Abstract
Acquired aplastic anemia (AA) is a potentially fatal bone marrow (BM) failure syndrome. IFN-γ-producing T helper (Th)1 CD4+ T cells mediate the immune destruction of hematopoietic cells, and are central to the pathogenesis. However, the molecular events that control the development of BM-destructive Th1 cells remain largely unknown. Ezh2 is a chromatin-modifying enzyme that regulates multiple cellular processes primarily by silencing gene expression. We recently reported that Ezh2 is crucial for inflammatory T cell responses after allogeneic BM transplantation. To elucidate whether Ezh2 mediates pathogenic Th1 responses in AA and the mechanism of Ezh2 action in regulating Th1 cells, we studied the effects of Ezh2 inhibition in CD4+ T cells using a mouse model of human AA. Conditionally deleting Ezh2 in mature T cells dramatically reduced the production of BM-destructive Th1 cells in vivo, decreased BM-infiltrating Th1 cells, and rescued mice from BM failure. Ezh2 inhibition resulted in significant decrease in the expression of Tbx21 and Stat4 (which encode transcription factors T-bet and STAT4, respectively). Introduction of T-bet but not STAT4 into Ezh2-deficient T cells fully rescued their differentiation into Th1 cells mediating AA. Ezh2 bound to the Tbx21 promoter in Th1 cells, and directly activated Tbx21 transcription. Unexpectedly, Ezh2 was also required to prevent proteasome-mediated degradation of T-bet protein in Th1 cells. Our results identify T-bet as the transcriptional and post-translational Ezh2 target that acts together to generate BM-destructive Th1 cells, and highlight the therapeutic potential of Ezh2 inhibition in reducing AA and other autoimmune diseases.
Introduction
Acquired aplastic anemia (AA) in humans is a fatal disorder characterized by bone marrow (BM) hypoplasia and blood pancytopenia.(40,57) Clinical studies indicate that in most cases, AA is a disease caused by immune-mediated destruction of hematopoietic stem cells and hematopoietic progenitor cells.(40,57) A role for T cells in AA was first suggested by their inhibition of hematopoietic cell colony formation in cultures in vitro.(57) Furthermore, CD4+ T cell clones isolated from the patients with AA have potent ability to lyse autologous CD34+ hematopoietic cells and inhibit formation of hematopoietic cell colonies.(59) Accumulating evidence indicate that CD4+ Th1 cells, which are characterized by production of high levels of IFN-γ, play important roles in mediating bone marrow failure (BMF).(38,42,47,55-57) IFN-γ displays potent effects on suppressing hematopoiesis in vitro.(57,59) Immunosuppressive therapy and allogeneic BM transplantation (BMT) have significantly improved the survival of severe AA. However, relapse still occurs in about 35% of AA patients when the immunosuppressive therapy is withdrawn.(40,57,58) Furthermore, graft-versus-host disease (GVHD) remains a major barrier to the success of allogeneic BMT.(4,13) Novel approaches are needed to improve the outcome of treatments for AA.
The transcription factor T-bet (encoded by Tbx21) is crucial for Th1 cell differentiation.(46,53,61,62) T-bet is induced by T cell receptor (TCR) signaling and strongly upregulated by activation of the STAT1 transcription factor.(29) T-bet binds to several enhancers and promoter of the Ifng genes, activating its transcription.(29,46) T-bet also promotes expression of the IL-12 receptor β2 chain (IL12Rβ2), resulting in greater IL-12 responsiveness and further elevated production of IFN-γ.(29) In addition, T-bet prevents Th2 differentiation by inhibiting Gata3.(29) T-bet is upregulated in peripheral blood T cells from patients with AA and is a useful marker predicting the responsiveness of AA patients to immunosuppressive therapy.(43) Furthermore, experimental studies suggested that T cells lacking T-bet were defective in induction of AA in mice.(47) These observations suggest that T-bet can be an attractive target for modulating Th1 cell-mediated AA. However, transcription factors are difficult drug targets.(11) Thus, identifying the molecular pathway(s) that control T-bet expression in Th1 cells may lead to new strategies to control AA.
Ezh2 is a histone methyltransferase that specifically catalyzes trimethylation of histone H3 at lysine 27 (H3K27me3).(27) Ezh2 forms Polycomb Repressive Complex 2 together with other Polycomb Group proteins Suz12 and Eed,(27) which is crucial for maintaining the cellular memory and transcriptional patterns primarily through a mechanism of silencing genes.(2,41) Several studies point to an important role of Ezh2 and H3K27me3 in multiple lineages of effector T cells.(14,17,20,25) Genome-wide mapping analysis revealed that repressive H3K27me3 marked genes associated with differentiation and maintenance of effector and memory T cells.(1,51) Most recently, we have demonstrated new and essential roles of Ezh2 in regulating inflammatory T cell responses in mice after allogeneic BMT.(15) Loss of Ezh2 led to impaired production of alloreactive T cells that induce damage to epithelial organs.(15) However, whether Ezh2 mediates pathogenic Th1 responses in AA and the mechanism of Ezh2 action in regulating Th1 cells remain unknown.
Mouse models of human AA have been successfully established.(8,38) Transfer of parent lymph node (LN) cells into haplo-identical daughter recipients caused BM hypoplasia and blood pancytopenia, typical features of clinical AA. These AA mouse models have proven to be a unique approach studying pathophysiology of immune cell-mediated BMF.(9,10,38,47) In this report, we exploited the functional impact of Ezh2 on Th1 cell responses in vitro and in vivo. Using genetic approaches and a mouse model of human BMF, we identified a novel and critical role of Ezh2 in regulating Th1 cells mediating AA.
Materials and Methods
Mice
C57BL/6 (B6, H-2b) and B6xDBA/2 F1 (BDF1, H-2b/d) mice were purchased from Taconic (Rockville, Maryland). Cd4-Cre mice were originally derived from the Jackson Laboratory. B6/129 mice with floxed alleles of Ezh2 (Ezh2fl/fl)(44) were crossed to Cd4-Cre mice, before backcrossing to the B6 background (>8 generations). Age-matched and sex matched controls were used. Experimental protocols were approved by the University of Michigan's Committee on Use and Care of Animals.
Antibodies and flow cytometry analysis
Abs used for immunofluorescence staining were purchased from eBioscience, BioLegend, or BD Biosciences. Flow cytometry analyses were performed using FACScan and Canto cytometer (Becton Dickinson) as described.(60)
Induction of AA
The mouse AA model was induced as previously described.(9,10) In brief, B6 LN cells (10 × 106/mouse) isolated from B6 mice were transplanted into irradiated BDF1 mice (6.5Gy). Peripheral blood was collected from the heart and the lateral tail vein. Complete blood counts were performed using a Hemavet 950 analyzer (Drew Scientific). BM cells were extracted from bilateral tibiae and femurs. Lymphocytes were isolated from inguinal, axillary and lateral axillary LNs.
Histological examination
The histological examination was performed on paraffin embedded femur pieces, fixed with 10% formalin and colored with Hematoxylin-Eosin. The histology picture was photographed by OlympusBX41 microscope (10/0.3 NA lens, 100× magnification, digital DP70 camera)
Real time RT- PCR
Total RNA was extracted from the indicated CD4+ T cell subsets using TRIzol (Invitrogen Life Technologies). cDNA was quantified through the quantitative real-time polymerase chain reaction (PCR) technique. Real-time PCR was performed with a SYBR Green PCR mix on a Mastercycler realplex (Eppendorf). Thermocycler conditions included an initial holding at 95°C for 2 min; this was followed by a three-step PCR program, as follows: 95°C for 30s, 55°C for 30s, and 72°C for 30s for 45 cycles. Transcript abundance was calculated using the ΔΔCt method (normalization with 18s). The primer sequences are listed in Supplemental Table 1.
Cell Culture
Splenic and LN CD4+ T cells were prepared by MACS purification with CD4 microbeads (Miltenyi Biotec), and the purity was usually 90%-95%. Th1-skewing culture conditions were set up as previously described.(12,48,64) In brief, CD44loCD4+ naïve T cells (Tn) were cultured in the presence of anti-CD3 (2μg/ml, Biolegend) and anti-CD28 (2μg/ml, Biolegend) Abs together with recombinant human IL-2 (10ng/ml, R&D Systems), recombinant mouse IL-12 (5ng/ml, R&D Systems) and BM-derived dendritic cells (DCs) at a ratio of 1:16. Cultured T cells were restimulated with anti-CD3 Ab (1μg/ml, Biolegend) 5 hours before intracellular staining.
Retroviral Construction and T cell infection
The MigR1 retroviral vector system was described previously.(36,48) MigR1 vector was kindly provided by Warren Pear (University of Pennsylvania), and MigR1 vector encoding T-bet or STAT4 was kindly provided by Steve Reiner (Columbia University, USA) and Takashi Usui (Kyoto University, Japan) respectively. MigR1 virus was produced as described.(35) For retroviral infection, CD4+ T cells were pre-stimulated with anti-CD3 and anti-CD28 Abs for 24 hours, and then the retrovirus supernatant was added in the presence of 8μg/ml polybrene (Sigma). Cells were spinoculated at 3000rpm, 32°C for 3 hours. The same retroviral infection procedure was repeated 24 hours later.
Western blot analysis
Western blot was performed as described.(16) Cell lysates were examined by routine western blotting. The blots were incubated with anti-Ezh2 (612667. BD), Stat4 (sc-486, Santa Cruz), T-bet (sc-21749, Santa Cruz), GATA-3 (558686. BD) or actin (ab3280, Abcam) Abs, and subsequently incubated with HRP-conjugated anti-rabbit or mouse IgG (Vector Laboratories) in TBS containing 5% nonfat dry milk and 0.05% Tween 20. The final reaction was developed with a chemiluminescent system (Pierce).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed as described by using EZ-Magna ChIP (17-10086, Millipore).(51,63) Sonicated extracts were precleared and incubated with Abs specific to Ezh2 (39901, active motif), H3K27me3 (PAb-069-050, Diagenode) or H3K4me3 (9751S, Cell Signaling) at 4°C overnight on a 360°C rotator. The immunoprecipitated DNA was quantitated by real-time quantitative PCR. The primer sequences are listed in Supplemental Table 2.
Enzyme-linked immunosorbent assay (ELISA)
CD4+ Tn isolated from WT B6 and T-KO mice were stimulated with anti-CD3 (2μg/ml, Biolegend) and anti-CD28 (2μg/ml, Biolegend) Abs together with recombinant human IL-2 (10ng/ml, R&D Systems) and recombinant mouse IL-12 (5ng/ml, R&D Systems). On Day 7, cells were restimulated in 96-well plates with plate-bound anti CD3 Abs (1μg/ml, Biolegend) for 5 hours before collecting the supernatants. Each group contained equal number of cells (1×106 cells/ml). The concentration of IFN-γ and IL-4 were measured in triplicate by using recombinant mouse IFN-γ and IL-4 ELISA kits in accordance with the manufacturer's instruction (Biolegend).
Luciferase Reporter assay
The Tbx21 promoter region ranging from +0.3kb to −1.0kb of the transcription start site (TSS) was cloned to pGL3 luciferase reporter vector to generate Tbx21-specific reporter (named pGL3-Tbx21 reporter). 3T3 cells were co-transfected with pGL3-Tbx21 reporter plasmid and MigR1 viral plasmid encoding Ezh2 or empty MigR1 plasmid. The cells were harvested 48 hours after transfection and analyzed with the Dual Luciferase system (Promega).
Statistical analysis
Survival in different groups was compared using the log-rank test. Comparison of 2 means was analyzed using the 2-tailed unpaired Student t test.
Results
In the absence of Ezh2, LN cells are defective in mediating AA in mice
We used genetic approach to determine the role of Ezh2 in the regulation of T cell-mediated AA. Mice with floxed alleles of Ezh2 (Ezh2fl/fl)(44) were crossed to B6 mice expressing Cre recombinase under control of the CD4 promoter to generate T cell-specific Ezh2 conditional knockout B6 mice (named T-KO). These T-KO mice were further backcrossed to B6 mice for 10 generations. The development of mature thymocytes and T cells in peripheral lymphoid tissues were normal in these T-KO mice, which is in agreement with previous observations.(15,44)
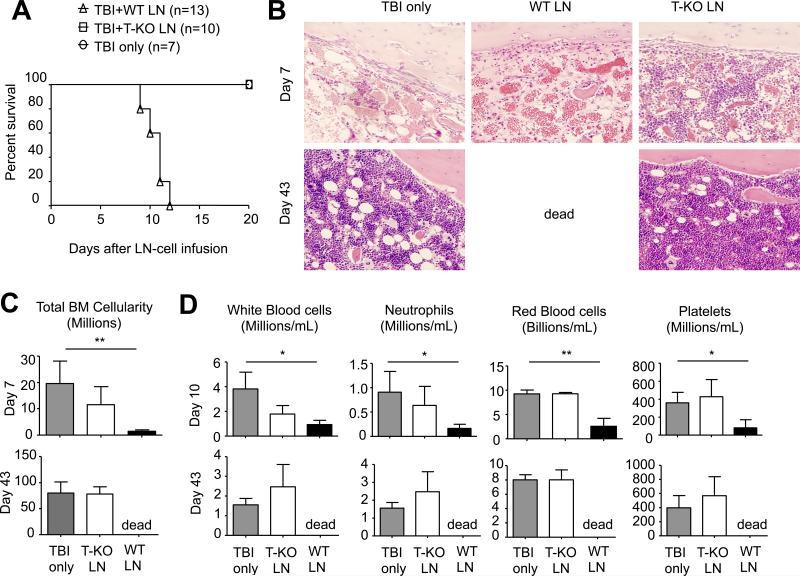
To assess whether conditionally deleting Ezh2 in T cells affected their ability to mediate AA, we transferred donor LN cells derived from WT and T-KO B6 mice into irradiated (6.5Gy) BDF1 recipients. In this setting, transfer of donor LN cells causes severe BM destruction and blood pancytopenia in these haplo-identical recipients, which closely reflects the pathogenesis of human AA.(9,38,47) As expected, BDF1 mice receiving WT B6 LN cells developed BM hypoplasia and severe blood pancytopenia after LN cell infusion compared to control total body irradiation (TBI) only mice, with all of them dying from the disease within 12 days after transfer (Fig.1A). Histological examination showed the destruction of BM and lack of hematopoietic cell islands in the BM of these recipients (Fig.1B). In contrast, transfer of T-KO LN cells did not cause severe AA in these BDF1 recipients (Fig.1A, B). As compared to control TBI mice, there was no significant reduction of BM cellularity and peripheral blood WBCs in these BDF1 mice receiving T-KO LN cells (Fig.1C, D). Importantly, all T-KO LN cell recipients survived without clinical signs and histological evidence of AA (Fig.1A, B). Thus, T cells required Ezh2 to mediate AA.
Fig.1. In the absence of Ezh2, LN cells are defective in mediating AA in mice.
WT B6 LN (10×106) or T-KO LN (10×106) cells were injected into irradiated BDF1 mice (6.5Gy). (A) The survival was monitored over time. (B) Histology analysis of BM from each group 7 or 43 days after LN-cell infusion. (C) Total BM cellularity was calculated assuming that bilateral tibia and femurs contain 25% of total marrow cells (mean ± SD, n=6 to 8 mice per group). (D) Peripheral blood was collected 10 or 43 days after LN-cell infusion and the cell number of RBC, WBC, platelets and neutrophils were calculated (mean ± SD, n=6 to 8 mice per group). Data are representative of two independent experiments. *, P<0.05, **, P<0.01.
Ezh2 is required for the development of Th1 cells inducing AA in mice
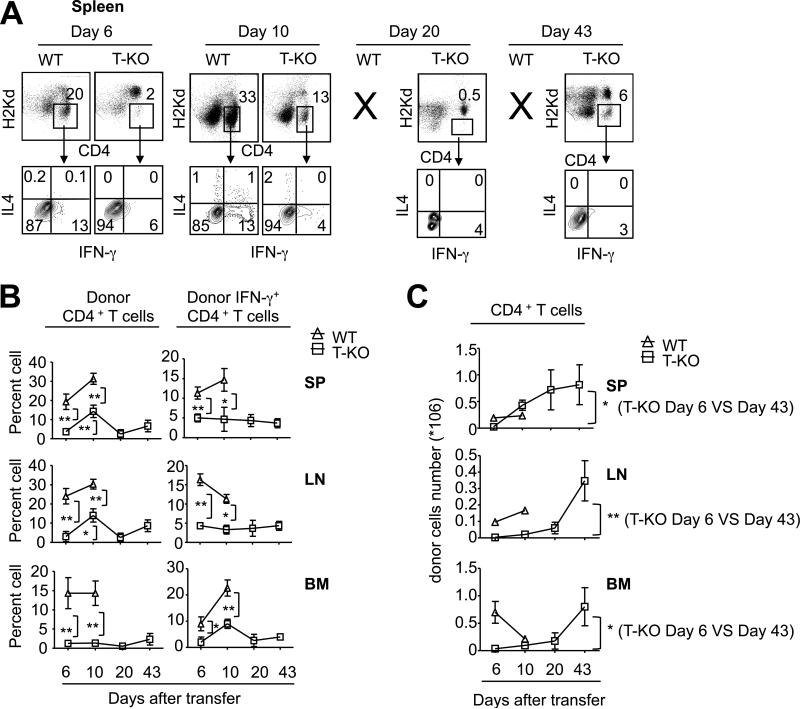
Previous studies have demonstrated that Th1 cells are crucial for inducing BMF in this model of experimental AA.(57,59) To examine the impact of Ezh2 deficiency on Th1 cell development in vivo, we harvested donor T cells from the spleen, LN and BM of these mice 10 days after transfer of WT or T-KO LN cells. We found that loss of Ezh2 led to a significant reduction in percentage and number of Th1 cells in the spleen, LN and BM (Fig.2A, B). Real time RT-PCR analysis showed that alloantigen-activated T-KO T cells expressed dramatically lower levels of Ifng (5-fold), Stat4 (2.5-fold) and Tbx21 (4-fold) transcripts than their WT counterparts (Fig.2C).
Fig.2. Loss of Ezh2 results in selective impairment of Th1 cell development in vivo during AA induction.
WT B6 LN (10×106) or T-KO LN (10×106) were injected into irradiated BDF1 mice (6.5Gy). 7 days after transfer, spleen, LN and BM were isolated, and donor derived CD4+ T cells were analyzed for the expression of IFN-γ and IL-4 (A-B). (A) Dot plots and graphs show the percentage and the mean fluorescence intensity (MFI) of donor CD4+ T cells producing cytokines. (B) Bar graphs show the number of donor CD4+ IFN-γ+ T cells. (C) Seven days after transfer, donor derived CD4+ T cells were sorted for the analysis of gene expression. Data are representative of two independent experiments with each group containing 4 to 6 mice. Error bars indicate mean ± SD. *, P<0.05, **, P<0.01, ***, P<0.001.
In this BMF model, only a small percentage of IL-4-producing CD4+ T cells occurred in the spleen, LN and BM of BDF1 mice receiving WT LN cells (Fig.2A, B), suggesting that the inflammatory stimuli produced during the AA process predominantly induces Th1 cell differentiation in vivo. Ezh2 deficiency did not result in significant changes in the ability of CD4+ T cells to produce IL-4 protein as assessed by flow cytometry (Fig.2A). Furthermore, loss of Ezh2 had no significant impact on the expression of Th2 cell genes (e.g., Il4, Il5, Il13, Stat6 and Gata3) (Fig.2C). There was a moderate reduction of Rorγt and increase of Foxp3 (Fig.2C). However, these moderate changes in genes expression did not lead to the reduction of Il17 expression (Fig.2C) and increase of regulatory T cells (Tregs, data not shown).
Taken together, these results suggest that during AA process Ezh2 is critically involved in regulating the development of Th1 cells, but has little impact on Th2 or Th17 cells.
Data from our previous studies and others indicate that Ezh2 deficiency may lead to impaired expansion and survival of activated T cells.(15,45) It is possible that the inability of T-KO LN cells to cause AA could result from decreased expansion and survival of BM-destructive Th1 cells. To test it, we tracked the longitudinal proliferation and differentiation of WT and T-KO LN cells that were injected into sublethally irradiated BDF1 recipients. As compared to WT LN cells, T-KO LN cells produced significantly less in frequency of both CD4+ T cells and IFN-γ-producing CD4+ T cells in the spleen, LN and BM 6 and 10 days after transfer (Fig.3A, B). Since WT LN cells caused lethal AA in all BDF1 recipients by day 12 (data not shown), it prevented us from further comparing the difference between WT and T-KO LN cells at later time points. However, in BDF1 mice receiving T-KO LN cells, although there was a marked increase in numbers of total donor CD4+ T cells in the spleen, LN and BM at day 43 after transfer compared to that at day 6 (Fig.3C), the frequency of IFN-γ-producing CD4+ T cells was not increased in parallel (Fig.3B). These data suggest that the lack of both differentiation and expansion of Th1 cells may be responsible for the inability of T-KO T cells to mediate AA early during disease process, whereas impaired Th1 differentiation of T-KO T cells could account for reduced AA during later stage.
Fig.3. Tracking the longitudinal proliferation and differentiation of transferred WT and T-KO LN cells in vivo.
WT B6 LN (10×106) or T-KO LN (10×106) cells were injected into irradiated BDF1 mice (6.5Gy). Spleen, LN and BM were isolated to measure cytokine production in donor derived CD4+ T cells at day 6, day 10, day 20 and day 43 after transfer, respectively (A-C). (A) Dot plots show the percentage of cytokine-producing donor CD4+ T cells in the spleen. (B) Graphs show the percentage of donor CD4+ T cells (left panel) and donor IFN-γ+CD4+ T cells (right panel). (C) Graphs show the total number of donor CD4+ T cells. Data are representative of two independent experiments with each group containing 4 to 6 mice. Error bars indicate mean ± SD. *, P<0.05, **, P<0.01.
Ezh2 promotes in vitro Th1 cell differentiation in cultures under Th1-skewing conditions
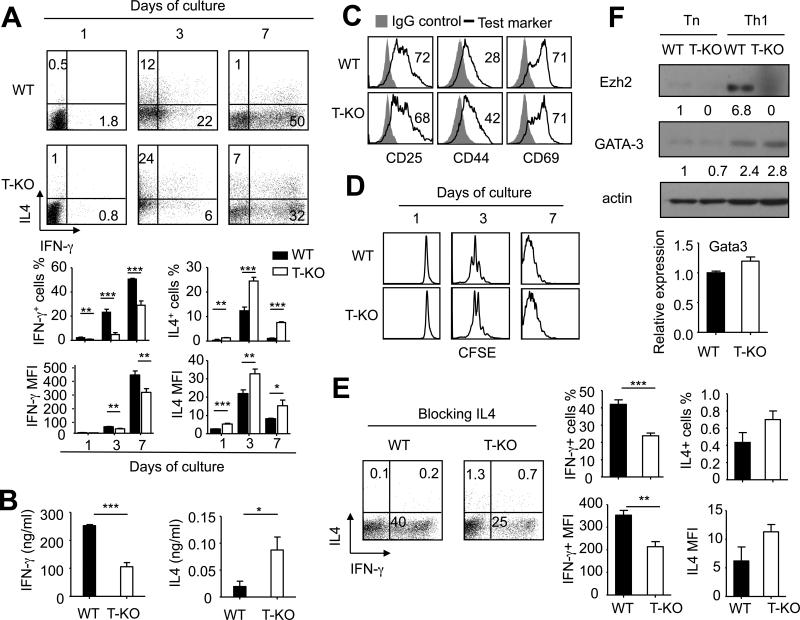
To further examine the importance of Ezh2 in regulating Th1 cell differentiation, we highly purified CD4+ Tn from WT and T-KO B6 mice and cultured them under Th1-skewing conditions without anti-IL-4 Ab. One day after activation in Th1-skewing cultures, both WT and T-KO CD4+ Tn produced barely detectable IFN-γ as assessed by intracellular cytokine staining (Fig. 4A). Three days after culture, there were 3.7-fold less in frequency of IFN-γ-producing CD4+ T cells in the culture of T-KO CD4+ T cells than that of WT CD4+ T cells (Fig.4A). This impaired ability of T-KO CD4+ T cells to produce IFN-γ persisted throughout a period of 7 days during culture (Fig.4A). ELISA further confirmed that T-KO CD4+ T cells secreted approximately 2.5-fold less IFN-γ than WT CD4+ T cells 7 days after culture (Fig.4B), when Th1 cells fully developed.(46,48) These data confirmed the observations in our preceding experiments in vivo (Fig.2) that Th1 cell differentiation was impaired in the absence of Ezh2.
Fig.4. Ezh2 promotes in vitro Th1 cell differentiation in cultures under Th1-skewing conditions.
Naive CD4+ T cells isolated from WT B6 and T-KO mice were stimulated with anti-CD3 and anti-CD28 Abs under Th1-skewing condition. Cells were collected at the indicated time for analysis. (A) Dot plots (upper panel) and graphs (lower panel) show the fraction and MFI of IFN-γ- and/or IL4-producing cells. (B) ELISA assays show the level of IFN-γ and IL-4 in the culture medium at day 7 after restimulation with anti-CD3 Ab. Each group contained equal number of T cells (1×106 cells/ml). (C) Naïve CD4+ T cells isolated from WT B6 or T-KO mice were pre-stained with CFSE and stimulated with anti-CD3 and anti-CD28 Abs. Two days after culture, cells were collected for the analysis of T cell activation markers. (D) Histograms show the cell divisions at the indicated time after culture. (E) Naive CD4+ T cells isolated from WT B6 and T-KO mice were stimulated with anti-CD3 and anti-CD28 Abs under Th1-skewing condition together with anti-IL-4 Ab (10μg/ml) for 7 days. Dot plots (left panel) and graphs (right panel) show the fraction of IFN-γ- and/or IL4-producing cells. (F) Naive CD4+ T cells before (Tn) and after cultured under Th1-skewing condition for 7 days (Th1) were harvested and lysed for western blot analysis (upper panel). The relative expression level of each protein was indicated under the band, which was determined by densitometry analysis. Seven days after culture, the indicated gene expression was analyzed in WT and T-KO cells (lower panel). Data are representative of two independent experiments. Error bars indicate mean ± SD. *, P<0.05, **, P<0.01, ***, P<0.001.
It has been shown that Ezh2 is associated with Vav1 protein,(45) which is a protein important for mediating proximal TCR signaling in T cell activation.(6) To rule out the possibility that the decreased induction of Th1 cells by Ezh2 inhibition might be associated with impaired activation of T cells in cultures, we examined the impact of Ezh2 deficiency on the activation and division of T-KO CD4+ T cells cultured under Th1-skewing conditions. We found that in the absence of Ezh2, CD4+ Tn were normally activated as evidenced by the upregulation of T cell activation markers CD25, CD44 and CD69 (Fig.4C). Furthermore, like WT CD4+ T cells, T-KO CD4+ T cells underwent extensive cell division as evidenced by their dilution of fluorescence dye CFSE in cultures (Fig. 4D). These data suggest that in the absence of Ezh2, CD4+ T cells can be normally activated to undergo cell division and expansion upon stimulation with Th1-skewing conditions.
Previous studies suggest that IL-4 can reduce Th1 cell differentiation.(34,48) We found that in vitro activated T-KO T cells produced more IL-4-producing T cells in frequency than their WT counterparts (Fig.4A). It is possible that increased production of IL-4 in T-KO T cells might account for their impaired Th1 cell differentiation. To test it, we added neutralizing anti-IL4 Ab into the cultures. Indeed, blockade of IL-4 efficiently inhibited the production of IL-4 by both WT and T-KO CD4+ T cells (Fig.4E). Seven days after cultures, there was no marked difference in frequency of IL-4-producing T cells between activated WT and T-KO CD4+ T cells (Fig.4E). In contrast, neutralizing IL-4 in cultures did not improve the ability of T-KO CD4+ T cells to produce IFN-γ (Fig.4E). Furthermore, we confirmed that both WT and T-KO CD4+ T cells derived from Th1-skewing cultures expressed similar levels of GATA3 protein and mRNA (Fig.4F). All these results suggest that Ezh2 promotes Th1 cell differentiation through a mechanism independent of IL-4 and GATA3.
Ezh2 associates with Th1 gene loci
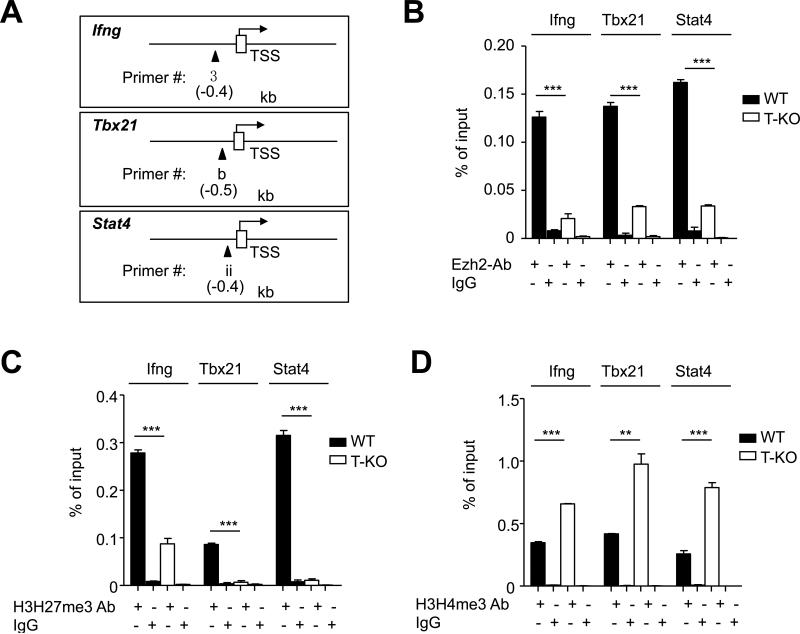
Development of Th1 cells involves a complex mechanism.(62) Th1 cell differentiation is initiated by IFN-γ upregulation of T-bet, which specifically activates Ifng transcription.(62) IL-12 activation of STAT4 is also important to promote Th1 cell differentiation.(53,61,62) To understand the mechanism by which Ezh2 promoted the development of Th1 cells, we first used ChIP assay to examine the presence of Ezh2 and histone methylation markers on Th1 gene loci (e.g., Ifng, Tbx21 and Stat4). CD4+ Tn were isolated from normal WT B6 mice and cultured under Th1-skewing conditions for 7 days. Chromatin was prepared from both CD4+ Tn and Th1 cells. As expected, the amount of H3K27me3 was markedly reduced at the promoter regions of Ifng, Tbx21 and Stat4 gene loci (Fig. 5A). In contrast, H3K4me3, which is a permissive histone methylation marker associated with gene activation,(3,51) was increased at the promoter region of these genes (Fig.5A). These data are in agreement with previous observations that H3K27me3 strongly marked the promoter, intergenic and 3’UTR regions of TBX21 in CD4+ Tn whereas differentiated Th1 cells had reduced H3K27me3 but increased H3K4me3 at the TBX21 regulatory regions.(14,51)
Fig.5. Ezh2 associates with the promoter regions of type-1 gene loci.
Freshly isolated WT CD4+ T cells (Tn) and WT CD4+ T cells after 7 days culture in Th1-skewing conditions (Th1) were processed for ChIP using Abs specific for H3K27me3, H3K4me3, Ezh2 and control IgG (A-B). (A) The graphs show the ChIP-qPCR for H3K27me3 or H3K4me3 binding to the promoter region of Ifng, Tbx21 or Stat4. (B) Schematic representation of the mouse Ifng, Tbx21 or Stat4 locus (upper panel). Open rectangles indicate the transcriptional start site. Closed triangles show the locations of real-time quantitative PCR primer pairs used in the ChIP assay, which are indicated in relative kilobases to the transcriptional start site. The graphs (lower panel) show the ChIP-qPCR for Ezh2 binding, with a serious of primer pairs shown above covering the regulatory and promoter region of Ifng, Tbx21 or Stat4 locus. (C) Graphs show the relative expression of indicated genes measured by realtime PCR in freshly isolated WT CD4+ T cells (Tn) and WT CD4+ T cells after 7 days culture in Th1-skewing conditions (Th1). (D) Dot plots show the fraction of IFN-γ- and/or Ezh2-expressing cells in freshly isolated WT CD4+ T cells (Tn) and WT CD4+ T cells after 7 days culture in Th1-skewing conditions (Th1). Data are representative of three independent experiments. Error bars indicate mean ± s.d. **, P <0.01, ***, P <0.001.
Interestingly, there was no significant reduction in the amount of Ezh2 at both regulatory and promoter regions of these Th1 genes in differentiated Th1 cells compared to CD4+ Tn, with moderately increased amount of Ezh2 at the promoter region of Stat4 (Fig.5B). Real-time RTPCR showed that Ezh2 was positively correlated with expression of these Th1 genes (Fig.5C). All CD4+ T cells producing IFN-γ also expressed high levels of Ezh2 (Fig.5D).
To further confirm that Ezh2 associated with Th1 gene loci, we prepared chromatin from WT and T-KO CD4+ T cells activated under Th1-skewing conditions for 7 days. ChIP analysis revealed significantly reduced amount of Ezh2 and H3K27me3 at the promoter regions of these gene loci in T-KO Th1 cells compared to WT Th1 cells (Fig.6A-C). Thus, despite the reduction of H3K27me3 at the promoter regions of these Th1 genes, its catalyzing enzyme Ezh2 remains associated with these regions. Notably, T-KO Th1 cells showed higher amount of H3K4me3 at the promoter regions of these Th1 gene loci than WT Th1 cells (Fig.6D). This suggests that loss of Ezh2 leads to the switch to a permissive histone methylation signature for Th1 genes, which is favorable for their activation. However, our results showed that loss of Ezh2 impaired Th1 cell development (Fig.2C).
Fig.6. Ezh2 specifically binds to promoter regions of Th1 type gene loci.
WT and T-KO CD4+ T cells collected 7 days after culture under Th1-skewing conditions and processed for ChIP using antibodies specific for Ezh2, H3K27me3, and H3K4me3. (A) A schematic of the primer regions in the mouse Ifng, Tbx21 or Stat4 locus. Open rectangles indicate the transcriptional start site (TSS). Closed triangles show the locations of real-time PCR primer pairs used for ChIP assay. (B-D) The graphs show the relative amount of Ezh2, H3K27me3, H3K4me3 and IgG at the regions of Ifng, Tbx21 or Stat4 locus.
Ezh2 regulates T-bet at both the transcriptional level and post-translational level
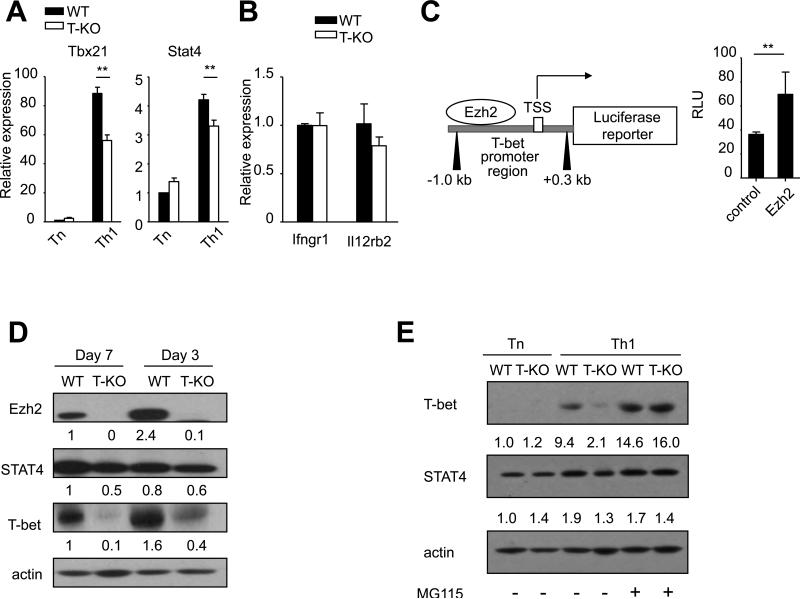
To determine how Ezh2 regulated the expression of T-bet and STAT4 in Th1 cells, we examined the expression of T-bet and STAT4 mRNA in WT and T-KO CD4+ T cell 7 days after culture under Th1-skewing conditions. CD4+ Tn were assessed as controls. Seven days after culture under Th1-skewing conditions, T-KO CD4+ T cells expressed approximately 1.8-fold less T-bet mRNA than their WT counterparts (Fig. 6A). STAT4 mRNA was slightly decreased in activated T-KO CD4+ T cells (Fig.7A). In addition, Ezh2 deficiency had no effect on the expression of Th1-related Ifngr1 and Il12rb2 genes (Fig.7B), two critical signaling molecules upstream of T-bet and STAT4, respectively.(62) These data together with our observations that Ezh2 was positively associated with Tbx21 gene (Fig.5 and Fig.6) indicate that Ezh2 promotes the expression of T-bet at the transcriptional level.
Fig.7. Ezh2 regulates T-bet at both the transcriptional level and post-translational level.
(A) Graphs show the gene expression of Stat4 and Tbx21 in freshly isolated WT or T-KO CD4+ T cells (Tn) and cells after 7 days culture in Th1-skewing conditions (Th1). (B) The Graph shows the relative expression of indicated genes measured by realtime PCR in WT or T-KO CD4+ T cells after 7 days culture in Th1-skewing conditions. (C) The schematic (left panel) shows the construction of Tbx21 promoter region ranging from +0.3kb to -1.0kb of the TSS. 3T3 cells were co-transfected with pGL3-Tbx21 reporter plasmid and an empty vector control or Ezh2 in combination. Luciferase reporter activity was normalized to the activity obtained for the cotransfected renilla control. The graph (right panel) represents the relative light units (RLU). (D) Western blot show the expression of Ezh2, Stat4 and T-bet after 3 or 7 days of Th1-skewing culture conditions. The relative expression level of each protein was indicated under the band, which was determined by densitometry analysis. (E) Western blots show the expression of T-bet and STAT4 in freshly isolated WT or T-KO CD4+ T cells (Tn) and cells after 7 days culture in Th1-skewing conditions (Th1) with or without the treatment of MG115 (2μM) for 6 hours. Data are representative of two independent experiments. Error bars indicate mean ± SD **, P <0.01.
To assess whether Ezh2 directly activated Tbx21 transcription, we co-transfected 3T3 cells with pGL3-Tbx21 reporter and MigR1 viral plasmid encoding Ezh2 or empty MigR1 plasmid. These 3T3 cells were harvested 48 hours after transfection and analyzed with the Dual Luciferase system. Overexpression of Ezh2 resulted in moderate induction of Tbx21 reporter activity (Fig.7C). This indicates that Ezh2 can directly activate Tbx21 transcription.
We further verified whether loss of Ezh2 led to reduction of STAT4 and T-bet protein in Th1 cells. As compared to activated WT CD4+ T cells, there was only minimal reduction of STAT4 protein in these activated T-KO CD4+ T cells (Fig.7D). Most notably, T-KO CD4+ T cells showed approximately 4-fold and 10-fold less T-bet protein at day 3 and day 7 after culture, respectively, than their WT counterparts (Fig.7D). This dramatic reduction of T-bet protein in T-KO Th1 cells appeared not to be completely supported by moderate reduction of T-bet mRNA in these cells. We reasoned that loss of Ezh2 might lead to increased degradation of T-bet protein in Th1 cells. To test it, we treated WT and T-KO Th1 cells with the proteasome inhibitor MG115.(18) Addition of the proteasome inhibitor MG-115 restored the expression of T-bet protein, but not STAT4 protein, in Ezh2-deficient Th1 cells (Fig.7E), suggesting that T-bet protein may be more susceptible than STAT4 to proteasome-mediated degradation in Th1 cells lacking Ezh2. Altogether, Ezh2 promotes T-bet expression at both transcriptional and post-translational levels, with the later the most extent. These results identify a novel and important role for Ezh2 to regulate Th1 cells.
Introduction of T-bet into T-KO CD4+ T cells fully rescues their differentiation into Th1 cells
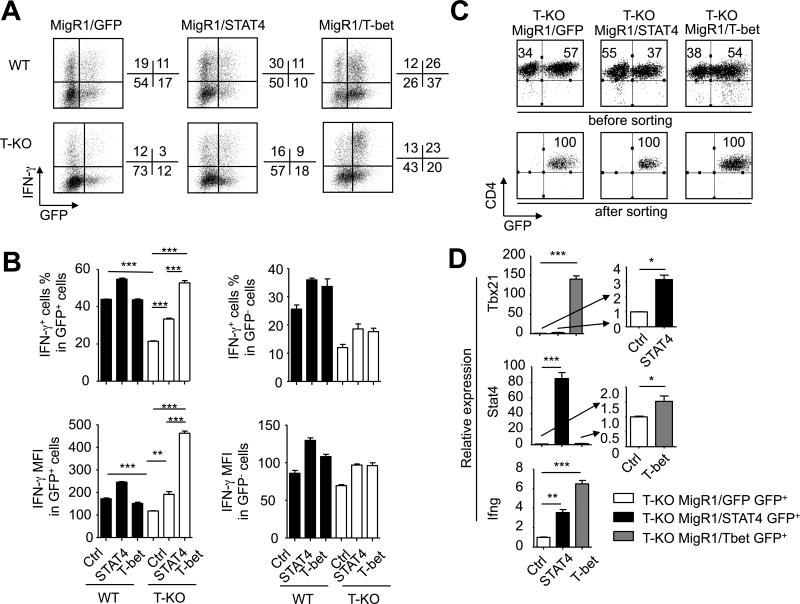
To determine whether the down-regulation of T-bet caused the impairment of Th1 cell development, we used MigR1 virus bicistronically encoding T-bet and GFP (named MigR1/T-bet) to infect T-KO CD4+ T cells cultured under Th1-skewing conditions. T-KO CD4+ T cells infected with MigR1 encoding STAT4 and GFP (named MigR1/STAT4) or GFP alone (named MigR1/GFP) were assessed in parallel. Expression of GFP allowed us to track cells expressing T-bet or STAT4. WT CD4+ T cells were also infected with each of these viruses as controls.
We found that T-KO GFP-positive (GFP+) CD4+ T cells that were derived from cultures infected by MigR1/T-bet, which over-expressed T-bet, produced similar percentage of IFN-γ+ T cells to WT GFP+ CD4+ T cells derived from cultures infected by either MigR1/T-bet or MigR1/GFP (Fig.8A,B). Interestingly, as compared to WT GFP+CD4+ T cells infected with MigR1/GFP or MigR1/STAT4, T-KO GFP+CD4+ T cells expressing STAT4 had significantly lower frequency of IFN-γ+ T cells (Fig.8A,B). Furthermore, T-KO CD4+ T cells expressing STAT4 contained about 40% less IFN-γ+ T cells than T-KO CD4+ T cells expressing T-bet (Fig.8A,B). These data suggest that viral expression of T-bet fully rescues the ability of Ezh2-deficient CD4+ T cells to differentiate into Th1 cells, whereas overexpression of STAT4 only partially improves Th1 cell differentiation of activated T-KO T cells.
Fig.8. Overexpression of T-bet fully rescues T-KO CD4+ T cells to differentiate into Th1 cells.
CD4+ Tn isolated from WT B6 and T-KO mice were stimulated with anti-CD3 and anti-CD28 antibodies under Th1-skewing conditions. One day after culture, T cells were infected with retrovirus encoding GFP-STAT4, GFP-T-bet or GFP-control, and then cultured for another 6 days. (A) Dot plots show the fraction of IFN-γ- and/or GFP-expressing cells 7 days after culture. (B) Bar graphs show the percent and MFI of IFN-γ+ cells in GFP+ (left panel) and GFP− (right panel) cells after infection. (C) GFP+CD4+ T-KO cells were sorted, and total RNAs were isolated and subjected to real-time PCR analysis. Dot plots show the purity of sorted GFP+ cells. (D) Graphs show the relative expression of indicated genes in sorted donor GFP+CD4+ T cells. Data are representative of two independent experiments. Error bars indicate mean ± SD. *, P<0.05, **, P <0.01, ***, P<0.001.
To validate these observations, we highly purified GFP-positive T-KO T cells from these cultures (Fig.8C) and confirmed the overexpression of T-bet and STAT4 in these T-KO CD4+ T cells, respectively, using real-time RT-PCR (Fig.8D). Furthermore, overexpression of T-bet in T-KO T cells induced significantly more Ifng transcripts than did overexpression of STAT4 (Fig.8D). Phosphorylation of STAT4 is critical for IFN-γ production and Th1 cell differentiation.(33,53) Western blot analysis showed that MigR1/STAT4-infected T-KO T cells expressed 7.5-fold more STAT4 protein and 1.2-fold more phosphorylated STAT4 than their WT counterparts (data not shown). Thus, the reduction of T-bet in activated T-KO CD4+ T cells is a major contributor to their impaired development of Th1 cells.
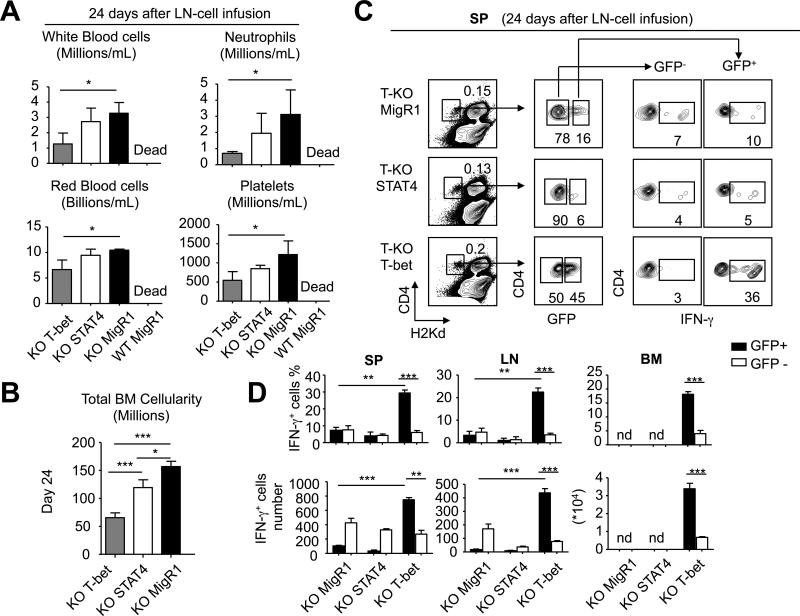
We finally examined whether retroviral expression of T-bet could rescue the ability of Ezh2-deficient T cells to induce AA. Both CD4+ and CD8+ T cells were highly purified from T-KO B6 mice and activated in vitro for 24 hours, followed by infection with MigR1/T-bet, MigR1/STAT4 and MigR1/GFP, respectively. Thirty-six hours later, these infected T cells were harvested, assessed using flow cytometry for the expression of GFP and transferred into sublethally irradiated BDF1 mice. Each mouse received unfractionated T cells that contained similar numbers of GFP+ cells (0.8×106 CD4+ T cells + 0.6×106 CD8+ T cells). BDF1 mice receiving T-KO T cells transduced by either T-bet or STAT4 survived over 24 days after transfer, whereas all BDF1 mice receiving WT T cells infected by control MigR1/GFP died from AA within 14 days (Fig.9A and data not shown). This suggests that transfer of this amount of T-KO T cells expressing T-bet or STAT4 did not cause lethal AA. However, as compared to control T-KO T cells infected by MigR1/GFP, MigR1/T-bet-infected T-KO T cells caused marked reduction of BM cells and peripheral blood cells (i.e., platelets, RBC, WBC and neutrophils) 24 days after transfer (Fig.9A,B). Furthermore, GFP+ MigR1/T-bet-infected KO T cells (which expressed T-bet) had about 4-fold more in frequency of IFN-γ-producing T cells than control GFP+ T-KO T cells infected by MigR1/GFP (Fig.9D). In contrast, transfer of MigR1/STAT4-infected T-KO T cells resulted in moderate decrease of BM cells, but not blood pancytopenia (Fig.9A,B). Notably, GFP+ MigR1/STAT4-infected T-KO T cells (which expressed STAT4) failed to produce high levels of IFN-γ (Fig.9C,D). Thus, Ezh2 regulation of T-bet is important for production of Th1 cells mediating AA.
Fig.9. Introduction of T-bet to T-KO T cells rescues their ability to mediate AA in mice.
CD4+ Tn and CD8+ Tn were isolated from WT and T-KO mice and separately cultured in the presence of anti-CD3 Ab (2μg/ml), anti-CD28 Ab (2μg/ml) and IL-2 (10ng/ml). Twenty-four hours later, T-KO T cell subsets were infected with MigR1/T-bet, MigR1/STAT4 and MigR1/GFP, respectively. WT T cell subsets were infected with MigR1/GFP as controls. Thirty-six hours after infection, these T cells were harvested and transferred into sublethally irradiated (6.5Gy) BDF1 mice. Each mouse received unfractionated T cells that contained similar numbers of GFP+ cells (0.8×106 CD4+ T cells + 0.6×106 CD8+ T cells). (A) Peripheral blood was collected 24 days after LN-cell infusion and the cell number of RBC, WBC, platelets and neutrophils were calculated (mean ± SD, n=6 to 8 mice per group). (B) Total BM cellularity was calculated assuming that bilateral tibia and femurs contain 25% of total marrow cells (mean ± SD, n=6 to 8 mice per group). (C) Twenty-four days after LN-cell infusion, donor CD4+ T cells were isolated from the spleen, LN and BM to measure the production of IFN-γ. The plots show the percentage of donor CD4+ T cells in spleen, the percentage of GFP− and GFP+ cells, and the fraction of IFN-γ-producing cells in GFP− and GFP+ cells. (D) Graphs show the percentage (upper panel) and the number (lower panel) of donor IFN-γ+CD4+ T cells in spleen, LN and BM. *, P<0.05, **, P<0.01, ***, P<0.001.
Discussion
Our findings elucidate a new and critical role of Ezh2 in controlling pathogenic Th1 cell responses during AA process, which has not been previously identified. Ezh2 is absolutely required for CD4+ Th1 cells to mediate fatal AA. Ezh2 inhibition led to dramatic reduction of BM-destructive Th1 cells in vivo, decreasing BM-infiltrating Th1 cells, and protecting mice from BMF. Decreased capability of Ezh2-deficient T cells to differentiate into Th1 cells resulted from reduced expression of T-bet mRNA and protein, as retroviral expression of T-bet in Ezh2-deficient CD4+ T cells fully rescued their differentiation into Th1 cells. In contrast, ectopic expression of STAT4 only partially restored the ability of Ezh2-deficient CD4+ T cells to produce IFN-γ. Interestingly, although Ezh2 is known to act primarily as a gene silencer, it promoted the expression of T-bet gene in Th1 cells via a mechanism of directly activating Tbx21 gene promoter. Furthermore, Ezh2 was required to prevent proteasome-mediated degradation of T-bet protein in Th1 cells. This constellation of Ezh2 actions induced the optimal production of Th1 cells destructing BM cells, highlighting the therapeutic potential of Ezh2 inhibition in controlling AA.
Since Ezh2 specifically catalyzes the repressive maker H3K27me3, a corollary believeth is that Ezh2 may be required for repressing cytokine gene expression.(7,14,52) CD4+ Tn showed moderate levels of H3K27me3 in the promoter regions of Ifng and Tbx21 gene loci.(5,7,39,51) Upon Th1 differentiation, H3K27me3 was reduced in the promoter regions of Th1 genes (e.g., Ifng, Tbx21 and Stat4), whereas H3K4me3 was upregulated in these gene loci.(51) We confirmed and further extended these observations that loss of Ezh2 caused the reduction of H3K27me3 at the Ifng, Tbx21 and Stat4 gene loci in Th1 cells, while increasing the amount of H3K4me3 in these gene loci. However, this skewed balance of histone methylation signature toward the transcription permission state at these Th1 gene loci did not lead to increased expression of these genes in Ezh2-deficient CD4+ T cells. In contrast, Ezh2 deficiency caused down-regulation of Tbx21 mRNA. ChIP assay and reporter analysis verified that Ezh2 possessed the ability to directly activate Tbx21 transcription. This result is in sharp contrast to conventional thoughts that Ezh2 might repress the expression of genes associated with effector differentiation of antigen-driven T cells during immune response.(7,14,51) Past studies have shown that Ezh2 and other polycomb proteins were unconventionally associated with effector cytokine genes (e.g., Ifng and Il4).(19,25) However, these studies did not report whether Ezh2 influenced the expression of Tbx21 and the development of Th1 cells in vivo during inflammatory responses.(19,25) Our data document for the first time that Ezh2 acted as an activator for inducing optimal expression of Tbx21 during Th1 cell differentiation. These findings are in agreement with observations by others that Ezh2 can activate some critical transcription factors in cancer cells in a context-dependent manner.(27,54)
Intriguingly, we identified a previously unknown role for Ezh2 in protecting proteasome-mediated degradation of T-bet protein in Th1 cells. Ezh2 deficiency led to increased degradation of T-bet protein in Th1 cells. Addition of the proteasome inhibitor MG-115 restored the expression of T-bet protein in Ezh2-deficient Th1 cells. This effect of Ezh2 on protecting T-bet from proteasome-mediated degradation differs from the effect of Ezh2 on repressing gene transcription.(27) T-bet is crucial for controlling appropriate Th1 cell differentiation through reversing and establishing new epigenetic states in T cells.(26,31) T-bet recruited the histone methyltranferase SET7/9 to catalyze permissive H3K4me2 at the Ifng promoter in Th1 cells.(26,31) Notably, T-bet recruited Jumonji domain-containing protein histone demethylase 3 (Jmjd3) to remove H3K27me3 across the Ifng locus.(30) These data suggest that epigenetic regulation of Th1 cell differentiation involves a complex and functional cooperation of histone methyltransferase, demethylase and transcription factors.(26,53) In the present study, we show T-bet as a post-translational Ezh2 target that acts together to regulate optimal Th1 cell responses. This effect of Ezh2 appeared to be independent of its activity of methylating histone proteins. Data from several studies indicate that Ezh2 can directly methylate signaling proteins to regulate gene transcription.(22,45) For example, Ezh2 methylates STAT3 in cancer cells, leading to enhanced STAT3 activity.(22) Given the presence of high amounts of Ezh2 at the promoter regions of Ifng gene locus in Th1 cells, we speculated that Ezh2 could play critical roles in stabilizing T-bet protein at this gene locus, thereby facilitating the T-bet regulation of Th1 cell differentiation. Formal experiments are needed to address this point and to determine the mechanisms whereby Ezh2 protects T-bet from proteasome-mediated degradation in Th1 cells.
Accumulating evidence indicates that T-bet can be an effective target for the prevention and treatment of immune-mediated AA. Abnormal expression of T-BET has been shown in human patients with AA.(43) T cells from AA patients who were refractory to immunosuppressive treatment expressed high levels of T-BET. In contrast, AA patients who had responded to the therapy showed low levels of T-BET in their circulating blood cells.(43) Experimental studies further indicate that genetic inhibition of T-bet leads to decreased induction of AA in mice.(47) However, so far, there is no pharmacological approach available to modulate T-bet. Furthermore, transcription factors are difficult drug targets.(11) Given that Ezh2 plays critical roles in the regulation of T-bet mRNA and protein in Th1 cells, we proposed that targeting Ezh2 may prove advantageous to modulate T-bet and its-controlled pathogenic T-cell inflammation.
Aberrant expression of Ezh2 has been associated with many types of malignant diseases such as prostate cancer, breast cancer and lymphomas.(27,32,49) Data from our previous studies indicate that Ezh2 is markedly increased in activated T cells.(16,21) Inhibition of Ezh2 leads to the reduction of GVHD in mice of allogeneic BMT.(15) In these mouse models of allogeneic BMT, infusion of donor BM cells fully rescues hematopoiesis and thymopoiesis in lethally irradiated recipients,(4,13) which prevents us from precisely assessing the impact of T cells in mediating BMF. Our experiments using the AA mouse model and in vitro cultures provide compelling evidence for a T cell-intrinsic contribution of Ezh2 to the expression of Th1-associated transcription factors and cytokines. Inhibition of Ezh2 in T cells through conditional deletion suppressed Th1 cell differentiation and development of BMF. As several Ezh2-specific inhibitors have recently been discovered for experimental treatment of cancer,(23,24,28,37) it will be important to test whether these Ezh2-specific inhibitors may control AA in experimental models.
In summary, we have identified the critical role of Ezh2 in regulating Th1 responses during AA and the mechanism by which Ezh2 promotes Th1 cell responses mediating BMF in mice. Our findings open new perspectives to study the importance of Ezh2 in regulating the development of other lineages of CD4+ T cells (e.g., Th2, Th17, Th9 and Tregs). Indeed, a most recent study has demonstrated that Ezh2 is involved in repressing the development of Th9 cells.(50) Furthermore, it will be important to investigate whether deregulated Ezh2 expression and activity may be associated with inflammatory disorders such as autoimmune diseases and chronic infections in humans.
Supplementary Material
Acknowledgments
We thank Jinfang Zhu (Molecular and Cellular Immunoregulation Unit Laboratory of Immunology, National Institute of Allergy and Infectious Diseases, National Institutes of Health, DHHS) and Cheong-hee Chang (Department of Microbiology & Immunology, University of Michigan) for the thoughtful discussion and critical reading of the manuscript.
Footnotes
This work was supported by the American Cancer Society (YZ), Department of Defense (YZ) and NIH (R01-CA-172106-01, YZ).
Author contributions
Q. T. and Y. Z. conceived and designed the project. Q.T., S.H., F.X., K.M., Y.L., I.M., L.M., H.S., Y.Z., Y.G., E.H. and Y.Z. performed experiments and analyzed the data. Q. T. and Y. Z. wrote the manuscript.
Conflict of interest disclosure
The authors declare that they have no conflicts of interest in relation to this manuscript.
Reference
- 1.Araki Y, Wang Z, Zang C, Wood WH, 3rd, Schones D, Cui K, Roh TY, Lhotsky B, Wersto RP, Peng W, Becker KG, Zhao K, Weng NP. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr Opin Cell Biol. 2006;18:275–283. doi: 10.1016/j.ceb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 4.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012;12:443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burbach BJ, Medeiros RB, Mueller KL, Shimizu Y. T-cell receptor signaling to integrins. Immunol Rev. 2007;218:65–81. doi: 10.1111/j.1600-065X.2007.00527.x. [DOI] [PubMed] [Google Scholar]
- 7.Chang S, Aune TM. Dynamic changes in histone-methylation 'marks' across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Lipovsky K, Ellison FM, Calado RT, Young NS. Bystander destruction of hematopoietic progenitor and stem cells in a mouse model of infusion-induced bone marrow failure. Blood. 2004;104:1671–1678. doi: 10.1182/blood-2004-03-1115. [DOI] [PubMed] [Google Scholar]
- 9.Chen J, Brandt JS, Ellison FM, Calado RT, Young NS. Defective stromal cell function in a mouse model of infusion-induced bone marrow failure. Exp Hematol. 2005;33:901–908. doi: 10.1016/j.exphem.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Chen J, Ellison FM, Eckhaus MA, Smith AL, Keyvanfar K, Calado RT, Young NS. Minor antigen h60-mediated aplastic anemia is ameliorated by immunosuppression and the infusion of regulatory T cells. J Immunol. 2007;178:4159–4168. doi: 10.4049/jimmunol.178.7.4159. [DOI] [PubMed] [Google Scholar]
- 11.Dunker AK, Uversky VN. Drugs for ‘protein clouds’: targeting intrinsically disordered transcription factors. Curr Opin Pharmacol. 2010;10:782–788. doi: 10.1016/j.coph.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Fang TC, Yashiro-Ohtani Y, Del Bianco C, Knoblock DM, Blacklow SC, Pear WS. Notch directly regulates Gata3 expression during T helper 2 cell differentiation. Immunity. 2007;27:100–110. doi: 10.1016/j.immuni.2007.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373:1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He S, Tong Q, Bishop DK, Zhang Y. Histone methyltransferase and histone methylation in inflammatory T-cell responses. Immunotherapy. 2013;5:989–1004. doi: 10.2217/imt.13.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He S, Xie F, Liu Y, Tong Q, Mochizuki K, Lapinski PE, Mani RS, Reddy P, Mochizuki I, Chinnaiyan AM, Mineishi S, King PD, Zhang Y. The histone methyltransferase Ezh2 is a crucial epigenetic regulator of allogeneic T cell responses mediating graft-versus-host disease. Blood. 2013;122:4119–4128. doi: 10.1182/blood-2013-05-505180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He S, Wang J, Kato K, Xie F, Varambally S, Mineishi S, Kuick R, Mochizuki K, Liu Y, Nieves E, Mani RS, Chinnaiyan AM, Marquez VE, Zhang Y. Inhibition of histone methylation arrests ongoing graft-versus-host disease in mice by selectively inducing apoptosis of alloreactive effector T cells. Blood. 2012;119:1274–1282. doi: 10.1182/blood-2011-06-364422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hod-Dvorai R, Jacob E, Boyko Y, Avni O. The binding activity of Mel-18 at the Il17a promoter is regulated by the integrated signals of the TCR and polarizing cytokines. Eur J Immunol. 2011;41:2424–2435. doi: 10.1002/eji.201141620. [DOI] [PubMed] [Google Scholar]
- 18.Imamura T, Haruta T, Takata Y, Usui I, Iwata M, Ishihara H, Ishiki M, Ishibashi O, Ueno E, Sasaoka T, Kobayashi M. Involvement of heat shock protein 90 in the degradation of mutant insulin receptors by the proteasome. J Biol Chem. 1998;273:11183–11188. doi: 10.1074/jbc.273.18.11183. [DOI] [PubMed] [Google Scholar]
- 19.Jacob E, Hod-Dvorai R, Schif-Zuck S, Avni O. Unconventional association of the polycomb group proteins with cytokine genes in differentiated T helper cells. J Biol Chem. 2008;283:13471–13481. doi: 10.1074/jbc.M709886200. [DOI] [PubMed] [Google Scholar]
- 20.Jacob E, Hod-Dvorai R, Ben-Mordechai OL, Boyko Y, Avni O. Dual function of polycomb group proteins in differentiated murine T helper (CD4+) cells. J Mol Signal. 2011;6:5. doi: 10.1186/1750-2187-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kato K, Cui S, Kuick R, Mineishi S, Hexner E, Ferrara JL, Emerson SG, Zhang Y. Identification of stem cell transcriptional programs normally expressed in embryonic and neural stem cells in alloreactive CD8+ T cells mediating graft-versus-host disease. Biol Blood Marrow Transplant. 2010;16:751–771. doi: 10.1016/j.bbmt.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim E, Kim M, Woo DH, Shin Y, Shin J, Chang N, Oh YT, Kim H, Rheey J, Nakano I, Lee C, Joo KM, Rich JN, Nam DH, Lee J. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell. 2013;23:839–852. doi: 10.1016/j.ccr.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knutson SK, Wigle TJ, Warholic NM, Sneeringer CJ, Allain CJ, Klaus CR, Sacks JD, Raimondi A, Majer CR, Song J, Scott MP, Jin L, Smith JJ, Olhava EJ, Chesworth R, Moyer MP, Richon VM, Copeland RA, Keilhack H, Pollock RM, Kuntz KW. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8:890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 24.Konze KD, Ma A, Li F, Barsyte-Lovejoy D, Parton T, Macnevin CJ, Liu F, Gao C, Huang XP, Kuznetsova E, Rougie M, Jiang A, Pattenden SG, Norris JL, James LI, Roth BL, Brown PJ, Frye SV, Arrowsmith CH, Hahn KM, Wang GG, Vedadi M, Jin J. An Orally Bioavailable Chemical Probe of the Lysine Methyltransferases EZH2 and EZH1. ACS Chem Biol. 2013;8:1324–1334. doi: 10.1021/cb400133j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koyanagi M, Baguet A, Martens J, Margueron R, Jenuwein T, Bix M. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J Biol Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 26.Lewis MD, Miller SA, Miazgowicz MM, Beima KM, Weinmann AS. T-bet's ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol Cell Biol. 2007;27:8510–8521. doi: 10.1128/MCB.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343–349. doi: 10.1038/nature09784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, 3rd, Diaz E, LaFrance LV, Mellinger M, Duquenne C, Tian X, Kruger RG, McHugh CF, Brandt M, Miller WH, Dhanak D, Verma SK, Tummino PJ, Creasy CL. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492:108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 29.Miller SA, Weinmann AS. Molecular mechanisms by which T-bet regulates T-helper cell commitment. Immunol Rev. 238:233–246. doi: 10.1111/j.1600-065X.2010.00952.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller SA, Mohn SE, Weinmann AS. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol Cell. 2013;40:594–605. doi: 10.1016/j.molcel.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller SA, Huang AC, Miazgowicz MM, Brassil MM, Weinmann AS. Coordinated but physically separable interaction with H3K27-demethylase and H3K4-methyltransferase activities are required for T-box protein-mediated activation of developmental gene expression. Genes Dev. 2008;22:2980–2993. doi: 10.1101/gad.1689708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–195. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, Feldman GM, Nishikomori R, O'Shea JJ. STAT4 serine phosphorylation is critical for IL-12-induced IFN-gamma production but not for cell proliferation. Proc Natl Acad Sci U S A. 2002;99:12281–12286. doi: 10.1073/pnas.182618999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745–755. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 35.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 36.Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, Banica M, DiCioccio CB, Gross DA, Mao CA, Shen H, Cereb N, Yang SY, Lindsten T, Rossant J, Hunter CA, Reiner SL. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 2003;302:1041–1043. doi: 10.1126/science.1090148. [DOI] [PubMed] [Google Scholar]
- 37.Qi W, Chan H, Teng L, Li L, Chuai S, Zhang R, Zeng J, Li M, Fan H, Lin Y, Gu J, Ardayfio O, Zhang JH, Yan X, Fang J, Mi Y, Zhang M, Zhou T, Feng G, Chen Z, Li G, Yang T, Zhao K, Liu X, Yu Z, Lu CX, Atadja P, Li E. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci U S A. 2012;109:21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roderick JE, Gonzalez-Perez G, Kuksin CA, Dongre A, Roberts ER, Srinivasan J, Andrzejewski C, Jr., Fauq AH, Golde TE, Miele L, Minter LM. Therapeutic targeting of NOTCH signaling ameliorates immune-mediated bone marrow failure of aplastic anemia. J Exp Med. 2013;210:1311–1329. doi: 10.1084/jem.20112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheinberg P, Young NS. How I treat acquired aplastic anemia. Blood. 2012;120:1185–1196. doi: 10.1182/blood-2011-12-274019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Sloand E, Kim S, Maciejewski JP, Tisdale J, Follmann D, Young NS. Intracellular interferon-gamma in circulating and marrow T cells detected by flow cytometry and the response to immunosuppressive therapy in patients with aplastic anemia. Blood. 2002;100:1185–1191. doi: 10.1182/blood-2002-01-0035. [DOI] [PubMed] [Google Scholar]
- 43.Solomou EE, Keyvanfar K, Young NS. T-bet, a Th1 transcription factor, is up-regulated in T cells from patients with aplastic anemia. Blood. 2006;107:3983–3991. doi: 10.1182/blood-2005-10-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 45.Su IH, Dobenecker MW, Dickinson E, Oser M, Basavaraj A, Marqueron R, Viale A, Reinberg D, Wulfing C, Tarakhovsky A. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 2005;121:425–436. doi: 10.1016/j.cell.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 46.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 47.Tang Y, Desierto MJ, Chen J, Young NS. The role of the Th1 transcription factor T-bet in a mouse model of immune-mediated bone-marrow failure. Blood. 2010;115:541–548. doi: 10.1182/blood-2009-03-211383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usui T, Nishikomori R, Kitani A, Strober W. GATA-3 suppresses Th1 development by downregulation of Stat4 and not through effects on IL-12Rbeta2 chain or T-bet. Immunity. 2003;18:415–428. doi: 10.1016/s1074-7613(03)00057-8. [DOI] [PubMed] [Google Scholar]
- 49.Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 50.Wang A, Pan D, Lee YH, Martinez GJ, Feng XH, Dong C. Cutting Edge: Smad2 and Smad4 Regulate TGF-beta-Mediated Il9 Gene Expression via EZH2 Displacement. J Immunol. 2013;191:4908–4912. doi: 10.4049/jimmunol.1300433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O'Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei J, Duramad O, Perng OA, Reiner SL, Liu YJ, Qin FX. Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A. 2007;104:18169–18174. doi: 10.1073/pnas.0703642104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 54.Xu K, Wu ZJ, Groner AC, He HH, Cai C, Lis RT, Wu X, Stack EC, Loda M, Liu T, Xu H, Cato L, Thornton JE, Gregory RI, Morrissey C, Vessella RL, Montironi R, Magi-Galluzzi C, Kantoff PW, Balk SP, Liu XS, Brown M. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338:1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young NS. Hematopoietic cell destruction by immune mechanisms in acquired aplastic anemia. Semin Hematol. 2000;37:3–14. doi: 10.1016/s0037-1963(00)90026-x. [DOI] [PubMed] [Google Scholar]
- 56.Young NS, Maciejewski J. The pathophysiology of acquired aplastic anemia. N Engl J Med. 1997;336:1365–1372. doi: 10.1056/NEJM199705083361906. [DOI] [PubMed] [Google Scholar]
- 57.Young NS, Scheinberg P, Calado RT. Aplastic anemia. Curr Opin Hematol. 2008;15:162–168. doi: 10.1097/MOH.0b013e3282fa7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young NS, Bacigalupo A, Marsh JC. Aplastic anemia: pathophysiology and treatment. Biol Blood Marrow Transplant. 2010;16:S119–125. doi: 10.1016/j.bbmt.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng W, Maciejewski JP, Chen G, Young NS. Limited heterogeneity of T cell receptor BV usage in aplastic anemia. J Clin Invest. 2001;108:765–773. doi: 10.1172/JCI12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang Y, Louboutin JP, Zhu J, Rivera AJ, Emerson SG. Preterminal host dendritic cells in irradiated mice prime CD8+ T cell-mediated acute graft-versus-host disease. J Clin Invest. 2002;109:1335–1344. doi: 10.1172/JCI14989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 62.Zhu J, Yamane H, Paul WE. Differentiation of effector CD4 T cell populations (*). Annu Rev Immunol. 2010;28:445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu J, Giannola DM, Zhang Y, Rivera AJ, Emerson SG. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood. 2003;102:2420–2427. doi: 10.1182/blood-2003-01-0251. [DOI] [PubMed] [Google Scholar]
- 64.Zhu J, Min B, Hu-Li J, Watson CJ, Grinberg A, Wang Q, Killeen N, Urban JF, Jr., Guo L, Paul WE. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.