Abstract
The initial stage of CRISPR–Cas immunity involves the acquisition of foreign DNA spacer segments into the host genomic CRISPR locus. The nucleases Cas1 and Cas2 are the only proteins conserved amongst all CRISPR–Cas systems, yet the molecular functions of these proteins during immunity are unknown. Here we show that Cas1 and Cas2 from Escherichia coli form a stable complex that is essential for spacer acquisition and determine the 2.3-Å resolution crystal structure of the Cas1–Cas2 complex. Mutations that perturb Cas1–Cas2 complex formation disrupt CRISPR DNA recognition and spacer acquisition in vivo. Unlike Cas1, active site mutants of Cas2 can still acquire new spacers indicating a non-enzymatic role of Cas2 during immunity. These results reveal the universal roles of Cas1 and Cas2 and suggest a mechanism by which Cas1–Cas2 complexes specify sites of CRISPR spacer integration.
INTRODUCTION
The clustered regularly interspaced short palindromic repeats (CRISPR) – CRISPR associated proteins (Cas) immune system is an RNA-guided defense mechanism against foreign genetic elements. CRISPR–Cas systems are present in approximately 40% of bacteria and almost all archaea1. CRISPR genomic loci consist of repeat sequences, typically 20–50 base pairs (bp) in length, separated by variable “spacer” sequences of similar length that frequently match a segment of foreign DNA2,3. Directly upstream of these repeat-spacer arrays is an AT-rich leader region. During the poorly understood process of adaptation, a segment of the foreign DNA, termed a protospacer, is site-specifically incorporated into the host CRISPR locus as a new spacer at the leader-proximal end, where it serves as a molecular memory of prior infection4–7. To generate immunity, CRISPR RNAs (crRNAs) derived from the CRISPR array8–12 are used as molecular guides by Cas proteins for base pairing with complementary sequences in foreign DNAs to trigger their degradation during the interference stage4,8,13–16.
The cas genes flanking CRISPR arrays encode proteins that play critical roles in the various steps of immunity. Although most cas genes are highly divergent and occur only in certain CRISPR loci, cas1 and cas2 are notably conserved across the three major types of CRISPR systems17. Genetic experiments, as well as spacer acquisition assays in Escherichia coli K12, demonstrate that Cas1 and Cas2 are the only Cas proteins required for new spacer acquisition into the host CRISPR locus5,7. Bioinformatic analyses indicate that spacer sequences are highly variable and can derive from both coding and non-coding regions of the foreign DNA5–7,18,19. However, their selection requires proximity to a protospacer adjacent motif (PAM) of ~2–4 base pairs that is also critical for correct target DNA binding, cleavage and self versus non-self discrimination20,21. The conserved presence of cas1 and cas2 suggest a common mechanism of spacer acquisition across the three CRISPR types. Despite these findings, along with previous biochemical studies identifying Cas1 and Cas2 as metal-dependent nucleases22–26, the molecular functions of Cas1 and Cas2 during CRISPR–Cas immunity remain elusive.
Here we show that Cas1 and Cas2 form a stable complex in vitro and present a crystal structure of the E. coli Cas1–Cas2 complex. With the Cas1–Cas2 complex as a structural guide, we set out to determine if heterocomplex formation is essential for new spacer acquisition in vivo. We combine an in vivo spacer acquisition assay with mutagenesis and immunoprecipitation experiments to show that physical disruption of complex formation abrogates spacer acquisition. While active site mutations in Cas1 inhibit spacer acquisition, the catalytic activity of Cas2 is not required for either Cas1–Cas2 complex formation or new spacer acquisition. The Cas1–Cas2 complex is uniquely capable of recognizing the CRISPR leader-repeat sequence, a property not shared by either protein alone. Together, these results provide the first functional insights into a Cas1–Cas2 complex that are likely to be shared across all three CRISPR systems.
RESULTS
Cas1 and Cas2 form a specific complex in vitro and in vivo
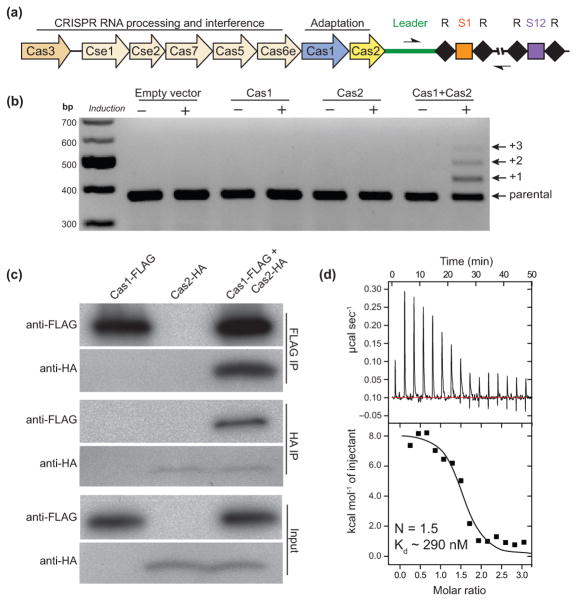
The E. coli K12 (MG1655) strain has two endogenous CRISPR loci, one of which is flanked by eight cas genes27 (Fig. 1a). In agreement with a previously developed assay5, when Cas1 and Cas2 from K12 are co-overexpressed in E. coli BL21-AI cells, which lack all cas genes, new spacer acquisition can be detected by PCR amplification of the CRISPR locus (Fig. 1b). We sequenced newly acquired spacers and verified that spacer acquisition in this model system retains accurate insertion of 33 base-pair (bp) spacers that are mostly derived from the foreign plasmid used for protein overexpression (Supplementary Table 1). In addition to the 33 bp spacer, each acquisition event duplicates the first repeat (28 bp), thereby expanding the parental locus by 61 bp5,28. Although these results demonstrate that spacer acquisition requires only the proteins Cas1 and Cas2, we observed variable PAM sequences adjacent to the protospacer in the foreign DNA. These results support the conclusion that the E. coli CRISPR interference machinery, the Cascade complex and Cas3 nuclease, are required for an accurate “priming” process where the interference stage is coupled to spacer acquisition to yield strict AAG PAM selection6,7,18,19.
Figure 1. Cas1 and Cas2 associate to form a complex.
(a) Representation of the CRISPR–Cas locus of E. coli K12. The 33-bp spacers (squares) are separated by 28-bp repeats (black diamonds). The half arrows flanking the leader and repeat-spacer arrays represent the positions of the primers used for PCR amplification in the spacer acquisition assays in BL21-AI cells. (b) Agarose gel of the PCR amplified CRISPR-I locus of BL21-AI cells after induced expression of empty vector, Cas1, Cas2 or Cas1+Cas2. Distinct bands represent the number of repeat-spacer arrays additions into the genomic parental CRISPR locus. (c) FLAG- and HA-immunoprecipitations in lysates overexpressing Cas1 only, Cas2 only or both. (d) ITC trace of Cas1 injection into a Cas2-containing cell. The reported N and Kd values are averages of three independent experiments.
With the finding that Cas1 and Cas2 are the only Cas proteins required for spacer acquisition, we tested whether Cas1 and Cas2 form a stable complex in vivo. We overexpressed Cas1-FLAG and Cas2-HA fusion proteins in BL21-AI cells and conducted immunoprecipitation experiments in cell lysates. We confirmed that the epitope-tagged proteins are active in acquiring new spacers (Supplementary Fig. 1a). Selective elution from FLAG or HA affinity beads with either a 3×-FLAG or an HA peptide resulted in the co-elution of Cas1 and Cas2 (Fig. 1c). To verify that this interaction is direct, we separately purified the untagged construct of each protein and determined the dissociation constant (Kd) of the interaction to be ~290 nM as measured by isothermal titration calorimetry (Fig. 1d). The calculated stoichiometry of Cas1 to Cas2 from the ITC experiments was ~1.5. To further probe for the stoichiometry of the complex, we conducted sedimentation velocity analytical ultracentrifugation (AUC) experiments and detected a strong peak at 5.2S with an apparent molecular weight of ~78.1 kDa (Supplementary Fig. 1b,c). This is consistent with a complex composed of one Cas1 dimer (66 kDa) and one Cas2 dimer (22 kDa). The retention time of the complex on a gel filtration column is also consistent with the AUC experiments (Supplementary Fig. 1d). Thus, we conclude that one dimer of Cas1 and one dimer of Cas2 interact to form a heterotetramer in solution.
Crystal structure of the Cas1–Cas2 complex
To gain insights into the structural organization of the Cas1–Cas2 complex, we determined the crystal structure of the complex. Crystal structures of Cas1 and Cas2 alone from various organisms, including E. coli K12, have been reported22–26,29,30. Cas1 proteins are asymmetrical homodimers with each monomer having an N-terminal β-sheet domain and C-terminal α-helical domain23,24,26. Cas2 proteins are symmetrical homodimers with a core ferredoxin fold22,25,29,30. We purified each protein and reconstituted the complex in vitro. Gel filtration chromatography showed the co-migration of both proteins as one peak, confirmed by SDS-PAGE analysis of the peak fractions (Supplementary Fig. 1d,e). The Cas1–Cas2 complex yielded crystals that diffracted X-rays to 2.3-Å resolution. We determined the structure by single-wavelength anomalous dispersion (SAD) using selenomethionine-derivatized crystals and refined the resulting model to an Rwork/Rfree of 22%/24% (Table 1).
Table 1.
Data collection and refinement statistics.
| Native | Se derivative | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 94.875, 125.70, 99.31 | 93.70, 127.70, 99.32 |
| a, b, g (°) | 90, 102.74, 90 | 90, 102.32, 90 |
| Resolution (Å) | 62.85–2.3 (2.383 – 2.3) | 48.63 – 2.89 |
| Rsym or Rmerge | 0.1301 (1.088) | 0.2045 (1.576) |
| I / sI | 9.19 (1.00) | 10.32 (1.37) |
| Completeness (%) | 97.22 (91.21) | 91.87 |
| Redundancy | 3.1 (2.9) | 7.8 (7.2) |
| Refinement | ||
| Resolution (Å) | 62.85 – 2.3 (2.383 – 2.3) | |
| No. reflections | 97,929 (9,271) | |
| Rwork / Rfree | 0.225/0.245 | |
| No. atoms | 10,451 | |
| Protein | 9,926 | |
| Water | 525 | |
| B factors | ||
| Protein | 55.60 | |
| Water | 53.90 | |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.003 | |
| Bond angles (°) | 0.62 | |
One crystal was used per data set. Values in parentheses denote highest-resolution shell.
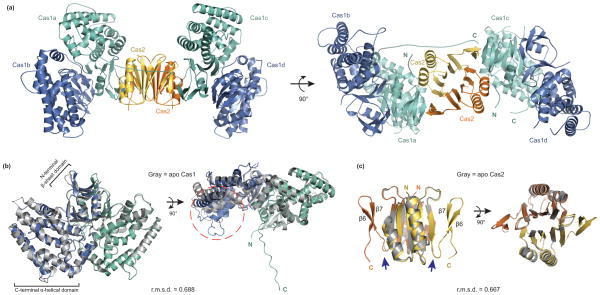
The overall architecture of the asymmetric unit is a heterohexameric complex consisting of two Cas1 dimers (Cas1a-b and Cas1c-d) that sandwich one Cas2 dimer (Fig. 2a). Cas1a and Cas1c make contacts with the Cas2 dimer and no contacts are observed between Cas1b or Cas1d and the Cas2 dimer. The Cas1c–Cas2 protein-protein interface buries a large surface area of ~3,100 Å2, whereas the Cas1a–Cas2 interface buries an additional 800 Å2 contributed by the C-terminus of Cas1a, as described further below. Superposition of the two Cas1 dimers (a-b dimer with c-d dimer) shows high structural similarity, with a root mean square deviation (r.m.s.d.) of 0.394 Å for the C-alpha atoms (Supplementary Fig. 2a,b). Similar contacts are present between Cas1a and Cas1c with Cas2 on opposite sides, creating a symmetrical complex. While Cas1 and Cas2 predominantly form a heterotetrameric complex in solution, our crystal structure suggests the complex may also be capable of accessing a hexameric state during acquisition.
Figure 2. Crystal structure of the Cas1–Cas2 complex.
(a) The overall structure consists of a Cas2 dimer (yellow and orange) and two Cas1 dimers (a-d, blue and teal). (b) Superposition of the Cas1a-b dimer with the previously determined E. coli Cas1 structure (gray, PDB 3NKD24). The dashed orange circle highlights the conformational change observed in the a-helical domain of Cas1a. (c) Superposition of the Cas2 dimer in the complex with the previously determined E. coli Cas2 structure (gray, PDB 4MAK30). The arrows point to the last resolved residue in the 4MAK structure. The N and C indicate the termini of each monomer; root-mean-square deviations (r.m.s.d.) of the superpositions are indicated.
Conformational changes and contacts within the complex
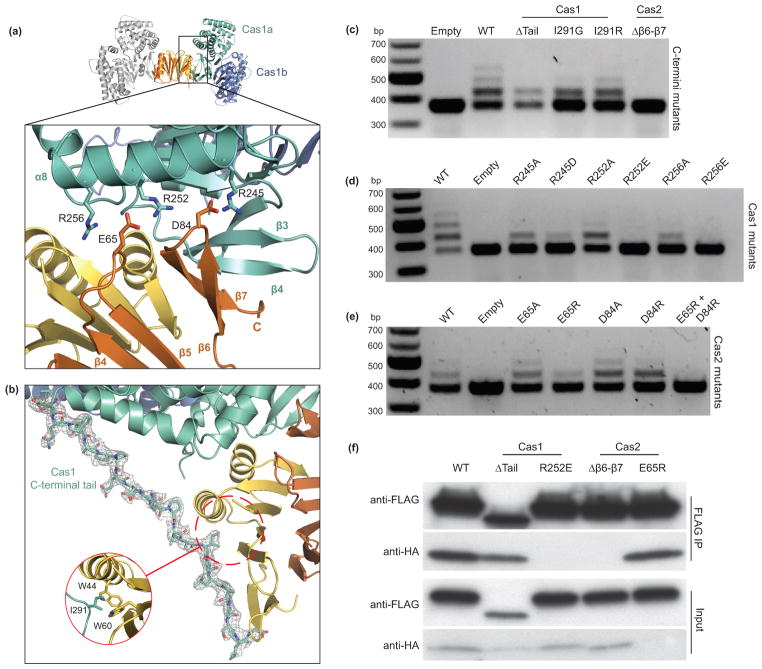
The interface between Cas1 and Cas2 consists of hydrogen bonding, electrostatic and hydrophobic interactions. We observe extensive electrostatic contacts between three arginine residues (R245, R252, R256) in α8 of Cas1 with two acidic residues (E65 and D84) of Cas2 (Fig. 3a and Supplementary Fig. 2c). The R252 residue is positioned between E65 and D84 and may sample salt bridges between the two acidic residues, although we observe continuous density between R252 and E65 at the Cas1a–Cas2 interface. In the same region, backbone hydrogen-bond contacts are present between the newly resolved Cas2 β7 C-terminus and β4 of Cas1 as discussed further below.
Figure 3. Disruption of complex formation affects spacer acquisition in vivo.
(a) A close-up view of the Cas1a–Cas2 protein-protein interface, with annotations for the residues involved in electrostatic interactions. (b) View of the ordered C-terminal tail of Cas1a with the electron density mesh contoured at 1.0 sigma. (c–e) Agarose gels of in vivo acquisition assays with mutations of Cas1 and Cas2 at the C-termini (c) and the electrostatic interface (d,e). (f) Western blot of FLAG immunoprecipitations in BL21-AI cells expressing Cas1-FLAG and Cas2-HA or various mutations of Cas1 and Cas2. Despite the low expression of Cas2 E65R, we still detect its co-elution with Cas1.
To identify Cas1 and Cas2 conformational changes that occur upon complex formation, we superimposed previously determined structures of apo Cas1 (PDB 3NKD)24 and Cas2 (PDB 4MAK)30 from E. coli with the Cas1–Cas2 complex structure (Fig. 2b, c). In addition to minor conformational changes present in the canonical βαββαβ ferredoxin fold of Cas2, the C-terminus forms two antiparallel β-sheets (β6–β7) that contact β4 of Cas1 (Fig. 2c and Fig. 3a). This region is unresolved in the apo-Cas2 structure, which terminates at the C-terminus of β5. Presumably, the β6–β7 region is flexible prior to complex formation with Cas1.
Although Cas1 does not undergo major conformational changes upon Cas2 binding (0.69 Å backbone r.m.s.d.), the proline-rich C-terminal “tail” of Cas1a is distinctively ordered in only the bound state and is stabilized by hydrophobic and electrostatic contacts (Fig. 3b). At the middle of the tail, I291 from Cas1 is positioned in a hydrophobic pocket of Cas2 that includes W44 and W60 (Fig. 3b). The C-terminus of Cas1c is likely to span the opposite face of Cas2 to fully encapsulate the dimer, although electron density for this tail was not observed due to crystal packing of an adjacent complex on this face. We also observe structural rearrangements in the C-terminal α-helical domains of Cas1b and Cas1d as compared to apo Cas1 (Fig. 2b), however it is unclear whether this conformational change is due to complex formation or crystallographic packing of another complex next to these monomers.
Cas1–Cas2 complex formation is required in vivo
To determine the function of Cas1–Cas2 complex formation, we conducted spacer acquisition assays in cells expressing Cas1 and Cas2 bearing mutations at the inter-protein interfaces. We found that a structured Cas2 C-terminus is critical for function as its deletion (Δβ6–β7) prevented detectable spacer acquisition (Fig. 3c). This deletion removes the backbone interaction of Cas2 with β4 of Cas1, as well as the D84 residue at the electrostatic interface. In contrast, deletion of the Cas1 tail (ΔP282–S305) did not abolish spacer acquisition. Furthermore, mutation of Cas1 I291, which binds in a hydrophobic pocket with W44 and W60 of Cas2, had no effect in spacer acquisition (Fig. 3a, c). Thus, the C-terminal tail of Cas1 is not essential for spacer acquisition, although it may supplement the critical interactions at the interface described below.
Mutations of residues involved in the electrostatic interactions between subunits have drastic effects on spacer acquisition. Cas1 constructs with mutations at the arginines of α8 to alanine (R245, R252 or R256) could still acquire spacers. However, constructs with mutations of the same residues to the opposite charge (R245D, R252E or R256E) supported little or no detectable spacer acquisition compared to wild-type Cas1 (Fig. 3d). To show that these mutations have little or no effect on Cas1 stability, we purified the R252E mutant to homogeneity and the mutant eluted at the expected retention time for wild-type Cas1 dimer (Supplementary Fig. 2d). In comparison, single mutations of either of the two acidic Cas2 interface residues E65 or D84 to alanine or arginine had little or no effect on spacer acquisition compared to wild-type Cas2. A double mutation of both residues to arginine (E65R and D84R) abolished spacer acquisition in vivo (Fig. 3e). Thus, while mutations in Cas1 at the electrostatic interface are more deleterious than those in Cas2, complementary charges in this interface permit acquisition.
To confirm that the observed in vivo effects are due to disruption of Cas1–Cas2 complex formation, we conducted FLAG immunoprecipitation experiments in lysates of BL21-AI cells overexpressing Cas1-FLAG and Cas2-HA mutants. Mutations that had little effect on spacer acquisition (Cas1 Δtail and Cas2 E65R) did not perturb co-precipitation of Cas1 and Cas2, indicating that the mutants are still able to form the complex (Fig. 3f). In contrast, acquisition-defective mutants (Cas1 R252E and Cas2 Δβ6–β7) can no longer form a stable Cas1–Cas2 complex. This result highlights the importance of the structured Cas2 C-terminus and the positioning of R252 between the two Cas2 acidic residues in the electrostatic interaction interface. Together, these findings support the conclusion that Cas1–Cas2 complex formation is required for spacer acquisition in vivo.
The catalytic activity of Cas2 is dispensable in vivo
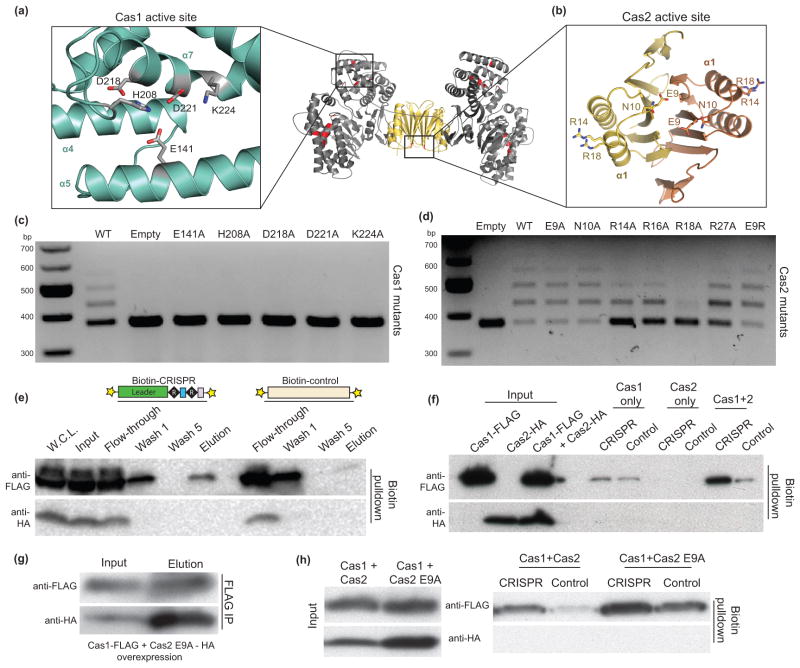
Despite the available literature on the biochemical activities of Cas1 and Cas2, the functional roles of these proteins during spacer acquisition are unknown. Cas1 is reported to be a sequence-independent, metal-dependent nuclease that can cleave ssDNA, linear and plasmid dsDNA, ssRNA and various DNA repair intermediates such as Holliday junctions23,24,26,31. Three different Cas2 homologs were found to have metal-dependent nuclease activity with a preference for ssRNA22 or dsDNA25 or to lack any detectable nuclease activity29. The heterohexameric Cas1-Cas2 complex has five potential active sites-one for each Cas1 monomer and one at the Cas2 homodimer interface (Fig. 4a, b). We conducted spacer acquisition assays with active site residue mutations in Cas1 and Cas2 to determine if the nuclease activities of both proteins are required.
Figure 4. Complex formation is required for CRISPR DNA recognition.
(a,b) Close-up views of the Cas1 and Cas2 active sites with stick representations for the conserved residues mutated in vivo. In the middle is a general view of the active sites in the complex, highlighted in red. (c,d) Acquisition assays of active site residue mutations of Cas1 and Cas2. (e) Western blot of fractions in the biotinylated DNA affinity precipitations. The cartoon representations are the DNA constructs used with the 5′-biotin labels (stars). W.C.L. refers to the whole cell lysate. (f) Western blot of the DNA affinity precipitations in BL21-AI lysates from overexpression of Cas1-FLAG only, Cas2-HA only or both. (g) FLAG IP in lysates from overexpression of Cas1-FLAG and Cas2 E9A-HA. (h) DNA affinity precipitations in the same lysates as in (g) using the same DNA constructs as in (e).
Alanine substitution of the conserved Cas1 active site residues abolished spacer acquisition, demonstrating its critical role in metal-dependent DNA cleavage during the adaptation stage (Fig. 4c). Despite the low protein sequence conservation of Cas2 proteins, a conserved acidic residue from each monomer is positioned in the active site to coordinate a metal ion during catalysis in vitro22,25 (Fig. 4b). Surprisingly, Cas2 mutated in the signature catalytic E9 residue to alanine or arginine supported spacer acquisition at frequencies similar to those observed in the presence of wild-type Cas2 (Fig. 4d). A mutation of this acidic residue was previously shown to have drastic effects on nucleic acid substrate cleavage in vitro22,25. Cas2 with an R14A mutation, which was also shown to be catalytically inactive in the Sulfolobus solfataricus Cas2 in vitro22, was still active in acquiring spacers. Of the nearby arginine residues, only the R18A construct had low spacer acquisition levels. This residue interacts with Cas1 at the inter-protein interface, as supported by its continuous electron density with the Cas1 backbone. These results support the notion that Cas1 is the likely nuclease that catalyzes the integration reaction, whereas the function of Cas2 during CRISPR-Cas immunity may not be nucleic acid cleavage.
The Cas1–Cas2 complex is essential for CRISPR locus binding
The molecular basis for new CRISPR spacer acquisition at the leader-proximal end of the CRISPR locus has been unknown. Although it is hypothesized that Cas1 or Cas2 might provide such spacer acquisition specificity, previous studies focused on the individual activities of these two proteins. These studies reported sequence-nonspecific DNA binding properties of purified Cas1 and Cas223–26,31. Our discovery that Cas1 and Cas2 form an essential complex that is required for CRISPR spacer acquisition in vivo led us to test for CRISPR DNA binding by Cas1 and Cas2. We initially conducted electrophoretic mobility shift assays (EMSA) of purified Cas1 and Cas2, either alone or as a complex, with various DNA substrates. Consistent with previous findings using either protein alone, purified Cas1–Cas2 complex has no sequence-specific DNA binding activity. These results suggest that other host factors may be required to stimulate loading of Cas1 and/or Cas2 on the CRISPR locus.
To alternatively probe for CRISPR locus binding specificity, we conducted biotinylated DNA affinity precipitation assays in lysates of BL21-AI cells overexpressing Cas1-FLAG and Cas2-HA (Supplementary Fig. 3a). We first tested the ability of Cas1 and/or Cas2 to bind a 186 bp 5′-biotinylated double-stranded DNA (dsDNA) containing two CRISPR repeats, two spacers and the minimal 60-bp leader sequence shown to be required for spacer acquisition in vivo5. As a control, a DNA of similar length with no CRISPR sequence was used. After a series of washes to remove non-specific binders, Western blot analysis of the elution samples confirmed the preferential binding of Cas1 to the CRISPR DNA compared to the control DNA (Fig. 4e and Supplementary Fig. 3b). Surprisingly, we did not detect Cas2 in the elution samples, which could be due to the washing conditions removing the weakly-bound Cas2. To determine whether Cas2 is required for the preferential binding of Cas1 to the CRISPR DNA, we conducted the affinity precipitation experiment in BL21-AI cell lysates containing over-expressed Cas1 only, Cas2 only or Cas1 and Cas2. Although we do not detect the presence of Cas2 in any of the elution samples, we found that Cas1 loses preference for CRISPR DNA binding in the absence of Cas2 (Fig. 4f). Cas1 is no longer able to recognize a DNA substrate when the conserved CRISPR leader sequence is replaced with random DNA (Supplementary Fig. 3c, d), indicating that in agreement with previous in vivo results, sequence or structural specific interactions with the leader DNA may be required to direct spacer acquisition5. To determine if a linear motif accounts for sequence-specific recognition of CRISPR DNA, we conducted DNA affinity precipitation experiments using dsDNA substrates of equal length that harbor scrambled portions of the CRISPR leader sequence (Supplementary Fig. 3c, d). In contrast to the severe binding defect resulting from complete removal of the CRISPR leader sequence, shorter scrambled stretches have a much less pronounced effect on the ability of Cas1 to recognize the DNA substrate. These results suggest that Cas1 recognition of the CRISPR leader sequence occurs through a yet unknown nonlinear sequence or structural basis.
Upon finding that disruption of the Cas1–Cas2 complex formation negatively affects spacer acquisition, we tested whether this defect is due in part to the inability of the complex to recognize the CRISPR locus. We conducted the DNA affinity purifications in lysates of cells expressing Cas1 and Cas2 mutants that were tested previously for in vitro complex formation (Fig. 3). Mutants that support spacer acquisition and form a complex (Cas1 Δtail and Cas2 E65R) retain the ability to bind the CRISPR DNA (Supplementary Fig. 3e). In contrast, mutants that do not support spacer acquisition (Cas1 R252E and Cas2 Δβ6–β7) lose the preference for CRISPR DNA recognition. A mutation of the active site E9 residue of Cas2 has no effect on complex formation or CRISPR DNA binding (Fig. 4g, h). Thus, mutations that disrupt complex formation may have lost the ability to support spacer acquisition due to the inability to recognize the leader-repeat sequence of the CRISPR locus.
DISCUSSION
The acquisition of new spacer sequences into the CRISPR locus as part of the adaptive immune response in bacteria requires the two conserved CRISPR-associated proteins Cas1 and Cas2. Our findings show that Cas1 and Cas2 assemble into a stable complex whose formation is essential for the incorporation of foreign DNA spacers into the host CRISPR locus in vivo. The 2.3-Å crystal structure of the Cas1–Cas2 complex reveals a 2:1 stoichiometry in which a Cas2 dimer binds two Cas1 dimers to form a crab-like architecture that specifies the site of integration at the leader end of the CRISPR locus. In solution, the complex is stable as a heterotetramer containing one dimer each of Cas1 and Cas2, leaving open the possibility that the tetrameric form is a functional unit during integration.
Our findings point to likely biochemical functions of Cas1 and Cas2 within the complex. Both proteins have been investigated independently and shown to possess non-specific nuclease activity in vitro22–25. Based on our active site mutational studies in vivo (Fig. 4), in which catalytically defective Cas1 mutants were incapable of supporting spacer acquisition, Cas1 functions as a bona fide nuclease involved in the adaptation stage of CRISPR–Cas immunity. In contrast, the catalytic activity of Cas2 is unnecessary for integration of sequences into the CRISPR locus in vivo. Furthermore, the observation that Cas2 does not co-precipitate with the biotinylated DNA probes suggests that Cas2 may bind weakly to the Cas1–DNA complex or may not bind directly to DNA within the Cas1–Cas2 complex. Together with the finding that Cas1–Cas2 complexes have a marked preference for binding to the CRISPR locus, which serves as the target site for spacer integration (Fig. 4), these results suggest that Cas2 recruits Cas1 to the leader sequence through an indirect mechanism. It remains possible that the nuclease activity of Cas2 contributes to a CRISPR-independent process, as suggested by the structural homology between Cas2 and the VapDHi toxin of the VapDHI-VapX toxin-antitoxin system in Haemophilus influenza32.
In addition to the catalytic function of Cas1, its ability to assemble with Cas2 is also essential for spacer acquisition. Mutations in either Cas1 or Cas2 that disrupt Cas1–Cas2 complex formation in vitro also interfere with spacer acquisition in vivo. Furthermore, this functionally critical interaction is conserved across divergent CRISPR systems. Recent experiments provided evidence for Cas1–Cas2 containing complexes in the Type I-A CRISPR system in the crenarchaeon Thermoproteus tenax-where Cas1 and Cas2 exist as a fusion protein-and in the Type I-F system in the plant pathogen Pectobacterium atrosepticum33,34. Despite the essential nature of the Cas1–Cas2 interaction in E.coli, we note that the observed inter-proteininterface contacts may not be conserved in other CRISPR–Cas systems. Structural alignment of available Cas1 and Cas2 crystal structures shows poor conservation of the three critical arginines that form salt bridges with E65 and D84 of Cas2 (Supplementary Fig. 4a,b). These residues may co-vary in other Cas1–Cas2 protein complexes, or they may be replaced by different interactions that ensure Cas1–Cas2 assembly in divergent CRISPR–Cas systems.
Alignment of Cas2 crystal structures also reveals significant structural flexibility outside of the core βαββαβ ferredoxin fold (Supplementary Fig. 4c). In particular, the C-terminus of the E. coli Cas2 in the Cas1–Cas2 complex is positioned ~90° from its position in the other Cas2 structures. The structural changes in the Cas2 C-terminal β6–β7 strands that we observe in the Cas1–Cas2 complex, and their requirement for both complex stability and in vivo spacer acquisition (Fig. 3c), underscore the role of Cas2 as a central structural component of the Cas1–Cas2 integration complex.
Together with previous work, our findings establish that at least two multi-protein complexes are fundamental for a fully functioning Type I CRISPR–Cas system-a Cas1–Cas2 spacer acquisition complex and an RNA-guided DNA interference complex. Whether or not these complexes interact to form a multi-functional super complex is not yet known. However, an interesting hint about this possibility comes from P. atrosepticum, where Cas2 exists as an N-terminal fusion with Cas3, the foreign DNA-targeting nuclease recruited by the DNA interference complex. The Cas2–Cas3 fusion protein in this organism was also shown to associate with Cas134. It is thus possible that at least some Type I CRISPR–Cas systems employ the Cas1–Cas2 complex not only for new spacer acquisition but also to couple this process to that of target recognition and destruction.
ONLINE METHODS
Protein purification
The cas1 and cas2 genes were PCR amplified from E. coli K12 (MG1655) genomic DNA. The cas1 gene was cloned into a Gateway compatible expression vector (pHMGWA) containing an N-terminal His6xMBP tag and cas2 was cloned into pET16b (Novagen) containing a C-terminal MBPHis6x tag35. The proteins were purified separately using the same protocol. The constructs were expressed in BL21(DE3) cells, grown to 0.6–0.8 OD600, and induced overnight at 16°C with 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). The cells were harvested and re-suspended in buffer A (500 mM KCl, 20 mM HEPES-KOH, pH 7.4, 10 mM imidazole, 0.1% Triton X-100, 2 mM TCEP, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], “Complete, EDTA-free” protease inhibitor [Roche] and 10% glycerol). After lysis by sonication, the lysates were cleared by centrifugation and incubated with Ni-NTA affinity resin in batch (QIAGEN). The resin was washed in buffer B (with 500 mM KCl, 20 mM HEPES-KOH, pH 7.4, 10 mM imidazole, 5% glycerol, 1 mM TCEP) and the protein was eluted with buffer B supplemented with 300 mM imidazole. The eluted protein was dialyzed against buffer B at 4°C in the presence of TEV protease to remove the affinity tags. The protein was concentrated and further purified on tandem MBPTrap HP (GE Healthcare) and Superdex 75 (16/60) size exclusion chromatography column using buffer B in the absence of imidazole. The selenomethionine proteins were overexpressed in minimal media as previously described36 and subsequently purified using the same purification protocol for the native proteins.
In vitro complex formation, crystallization and structure determination
Purified Cas1 and Cas2 were separately dialyzed against 150 mM KCl, 20 mM HEPES-KOH, pH 7.4, 5% glycerol and 1 mM TCEP at 4°C overnight. The proteins were incubated together at a 1:3 Cas1:Cas2 molar ratio for one hour on ice. The sample was loaded on a Superdex 75 (16/60) size exclusion column and the peak fractions corresponding to the complex were pooled and concentrated for crystallization. We note that the molar ratio of Cas1:Cas2 for pre-incubation was chosen to obtain clear separation between the Cas1–Cas2 complex and Cas1 only peaks on gel filtration. There is no difference in the retention time of the complex when the proteins were pre-incubated at a 1:1 ratio.
The selenomethionine-derivatized complex was concentrated to ~4 mg mL−1 and crystallized by hanging drop vapor diffusion at room temperature in 120 mM calcium acetate, 10% (w/v) PEG 8000 and 50 mM sodium cacodylate, pH 6.50. The crystals were briefly transferred into a drop containing 20% glycerol for cryoprotection and frozen in liquid nitrogen until data collection. The native protein was crystallized in 150 mM NaCl, 6% (w/v) PEG 8000 and 100 mM Tris, pH 8.0. The crystals were frozen in the presence of 20% PEG 8000 as cryoprotectant.
The diffraction data were collected under cryogenic conditions at the Lawrence Berkeley National Laboratory Advanced Light Source (beamline 8.3.1). The selenomethionine-derivative crystals were scanned for fluorescence to determine the selenium absorption edge and diffraction data was collected at the peak wavelength.. The data were processed with XDS37 and SCALA38. The crystals belonged to a monoclinic space group (P 21) with four copies of Cas1 and two copies of Cas2 in the asymmetric unit. Autosol was used for experimental phasing and AutoBuild within PHENIX39 to obtain an initial model. Iterative rounds of model building in Coot40 and refinement using PHENIX was performed to obtain a selenomethionine-derivative model. The 2.3-Å native model was built using Phaser–Molecular Replacement in PHENIX for phasing, followed by Coot and PHENIX for model building and refinement, respectively.
Isothermal titration calorimetry
ITC experiments were conducted on a MicroCal Auto-iTC200 system (GE Healthcare). Purified Cas1 and Cas2 were separately dialyzed at 4°C against 200 mM KCl, 20 mM HEPES-KOH, pH 7.5, 5% glycerol and 1 mM TCEP. Cas1 (150 μM) was titrated into the cell containing 15 μM Cas2 with 10–15 3.1 μl injections at 4°C. The Origin software (OriginLab) was used for baseline correction, integration and curve fitting. The Kd reported is an average of three separate experiments (308, 212 and 351 nM, standard deviation of 58 nM). The calculated N values of each independent run were 1.52, 1.51 and 1.50.
Analytical ultracentrifugation
Sedimentation velocity experiments were conducted at 50,000 rpm using the Beckman Coulter XLI (Beckman Coulter, Fullerton, CA, USA). The samples were monitored by absorbance optics at 280 nm. The proteins were dialyzed against 20 mM HEPES-KOH pH 7.5, 150 mM KCl and 1 mM TCEP. Three concentration series for Cas1 at 7, 14 and 28 μM and two concentrations of Cas2 at 20 and 40 μM were conducted to evaluate the formation of higher-order species. The Cas1–Cas2 complex was characterized using a constant concentration of 5 μM of Cas1 and two concentrations of Cas2 at 5 and 10 μM. The solvent density (1.00688 g.ml−1), viscosity (0.01022 poise), and the partial specific volumes that were used for the analyses, 0.7453 ml.g−1 (Cas1), 0.7443 ml.g−1 (Cas2), 0.7451 ml.g−1 (Cas1-Cas2), were calculated by SEDNTERP v. 2012082841. The sedimentation coefficients and apparent molecular weights were calculated from size distribution analyses [c(s)] using SEDFIT v. 14.3e 42,43. The figures were prepared using Origin v. 6.0 (Microcal Software Inc.).
Spacer acquisition assays
The assays were conducted as previously described5 with slight modifications. Briefly, cas1 and cas2 were both cloned into pCDF-1b (Novagen) and transformed into E. coli BL21-AI (Invitrogen). After preparation of an overnight culture in LB medium containing 50 μg ml−1 streptomycin, a sample was transferred (1:300) into a 10 ml culture containing 0.2% L-arabinose, 0.1 mM IPTG and 50 μg ml−1 streptomycin to induce protein expression. After 20–24 hours, a sample of the culture was diluted in water, boiled at 95°C for 5 min and centrifuged (16,100 × g). A sample of the supernatant was used as template for PCR amplification of the CRISPR locus using the same primers as previously described5. The PCR reactions were analyzed on 1.5% agarose gels. For comparison in spacer acquisition efficacy of mutant proteins, the OD600 of each culture was measured and the amount of culture obtained for PCR amplification was normalized accordingly. The newly acquired spacers reported in Supplementary Table 1 were obtained by plating a sample of the culture on LB agar plates and amplifying the CRISPR-I locus of single clones to detect locus expansion. The PCR products of clones with expanded loci were submitted for sequencing. The gene annotations were obtained from the NCBI Basic Local Alignment Search Tool (BLAST)44 using the Escherichia coli BL21 (taxid:469008) genomic sequence. All of the acquisition assays reported in this study have been replicated at least three times.
Immunoprecipitation assays
The Cas1-FLAG and Cas2-HA constructs were both cloned into pCDF-1b and co-expressed in BL21-AI cells for 20–24 hours in spacer acquisition-inducing conditions, as described above. The amount of cells used was normalized to the OD600 measurements of the cultures. The cells were pelleted and re-suspended in lysis buffer (150 mM KCl, 50 mM Tris, pH 7.50, 1 mM TCEP, 1% Triton X-100, 0.5 mM PMSF and protease inhibitors). After sonication on ice, the lysates were cleared and rocked for 1.5–2 hours at 4°C with either anti-FLAG M2 or anti-HA affinity resin (Sigma-Aldrich). The resin was washed five times with 400 mM KCl, 50 mM Tris, pH 7.50, 1 mM TCEP and 1% Triton X-100. The proteins were eluted with either 100 ng μl−1 3X FLAG peptide (DYKDDDDK) or HA peptide (YPYDVPDYA), synthesized by David King (HHMI, UC Berkeley). The epitope-tagged proteins were detected with monoclonal anti-FLAG M2-peroxidase (HRP) mouse antibody (Sigma-Aldrich A8592, 1:22,000) or HRP-conjugated anti-HA mouse antibody (Cell Signaling 2999S, 1:10,000). All of the FLAG immunoprecipitation experiments reported in this study have been replicated at least three times.
DNA affinity precipitation assays
The 186-bp CRISPR DNA bait was generated by PCR amplification of the BL21-AI CRISPR-I locus using 5′ biotin-conjugated forward and reverse primers (synthesized by Integrated Device Technology). The 186-bp control DNA was PCR amplified from the ori sequence of the pUC19 vector. The input lysates were prepared as described above for IP assays. The amount of biotinylated DNA probe was normalized to 100 nM and rocked with the lysate at 4°C for 30 min. Avidin agarose (Pierce, Fig. 4e) or streptavidin magnetic beads (NEB, Supplementary Fig. 4b) was added to the reaction and rocked for an additional 1.5–2 hours. The samples were washed five times with lysis buffer and the proteins were eluted with Laemmli buffer by boiling at 95°C for 5 minutes. Western blotting was conducted to detect Cas1-FLAG and Cas2-HA in the samples as described in the IP procedure.
Structure-based sequence alignments
The amino acid sequences of the Cas1 and Cas2 proteins were obtained from the RCSB Protein Data Bank. The alignment was generated by PROMALS3D45 and the output was analyzed on Jalview46. The BLOSUM62 score threshold on Jalview was set to 50% to generate the conservation colors. The PDB IDs of the Cas1 structures are: 3LFX (Thermotoga maritima), 3PV9 (Pyrococcus horikoshii), 2YZS (Aquifex aeolicus) and 3GOD (Pseudomonas aeruginosa). The PDB IDs of the Cas2 structures are: 3OQ2 (Desulfovibrio vulgaris), 4ES2 (Bacillus halodurans), 1ZPW (Thermus thermophilus), 2I0X (Pyrococcus furious) and 2I8E (Sulfolobus solfataricus).
Supplementary Material
Acknowledgments
We are grateful for the input on this work provided by members of the Doudna lab. We thank S. Floor, A.S. Lee, H.Y. Lee, R. Wilson, R. Wu and K. Zhou for technical assistance, the 8.3.1 beamline staff at the Advanced Light Source and A. Iavarone (UC Berkeley) for mass spectrometry. This project was funded by a US National Science Foundation grant to J.A.D (No. 1244557). J.K.N. and A.V.W. are supported by US National Science Foundation Graduate Research Fellowships and J.K.N. by a UC Berkeley Chancellor’s Fellowship. P.J.K. is a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation. J.N. is supported by a Long-Term Postdoctoral Fellowship from the Human Frontier Science Program Organization. J.A.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
ACCESSION CODES
Coordinates and structure factors for the Cas1–Cas2 complex have been deposited in the Protein Data Bank under accession code 4P6I.
AUTHOR CONTRIBUTIONS
J.K.N. performed the protein purification, biochemical and crystallography experiments. X-ray diffraction data were collected by J.K.N., P.J.K. and J.N., and structure determination was performed by J.K.N and P.J.K. A.V.W. assisted J.K.N. with in vivo acquisition and immunoprecipitation assays. C.W.D. performed and analyzed analytical ultracentrifugation experiments. J.K.N. and J.A.D. designed the study, analyzed all data and wrote the manuscript.
References
- 1.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–66. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 2.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–82. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 3.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–61. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 4.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 5.Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. doi: 10.1093/nar/gks216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swarts DC, Mosterd C, van Passel MW, Brouns SJ. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. doi: 10.1371/journal.pone.0035888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Datsenko KA, et al. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. doi: 10.1038/ncomms1937. [DOI] [PubMed] [Google Scholar]
- 8.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–96. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–8. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol. 2011;18:680–7. doi: 10.1038/nsmb.2043. [DOI] [PubMed] [Google Scholar]
- 13.Wiedenheft B, et al. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–9. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 16.Jore MM, et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–36. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 17.Makarova KS, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–77. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savitskaya E, Semenova E, Dedkov V, Metlitskaya A, Severinov K. High-throughput analysis of type I-E CRISPR/Cas spacer acquisition in E. coli. RNA Biol. 2013;10:716–25. doi: 10.4161/rna.24325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diez-Villasenor C, Guzman NM, Almendros C, Garcia-Martinez J, Mojica FJ. CRISPR-spacer integration reporter plasmids reveal distinct genuine acquisition specificities among CRISPR-Cas I-E variants of Escherichia coli. RNA Biol. 2013;10:792–802. doi: 10.4161/rna.24023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–40. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 21.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–15. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beloglazova N, et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J Biol Chem. 2008;283:20361–71. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiedenheft B, et al. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–12. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 24.Babu M, et al. A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol. 2011;79:484–502. doi: 10.1111/j.1365-2958.2010.07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nam KH, et al. Double-stranded endonuclease activity in Bacillus halodurans clustered regularly interspaced short palindromic repeats (CRISPR)-associated Cas2 protein. J Biol Chem. 2012;287:35943–52. doi: 10.1074/jbc.M112.382598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim TY, Shin M, Huynh Thi Yen L, Kim JS. Crystal structure of Cas1 from Archaeoglobus fulgidus and characterization of its nucleolytic activity. Biochem Biophys Res Commun. 2013 doi: 10.1016/j.bbrc.2013.10.122. [DOI] [PubMed] [Google Scholar]
- 27.Diez-Villasenor C, Almendros C, Garcia-Martinez J, Mojica FJ. Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010;156:1351–61. doi: 10.1099/mic.0.036046-0. [DOI] [PubMed] [Google Scholar]
- 28.Goren MG, Yosef I, Auster O, Qimron U. Experimental definition of a clustered regularly interspaced short palindromic duplicon in Escherichia coli. J Mol Biol. 2012;423:14–6. doi: 10.1016/j.jmb.2012.06.037. [DOI] [PubMed] [Google Scholar]
- 29.Samai P, Smith P, Shuman S. Structure of a CRISPR-associated protein Cas2 from Desulfovibrio vulgaris. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1552–6. doi: 10.1107/S1744309110039801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nocek B, Skarina T, Brown G, Yakunin AF, Joachimiak A. RCSB Protein Data Bank. 2013. 4MAK: Crystal structure of a putative ssRNA endonuclease Cas2, CRISPR adaptation protein from E. coli. [Google Scholar]
- 31.Han D, Lehmann K, Krauss G. SSO1450--a CAS1 protein from Sulfolobus solfataricus P2 with high affinity for RNA and DNA. FEBS Lett. 2009;583:1928–32. doi: 10.1016/j.febslet.2009.04.047. [DOI] [PubMed] [Google Scholar]
- 32.Makarova KS, Anantharaman V, Aravind L, Koonin EV. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plagens A, Tjaden B, Hagemann A, Randau L, Hensel R. Characterization of the CRISPR/Cas subtype I-A system of the hyperthermophilic crenarchaeon Thermoproteus tenax. J Bacteriol. 2012;194:2491–500. doi: 10.1128/JB.00206-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richter C, Gristwood T, Clulow JS, Fineran PC. In vivo protein interactions and complex formation in the Pectobacterium atrosepticum subtype I-F CRISPR/Cas System. PLoS One. 2012;7:e49549. doi: 10.1371/journal.pone.0049549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kranzusch PJ, Lee AS, Berger JM, Doudna JA. Structure of human cGAS reveals a conserved family of second-messenger enzymes in innate immunity. Cell Rep. 2013;3:1362–8. doi: 10.1016/j.celrep.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J Mol Biol. 1993;229:105–24. doi: 10.1006/jmbi.1993.1012. [DOI] [PubMed] [Google Scholar]
- 37.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 39.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society of Chemistry. 1992:90–125. [Google Scholar]
- 42.Brown PH, Schuck P. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys J. 2006;90:4651–61. doi: 10.1529/biophysj.106.081372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–19. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 45.Pei J, Kim BH, Grishin NV. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res. 2008;36:2295–300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.