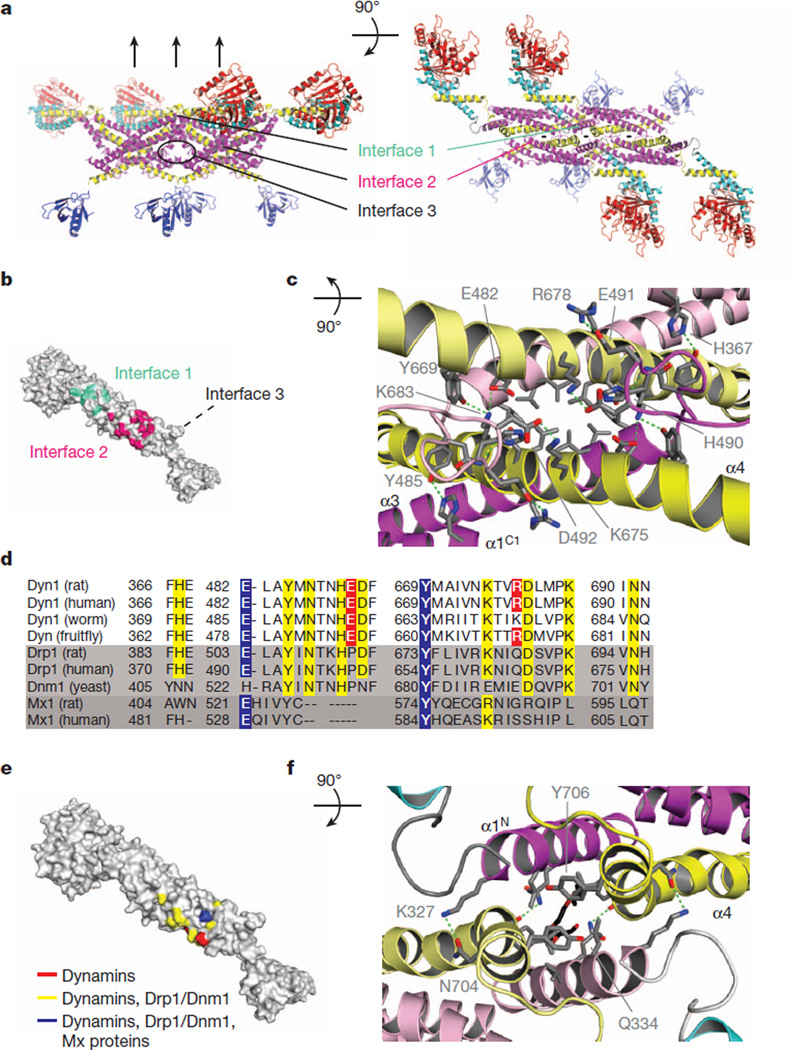
The crystal lattice contains linear filaments of dynamin assembled via three stalk interfaces, similar to MxA18, resulting in layers of interacting stalks separated by GTPase and PH domains (Fig. 3a, b). Interface 2, the largest with a buried surface of 1,339 Å 2, has two-fold symmetry and is formed by residues from stalk helices α4 and α3 (Fig. 3c), with an additional residue from α1C1 (H367). Each protomer in the interface contributes seven direct hydrogen bonds and eight hydrophobic residues line the site of contact between α3 and α4 (Fig. 3c).
Figure 3. Dyn1 G397D ΔPRD stalk interfaces mediate self-assembly.
a, Schematic diagramof fourmonomers showing interfaces in the crystal lattice. b, Surface representation showing the locations of interfaces 1 and 2. c, Detail of interface 2. Protomers are shown in lighter and darker hues. Green dotted lines are hydrogen bonds. d, Conservation of interface 2 residues involved in hydrogen bonding in dyamins, Drp1/Dnm1 and Mx proteins. Blue, conserved in dynamins, Drp1/Dnm1 and Mx proteins; yellow, conserved in dynamins and Drps; red, conserved in dynamins. Alignment shows a subset of sequences used to determine the conservation. Sequences are identical to Supplementary Fig. 1, with the addition of Rattus norvegicus Mx1 (P18588.1) and Homo sapiens Mx1 (P20591.4). Fruitfly, Drosophila melanogaster; human, Homo sapiens; rat, Rattus norvegicus; worm, Caenorhabditis elegans; yeast, Saccharomyces cerevisiae. e, Surface representation of conservation data shown in (d). f, Interface 1. Density interpreted as a PEG400 molecule is shown in black.
Interface 2 sequence conservation indicates a mechanism for dimer specificity within the DRP superfamily. Phylogenetic analysis of the hydrogen bonding partners within this region in dynamins, Drp1s and Dnm1, and Mx proteins allowed us to categorize three classes of residues: conserved in dynamins, Drp1/Dnm1, and Mx proteins (in blue), conserved only within dynamins and Drp1/Dnm1 (in yellow), and conserved only in the dynamins (in red) (Fig. 3d). Plotting these classes onto a surface representation of the dynamin monomer (Fig. 3e) indicates that dimerization specificity is controlled by a spatial combinatorial code. As predicted from this code, hetero-oligomers consisting of dynamin 1 and dynamin 2 are observed35. Several residues contributing to the apparent specificity are localized in the strongly conserved coil of dynamin, Drp1 and Dnm1 that follows α3.
Dynamin stalk interfaces drive self assembly
Interfaces 1 and 3 are also stalk-localized and mediate higher order assembly of dynamin dimers (Fig. 3a). Interface 1 is at the tips of interacting stalks, proximal to the GTPase domain and BSE (Fig. 3f) and is formed through interactions between protomer helices α4 and α1N. The interface is capped by a stacking interaction between opposing and flexible Y706 residues, conserved in dynamins and Drp1s. Interface 1 includes four hydrogen bonds (Fig. 3f, dashed green lines), with the remainder consisting of hydrophobic residues. The buried surface area of interface 1 is relatively small (624 Å2) and is likely to be dynamic to tolerate protomer interactions in a range of orientations.
Interface 3 is at the distal end of the stalk, where L2 loops from symmetry-related molecules are in close proximity (Supplementary Fig. 4b). The G397D mutation is within L2, which could not be traced due to poor density. The N-terminal region of L2 is also in close proximity to a symmetry-related L1, which features a highly conserved glycine, G346, that when mutated in Dnm1 (G385D) blocks assembly beyond the dimer16. We therefore predict that G397 and G346 are near one another in assembled dynamin. In addition, mutations at two dynamin arginine residues, R399 (in L2) and R361 (in α1C1, following L1), and the corresponding interface 3 mutations in MxA also inhibit assembly and stabilize dimeric forms18,27. In contrast, disruption of MxA interface 1 yields a mixture of dimers and tetramers18. Thus, our data indicate that interface 3 is critical at an early step in the dynamin assembly pathway as its disruption stabilizes dimeric dynamin and allows linear filament formation. In contrast, interface 1 interactions probably function only to stabilize oligomerization.
From our structure, we propose that the dynamin dimer interface 2 is constitutive and relatively rigid. As compared to linear arrangement in the crystal, in helical assemblies we propose that the necessary rotational and translational shifts occur between adjacent dimers at interfaces 1 and 3 (Fig. 4a). Indeed, stalk dimers fit into the GMPPCP-bound helical electron microscopic reconstruction of dynamin25 (Fig. 4b) possess a more tightly packed interface 3 and interface 1. These differences can be attributed to the disordered interface 3 in the crystal that is probably due to steric hindrance from the G397D mutation, and the dynamic nature of the hydrophobic interface 1.
Figure 4. Oligomerization of dynamin into helical structures.
a, Dynamin helices derived from the linear arrangement in our crystal structure. Two stalk dimers (green and magenta) that engage in interface 1 and 3 are related by crystallographic translation. Experimentally determined helical parameters for dynamin assembled into helices in the GMPPCP-bound state25 were matched by applying a small shift and tilt of one stalk dimer with respect to the other. b, Placement of oligomerized dynamin model into the electron microscopy density map contoured at 1.2s. In side view: the fit of the GTPase domain as a GTPase–GTPase dimer with the BSE in open conformation to connect to interface 1 of the stalk helix (solid density is contoured at 3.6σ). c, Observed conformational flexibility of the BSE. Model fitted into the helical reconstruction is shown as black superimposed ribbon on the crystal structure of the GTPase–CGED fusion dimer (PDB accession no. 2×2E)10.
Regulation of DRPs
The dynamin PH is essential for endocytosis and interacts with inositol phospholipids with low affinity12,36. Centronuclear myopathy (CNM) disease mutations cluster at the C terminus of the PH α-helix (Supplementary Fig. 5a), underscoring its importance. They cause an increase in basal GTPase activity, without altering interactions with inositol phospholipids33. In addition, SAXS analysis of an assembly-deficient dynamin indicates that the CNM mutants have a different conformation compared to wild type33,37. In our lattice, three PH domains related by crystallographic symmetry lie close to interface 3 and the L1 loop (Fig. 2d), suggesting an interaction. Thus, the PH may serve to regulate access to this key multimerization interface to couple dynamin membrane interactions to dynamin assembly. Phosphoinositide binding by the PH variable loops and penetration of the membrane by variable loop 114 could help to expose dynamin interface 3 and/or L1 and thus promote multimerization.
Alignment of the dynamin PH and corresponding sequences from Drp1s shows conservation of key residues (Supplementary Figs 1 and 5b), indicating that lipids may similarly regulate mammalian mitochondrial division. However, mammalian Drp1s lack most of the C-terminal PH α-helix, including residues mutated in CNM. The details of Drp1 interface 3 regulation may therefore be different. Consistently, Drp1 splice variants have deletions in this region, pointing to a potential regulatory role. By contrast, Danio rerio Drp1 and fungal Dnm1 have an insert B of unknown structure in this region. Mitochondrial division DRPs can therefore be subdivided on this basis, which correlates with the divergence of their corresponding effectors, yeast Mdv1 and mammalian Mff38,39.
Discussion
Our structure provides insight into the mechanism and regulation of dynamin assembly and into how the dynamin GTPase cycle is harnessed for function. Several observations point to a key role of the dynamin BSE in the formation of the GTPase–GTPase dimer interface observed in the GTPase–CGED GDP•AlF4 − crystal structure10. This interface is likely to form from adjacent rungs of a dynamin helix and is critical to dynamin function as it mediates assemblystimulated GTP hydrolysis. Comparison of the GTPase–GTPase dimer with our nucleotide-free structure indicates that in addition to expected differences, the BSE is flexible. Dynamin genetic data also support a role for the BSE in the regulation of GTPase–GTPase interface formation. Specifically, the dynamin switch 2 shibire ts2 mutation, G146S, which causes endocytic intermediates with ‘collared’ dynamin necks to accumulate40, is suppressed by the sushi mutation A738T41,42, located in the CGED peptide, facing the hydrophobic groove (Fig. 5a). Insight into how dynamin undergoes conformational changes also comes from distantly related DRP structures (Supplementary Fig. 2). GTP-dependent GTPase domain dimerization is also observed for guanylate binding protein, indicating that this may be a common feature of the DRP superfamily22 (Supplementary Fig. 2c). In addition, two recent structures of the atlastin 1 cytosolic domain, thought to represent pre- and post-endoplasmic reticulummembrane fusion conformations23,24, indicate that large changes occur in the position of the 3-helix bundle ‘middle domain’ relative to the GTPase domain (Supplementary Fig. 2b).
Figure 5. Model for dynamin GTP cycle conformational changes.
a, Mapping of dynamin shibire and sushi mutations. b, Nucleotide-dependent dynamin conformations. The GTPase core domains (red) are in the same orientation. Left, GTP-bound state with open BSE conformation of dynamin as fitted into the GMPPCP-bound electron microscopic reconstruction shown in Fig. 4. Right, transition state of dynamin obtained by superposition of the BSE residues 291–312 and 727–743 of our structure on the corresponding residues of the GDP•AlF4 −-bound GTPase–CGED fusion dimer (PDB accession no. 2×2E)10. Transition from open to closed BSE conformation results in movement of stalk domains. c, Model for Dyn1 ΔPRD GTP-bound helix. The BSE is opened to allow GTPase–GTPase dimer formation. d, GTP hydrolysis closes the BSE and adopts the conformation of the GDP•AlF4 −-bound transition state. This results in a substantial global constriction of the helical oligomeric assembly causing membrane deformation and scission. e, Schematic of how the proposed GTP hydrolysis triggered BSE conformational change is transmitted to oligomerized stalk domains.
We propose that GTP binding and self-assembly promote dynamin GTPase–GTPase dimer formation via an opening of the BSE relative to the GTPase domain. In support, a modified version of our structure can be fit into the cryo-electron microscopic reconstruction of GMPPCP-bound dynamin with the BSE in a substantially more open conformation (Fig. 4b). We obtained a good fit of the GTPase– GTPase dimer domains and stalk interface 1 using constraints dictated by the helical fit of the oligomeric stalk and the strong GTPase domain-derived density (Fig. 4b). The BSE α-helices fit into the density stretching from the GTPase domain to the oligomeric stalk, indicating that a BSE flipping-out motion occurs at two hinge regions: the α-helical kink at P294 and in the loop connecting the NGTPase and the GTPase domain, at P32 (Fig. 4c). Consistently, superposition of GDP•AlF4 − GTPase–CGED fusion GTPase–GTPase dimer partners indicates a smaller yet directionally equivalent opening of the BSE in one protomer10. This proposed conformational change is feasible as there are relatively few contacts that hold the NGTPase helix to the body of the GTPase domain in our structure. In addition, the BSE links the GTPase domain to the stalk and interface 1 via a short turn, making it well placed to transmit conformational alterations. As previously noted, comparison of the nucleotide-free and GMPPCP-bound electron microscopic reconstructions indicates that the GMPPCP-bound helix has a relatively smaller diameter17. In addition, the stalk density is more ‘kinked’ in the GMPPCP-bound form17. When we dock our structure, we observed that the bases of the stalks do not fit the density of the GMPPCP-bound helix. However, where the fit becomes poor there are strongly conserved prolines in α2 and α4 and a partially conserved proline in α1C1 clustered in the stalk. We propose that these proline residues facilitate the formation of a kink at the stalk base in the GMPPCP-bound helix, which might allow interface 1 to form more fully in the assembled helix.
As predicted by our model, GTP hydrolysis would induce closure of the BSE via a transition state represented by the structure of the GTPase domain–BSE fusion in the presence of GDP•AlF4 −, towards the conformation observed in our nucleotide-free structure (Fig. 5b). As GTP binding to dynamin is not rate limiting and GTP hydrolysis is stimulated by the formation of the GTPase–GTPase interface between adjacent rungs, it is likely that the BSE-dependent conformational change occurs in the context of short dynamin helical assemblies rather than within a helix consisting of many rungs8,32. Short assemblies could also result in an approximate temporally coordinated conformational change (Fig. 5c–e). The conformational changes we propose would cause disruption to the assembled helix and the underlying membrane via local rung shifts. Interface 1 in our structure, which buries a relatively small surface area in the assembled stalk lattice, will be especially susceptible to change by inter-rung GTPase–GTPase dimer formation and its subsequent disruption by GTP hydrolysis, and these changes will be transmitted to interface 3 and the PH. The combined effects of curvature stress imposed by a short dynamin helical assembly coupled with PH insertion into the membrane is likely to destabilize the membrane and result in membrane fission14. Given the strong similarities between dynamins and other DRP family members, the structure of dynamin and our proposed model will serve to guide studies on the mechanisms of action of DRPs in diverse cellular functions.
METHODS SUMMARY
The conserved Dyn1 ΔPRD assembly-defective mutant G397D was identified by a cytological assay using the yeast mitochondrial fission DRP Dnm1–GFP. Dyn1 ΔPRD and Dyn1 G397D ΔPRD were expressed in E. coli and were purified as described in Methods. Light scattering, sucrose density gradient centrifugation, mass determination, continuous GTPase assays and electron microscopy were performed as described in Methods. Crystals were grown by microbatch from 3.2 µl droplets containing 52.5mM Tris/Cl pH 7.7, 175mM NaCl, 32.5mM NaNO3, 20% v/v PEG 400, 0.97mM β-mercaptoethanol and 31.9 µM Dyn1 G397D ΔPRD and cryo-protected with Paretone-N. Reflection data were collected at 100K at Beamline 8.3.1 at the Advanced Light Source (Berkeley, California, USA) at a wavelength of 0.9488Å. Data collection and processing are described in Methods. The structure was determined by molecular replacement, using known structures of the nucleotide-free rat dynamin 1 GTPase domain (PDB accession no. 2AKA chain B), the human dynamin 1 PH domain (PDB accession no. 2DYNchain B) and a truncated form of the human MxA stalk (PDB accession no. 3LJB chain B) as sequential search models. Structure refinement is described in Methods. The stalk interface 2 dimer was fit into the previously described cryo-electron microscopic reconstruction of GMPPCP-bound dynamin by applying tilt and twist to the sequential dimers from the linear filaments observed in the crystal to match the helical parameters described for the reconstruction. The previously described GTPase domain dimer formed in the presence of GDP•AlF4 − was subsequently fit into the density. Connection of the GTPase domain to the stalk required a conformational rearrangement of the BSE, which was independently fit into the visible density. The fit was subjected to rigid body refinement as described in Methods.
Supplementary Material
Acknowledgements
The authors would like to express thanks to I. Stokes-Rees for assistance with the Wide Space Molecular Replacement, J. Holton for advice and assistance with data collection and H. McMahon for the kind gift of the dynamin clone. We would also like to thank J. Al-Bassam, J. Chappie, A. McCoy, S. Harrison, D. Owen, A. Fisher, E. Baldwin, L. Lackner, G. Adamson, N. Varlakhanova and members of the Nunnari lab for extensive discussions. S.J. is a Damon Runyon Cancer Research Foundation Fellow supported by the Howard Hughes Medical Institute (DRG-2004-09). This work was supported by an American Heart Innovator Award and NIH grants (R01GM062942S1 and R01GM097432) to J.N. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions M.G.J.F. purified, biochemically characterized and crystallized the Dyn1 G397D ΔPRD. M.G.J.F. collected X-ray diffraction data and M.G.J.F. and S.J. solved the structure. M.G.J.F. and J.N. designed experiments and M.G.J.F., S.J. and J.N. interpreted data and prepared the manuscript.
Author Information Atomic coordinates and structure factors for Dyn1 G397D ΔPRD have been deposited with the Protein Data Bank under accession code 3ZVR.
The authors declare no competing financial interests.
References
- 1.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nature Rev. Mol. Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 2.Hoppins S, Lackner L, Nunnari J. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 2007;76:751–780. doi: 10.1146/annurev.biochem.76.071905.090048. [DOI] [PubMed] [Google Scholar]
- 3.Marks B, et al. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- 4.Stowell MHB, Marks B, Wigge P, McMahon HT. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nature Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- 5.Mears JA, et al. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nature Struct. Mol. Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. J. Biol. Chem. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- 7.Danino D, Moon KH, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J. Struct. Biol. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chappie JS, et al. An intramolecular signaling element that modulates dynamin function in vitro and in vivo. Mol. Biol. Cell. 2009;20:3561–3571. doi: 10.1091/mbc.E09-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Timm D, et al. Crystal structure of the pleckstrin homology domain from dynamin. Nature Struct. Biol. 1994;1:782–788. doi: 10.1038/nsb1194-782. [DOI] [PubMed] [Google Scholar]
- 12.Achiriloaie M, Barylko B, Albanesi JP. Essential role of the dynamin pleckstrin homology domain in receptor-mediated endocytosis. Mol. Cell. Biol. 1999;19:1410–1415. doi: 10.1128/mcb.19.2.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Crystal structure at 2.2 Å resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 14.Ramachandran R, et al. Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol. Biol. Cell. 2009;20:4630–4639. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 16.Ingerman E, et al. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J. Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao S, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 19.Low HH, Sachse C, Amos LA, Lowe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 21.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature. 2006;440:101–104. doi: 10.1038/nature04510. [DOI] [PubMed] [Google Scholar]
- 23.Bian X, et al. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl Acad. Sci. USA. 2011;108:3976–3981. doi: 10.1073/pnas.1101643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrnes LJ, Sondermann H. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl Acad. Sci.USA. 2011;108:2216–2221. doi: 10.1073/pnas.1012792108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nature Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 26.Siegel LM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim. Biophys. Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- 27.Ramachandran R, et al. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao S, et al. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- 29.Song BD, Yarar D, Schmid SL. An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol. Biol. Cell. 2004;15:2243–2252. doi: 10.1091/mbc.E04-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reubold TF, et al. Crystal structure of the GTPase domain of rat dynamin 1. Proc. Natl Acad. Sci. USA. 2005;102:13093–13098. doi: 10.1073/pnas.0506491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 32.Bashkirov PV, et al. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niemann HH, Knetsch ML, Scherer A, Manstein DJ, Kull FJ. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barylko B, et al. The proline/arginine-rich domain is a major determinant of dynamin self-activation. Biochemistry. 2010;49:10592–10594. doi: 10.1021/bi101343p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vallis Y, Wigge P, Marks B, Evans PR, McMahon HT. Importance of the pleckstrin homology domain of dynamin in clathrin-mediated endocytosis. Curr. Biol. 1999;9:257–263. doi: 10.1016/s0960-9822(99)80114-6. [DOI] [PubMed] [Google Scholar]
- 37.Durieux AC, Prudhon B, Guicheney P, Bitoun M. Dynamin 2 and human diseases. J. Mol. Med. 2010;88:339–350. doi: 10.1007/s00109-009-0587-4. [DOI] [PubMed] [Google Scholar]
- 38.Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single genemutation in Drosophila. J. Neurobiol. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- 41.Ramaswami M, Rao S, van der Bliek A, Kelly RB, Krishnan KS. Genetic studies on dynamin function in Drosophila. J. Neurogenet. 1993;9:73–87. doi: 10.3109/01677069309083451. [DOI] [PubMed] [Google Scholar]
- 42.Narayanan R, Leonard M, Song BD, Schmid SL, Ramaswami M. An internal GAP domain negatively regulates presynaptic dynamin in vivo: a two-step model for dynamin function. J. Cell Biol. 2005;169:117–126. doi: 10.1083/jcb.200502042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ingerman E, Nunnari J. A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol. 2005;404:611–619. doi: 10.1016/S0076-6879(05)04053-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.