Abstract
The majority of disease-associated variants lie outside protein-coding regions, suggesting a link between variation in regulatory regions and disease predisposition. We studied differences in chromatin states using five histone modifications, cohesin, and CTCF in lymphoblastoid lines from 19 individuals of diverse ancestry. We found extensive signal variation in regulatory regions, which often switch between active and repressed states across individuals. Enhancer activity is particularly diverse among individuals, whereas gene expression remains relatively stable. Chromatin variability shows genetic inheritance in trios, correlates with genetic variation and population divergence, and is associated with disruptions of transcription factor binding motifs. Overall, our results provide insights into chromatin variation among humans.
Association and gene expression studies have linked disease predisposition to specific alleles (1–3) and identified intermediate molecular phenotypes that may be responsible for organismal differences (4–7). However, the underlying mechanisms by which genetic variation drives either disease or expression differences remain poorly understood. Interindividual variability has been reported for transcription factor (TF) binding (8–10) and deoxyribonuclease I (DNase I) accessibility (11, 12). However, TF studies assay a very small fraction of regulatory elements, whereas DNase I hypersensitivity does not distinguish between different types of regulatory elements (e.g., enhancers versus promoters), is biased toward active elements, and provides little information on domain-level features (e.g., Polycomb-repressed domains).
To further characterize human variation in diverse types of regulatory elements, we studied the chromatin state of lymphoblastoid cell lines (LCLs) derived from 19 individuals: five European (CEU), seven Yoruban (YRI), and two Asian individuals from the 1000 Genomes Project (including two mother-father-daughter trios) (13), an additional Caucasian individual (14), and four deeply sequenced individuals from the San population (15) (table S1). We used RNA sequencing (RNA-seq) to measure expression and chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq) to map five histone modifications (H3K27ac, H3K4me1, H3K4me3, H3K36me3, and H3K27me3) and two general factors (CTCF and SA1, a subunit of cohesin) (figs. S1 and S2 and tables S1 to S4). ChIP-seq reads were mapped to each line’s phased genome to reduce mapping biases.
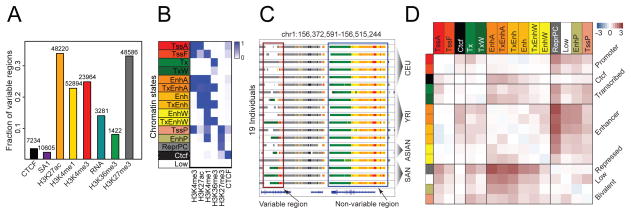
To systematically identify variable regions across individuals, we used analysis of variance (ANOVA) as well as DESeq (figs. S3 and S4) (16). Active chromatin marks H3K27ac, H3K4me1, and H3K4me3, and the repressive mark H3K27me3 show the highest fraction of variable regions, in contrast to gene-body marks and RNA expression levels (Fig. 1A and fig. S4).
Fig. 1.
Variation in chromatin, factors, and expression across individuals. (A) Number and fraction of enriched regions (for chromatin marks and factors) and expressed genes (for RNA) that are variable across individuals. (B) Composition (emission probability) of five chromatin marks and CTCF in 15 chromatin states: TssA (active promoters), TssF (flanking active promoters), Tx (strong transcription), TxW (weak transcription), EnhA (active enhancers with H3K4me3), TxEnhA (active enhancers in transcribed regions), Enh (active enhancers without H3K4me3), TxEnh (active enhancers without H3K4me3 in transcribed regions), EnhW (weak enhancers), TxEnhW (weak enhancers in transcribed regions), TssP (poised promoters), EnhP (poised enhancers), ReprPC (Polycomb repressed), Ctcf (CTCF enriched regions), and Low (low signal). (C) Examples of a nonvariable and a variable region. Coordinates are in build hg19 of the human reference sequence. State colors are as in (B). (D) log10 ratio of the observed probability that a region switches from one state (row) to another (column) in any pair of individuals relative to background switching across pairs of replicates.
We additionally used ChromHMM (17) to segment the genome of each individual into 15 chromatin states based on the combinatorial patterns of the chromatin marks and CTCF (Fig. 1, B and C, figs. S5 to S8, and table S5). We found that enhancer states exhibit the most variability (fig. S9), with bivalent (poised) enhancers having the highest fraction of individual-specific regions, followed by weak enhancers, followed by strong active enhancers. Similarly, bivalent promoters are more variable than active promoters and strongly transcribed states.
The variability of chromatin marks is often dependent on functional context defined by combinatorial chromatin patterns. H3K27ac and H3K4me3 show significantly higher variability at enhancers compared with promoters (fig. S10). This could explain the apparent discrepancy between the high variability of H3K4me3 and the low variability of expression and gene body marks (Fig. 1A). Furthermore, the repressive mark H3K27me3 is significantly more variable when co-enriched with other marks in bivalent states—such as poised enhancers and poised promoters—than in stable Polycomb-repressed domains (Fig. 1B and fig. S10).
Investigating the dynamics of chromatin state conversions among individuals, we found that most significant state switches are between active states, such as enhancers, promoters, or transcribed regions, and repressed or weakly active states (Fig. 1D and fig. S11). Although changes in activity are common, switching between enhancers and core promoter states is rare, highlighting that these are distinct types of regulatory elements.
We examined the effects of enhancer variability on gene expression and found no significant difference in expression variability between genes with one variable enhancer and those lacking variable enhancers. However, there is a significant increase in expression variability when more than 60% of the gene’s enhancers vary (fig. S12A; Wilcoxon’s rank sum test, P < 0.05), indicating that changes in multiple enhancers are often required to alter gene expression. We also found that 74% of nonvariable genes and 99% of variable genes are associated with at least one variable enhancer (24-fold enrichment, Fisher’s exact test, P < 2.2 × 10−16 ) (15, 18) and that enhancer-gene expression correlations are stronger for genes with a single enhancer than for genes linked to multiple enhancers (fig. S12B; Wilcoxon, P = 7 × 10−5). Thus, a substantial fraction of the enhancers that are variable across individuals do not result in detectable differences in gene expression, suggesting that compensatory regulatory effects, enhancer redundancy, subtle gene expression variation, or nonconsequential enhancer variation exist under the experimental conditions examined.
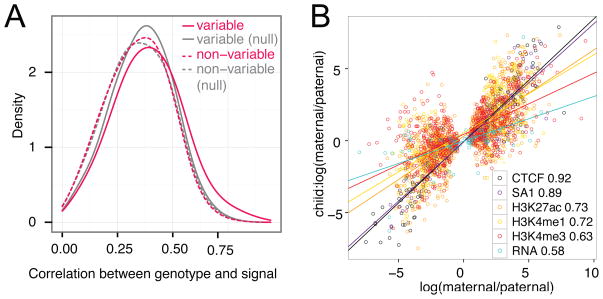
Variable regions are enriched in single-nucleotide polymorphisms (SNPs) relative to nonvariable regions (2.8-fold; P < 2.2 × 10−16; Fisher’s exact test), with an increased number of SNPs associated with higher variability (fig. S13). Signal variability also increases with nucleotide diversity (P < 1 × 10−10; Wilcoxon test) (15). Consistently, the correlation between genotype and signal is stronger for variable than nonvariable H3K27ac peaks (P < 1 × 10−15; Wilcoxon test) (Fig. 2A). Nonvariable H3K4me1 and H3K27ac F2 peaks have suppressed derived allele frequencies in both the Yoruban and Caucasian populations (P < 2 × 10−5; Wilcoxon test) and increased conservation scores (P < 0.005; binomial test) (15) compared with variable regions, suggesting stronger negative selection in nonvariable regions. Also, the fraction of heterozygous SNPs with allele-specific signal is highest for the active marks H3K27ac, H3K4me1, and H3K4me3 (fig. S14A), which is in agreement with cis effects on the variability of these marks. Finally, rare variants (allele frequency < 0.01 in the 1000 Genomes Project) are enriched in variable H3K27ac regions compared with nonvariable regions (P < 2.5 × 10−14; two-sample t test), indicating that rare variants may underlie enhancer variation.
Fig. 2.
Genetic basis of chromatin variation. (A) Spearman correlation between genotype and signal at variable and nonvariable H3K27ac regions after correcting for differences in length and signal strength. For the null sets, we shuffled the signal. (B) Correlation of allelic biases between the parents and the daughter of the YRI trio at allele-specific SNPs of the daughter that are homozygous in both parents (Pearson correlation coefficients are in the legend; linear fits are shown as lines). Only marks with at least 50 SNPs are shown.
We observed strong correlation of allele-specific signal between daughters and parents, especially for CTCF, SA1, and the enhancer and promoter marks, which suggests that the patterns of chromatin modifications and TF binding are heritable (Fig. 2B and fig. S14B). For the majority of marks, more than 75% of sites agree in the direction of allelic bias between daughters and parents (fig. S14, C and D). Gene expression is less heritable (Fig. 2B), in agreement with previous studies (19).
Next, we analyzed variation across individuals grouped by ancestry. For all marks, ancestry explains less than 20% of the variance at a majority of regions (fig. S15). The enhancer marks H3K27ac and H3K4me1 have the largest fraction of regions that discriminate ancestry groups [F-test corrected P < 0.01 (15)] (Fig. 3, A and B, and figs. S16 and S17, A and B), with signal divergence often correlating with genetic divergence (Fig. 3C). The expression of genes overlapping these regions shows a similar but weaker pattern (fig. S17C), suggesting that the impact of genetic variation at regulatory elements may be diluted at the level of downstream expression. Regions with divergent signal across ancestry groups are enriched for SNPs compared with other regions for the same marks (binomial P = 2 × 10−58 to 1.8 × 10−5 for all marks) (fig. S17D). They are also enriched for SNPs with high fixation index (FST) (20), a measure of genetic divergence across populations (binomial P = 1 × 10− to 0.01) (Fig. 3D). Although the observed signal patterns may not generalize to larger samples, they establish a link between chromatin variation and genetic divergence.
Fig. 3.
Correlation between chromatin signal and ancestry. (A) Fraction and number of regions where ancestry has a significant contribution to signal variation. (B) Row-standardized signal at H3K27ac peaks from (A), grouped into four clusters (C1 to C4). (C) Fraction of regions from (B) with SNPs characteristic of individuals in each ancestry group. Each column is divided by its maximum. The maximum genetic divergence for each ancestry group (squares with the value 1) is achieved in the cluster that shows the most divergent signal for that group [from (B)]. (D) Enrichment of regions from (A) for SNPs with high FST. Stars indicate P < 0.01 (binomial test) after accounting for the overall enrichment for SNPs.
One possible mechanism through which genetic variability leads to chromatin variation is the disruption of TF binding (8, 12). We found that variable regions of active chromatin marks are enriched for motif-disrupting SNPs (1.3- to 2.3-fold; Fisher’s exact test P < 5.1 × 10−46) (fig. S18A). Of the variable H3K27ac regions overlapping Encyclopedia of DNA Elements TF binding sites in GM12878 (21), 32% show significant associations between signal differences and motif disruptions (fig. S18B) (15). The most frequent motif disruptions involve cell-type–specific regulatory factors (Fig. 4A and fig. S19), some of which are differentially associated with H3K27ac variation at enhancer and promoter states (Fig. 4B). Finally, variable regions and allele-specific SNPs are enriched for DNase I sensitivity quantitative trait loci (dsQTLs), expression QTLs (eQTLs), and genome-wide association studies (GWAS) SNPs, providing further evidence of the functional implications of chromatin variability (Fig. 4C and fig. S20).
Fig. 4.
Mechanism and functional consequences of chromatin variation. (A) Correlation coefficients of TF motif disruption scores and H3K27ac signal across individuals. Motifs are sorted based on the number of associated peaks; peaks are sorted based on their associated motifs. (B) Log2 fold-enrichment of motifs in promoter (red) versus enhancer (orange) states. Only significant enrichments (Fisher’s exact test P < 0.05) are shown. (C) eQTLs and GWAS hits in variable regions. Stars indicate P < 0.05.
In summary, enhancers are highly variable and may contribute to phenotypic differences between individuals and ancestral groups through heritable variation in histone modifications arising from SNPs in TF binding sites.
Supplementary Material
Acknowledgments
Funded by grants from the NIH and Genetics Department, Stanford University, Vera Moulton Wall Center for Pulmonary Vascular Disease and NIH MSTP TG T32GM07205 (M.K.), the Siebel Scholars Foundation (S.-K.P.), the KAUST-Stanford Academic Excellence Alliance program (S.-K.P. and S.B.), the Swiss National Foundation, and the Janggen-Poehn Foundation (J.B.Z.). Data sets are available at the Gene Expression Omnibus (GEO) database with accession no. GSE50893. We thank S. Montgomery, W. Huber, C. Bustamante, H. Tang, S. Anders, G. Euskirchen, B. Altshuler, M. Eaton, and L. Ward. M.S. is a founder and member of the science advisory board for Personalis and a science advisory board member for Genapsys and AxioMx. S.B. is a founder and advisor for DNA nexus and serves on the advisory boards of 23 and Me and Eve Biomedical. Genotype calls and BAM files with mapped sequencing reads for the San individuals are available through a data access agreement for transfer of genetic data by contacting M.S.
References and Notes
- 1.Wellcome Trust Case Control Consortium. Nature. 2007;447:661–678. [Google Scholar]
- 2.McCarthy MI, et al. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.Hindorff LA, et al. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morley M, et al. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stranger BE, et al. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dixon AL, et al. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 7.Pickrell JK, et al. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasowski M, et al. Science. 2010;328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borneman AR, et al. Science. 2007;317:815–819. doi: 10.1126/science.1140748. [DOI] [PubMed] [Google Scholar]
- 10.Zheng W, Zhao H, Mancera E, Steinmetz LM, Snyder M. Nature. 2010;464:1187–1191. doi: 10.1038/nature08934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McDaniell R, et al. Science. 2010;328:235–239. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degner JF, et al. Nature. 2012;482:390–394. doi: 10.1038/nature10808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abecasis GR, et al. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen R, et al. Cell. 2012;148:1293–1307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Materials and methods and supporting data are available on Science Online.
- 16.Anders S, Huber W. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ernst J, Kellis M. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McLean CY, et al. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reddy TE, et al. Genome Res. 2012;22:860–869. doi: 10.1101/gr.131201.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duan S, Zhang W, Cox NJ, Dolan ME. Bioinformation. 2008;3:139–141. doi: 10.6026/97320630003139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerstein MB, et al. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.