Abstract
The actin cytoskeleton carries out cellular functions, including division, migration, adhesion, and intracellular transport, that require a variety of actin binding proteins, including myosins. Our focus here is on class II nonmuscle myosin isoforms, NMIIA, NMIIB, and NMIIC, and their regulation by the actin binding protein, tropomyosin. NMII myosins are localized to different populations of stress fibers and the contractile ring, structures involved in force generation required for cell migration, adhesion, and cytokinesis. The stress fibers and contractile ring that contain NMII myosins also contain tropomyosin. Four mammalian genes encode more than 40 tropomyosins. Tropomyosins inhibit or activate actomyosin MgATPase and motility depending on the myosin and tropomyosin isoform. In vivo, tropomyosins play a role in cell migration, adhesion, cytokinesis, and NMII isoform localization in an isoform-specific manner. We postulate that the isoform-specific tropomyosin localization and effect on NMII isoform localization reflect modulation of NMII actomyosin kinetics and motile function. In this study, we compare the ability of different tropomyosin isoforms to support actin filament motility with NMIIA, NMIIB, and NMIIC as well as skeletal muscle myosin. Tropomyosins activated, inhibited, or had no effect on motility depending on the myosin, indicating that the myosin isoform is the primary determinant of the isoform-specific effect of tropomyosin on actomyosin regulation. Activation of motility of nonmuscle tropomyosin–actin filaments by NMII myosin correlates with an increased Vmax of the myosin MgATPase, implying a direct effect on the myosin MgATPase, in contrast to the skeletal tropomyosin–actin filament that has no effect on the Vmax or maximal filament velocity.
Cells depend on the actin cytoskeleton for a variety of cellular functions, including determination of cellular shape and dynamic changes during motile events, intracellular transport of molecules, vesicles, and organelles, formation of adhesions to other cells and the extracellular matrix, and cytokinesis with formation and closure of the contractile ring. In the quest to understand these structures and processes in cells and their response to physiological stimuli in normal and diseased states, a multitude of proteins has been identified and analyzed. How they function in macromolecular assemblies remains to be understood in most cases.
In this work, we focus on class II myosins, the filament-forming myosins, and their regulation by the actin binding protein, tropomyosin (Tm). Myosin II is the founding member of the myosin family, being originally isolated from skeletal muscle.1 In striated muscle, the primary mechanism of regulation is via troponin–tropomyosin bound to actin in which Ca2+ binding to troponin (Tn) and the strong binding of myosin crossbridges together cooperatively activate the thin filament.2 Tropomyosin, a coiled-coil protein that binds along the length of four to seven actin subunits in the actin filament, depending on the species and isoform, is responsible for the cooperativity.3−5 Myosin and tropomyosin promote each other’s affinity for actin in a cooperative manner.6
While troponin is found only in cardiac and skeletal muscle cells, tropomyosin is ubiquitous.7 Tropomyosin is well-established in its cooperative regulation of smooth muscle myosin. Smooth muscle myosin, together with nonmuscle forms of myosin II, is primarily regulated by phosphorylation of the regulatory light chain by myosin light chain kinase and Rho kinase.8 Studies comparing the Tm-dependent regulation of smooth and striated muscle myosin II isoforms and their respective tissue-specific Tm isoforms first showed that both proteins influence the regulatory state of the actin filament such that Tm can be inhibitory or activating.9−12
The proposal that regulation of myosin is coordinated with specific Tms in a manner that translates to cellular localization and specific cytoskeletal functions has received support from both biochemical and cellular studies. Myosin I, localized in regions of the cell where actin filaments are dynamic and Tm is sparse,13−16 is inhibited by Tm.17−20 In contrast, Tm activates myosin V isoforms isolated from yeast, the class of myosins that engage in intracellular transport. Yeast myosin V selectively travels on actin filaments that contain Tm, and Tm makes the motor more processive.19,21,22 However, a vertebrate myosin V that is processive is unaffected by skeletal muscle Tm.23
In mammals, there are three nonmuscle myosin IIs, NMIIA, NMIIB, and NMIIC, encoded by separate genes. In addition, each of these genes is subject to alternative splicing giving rise to additional isoforms.8 Like the smooth muscle myosin II, NMIIs are activated by phosphorylation of the regulatory light chain.8 Kinetic analysis of NMII isoforms shows they are distinct from each other and from their sarcomeric counterparts.8,24−29 NMII myosins are much slower than muscle myosin IIs, with higher affinity for ADP and slower rates of ADP release. Of the three, NMIIA is the fastest and NMIIB the slowest. All three have duty ratios higher than those of muscle myosin IIs but fall short of being processive myosins that typically have duty ratios of ∼50%. The kinetic profile suits this class of myosins for being involved in processes that require maintenance of tension, rather than rapid movement, consistent with their cellular localizations and reported functions.8
The nonmuscle filament-forming myosin IIs are localized to different populations of stress fibers and regions of the contractile ring,30−32 structures that are involved in force generation required for cell migration, cell adhesion, and cytokinesis.8,33,34 NMIIA and NMIIB may be spatially segregated in the cells, localized in different populations of stress fibers that help define cellular polarity, as well as coassembled in heterofilaments in regions of spatial overlap.31,32,35−37 Stress fibers containing NMIIA are attached at one end to focal adhesions. Active myosin II (as defined by the need for phosphorylation of the regulatory light chain) is required in the stress fibers that form along adhesions for maturation of focal adhesions and adhesion-dependent cell migration.38,39
The stress fibers that contain myosin II also contain tropomyosin, as does the contractile ring.40−42 More than 40 Tm isoforms are encoded by the four mammalian genes as a consequence of alternate promotors and alternative exon splicing.7 Tropomyosin isoforms are enriched in the lamellum and in stress fibers that contain myosin II, but not in the lamellipodium of epithelial cells.13,15 Microinjection of skeletal muscle tropomyosin into cells resulted in the recruitment of myosin II, decreasing the extent of the lamellipodium while supporting rapid migration and the formation of focal adhesions closer to the leading edge of the cell.14 This study indicated Tm has a role in regulating regional specializations of the cytoskeleton for cellular migration.
Subsequent studies have studied the role of specific Tm isoforms. Overexpression of Tm5NM1 (a cytoskeletal Tm isoform) results in an increased number of stress fibers, increased spreading, and reduced cell migration velocity, compared to other Tm isoforms not typically associated with stress fibers.41,43 The associated stabilization of focal adhesions has led to the suggestion that overexpressing Tm5NM1 results in a tension-dependent decrease in focal adhesion turnover and consequent remodeling of the ECM.43 NMIIA is recruited to the Tm5NM1-containing stress fibers.41
Fission yeast Tm promotes myosin II (Myo2) activity,44 but the effect of Tm on mammalian cytosolic myosin II is unknown. We postulate that the isoform-specific Tm localization and the effect of overexpression on NMII isoform localization reflect the modulation of acto-NMII kinetics and motile function. In this study, we compared the ability of different Tm isoforms to support actin filament motility with nonmuscle HMMs IIA, IIB, and IIC and skeletal myosin. The Tm isoforms have differential effects on filament velocities depending on the myosin isoform, indicating that the primary determinant of the isoform-specific effect of Tm on actomyosin regulation is the myosin isoform. We also measured the steady-state MgATPase activity of NMIIA, NMIIB, and NMIIC HMMs in the presence of a Tm isoform (Tm5NM1) that increased the velocity of NMIIA and NMIIC HMMs in the motility assays. Tm5NM1 increased the ATPase rates of NMIIA and NMIIC, in support of the motility data, by increasing the Vmax and decreasing the KATPase.
Experimental Procedures
Protein Expression and Purification
The rat AS-Tmstα, Tm2, Tm5a, and Tm5NM1 and human Tm4 proteins were expressed in Escherichia coli BL21(DE3) cells and purified as previously described.45 Actin was purified from acetone powder of chicken pectoral skeletal muscle actin.46 Tropomyosin and actin concentrations were determined by measuring the difference spectrum of tyrosine.45 Skeletal myosin was prepared from chicken pectoral muscle.47 Monoclonal antibody 10F12.3 that reacts specifically with chicken skeletal muscle myosin subfragment 2 was prepared and characterized as described previously.47,48
Recombinant human nonmuscle heavy meromyosin IIA, MYH9 (NMIIA HMM), and IIB, MYH10 (NMIIB HMM), and mouse IIC, MYH14 (NMIIC HMM), with a C-terminal EGFP and Flag tag were co-expressed with bovine nonmuscle regulatory light chain (MYL9) and chicken essential (MYL6) light chains in the baculovirus/Sf9 system (Invitrogen, New York, NY).49 The infected Sf9 cells were harvested by sedimentation after they had grown for 24 h and then stored at −80 °C. The cell pellets were extracted and homogenized in a buffer containing 0.5 M NaCl, 10 mM MOPS (pH 7.3), 10 mM MgCl2, 1 mM EGTA, 3 mM NaN3, 2 mM ATP, 0.1 mM phenylmethanesulfonyl fluoride, 0.1 mM dithiothreitol, and a 5 μg/mL leupeptin/proteinase inhibitor mixture [2 μg/mL chymostatin (MP Biochemicals, Santa Ana, CA), 1 μg/mL pepstatin (MP Biochemicals), 1 μg/mL Nα-tosyl-Lys-chloromethylketone-HCl (TLCK) (Sigma, St. Louis, MO), and 1 μg/mL N-p-tosyl-l-phenylalanine chloromethyl ketone (TPCK) (Sigma)]. The HMM fragments were copurified with the light chains by Flag affinity chromatography using M2 Flag affinity gel (Sigma). The full-length rabbit smooth muscle myosin light chain kinase (MLCK, NP_001075775) was expressed in the baculovirus/Sf9 system and purified via an N-terminal Flag tag.
Cosedimentation Assays
Tropomyosin (0.1–8 μM) was combined with 5 μM F-actin and cosedimented at 20 °C in 200 mM NaCl, 10 mM Tris-HCl (pH 7.5), 2 mM MgCl2, and 0.5 mM DTT at 60000 rpm in a Beckman model TL-100 ultracentrifuge in a TLA-100 rotor.50 The pellets and supernatants were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis, stained with Coomassie blue, and scanned and analyzed using an ImageScanner III densitometer with Labscan version 6.0 and ImageQuant TL version 7.0. The free Tm in the supernatants was calculated from standard curves for each Tm. The binding constant Kapp and the Hill coefficient (αH) were determined by fitting the experimental data to the Hill equation (eq 1) using Kaleidagraph:
| 1 |
where ν is the maximal fraction of Tm binding to actin, n is the maximal Tm bound, and [Tm] = [Tm]free. The Tm:actin ratio was normalized to 1 by dividing the Tm:actin ratio obtained from densitometry by the Tm:actin ratio observed at saturation.
Circular Dichroism (CD) Measurements
Thermal stability measurements were taken by following the ellipticity of 0.1 mg/mL Tm at 222 nm as a function of temperature in 0.5 M NaCl, 10 mM sodium phosphate (pH 7.5), 1 mM EDTA, and 1 mM DTT in an Aviv model 400 spectropolarimeter at the Robert Wood Johnson Medical School CD facility. The observed melting temperature (TM) is defined as the temperature at which the ellipticity at 222 nm, normalized to a scale of 0–1, is equal to 0.5.51
In Vitro Motility Assays
The in vitro motility assays were conducted as described previously.52,53 The chicken skeletal myosin and NMII HMMs were flash-frozen, stored in liquid nitrogen in 25–150 μL aliquots, and thawed in ice followed by dead head removal. To remove dead heads, the skeletal myosin and NMII HMMs were combined with F-actin and 5 mM MgATP in high-salt buffer (HSB) [25 mM imidazole (pH 7.6), 0.3 M KCl, 4 mM MgCl2, and 5 mM DTT] and pelleted at 90000 rpm for 20 min at 4 °C in a Beckman TLA 100.2 rotor. This procedure pellets all of the actin and 5–10% of the total myosin, including inactive myosin that is no longer dissociated from actin by ATP (dead heads). The clear myosin supernatant was recovered and used within 0–16 h of dead head removal.
For the assays with skeletal myosin, nitrocellulose-coated glass coverslips were incubated with 0.15 mg/mL monoclonal antibody that reacts specifically with chicken pectoral muscle myosin subfragment 2 (10F12.3), followed by incubation with 1% BSA for blocking. Skeletal myosin was diluted in HSB supplemented with 1% BSA (HSB/BSA) to a final concentration of 40 μg/mL. The antibody-coated coverslips were incubated with 40 μg/mL myosin for ∼2 h in a humidified chamber at 4 °C, followed by washes with HSB/BSA and then motility buffer [25 mM imidazole (pH 7.6), 25 mM KCl, 4 mM MgCl2, 1 mM MgATP, and 5 mM DTT].
For the assays with nonmuscle HMMs, IIA, IIB, and IIC, nitrocellulose-coated glass coverslips were incubated with 60 μg/mL HMM, followed by incubation with 1% BSA for blocking at room temperature. Previous studies have shown that no antibody is needed to facilitate the attachment of this myosin fragment.25 The HMMs were then phosphorylated by incubating the coverslips with 0.1 μM calmodulin and 10 nM MLCK in buffer containing 10 mM MOPS (pH 7.3), 50 mM KCl, 5 mM MgCl2, 0.2 mM CaCl2, 0.1 mM EGTA, 1 mM DTT, and 0.2 mM ATP for 10 min at room temperature, followed by washes with motility buffer at 4 °C.
The myosin-coated coverslips were transferred to a 15 μL drop of 2 nM rhodamine-phalloidin-labeled actin (chicken skeletal muscle) or actin–Tm (Tm concentration of 1–2 μM for all motility assays, except where indicated) in a modified motility buffer [25 mM imidazole (pH 7.6), 25 mM KCl, 4 mM MgCl2, 7.6 mM MgATP, 50 mM DTT, 0.5% methyl cellulose, 0.1 mg/mL glucose oxidase, 0.018 mg/mL catalase, and 2.3 mg/mL glucose] in a small parafilm ring fixed on an alumina slide with vacuum grease. This chamber was observed with an Olympus BH-2 microscope. The movement of actin filaments from 2 to 5 min of continuous video (Movies S1 and S2 of the Supporting Information) was recorded from several fields for each assay condition and analyzed with semiautomated filament tracking programs as described below.54,55
Analysis of Motility Data
The distance versus time plot for each filament is divided into segments of uniform velocity by the tracking program, and the filament length and velocity determined for each segment of every track are saved. The list of all segments is then sorted in descending order of filament velocity. The minimal and maximal velocities are determined and divided into 20 bins between these limits of width = max – min/20. The data from the list are sorted into these bins, and a mean velocity and a mean count for each bin are determined. The trajectory of every filament with a lifetime of at least 10 frames is determined, and the instantaneous velocity of the filament moving along the trajectory, the filament length, and the distance of continuous motion are tabulated. When hundreds of filaments are tracked, a total distance moved by all the filaments can be determined from the tabulated data list. The question of interest is the average velocity at which the filaments move over this total distance. Unfortunately, defects arise in the preparation of surfaces that result in pinning of actin filaments and slowing and even stopping of the sliding movement for various periods. To minimize the influence of these defects in determining the filament velocity, the binned velocity data are weighted on the basis of the fraction of the total distance the filaments in each bin have moved. This weighting approach favors moving filaments over stationary filaments. As a consequence, the resulting velocity distribution reflects the mean velocity of smooth movement and is not weighted down by stationary filaments, thus minimizing the effect of surface defects that slow and pin filaments. The weighted probability of the filament velocity for hundreds of events was fit to a Gaussian distribution to determine a mean velocity ± standard deviation (SD) for each experiment (Figure S1 of the Supporting Information).
ATPase Assays
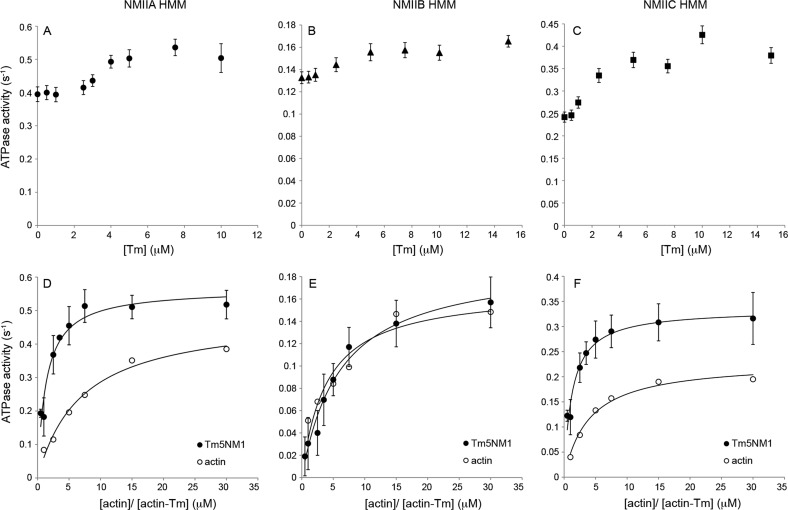
The HMM fragments at 2.2–3.4 μM were phosphorylated with MLCK (1–10 nM) for 15 min at room temperature in a reaction mixture containing 10 mM MOPS (pH 7.3), 50 mM KCl, 5 mM MgCl2, 0.2 mM CaCl2, 0.1 mM EGTA, 0.1 μM calmodulin, 1 mM DTT, and 0.2 mM ATP.28 The MgATPase activity was measured using an NADH-coupled assay at 25 °C in 10 mM MOPS (pH 7.0), 2 mM MgCl2, 1 mM ATP, 50 mM KCl, 0.15 mM EGTA, 40 units/mL l-lactic dehydrogenase, 200 units/mL pyruvate kinase, 200 μM NADH, and 1 mM phospho(enol)pyruvate. Fifteen micromolar actin (rabbit skeletal muscle) with varying concentrations of Tm5NM1 (from 0 to 15 μM) and varying concentrations of actin–Tm5NM1 (from 0.5 to 30 μM) combined at a 1:1 actin:Tm molar ratio was used for the assays shown in Figure 4. Each HMM II protein was at a concentration of 0.1 μM.
Figure 4.
Actin-activated MgATPase activity of myosins. The steady-state ATPase activity of phosphorylated NMIIA (A and D), NMIIB (B and E), and NMIIC (C and F) HMMs was measured in the presence of (A–C) 15 μM actin and varying concentrations of Tm5NM1 (0–15 μM) or (D–F) varying concentrations of actin and actin–Tm5NM1 (0.5–30 μM) combined at a 1:1 molar ratio. The data in panels D–F were fit to a hyperbolic equation (—) to obtain the values for Vmax and KATPase. Each HMM II protein was at a concentration of 0.1 μM. Each data point is shown with the standard deviation from three or four experiments, and the data without error bars are from a single experiment. The Vmax and KATPase values are listed in Table 2. Assay conditions: 10 mM MOPS (pH 7.0), 2 mM MgCl2, 1 mM ATP, 50 mM KCl, 0.15 mM EGTA, 40 units/mL l-lactic dehydrogenase, 200 units/mL pyruvate kinase, 200 μM NADH, and 1 mM phospho(enol)pyruvate. The temperature was 25 °C.
Results
Tropomyosin and Myosin Isoforms
The tropomyosins were purified from E. coli and were unacetylated. The striated muscle αTm has an Ala-Ser extension (AS-Tmstα) at the N-terminal end to increase actin affinity.56 There are four TPM genes (TPM1–TPM4) in vertebrates that express more than 40 isoforms through the use of alternate promoters (exons 1a/b) and alternative splicing of three exons: 2a/b, 6a/b, and 9a-d. The difference in the lengths of high-molecular weight (HMW, 284 a.a.) and low-molecular weight (LMW, 247 a.a.) forms of Tm arises from the use of alternate promoters for exons 1a and 2 or exon 1b (Figure 1).7 Rat striated αTm (1a-2b-6b-9a) and Tm2 (1a-2b-6b-9d) are HMW isoforms encoded by TPM1 that differ in the ninth C-terminal exon. Rat Tm5a (1b-6b-9d), Tm5NM1 (1b-6a-9d), and human Tm4 (1b-6b-9d) are LMW isoforms encoded by TPM1, TPM3, and TPM4 genes, respectively (Figure 1). Recombinant human nonmuscle heavy meromyosin IIA, MYH9 (NMIIA HMM), and IIB, MYH10 (NMIIB HMM), and mouse IIC, MYH14 (NMIIC HMM), with a C-terminal EGFP and Flag tag were co-expressed with bovine nonmuscle regulatory light chain (MYL9) and chicken essential (MYL6) light chains in the baculovirus/Sf9 system.49
Figure 1.
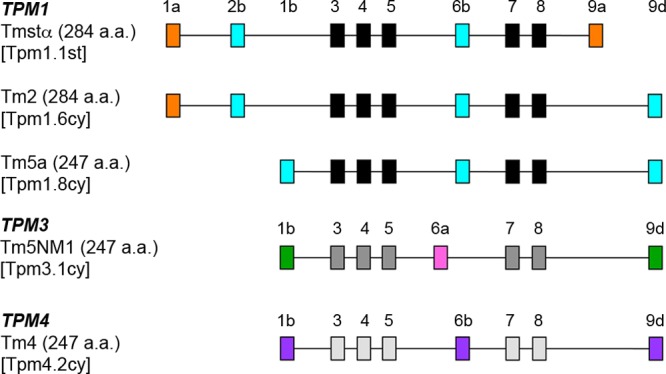
Tropomyosin gene structure in vertebrates. The arrangement of exons expressed in the Tm isoforms used in this study from TPM1, TPM3, and TPM4 genes is shown. The names of the Tms in parentheses correspond to the systematic nomenclature proposed for mammalian tropomyosins by Geeves, Hitchcock-DeGregori, and Gunning (manuscript submitted for publication). The full forms of the names in the parentheses are as follows: Tpm1.1st, Tpm1.1st(a.b.b.a); Tpm1.6cy, Tpm1.6cy(a.b.b.d); Tpm1.8cy, Tpm1.8cy(b.-.b.d.); Tpm3.1cy, Tpm3.1cy(b.-.a.d); Tpm4.2cy, Tpm4.2cy(b.-.b.d).
In Vitro Motility of Actin–Tropomyosin
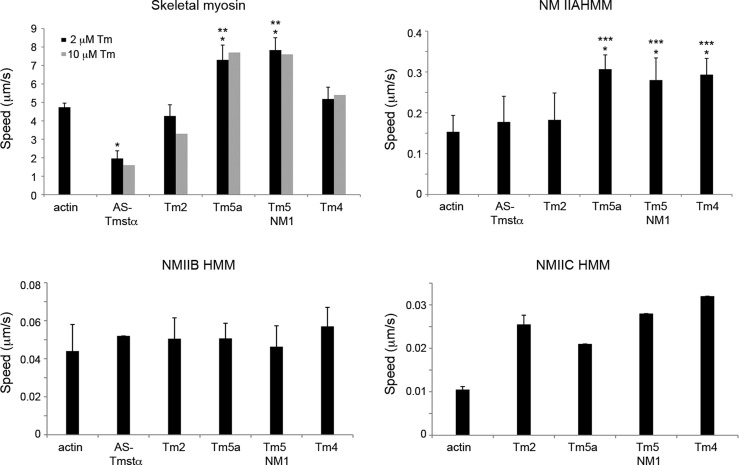
The velocity of actin–Tm filaments on skeletal myosin and nonmuscle IIA, IIB, and IIC HMMs was determined in in vitro motility assays (Figure 2 and Table 1). The motility assays were performed at 27 °C for skeletal myosin. Because of their slower velocities, the motility assays for the NMII HMMs were done at a higher temperature (32 °C) for ease of measurement. Of the NMII isoforms, NMIIC HMM was the slowest (0.01 μm/s) and NMIIA HMM the fastest (0.16 μm/s), in agreement with previous studies.25 The NMII HMMs were phosphorylated in the presence of Ca2+-calmodulin-MLCK prior to the motility assays. In the presence of Tm, the velocities of actin filaments were inhibited, activated, or remained unaffected depending on the Tm and myosin isoforms. The motility assays for actin–Tm were done in the presence of 2 μM Tm. Filament velocities were also determined at a higher Tm concentration, 10 μM, to ensure that the binding of Tm to actin is saturated with 2 μM Tm. The velocities of actin–Tm were not significantly different at 2 and 10 μM Tm (Figure 2), indicating that 2 μM Tm is sufficient to saturate the actin in the motility assays.
Figure 2.
Filament speeds of actin–tropomyosin in in vitro motility assays. Filament speeds were determined for actin and actin–Tm on skeletal myosin and phosphorylated NMIIA, NMIIB, and NMIIC HMMs. The assays with actin–Tm were conducted with 2 μM Tm (black bars). Assays were also conducted with 10 μM Tm for skeletal myosin (gray bars). The filament speeds are means ± SD from two to six experiments (Table 1). The data with no error bars are from a single experiment. *P < 0.01 compared with actin. **P < 0.01 compared with AS-Tmstα, Tm2, and Tm4. ***P < 0.05 compared with AS-Tmstα and Tm2 (unpaired Student’s t test). Assay conditions: 25 mM imidazole (pH 7.6), 25 mM KCl, 4 mM MgCl2, 7.6 mM MgATP, 50 mM DTT, 0.5% methyl cellulose, and an oxygen scavenger system (0.1 mg/mL glucose oxidase, 0.02 mg/mL catalase, and 2.5 mg/mL glucose). The temperature was 27 °C for skeletal myosin and 32 °C for HMMs IIA, IIB, and IIC.
Table 1. Filament Speeds of Actin and Actin–Tm with Skeletal and NMII Myosins.
| speed (μm/s)a (% moving,
total number) |
||||
|---|---|---|---|---|
| skeletal myosin | NMIIA HMM | NMIIB HMM | NMIIC HMM | |
| actin | 4.8 ± 0.2, n = 6 (97, 320) | 0.16 ± 0.03, n = 6 (55, 110) | 0.04 ± 0.01, n = 3 (36, 110) | 0.011 ± 0.001, n = 2 (11, 100) |
| AS-Tmstα | 2.0 ± 0.5,bn = 5 (75, 160) | 0.20 ± 0.10, n = 4 (56, 75) | 0.05, n = 1 (41, 65) | – |
| Tm2 | 4.4 ± 0.6, n = 5 (96, 330) | 0.18 ± 0.08, n = 4 (64, 120) | 0.05 ± 0.01, n = 2 (31, 90) | 0.026 ± 0.002, n = 2 (12, 85) |
| Tm5a | 7.4 ± 1.0,b,cn = 5 (99, 320) | 0.31 ± 0.04,b,dn = 3 (87, 130) | 0.05 ± 0.01, n = 3 (27, 120) | 0.021, n = 1 (10, 100) |
| Tm5NM1 | 7.9 ± 0.8,b,cn = 5 (99, 390) | 0.30 ± 0.04,b,dn = 5 (67, 150) | 0.05 ± 0.01, n = 3 (23, 110) | 0.028, n = 1 (24, 120) |
| Tm4 | 5.2 ± 0.7, n = 5 (96, 280) | 0.29 ± 0.04,b,dn = 3 (82, 200) | 0.06 ± 0.01, n = 3 (29, 90) | 0.032, n = 1 (18, 110) |
Filament speeds ± the standard deviation (SD) of actin and actin–Tm from in vitro motility assays from two to six experiments. The values with no SD are from a single experiment. The average values for the percent moving filaments and total number of filaments are reported in parentheses.
P < 0.01 compared with actin (unpaired Student’s t test) (Figure 2).
P < 0.01 compared with AS-Tmstα, Tm2, and Tm4 (unpaired Student’s t test) (Figure 2).
P < 0.05 compared with AS-Tmstα and Tm2 (unpaired Student’s t test) (Figure 2).
With skeletal myosin, filament velocities were inhibited by AS-Tmstα (∼60%) but activated by Tm5a and Tm5NM1 (∼60%) relative to the rate with actin alone (P < 0.01, unpaired Student’s t test), whereas Tm2 and Tm4 had no significant effect (Figure 2 and Table 1). In the case of NMIIA HMM, the LMW nonmuscle Tms (Tm5a, Tm5NM1, and Tm4) activated filament velocities by ∼2-fold (P < 0.01), whereas AS-Tmstα and Tm2 had no significant effect, relative to the rate with actin alone. None of the Tm isoforms significantly affected filament velocity with NMIIB HMM (P > 0.1). With NMIIC HMM, all nonmuscle Tms activated the velocities of actin filaments by 2–3-fold compared to that with actin alone. However, because of the extremely slow velocities of actin filaments translocated by NMIIC HMM, the velocities for this isoform were calculated from fewer filaments and the experiments were not repeated in most cases. The fraction of filaments moving for actin and actin–Tm ranged from 75 to 99% with skeletal myosin, from 55 to 90% with NMIIA HMM, from 20 to 40% with NMIIB HMM, and from 10 to 25% for NMIIC HMM (Table 1). The speed of individual filaments within each experiment was independent of filament length except for very short filaments, <1 μm (Figure S2 of the Supporting Information).
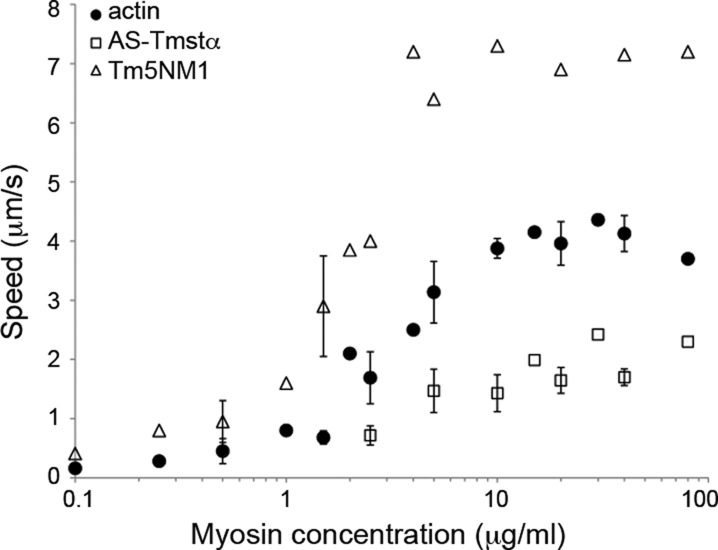
Dependence of Motility on Surface Myosin Density
We determined the velocities of actin–AS-Tmstα and actin–Tm5NM1 as a function of skeletal myosin loading concentration (Figure 3). The myosin surface density on the coverslips is directly proportional to the myosin loading concentration, and the surface is saturated at ∼10–20 μg/mL myosin.53 The velocity of actin is saturated above 5–10 μg/mL myosin and decreases at myosin concentrations of <5 μg/mL. The velocities of AS-Tmstα and Tm5NM1 have a myosin concentration dependence similar to that of actin. AS-Tmstα inhibited actin velocities, whereas Tm5NM1 activated actin velocities at all myosin concentrations tested (0.1–80 μg/mL). The fraction of moving filaments was ∼35% for actin and ∼65% for Tm5NM1 at the lowest myosin concentration (0.1 μg/mL) and increased to >80% at >1 μg/mL myosin.
Figure 3.
Filament speeds as a function of surface myosin density. Filament speeds were determined for actin, actin–AS-Tmstα, and actin–Tm5NM1 as a function of skeletal myosin loading concentration. The data with error bars show the standard deviation from two or three experiments, and the data with no error bars are from a single experiment. Assay conditions are the same as in Figure 2. The temperature was 27 °C.
ATPase Activity of Nonmuscle Forms of Myosin II with Actin–Tm5NM1
The actin-activated MgATPase activities of NMIIA, NMIIB, and NMIIC HMMs were measured to determine the mechanism of activation by Tm5NM1, a nonmuscle Tm that activated NMIIA and NMIIC in the motility assays. We focused on the Tm5NM1 isoform for further analysis because it is the most studied cytoskeletal Tm and plays an important role in cellular functions such as determining cell morphology, formation of stress fibers and focal adhesions during cell migration, and cytokinesis.41−43,57 Tm5NM1 is also a major form in transformed, metastatic cancer cells and has recently been shown to be a target for anticancer drugs.58
The ATPase rates of the myosin isoforms vary, with NMIIA HMM being the fastest (0.49 s–1) and the NMIIB and NMIIC HMMs being much slower (0.17 and 0.23 s–1, respectively).25 The ATPase activity of NMIIA and NMIIC HMMs increased as a function of Tm concentration and reached saturation when the Tm:actin molar ratio reached saturation (1:6 Tm5NM1:actin) (Figure 4A–C). The ATPase activity of NMIIA, NMIIB, and NMIIC HMMs was also determined as a function of actin–Tm concentration. The plots were fit to the Michaelis–Menten equation to give values for the kinetic constants, Vmax (maximal rate of ATP hydrolysis at a saturating actin concentration) and KATPase (concentration of actin required for the half-maximal rate of ATP hydrolysis) (Figure 4D–F and Table 2). Tm5NM1 increased the Vmax of NMIIA and NMIIC by ∼20–40%, with a smaller effect on IIB (12%) compared to that with actin alone, thus supporting the motility data. Tm5NM1 also decreased the KATPase of NMIIA and NMIIC by 3–5-fold but increased the KATPase of NMIIB by ∼2-fold. Therefore, Tm5NM1 may activate NMIIA and NMIIC by increasing the rate of the ATPase cycle and reaching the maximal rate at a lower actin concentration.
Table 2. Actin-Activated MgATPase Rates of NMII Myosins.
|
Vmax (s–1), KATPase (μM) (from steady-state
ATPasea) |
|||
|---|---|---|---|
| NMIIA HMM | NMIIB HMM | NMIIC HMM | |
| actin | 0.49 ± 0.03, 7.0 ± 1.2 | 0.17 ± 0.02, 3.9 ± 1.2 | 0.23 ± 0.01, 3.9 ± 0.6 |
| Tm5NM1 | 0.57 ± 0.03, 1.3 ± 0.3 | 0.19 ± 0.01, 6.2 ± 1.1 | 0.33 ± 0.01, 1.3 ± 0.2 |
The values of Vmax and KATPase from steady-state ATPase assays ± the standard error from three or four experiments for Tm5NM1 and one experiment for actin (Figure 4).
Actin Affinity and Thermal Stability of Tropomyosins
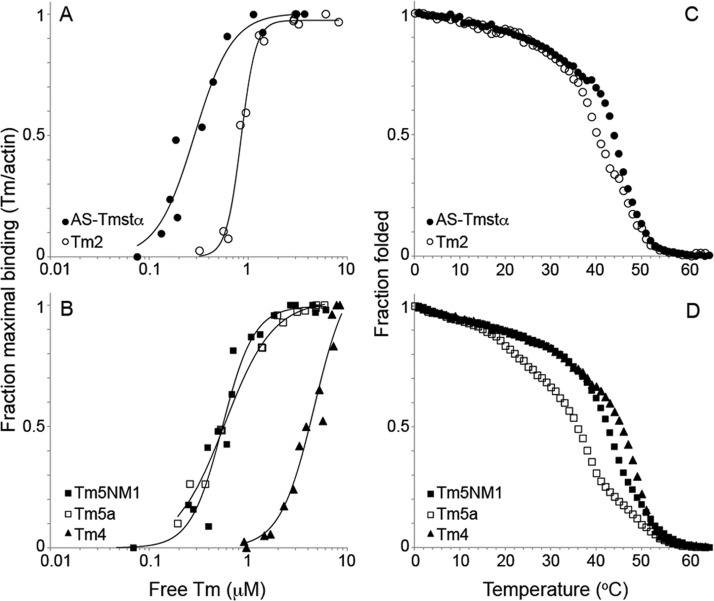
We conducted cosedimentation assays and thermal stability measurements of the Tm isoforms to ensure that the effects on actomyosin regulation are not related to actin affinity or stability. All isoforms bound to actin but with different affinities (Figure 5A,B and Table 3). AS-Tmstα has the highest actin affinity with a Kapp of 3.5 × 106 M–1. Tm2, Tm5a, and Tm5NM1 have ∼2–3-fold lower affinity, whereas Tm4 has an affinity >10-fold lower than that of AS-Tmstα. The differences in actin affinity between Tm isoforms are suggested to be primarily due to the presence of specific alternate exons at the N- and C-termini (1a vs 1b and 9a vs 9d) of the molecule that can affect the end-to-end overlap complex and hence the affinity.59 However, we did not observe any clear relationship between alternate exon usage and actin affinity for isoforms. For example, Tm5a (1b) and Tm2 (1a and 2b) are both expressed from the TPM1 gene with alternate promoters but have similar actin affinities.
Figure 5.
Actin affinity and thermal stability of tropomyosins. (A and B) Binding of Tm measured by cosedimentation with F-actin. Tropomyosin (0.1–8 μM) was combined with 5 μM F-actin and sedimented at 20 °C in 200 mM NaCl, 10 mM Tris-HCl (pH 7.5), 2 mM MgCl2, and 0.5 mM DTT. Stoichiometric binding of one Tm per seven actins is represented by a maximal fraction of binding of 1. The data for each isoform were obtained from two to four independent experiments. The Kapp and Hill coefficient (αH) values are listed in Table 3. (C and D) Fraction folded as measured by the relative ellipticity at 222 nm as a function of temperature (0–65 °C). The Tm concentration was 1.5 μM in 0.5 M NaCl, 10 mM sodium phosphate (pH 7.5), 1 mM EDTA, and 1 mM DTT. The fraction folded is relative to the mean residue ellipticity at 0 °C, where the proteins were fully folded. The TM values are listed in Table 3.
Table 3. Actin Affinities and Thermal Stabilities of Tropomyosins.
| Kappa (×106 M–1) | αHa | TMb (°C) | |
|---|---|---|---|
| AS-Tmstα | 3.5 ± 0.4 | 2.3 ± 0.5 | 44.0 |
| Tm2 | 1.2 ± 0.0 | 5.8 ± 0.7 | 40.0 |
| Tm5a | 1.7 ± 0.1 | 1.8 ± 0.1 | 36.0 |
| Tm5NM1 | 1.8 ± 0.1 | 2.7 ± 0.5 | 42.5 |
| Tm4 | 0.2 ± 0.0 | 2.5 ± 0.5 | 45.5 |
The values for the binding constant of Tm to actin, Kapp ± the standard error, and the Hill coefficient, αH ± the standard error, from two or three experiments (Figure 5). The data were fit to the Hill equation, and the Kapp and αH are those reported by Kaleidagraph.
TM is the temperature at which the ellipticity at 222 nm, normalized to a scale of 0–1, is equal to 0.5 (Figure 5).
The thermal stability of the Tm isoforms was determined by following the ellipticity at 222 nm as a function of temperature using CD spectroscopy (Figure 5C,D). The Tm isoforms had varying thermal stabilities, and the overall melting temperature, TM, ranged from 36 °C for Tm5a to 45.5 °C for Tm4 (Table 3), consistent with previous studies.59 Tropomyosin undergoes multiple thermal transitions, where the first and minor transition corresponds to unfolding of the middle of the molecule (residues ∼130–190), the second and major transition corresponds to unfolding of the rest of the molecule or the C-terminal region, and the third transition (when present) corresponds to unfolding of the N-terminal region.51 The thermal stability of the Tms does not correlate with actin affinity, allaying concerns that the differences in stability may produce global conformational changes that can affect actin affinity. For example, Tm5a and Tm5NM1 have an ∼6 °C difference in their thermal stabilities but similar actin affinities.
Discussion
Tropomyosins can regulate actomyosin by inhibiting or activating actomyosin MgATPase activity and motility depending on the myosin and Tm isoforms.10,16,19,20,44 In this study, we have compared the regulation of three nonmuscle myosin II isoforms, IIA, IIB, and IIC, by nonmuscle Tm isoforms Tm2, Tm5a, Tm5NM1, and Tm4 by determining the velocities of actin filaments in in vitro motility assays. We also looked at skeletal myosin and striated αTm (AS-Tmstα) isoforms, in addition to the nonmuscle isoforms mentioned above. The velocities of actin filaments were differentially regulated depending on the myosin and Tm isoform. With skeletal myosin II, filament velocities were inhibited in the presence of AS-Tmstα, were unaffected in the presence Tm2 and Tm4, and were activated in the presence Tm5NM1 and Tm5a. AS-Tmstα (1a-2b-6b-9a) and Tm2 (1a-2b-6b-9d) are HMW Tms encoded by the same (TPM1) gene and differ only in the sequence of the ninth exon. Tm5a (1b-6b-9d) is a LMW Tm also encoded by the TPM1 gene that has the same ninth exon as Tm2 but expresses an alternate first exon (1b) from AS-Tmstα and Tm2 (1a and 2b). This implies that inhibition is chiefly determined by the presence of exon 9a (AS-Tmstα), which is primarily a striated muscle-specific exon, whereas the presence of exon 9d does not cause inhibition (Tm2) and may cause activation (Tm5a). All three Tms are unacetylated, but AS-Tmstα has an N-terminal Ala-Ser. The presence of the N-terminal Ala-Ser may also be a contributing factor to the inhibition of velocity by AS-Tmsta in contrast to Tm2, which does not inhibit, with skeletal myosin. With NMIIA HMM, filament velocities in the presence of the LMW isoforms, Tm5a, Tm5NM1, and Tm4, were activated ∼2-fold (P < 0.05) compared to the velocities in the presence of HMW isoforms, AS-Tmstα and Tm2 (Figure 2 and Table 1). On the basis of these results, the primary determinant of activation of NMIIA is the presence of exon 1b (LMW Tms) as opposed to exon 1a and exon 2 (HMW Tms) or the size of the Tms (Figure 1). Tm isoforms did not significantly affect the filament velocity of NMIIB HMM. Inhibition of filament velocity was observed only for skeletal myosin in the presence of striated αTm, indicating that the inhibition of actomyosin activity by Tm is specific to striated muscle isoforms of Tm and myosin. Striated muscle myosin is regulated by Tm–troponin in a Ca2+-dependent manner, whereas smooth muscle and nonmuscle forms of myosins II are primarily regulated by phosphorylation of their regulatory light chains.2,8,60
The MgATPase activity of NMIIA and NMIIC HMMs was enhanced, while that of NMIIB HMM was unaffected in the presence of a nonmuscle Tm isoform, Tm5NM1, in support of the motility data. The enhancement of MgATPase rates was due to an ∼20–40% increase in Vmax accompanied by a 3–5-fold decrease in KATPase. This is similar to the activation of MgATPase activity of smooth muscle myosin by smooth muscle Tm, which is accompanied by an increase in Vmax and a decrease in KATPase,61 and in contrast to the regulation of skeletal myosin by striated αTm, where activation of the MgATPase is largely due to a decrease in KATPase with no effect on Vmax.62 Thus, nonmuscle II and smooth muscle myosins are regulated by Tms by a mechanism that infers a direct effect of Tm on myosin in contrast to skeletal myosin, which is thought to be regulated by Tm via a predominantly steric effect through the actin filament. Note, however, that the kinetic step that regulates the rate of in vitro motility is likely to be the release of ADP from the A·M·ADP complex, whereas the step that regulates actin-activated MgATPase activity is likely to be the release of Pi from the A·M·ADP·Pi complex. Thus, there is no reason that tropomyosin would have the same effect on the two measurements.63
A noteworthy outcome of our research is that various Tm isoforms have differing affects on the motility and ATPase activities of the three nonmuscle myosin HMM isoforms. While these isoforms do have the common property of having comparatively slow actin translocation rates and ATPase activities, there are significant kinetic differences among them. For example, NMIIB has a duty ratio significantly higher than those of NMIIA and NMIIC, and NMIIA is faster enzymatically than the other two.26,28,64 It is not possible to speculate about the structural basis for the differences in the motility or ATPase activities because there are no crystal structures available for any of these myosins.
In striated muscle, regulation of binding of myosin to the thin filament (actin–Tm–Tn) is described by a three-state model, in which the three kinetic and structural states are defined as (1) the blocked state, in which in the absence of Ca2+, Tm blocks the interaction of myosin with actin, (2) the closed state, in which in the presence of Ca2+, Tm shifts azimuthally on the actin filament to partially expose the myosin binding site and allow weak myosin binding, and (3) the open state, in which in the presence of myosin, Tm azimuthally shifts further on the actin filament, allowing strong myosin binding and force production.65−68 The binding of myosin to thin filaments shifts the equilibrium from the closed to open state in which both Tm and myosin bind to actin with higher affinity.3,6 The cooperative activation of the thin filament by myosin from the closed to open state can be produced by Tm alone, independent of Tn and Ca2+. In the absence of Tn and myosin, Tms occupy the blocked or closed positions on the actin filament, depending on the Tm isoform.69 The open position is occupied only in the presence of myosin. The structural transition of Tm from the blocked/closed position to the open position in the presence of myosin may modulate the regulation of actomyosin in an isoform-specific manner. Tropomyosins that occupy average positions closer to the open state may be easier to switch to the open state, thus producing activation of the actomyosin. However, the average position of Tm5a, a short Tm isoform that activates filament velocities with skeletal myosin, was reported to be in the closed state, similar to Tmstα, which inhibits velocities with skeletal myosin.69 In the study presented here, both Tm5NM1 and Tm5a activated filament velocities with skeletal myosin and NMIIA and NMIIC HMMs, suggesting that they may occupy similar positions on the actin filament. Additionally, Tm5NM1 activated the motility of actin filaments at all myosin concentrations ranging from 0.1 to 80 μg/mL. The differences in the position of Tms on the actin filament and cooperativity are most likely related to differences in sequence of the Tm isoforms as well as subtle differences in the structure of the ends in the overlap complex such as the extent of overlap and flexibility70−73 and may be responsible for the differential effect of Tm isoforms on the ATPase rate and motility of actomyosin.
Actomyosin bundles form the contractile component of stress fibers that are essential for the formation of focal adhesions and cellular movement during cell migration.8 Actomyosin filaments are also a major component of the contractile ring and play an important role in the constriction of the ring during cell division.8 The enhancement of ATPase rates as well as the translocation rates of nonmuscle myosin IIA by specific Tm isoforms introduces an additional way to modulate the actomyosin interaction, apart from phosphorylation of the regulatory light chain, while conducting these cellular functions. In addition to the biochemical regulation of actomyosin, Tm may also be involved in the mechanical regulation of actomyosin through an increase in force production. This is of particular interest in cancer, where tumor cells alter the mechanical properties of the extracellular matrix using the contractile properties of focal adhesions and stress fibers to aid cell proliferation and metastasis.74 Knowing the localization of Tm isoforms in relation to myosin isoforms at the level of single filaments and/or stress fibers would provide insight into the specificity of regulation.
Acknowledgments
We thank Prof. Don Winkelmann for use of the instrument and analysis software for in vitro motility assays and for discussions and help with motility data analysis. We thank Dr. Fang Zhang for technical assistance.
Glossary
Abbreviations
- Tm and Tpm
tropomyosin
- Tn
troponin
- HMM
heavy meromyosin
- CD
circular dichroism
- TM
melting temperature
- NM
nonmuscle
- MLCK
myosin light chain kinase
- Tmstα and Tpm1.1st
Tpm1.1st(a.b.b.a)
- Tm2 and Tpm1.6cy
Tpm1.6cy(a.b.b.d)
- Tm5a and Tpm1.8cy
Tpm1.8cy(b.-.b.d.)
- Tm5NM1 and Tpm3.1cy
Tpm3.1cy(b.-.a.d)
- Tm4 and Tpm4.2cy
Tpm4.2cy(b.-.b.d).
Supporting Information Available
Velocity probability distributions for actin, actin–AS-Tmstα, and actin–Tm5NM1 on skeletal myosin and NMIIA HMM (Figure S1), speeds of individual actin, actin–AS-Tmstα, and actin–Tm5NM1 filaments as a function of filament length on skeletal myosin and NMIIA HMM (Figure S2), and videos showing in vitro motility of actin, actin–AS-Tmstα, and actin–Tm5NM1 filaments on skeletal myosin and NMIIA HMM [Movies S1 (parts A–C) and S2 (parts A–C)]. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This work was supported by National Institutes of Health (NIH) Grant GM093065 to S.E.H.-D. and NIH Intramural Project HL001786 (J.R.S.).
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Engelhardt W. A.; Ljubimowa M. N. (1939) Myosine and adenosinetriphosphatase. Nature 144, 668–669. [Google Scholar]
- Gordon A. M.; Homsher E.; Regnier M. (2000) Regulation of contraction in striated muscle. Physiol. Rev. 80, 853–924. [DOI] [PubMed] [Google Scholar]
- Bremel R. D.; Weber A. (1972) Cooperation within actin filament in vertebrate skeletal muscle. Nat. New Biol. 238, 97–101. [DOI] [PubMed] [Google Scholar]
- Wang C. L.; Coluccio L. M. (2010) New insights into the regulation of the actin cytoskeleton by tropomyosin. Int. Rev. Cell Mol. Biol. 281, 91–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne D. (2008) Tropomyosin function in yeast. Adv. Exp. Med. Biol. 644, 168–186. [DOI] [PubMed] [Google Scholar]
- Eaton B. L. (1976) Tropomyosin binding to F-actin induced by myosin heads. Science 192, 1337–1339. [DOI] [PubMed] [Google Scholar]
- Gunning P.; O’Neill G.; Hardeman E. (2008) Tropomyosin-based regulation of the actin cytoskeleton in time and space. Physiol. Rev. 88, 1–35. [DOI] [PubMed] [Google Scholar]
- Heissler S. M.; Manstein D. J. (2013) Nonmuscle myosin-2: Mix and match. Cell. Mol. Life Sci. 70, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehrer S. S.; Morris E. P. (1982) Dual effects of tropomyosin and troponin-tropomyosin on actomyosin subfragment 1 ATPase. J. Biol. Chem. 257, 8073–8080. [PubMed] [Google Scholar]
- Lehrer S. S.; Morris E. P. (1984) Comparison of the effects of smooth and skeletal tropomyosin on skeletal actomyosin subfragment 1 ATPase. J. Biol. Chem. 259, 2070–2072. [PubMed] [Google Scholar]
- Williams D. L. Jr.; Greene L. E.; Eisenberg E. (1984) Comparison of effects of smooth and skeletal muscle tropomyosins on interactions of actin and myosin subfragment 1. Biochemistry 23, 4150–4155. [DOI] [PubMed] [Google Scholar]
- Miyata H.; Chacko S. (1986) Role of tropomyosin in smooth muscle contraction: Effect of tropomyosin binding to actin on actin activation of myosin ATPase. Biochemistry 25, 2725–2729. [DOI] [PubMed] [Google Scholar]
- DesMarais V.; Ichetovkin I.; Condeelis J.; Hitchcock-DeGregori S. E. (2002) Spatial regulation of actin dynamics: A tropomyosin-free, actin-rich compartment at the leading edge. J. Cell Sci. 115, 4649–4660. [DOI] [PubMed] [Google Scholar]
- Gupton S. L.; Anderson K. L.; Kole T. P.; Fischer R. S.; Ponti A.; Hitchcock-DeGregori S. E.; Danuser G.; Fowler V. M.; Wirtz D.; Hanein D.; Waterman-Storer C. M. (2005) Cell migration without a lamellipodium: Translation of actin dynamics into cell movement mediated by tropomyosin. J. Cell Biol. 168, 619–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg U.; Schutt C. E.; Goldman R. D.; Nyakern-Meazza M.; Hillberg L.; Rathje L. S.; Grenklo S. (2008) Tropomyosins regulate the impact of actin binding proteins on actin filaments. Adv. Exp. Med. Biol. 644, 223–231. [DOI] [PubMed] [Google Scholar]
- Ostap E. M. (2008) Tropomyosins as discriminators of myosin function. Adv. Exp. Med. Biol. 644, 273–282. [DOI] [PubMed] [Google Scholar]
- Tang N.; Ostap E. M. (2001) Motor domain-dependent localization of myo1b (myr-1). Curr. Biol. 11, 1131–1135. [DOI] [PubMed] [Google Scholar]
- Collins K.; Sellers J. R.; Matsudaira P. (1990) Calmodulin dissociation regulates brush border myosin I (110-kD-calmodulin) mechanochemical activity in vitro. J. Cell Biol. 110, 1137–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J. E.; Sammons M. R.; Stark B. C.; Hodges A. R.; Lord M. (2010) Differential regulation of unconventional fission yeast myosins via the actin track. Curr. Biol. 20, 1423–1431. [DOI] [PubMed] [Google Scholar]
- Fanning A. S.; Wolenski J. S.; Mooseker M. S.; Izant J. G. (1994) Differential regulation of skeletal muscle myosin-II and brush border myosin-I enzymology and mechanochemistry by bacterially produced tropomyosin isoforms. Cell Motil. Cytoskeleton 29, 29–45. [DOI] [PubMed] [Google Scholar]
- Clayton J. E.; Pollard L. W.; Sckolnick M.; Bookwalter C. S.; Hodges A. R.; Trybus K. M.; Lord M. (2013) Fission yeast tropomyosin specifies directed transport of myosin-V along actin cables. Mol. Biol. Cell 25, 66–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges A. R.; Krementsova E. B.; Bookwalter C. S.; Fagnant P. M.; Sladewski T. E.; Trybus K. M. (2012) Tropomyosin is essential for processive movement of a class V myosin from budding yeast. Curr. Biol. 22, 1410–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolenski J. S.; Cheney R. E.; Mooseker M. S.; Forscher P. (1995) In vitro motility of immunoadsorbed brain myosin-V using a Limulus acrosomal process and optical tweezer-based assay. J. Cell Sci. 108(Part 4), 1489–1496. [DOI] [PubMed] [Google Scholar]
- Golomb E.; Ma X.; Jana S. S.; Preston Y. A.; Kawamoto S.; Shoham N. G.; Goldin E.; Conti M. A.; Sellers J. R.; Adelstein R. S. (2004) Identification and characterization of nonmuscle myosin II-C, a new member of the myosin II family. J. Biol. Chem. 279, 2800–2808. [DOI] [PubMed] [Google Scholar]
- Kim K. Y.; Kovacs M.; Kawamoto S.; Sellers J. R.; Adelstein R. S. (2005) Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J. Biol. Chem. 280, 22769–22775. [DOI] [PubMed] [Google Scholar]
- Kovacs M.; Wang F.; Hu A.; Zhang Y.; Sellers J. R. (2003) Functional divergence of human cytoplasmic myosin II: Kinetic characterization of the non-muscle IIA isoform. J. Biol. Chem. 278, 38132–38140. [DOI] [PubMed] [Google Scholar]
- Nagy A.; Takagi Y.; Billington N.; Sun S. A.; Hong D. K.; Homsher E.; Wang A.; Sellers J. R. (2013) Kinetic characterization of nonmuscle myosin IIb at the single molecule level. J. Biol. Chem. 288, 709–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F.; Kovacs M.; Hu A.; Limouze J.; Harvey E. V.; Sellers J. R. (2003) Kinetic mechanism of non-muscle myosin IIB: Functional adaptations for tension generation and maintenance. J. Biol. Chem. 278, 27439–27448. [DOI] [PubMed] [Google Scholar]
- Rosenfeld S. S.; Xing J.; Chen L. Q.; Sweeney H. L. (2003) Myosin IIb is unconventionally conventional. J. Biol. Chem. 278, 27449–27455. [DOI] [PubMed] [Google Scholar]
- Jana S. S.; Kawamoto S.; Adelstein R. S. (2006) A specific isoform of nonmuscle myosin II-C is required for cytokinesis in a tumor cell line. J. Biol. Chem. 281, 24662–24670. [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M.; Koach M. A.; Whitmore L.; Lamers M. L.; Horwitz A. F. (2008) Segregation and activation of myosin IIB creates a rear in migrating cells. J. Cell Biol. 183, 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maupin P.; Phillips C. L.; Adelstein R. S.; Pollard T. D. (1994) Differential localization of myosin-II isozymes in human cultured cells and blood cells. J. Cell Sci. 107(Part 11), 3077–3090. [DOI] [PubMed] [Google Scholar]
- Chrzanowska-Wodnicka M.; Burridge K. (1996) Rho-stimulated contractility drives the formation of stress fibers and focal adhesions. J. Cell Biol. 133, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M.; Ma X.; Adelstein R. S.; Horwitz A. R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M.; Zareno J.; Whitmore L.; Choi C. K.; Horwitz A. F. (2007) Regulation of protrusion, adhesion dynamics, and polarity by myosins IIA and IIB in migrating cells. J. Cell Biol. 176, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betapudi V. (2010) Myosin II motor proteins with different functions determine the fate of lamellipodia extension during cell spreading. PLoS One 5, e8560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach J. R.; Shao L.; Remmert K.; Li D.; Betzig E.; Hammer J. A. (2014) Nonmuscle Myosin II Isoforms Coassemble in Living Cells. Curr. Biol. 24, 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente-Manzanares M.; Horwitz A. R. (2010) Myosin light chain mono- and di-phosphorylation differentially regulate adhesion and polarity in migrating cells. Biochem. Biophys. Res. Commun. 402, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubow K. E.; Conrad S. K.; Horwitz A. R. (2013) Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr. Biol. 23, 1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S.; Gateva G.; Schevzov G.; Hotulainen P.; Naumanen P.; Martin C.; Gunning P. W.; Lappalainen P. (2011) A molecular pathway for myosin II recruitment to stress fibers. Curr. Biol. 21, 539–550. [DOI] [PubMed] [Google Scholar]
- Bryce N. S.; Schevzov G.; Ferguson V.; Percival J. M.; Lin J. J.; Matsumura F.; Bamburg J. R.; Jeffrey P. L.; Hardeman E. C.; Gunning P.; Weinberger R. P. (2003) Specification of actin filament function and molecular composition by tropomyosin isoforms. Mol. Biol. Cell 14, 1002–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppinga R. D.; Li Y.; Lin J. L.; Lin J. J. (2006) Tropomyosin and caldesmon regulate cytokinesis speed and membrane stability during cell division. Arch. Biochem. Biophys. 456, 161–174. [DOI] [PubMed] [Google Scholar]
- Bach C. T.; Creed S.; Zhong J.; Mahmassani M.; Schevzov G.; Stehn J.; Cowell L. N.; Naumanen P.; Lappalainen P.; Gunning P. W.; O’Neill G. M. (2009) Tropomyosin isoform expression regulates the transition of adhesions to determine cell speed and direction. Mol. Cell. Biol. 29, 1506–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark B. C.; Sladewski T. E.; Pollard L. W.; Lord M. (2010) Tropomyosin and myosin-II cellular levels promote actomyosin ring assembly in fission yeast. Mol. Biol. Cell 21, 989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A.; Hitchcock-DeGregori S. E. (2003) Local destabilization of the tropomyosin coiled coil gives the molecular flexibility required for actin binding. Biochemistry 42, 14114–14121. [DOI] [PubMed] [Google Scholar]
- Hitchcock-De Gregori S. E.; Mandala S.; Sachs G. A. (1982) Changes in actin lysine reactivities during polymerization detected using a competitive labeling method. J. Biol. Chem. 257, 12573–12580. [PubMed] [Google Scholar]
- Winkelmann D. A.; Kinose F.; Chung A. L. (1993) Inhibition of actin filament movement by monoclonal antibodies against the motor domain of myosin. J. Muscle Res. Cell Motil. 14, 452–467. [DOI] [PubMed] [Google Scholar]
- Winkelmann D. A.; Lowey S.; Press J. L. (1983) Monoclonal antibodies localize changes on myosin heavy chain isozymes during avian myogenesis. Cell 34, 295–306. [DOI] [PubMed] [Google Scholar]
- Wang F.; Harvey E. V.; Conti M. A.; Wei D.; Sellers J. R. (2000) A conserved negatively charged amino acid modulates function in human nonmuscle myosin IIA. Biochemistry 39, 5555–5560. [DOI] [PubMed] [Google Scholar]
- Hammell R. L.; Hitchcock-DeGregori S. E. (1996) Mapping the functional domains within the carboxyl terminus of α-tropomyosin encoded by the alternatively spliced ninth exon. J. Biol. Chem. 271, 4236–4242. [DOI] [PubMed] [Google Scholar]
- Greenfield N. J.; Hitchcock-DeGregori S. E. (1995) The stability of tropomyosin, a two-stranded coiled-coil protein, is primarily a function of the hydrophobicity of residues at the helix-helix interface. Biochemistry 34, 16797–16805. [DOI] [PubMed] [Google Scholar]
- Barua B.; Winkelmann D. A.; White H. D.; Hitchcock-DeGregori S. E. (2012) Regulation of actin-myosin interaction by conserved periodic sites of tropomyosin. Proc. Natl. Acad. Sci. U.S.A. 109, 18425–18430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann D. A.; Bourdieu L.; Ott A.; Kinose F.; Libchaber A. (1995) Flexibility of myosin attachment to surfaces influences F-actin motion. Biophys. J. 68, 2444–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdieu L.; Magnasco M. O.; Winkelmann D. A.; Libchaber A. (1995) Actin filaments on myosin beds: The velocity distribution. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 52, 6573–6579. [DOI] [PubMed] [Google Scholar]
- Wang Q.; Moncman C. L.; Winkelmann D. A. (2003) Mutations in the motor domain modulate myosin activity and myofibril organization. J. Cell Sci. 116, 4227–4238. [DOI] [PubMed] [Google Scholar]
- Monteiro P. B.; Lataro R. C.; Ferro J. A.; Reinach F. C. (1994) Functional α-tropomyosin produced in Escherichia coli. A dipeptide extension can substitute the amino-terminal acetyl group. J. Biol. Chem. 269, 10461–10466. [PubMed] [Google Scholar]
- Schevzov G.; Bryce N. S.; Almonte-Baldonado R.; Joya J.; Lin J. J.; Hardeman E.; Weinberger R.; Gunning P. (2005) Specific features of neuronal size and shape are regulated by tropomyosin isoforms. Mol. Biol. Cell 16, 3425–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehn J. R.; Haass N. K.; Bonello T.; Desouza M.; Kottyan G.; Treutlein H.; Zeng J.; Nascimento P. R.; Sequeira V. B.; Butler T. L.; Allanson M.; Fath T.; Hill T. A.; McCluskey A.; Schevzov G.; Palmer S. J.; Hardeman E. C.; Winlaw D.; Reeve V. E.; Dixon I.; Weninger W.; Cripe T. P.; Gunning P. W. (2013) A novel class of anticancer compounds targets the actin cytoskeleton in tumor cells. Cancer Res. 73, 5169–5182. [DOI] [PubMed] [Google Scholar]
- Moraczewska J.; Nicholson-Flynn K.; Hitchcock-DeGregori S. E. (1999) The ends of tropomyosin are major determinants of actin affinity and myosin subfragment 1-induced binding to F-actin in the open state. Biochemistry 38, 15885–15892. [DOI] [PubMed] [Google Scholar]
- Sellers J. R.; Knight P. J. (2007) Folding and regulation in myosins II and V. J. Muscle Res. Cell Motil. 28, 363–370. [DOI] [PubMed] [Google Scholar]
- Chacko S.; Eisenberg E. (1990) Cooperativity of actin-activated ATPase of gizzard heavy meromyosin in the presence of gizzard tropomyosin. J. Biol. Chem. 265, 2105–2110. [PubMed] [Google Scholar]
- Williams D. L. Jr.; Greene L. E.; Eisenberg E. (1988) Cooperative turning on of myosin subfragment 1 adenosinetriphosphatase activity by the troponin-tropomyosin-actin complex. Biochemistry 27, 6987–6993. [DOI] [PubMed] [Google Scholar]
- Yengo C. M.; Takagi Y.; Sellers J. R. (2012) Temperature dependent measurements reveal similarities between muscle and non-muscle myosin motility. J. Muscle Res. Cell Motil. 33, 385–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heissler S. M.; Manstein D. J. (2011) Comparative kinetic and functional characterization of the motor domains of human nonmuscle myosin-2C isoforms. J. Biol. Chem. 286, 21191–21202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrmann E.; Muller M.; Penczek P. A.; Mannherz H. G.; Manstein D. J.; Raunser S. (2012) Structure of the rigor actin-tropomyosin-myosin complex. Cell 150, 327–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heeley D. H.; Belknap B.; White H. D. (2006) Maximal activation of skeletal muscle thin filaments requires both rigor myosin S1 and calcium. J. Biol. Chem. 281, 668–676. [DOI] [PubMed] [Google Scholar]
- Lehman W.; Craig R. (2008) Tropomyosin and the steric mechanism of muscle regulation. Adv. Exp. Med. Biol. 644, 95–109. [DOI] [PubMed] [Google Scholar]
- McKillop D. F.; Geeves M. A. (1993) Regulation of the interaction between actin and myosin subfragment 1: Evidence for three states of the thin filament. Biophys. J. 65, 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman W.; Hatch V.; Korman V.; Rosol M.; Thomas L.; Maytum R.; Geeves M. A.; Van Eyk J. E.; Tobacman L. S.; Craig R. (2000) Tropomyosin and actin isoforms modulate the localization of tropomyosin strands on actin filaments. J. Mol. Biol. 302, 593–606. [DOI] [PubMed] [Google Scholar]
- Frye J.; Klenchin V. A.; Rayment I. (2010) Structure of the tropomyosin overlap complex from chicken smooth muscle: Insight into the diversity of N-terminal recognition. Biochemistry 49, 4908–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N. J.; Huang Y. J.; Swapna G. V.; Bhattacharya A.; Rapp B.; Singh A.; Montelione G. T.; Hitchcock-DeGregori S. E. (2006) Solution NMR structure of the junction between tropomyosin molecules: Implications for actin binding and regulation. J. Mol. Biol. 364, 80–96. [DOI] [PubMed] [Google Scholar]
- Greenfield N. J.; Kotlyanskaya L.; Hitchcock-DeGregori S. E. (2009) Structure of the N terminus of a nonmuscle α-tropomyosin in complex with the C terminus: Implications for actin binding. Biochemistry 48, 1272–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. E.; Orzechowski M.; Lehman W.; Fischer S. (2014) Structure and flexibility of the tropomyosin overlap junction. Biochem. Biophys. Res. Commun. 446, 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar-Cuenca R.; Juanes-Garcia A.; Vicente-Manzanares M. (2013) Myosin II in mechanotransduction: Master and commander of cell migration, morphogenesis, and cancer. Cell. Mol. Life Sci. 71, 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.