Mitochondria are highly dynamic organelles that continuously divide and fuse. These dynamic processes regulate the size, shape, and distribution of the mitochondrial network. In addition, mitochondrial division and fusion play critical roles in cell physiology. This review will focus on the dynamic process of mitochondrial division, which is highly conserved from yeast to humans. We will discuss what is known about how the essential components of the division machinery function to mediate mitochondrial division and then focus on proteins that have been implicated in division but whose functions remain unclear. We will then briefly discuss the cellular functions of mitochondrial division and the problems that arise when division is disrupted.
A dynamin-related protein drives mitochondrial division
Mitochondrial division is driven by Dnm1 in yeast and its homolog Drp1 in higher eukaryotes. Consistent with a role in the scission of mitochondrial membranes, Dnm1 and Drp1 belong to a family of dynamin related proteins (DRPs), members of which are large self-assembling GTPases that function to mediate membrane dynamics in a variety of cellular processes [1]. The prototypic and most extensively characterized member of the DRP family is dynamin-1. Likely via the generation of mechanochemical forces, dynamin-1 functions in the scission of endocytic vesicles from the plasma membrane [2, 3].
Dnm1 and Drp1, like all DRPs, possess a highly conserved GTPase domain that adopts the core fold common to all regulatory GTPases [4–6]. However, DRPs are set apart from regulatory GTPases by their large size as well as kinetic and structural properties [7]. Two functionally unique and essential features of DRPs are GTP-driven self-assembly and assembly stimulated GTP-hydrolysis. Compared to regulatory GTPases, DRPs exhibit lower affinities for GTP and GDP and faster rates of GTP hydrolysis, especially in the assembled state, obviating the need for nucleotide exchange factors and GTPase activating proteins [8, 9].
In addition to the GTPase domain, DRPs are comprised of additional regions predicted to adopt coiled-coil structures, referred to as the middle domain and GTPase effector domain (GED) [3, 10]. Both regions participate in intra- and inter-molecular interactions that are required for self-assembly and assembly-stimulated hydrolysis [11, 12]. Interestingly, the high resolution structure of the full-length bacterial dynamin-like protein, BDLP, from N. punctiforme, suggests that the middle and GED regions of the protein do not exist as structurally distinct domains. Rather, in the BDLP structure, the helixes that comprise the middle and GED regions run in parallel forming a 4-helix bundle [13]. BDLP is most closely related to the more distant DRP mitofusin/fzo family, members of which are involved in mitochondrial outer membrane fusion. Thus, the relevance of the BDLP structure for the DRP family as a whole and especially for those members more evolutionarily removed, such as the membrane scission DRPs, awaits verification.
Interestingly, assembled BDLP forms a helical array of repeating T-shaped units similar to what has been observed in three-dimensional (cryo)-EM reconstructions of assembled dynamin [13, 14]. For dynamin this repeating T unit is suggested to be a dimer in which the GTPase domain and the middle and GED regions are predicted to form the head and stalk of the T, respectively. The helical dynamin cryo-EM reconstruction suggests that the strongest interactions within and between dimers are mediated by the middle and GED regions [14, 15]. In addition to middle and GED interactions, GTPase-GTPase and GTPase-middle/GED interactions also contribute to self-assembly, which is critical for DRP function [11, 14, 15].
In yeast, Dnm1 assembles into structures that localize in a dynamic fashion to the cytosol and to the mitochondrial surface [16–19]. In contrast, the majority of Drp1 in higher eukaryotes is diffusely dispersed throughout the cytosol, while a small fraction of the protein is found assembled on mitochondria [20, 21] (Fig. 1). The assembled Dnm1/Drp1 structures are highly dynamic as there is a continuous exchange of subunits between cytosolic and assembled Dnm1/Drp1 [18, 22]. Significantly, while division does not occur at all sites of mitochondrial-associated Dnm1/Drp1 assemblies, Dnm1/Drp1 assemblies are always found at sites of mitochondrial division [17–21]. What differentiates those structures that go on to mediate division from those that do not is currently unknown. At any given time, only a small fraction of Dnm1/Drp1 is participating in division, suggesting roles for these proteins outside of mitochondrial division.
Figure 1. The cellular distribution of Dnm1 and Drp1.
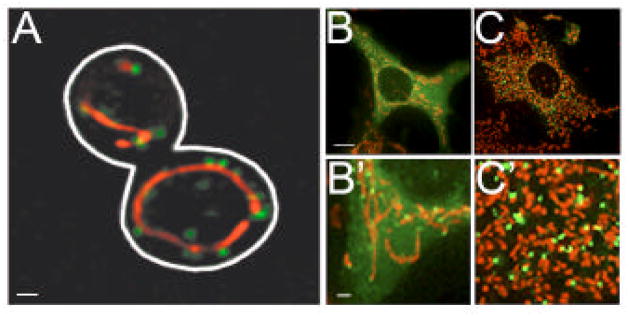
A) In yeast, Dnm1 is predominantly found in self-assembled structures, a majority of which are associated with mitochondria. B and C) Under normal growth conditions (B and B′), the bulk of Drp1 in COS cells is diffusely dispersed in the cytosol with only a small fraction of the protein found assembled on mitochondria. Following STS treatment (C and C′), the assembly and mitochondrial targeting of Drp1 are markedly increased resulting in increased mitochondrial division and fragmentation. Panels B′ and C′ are a representative region of each cell shown in panels B and C and are magnified 7-fold. Dnm1 and Drp1 are shown in green and mitochondria are shown in red. Panels B, B′, C and C′ are reproduced from Cassidy- Stone et al. 2008, Developmental Cell [95]. Scale bars: (A) 1 μm, (B and C) 10 μm, (B′and C′) 1 μm.
Assembly of Dnm1/Drp1 is required for its function [23, 24]. A thorough structural characterization of Dnm1 has provided mechanistic insight into the relationship between assembly and division [24]. In its GTP-bound form, Dnm1 assembles into extended helical structures that are markedly different than the slightly curved filaments formed by nucleotide-free and GDP-bound forms of the protein. While similar to the helices formed by dynamin [25], the Dnm1 helices possess a larger diameter that, remarkably, is equivalent to the diameter of mitochondrial constriction sites in vivo. Dnm1 can also assemble on and constrict artificial liposomes in vitro, forming tubules with diameters similar to mitochondrial constriction sites in vivo. Thus, these data indicate that GTP-driven Dnm1 assembly drives mitochondrial membrane constriction. Dnm1 GTPase mutants that are defective in GTP-binding do not assemble into helices in vitro and do not support division in vivo [19]. Thus, GTP-mediated structural changes are required for helix formation, which in turn is required for mitochondrial division.
Two regulatory features of GTP-driven Dnm1 assembly have been revealed by a detailed kinetic analysis of the hydrolysis activity of the protein, which is intimately tied to self-assembly [24]. In the early steps of the assembly pathway, Dnm1 assembly is positively regulated by GTP-dependent conformational changes and proceeds via a rate-limiting nucleation step. These kinetic features suggest that a rate-limiting event in division is the regulation of Dnm1 self-assembly. Together, the structural and kinetic characterizations of Dnm1 support a model in which nucleation-dependent self-assembly of Dnm1 into helices drives the constriction of the mitochondrial membranes during mitochondrial division.
Drp1 has also been shown to self-assemble in vitro, albeit with less efficiency than Dnm1, and only into simple rings [20, 26]. This suggests additional factors may be required to regulate Drp1 self-assembly. The difference in the assembled states of Dnm1 and Drp1 in vivo also suggest a higher degree of regulation for Drp1 assembly. While Dnm1 is predominantly found in self-assembled structures, the majority of which are mitochondrial associated, the bulk of Drp1 is diffusely dispersed in the cytosol [16, 17, 20, 21] (Fig. 1). A higher degree of regulation may be required to achieve the dramatic and physiologically relevant variation in mitochondrial structure and dynamics observed in various mammalian cell types [27]. In addition, a more fine tuned regulation of assembly is likely to be important in regulating the mitochondrial functions of Drp1 outside of division, such as in apoptosis. While regulation of assembly may be more complex for Drp1, purified Drp1, like Dnm1, can assemble on and tubulate liposomes in vitro [26]. Thus, the mechanism of assembly-driven constriction in mitochondrial division is conserved between yeast and higher eukaryotes.
While GTP binding is critical for assembly-driven constriction, GTP hydrolysis is likely to be required for subsequent membrane fission. When GTP is exchanged for a bound non-hydrolyzable GTP analog, GMPPCP, Dnm1 helices rapidly contort and disassemble [24]. This raises the possibility that structural changes associated with nucleotide hydrolysis may be used to facilitate fission. A role for nucleotide hydrolysis in membrane fission is highlighted in studies using the hydrolysis deficient mutant Dnm1K41A [19]. This Dnm1 mutant, which can bind but not hydrolyze GTP, assembles into helices in vitro and on mitochondria in vivo but does not support division. Thus, GTP binding is sufficient for assembly, but GTP hydrolysis is critical to complete division. For dynamin, nucleotide hydrolysis has been coupled to a decrease in the pitch and diameter of the helix [28, 29]. This constriction has been proposed to provide the mechanochemical force necessary for fission. Data suggests that the hydrolysis-driven constriction of the dynamin helix may be a consequence of a rotary movement of the helical turns relative to one another or a conformational change involving the sliding and twisting of individual subunits of the dynamin helix, which are not mutually exclusive possibilities. Such twisting movements would likely place a strain on the lipid bilayer that could result in fission [15, 30]. Indeed, dynamin can drive the scission of lipid tubules in a GTP hydrolysis-dependent manner [30]. Interestingly, however, hydrolysis-driven constriction alone is not sufficient for membrane scission; tension along the lipid tubule is also required [30].
Thus, the current working model of mitochondrial division is that after Dnm1/Drp1 is targeted to mitochondria, it undergoes GTP-driven assembly into helical structures, and this self-assembly drives membrane constriction. In turn, self-assembly also stimulates GTP hydrolysis, which is likely to evoke conformational changes within the Dnm1/Drp1 helix that complete membrane fission (Fig. 2). Although it is the master regulator of mitochondrial division, in vivo, Dnm1/Drp1 alone is not sufficient to mediate this process. A myriad of players have been identified that are required for or help to facilitate Dnm1/Drp1-driven mitochondrial division.
Figure 2. A model of Dnm1/Drp1-driven mitochondrial division.
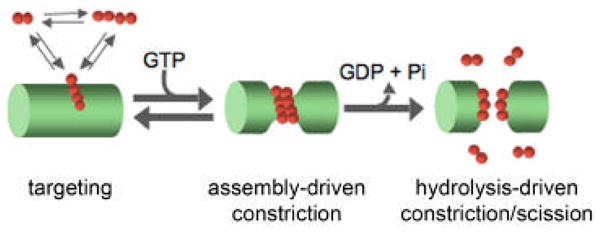
Dnm1/Drp1-driven mitochondrial division can be broken down into 3 stages: targeting, assembly-driven constriction, and hydrolysis-mediated constriction/scission. Briefly, Dnm1/Drp1 is first targeted to the mitochondrial surface. Once targeted, Dnm1/Drp1 undergoes GTP-driven assembly into a helical structure, which drives the constriction of the mitochondrial tubule. GTP-driven assembly also stimulates nucleotide hydrolysis which is likely to evoke additional conformational changes in the Dnm1 helix that are required for further constriction and subsequent scission of the mitochondrial membranes. Dnm1/Drp1 is shown in red and a portion of a mitochondrial tubule is shown in green.
Targeting of the mitochondrial division dynamin
Dnm1-dependent mitochondrial division requires additional players to target the mitochondrial division dynamin to the mitochondrial surface. In yeast, there are at least three ways in which Dnm1 can be targeted to the mitochondrial outer membrane; however, not all means of targeting result in efficient mitochondrial division.
Fis1/Mdv1-dependent targeting was the first targeting mechanism to be identified and to date is the only one known to be essential for division. Fis1 is anchored to the mitochondrial outer membrane via a C-terminal transmembrane domain [31]. The N-terminus of the protein, which is exposed to the cytosol, adopts a super helical tetratricopepetide-like (TPR) fold [32–34]. Consistent with the known role of TPR motifs in mediating protein-protein interactions, Fis1 interacts with and consequently recruits Mdv1 to the mitochondrial outer membrane [31, 35–40]. Mdv1 is comprised of 3 regions: an N-terminal extension that contains a helix-loop-helix motif that interacts with Fis1, a central coiled-coil region that mediates Mdv1 homo-oligomerization, and a C-terminal WD repeat predicted to form a seven-bladed propeller that interacts with Dnm1 [35, 36, 40–42]. Thus Mdv1 serves as a bridge between mitochondrial-associated Fis1 and soluble Dnm1.
Fis1 also functions to target Dnm1 to the mitochondrial outer membrane via its interaction with the Mdv1 paralog Caf4 [37]. Caf4 possesses the same domain architecture as Mdv1 and participates in analogous intra- and inter-molecular interactions. While structurally similar, Mdv1 and Caf4 are not functionally redundant as recruitment mediated by Caf4 alone is not productive and mitochondrial division is severely compromised in these cells. In contrast, cells lacking Caf4 exhibit wild type mitochondrial morphology and division rates [37]. At present, the role of Fis1/Caf4-dependent targeting of Dnm1 in mitochondrial division is unknown and clearly non-essential. There is evidence to suggest that the Fis1/Caf4 complex may play a role in establishing and/or maintaining a cell cortex-directed orientation of a subset of Dnm1 clusters that may have a physical attachment to the cell cortex [43]. Although the biological significance of such cortex tethering is unclear at this point, it is conceivable that tethering may serve a function in mitochondrial motility and retention during transport of the organelle to the incipient bud.
Interestingly, Num1-dependent targeting, the third mechanism by which Dnm1 is targeted to the mitochondria, is independent of Fis1 [44]. Num1 is a large 313 kD protein that is anchored to the cell cortex via a C-terminal pleckstrin homology domain [45, 46]. While Num1 has long been known to function in nuclear segregation [45], a function for Num1 in mitochondrial division and distribution has recently been uncovered [44]. In the absence of Num1, mitochondrial division is attenuated but not completely disrupted as is seen in the absence of the essential division proteins, Dnm1, Mdv1, and Fis1. Thus, while not essential for division, Num1 facilitates division by an unknown mechanism. Num1 colocalizes with a subset of mitochondrial associated Dnm1 assemblies and these are mutually exclusive of those that contain Mdv1. Interestingly, in a fraction of cells that lack both Dnm1 and Num1, a defect in mother cell mitochondrial retention is observed suggesting a role for both proteins in mitochondrial retention/inheritance. Num1, through its interaction with Dnm1, might therefore provide a link between mitochondrial division and inheritance. In addition, the interactions of Num1 with tubulin and dynein as well as with the actin nucleator Bni1 link cytoskeletal elements with mitochondrial division and distribution [47]. A link between division and the cytoskeleton may be functionally important, as interactions with the cytoskeleton could generate tension along mitochondrial tubules that may be critical for membrane scission [30]. Although targeting of Dnm1 via the Fis1/Caf4 or Num1 pathways alone is not sufficient for consequent fission of mitochondrial membranes, it is possible that Dnm1 recruited by these pathways could then go on to participate in Fis1/Mdv1-dependent fission.
The requirements for Drp1 targeting in higher eukaryotes, which possess only a Fis1 homolog, are less clear. It has been suggested that human Fis1 (hFis1) functions in part to directly target Drp1 to the mitochondrial outer membrane. Consistently, hFis1 and Drp1 have been shown to interact [48–50]. However, Drp1 recruitment is not altered in cells in which levels of hFis1 are depleted suggesting additional mechanisms to target Drp1 exist in higher eukaryotes [51]. Interestingly, targeting of Drp1 is dramatically increased following stimulation of apoptosis suggesting that Drp1 targeting even in homeostatic conditions may require factors whose activity and/or expression is increased during apoptosis [52–54]. In addition, the mitochondrial targeting of Drp1 may be facilitated by cytoskeletal elements; disruption of the actin cytoskeleton and inactivation of the dynein/dynactin complex both disrupt targeting of Drp1 to the mitochondrial surface [55, 56].
Post-targeting roles for essential mitochondrial division proteins
While hFis1 may not be required to target Drp1 to the mitochondrial surface, mitochondrial division is blocked when hFis1 levels are reduced suggesting that hFis1 also functions downstream of Drp1 targeting [32, 51, 57]. Interestingly, overexpression of hFis1 results in mitochondrial fragmentation suggesting hFis1 activity is rate limiting in division [48, 57, 58].
In yeast the post-targeting roles/requirements of Fis1 are less clear. Fis1 is suggested to be present in the division complex but is uniformly distributed along the mitochondrial surface; Fis1 does not appear to accumulate in clusters at sites of division as is seen for Dnm1 and Mdv1 [31, 59]. Therefore, if present in the division complex, it is there at substoichiometric levels. Interestingly, in vitro a direct interaction between Fis1 and Dnm1 in the absence of Mdv1 has been observed when residues that comprise the extreme N-terminal helix of Fis1 are removed, but the in vivo significance of this observation is not known [39]. This small helix, known as the N-terminal arm, binds to a groove in the concave surface of the Fis1 TPR motif suggesting that the N-terminal arm may regulate access to a Dnm1 binding site [33, 40]. While not likely to participate in targeting, as Fis1-dependent targeting requires Mdv1 or Caf4, a Fis1-Dnm1 interaction may function to regulate Dnm1 activity post-targeting.
While Mdv1 is also required to target Dnm1 to the mitochondrial surface, the protein’s function in mitochondrial division clearly extends beyond serving to simply bridge the Dnm1-Fis1 interaction. Interestingly, Mdv1 accumulates at nascent division sites following the targeting of Dnm1 and, importantly, Mdv1 remains present with Dnm1 at the time of division [19]. A mutation in the WD domain of Mdv1, Mdv1N544R, that strengthens the Dnm1-Mdv1 interaction supports division complex assembly but not membrane scission [19]. Thus targeting alone is not sufficient; a dynamic Dnm1-Mdv1 interaction is required to facilitate fission. Interestingly, cytological and yeast two hybrid studies indicate that Mdv1 preferentially interacts with the GTP-bound, assembled form of Dnm1 suggesting that Mdv1 may facilitate division by promoting/nucleating the assembly of Dnm1 into higher-order division competent structures [19, 36].
Additional regulators of mitochondrial division
While great progress has been made in dissecting the functions of the core components of the division machinery, the future of the field lies in understanding how the function of these proteins is regulated/modulated by other protein factors.
In yeast, elucidating the functional significance of the Dnm1-Caf4 and Dnm1-Num1 interactions will likely provide insight into potential roles for Dnm1 outside of its active role in mitochondrial division. Thus, although saturated genetic selections in yeast have likely identified all of the essential components of the division machinery, additional players that regulate/modulate the activities of these essential components will likely be identified.
In higher eukaryotes, numerous proteins have been implicated in regulating the process of mitochondrial division. In the mammalian system, the next challenge will be to place these players into a cohesive pathway for mitochondrial division and to identify their roles more precisely.
One potential division effector is endophilin B1 (Bif-1/SH3GLB1), which acts downstream of Drp1 in the maintenance of mitochondrial morphology. Depletion of endophilin B1 causes mitochondria to be more connected, a phenotype consistent with attenuated mitochondrial division [60]. Endophilin B1 interacts with the pro-apoptotic protein Bax, and like Bax, endophilin B1 translocates to and forms clusters on the mitochondrial surface that co-localize with Drp1 following the induction of apoptosis [60–62]. Interestingly, endophilin B1 belongs to an endophilin family of proteins that self-assemble to form filaments that remodel membranes and, as in the case of endophilin A, co-assemble with dynamin to promote endocytosis [63, 66]. Like other endophilins, endophilin B1 possesses an N-BAR domain; such domains are thought to sense and/or induce membrane curvature [64, 65], which is consistent with the ability of endophilin B1 to tubulate liposomes in vitro [66]. Thus, endophilin B1 may function to remodel mitochondrial membranes during mitochondrial division.
Mff (mitochondrial fission factor) was recently identified as a regulator of mitochondrial division [67]. Similar to hFis1, Mff is a C-tail anchored protein of the mitochondrial outer membrane whose depletion attenuates mitochondrial division. Interestingly, the two proteins exist in separate complexes suggesting independent roles in division. While the role of Mff in mitochondrial division is unknown, it has been postulated to function in the assembly and/or organization of division complexes.
GDAP1, ganglioside-induced differentiation associated protein 1, and MTP18 have also been proposed to function as effectors of mitochondrial division. Overexpression of GDAP1, an integral protein of the mitochondrial outer membrane, promotes mitochondrial fragmentation. Mutations in GDAP1 are associated with Charcot-Marie-Toothe disease type 4A. Overexpression of the disease alleles does not cause mitochondrial fragmentation suggesting a link between impaired mitochondrial division and disease [68]. Overexpression of MTP18, a resident of the inner membrane space, also promotes mitochondrial fragmentation. This coupled with the fact that depletion of MTP18 results in elongation of mitochondria as well as in a reduction of mitochondrial associated Drp1 assemblies suggests a regulatory role for MTP18 in mitochondrial division [69, 70]. Additional insight into the functions of both GDAP1 and MTP18 is required to establish whether these proteins are indeed bona fide components/regulators of mitochondrial division.
Various post-translation modifications, including ubiquitination, sumoylation, and phosphorylation also have been convincingly shown to play regulatory roles in mitochondrial division. MARCH5, an E3 ubiquitin ligase of the mitochondrial outer membrane, ubiquitinates both Drp1 and hFis1 [71, 72]. While the exact role of ubiquitination is unknown, it has been postulated that ubiquitination by MARCH5 may regulate the stability and/or activity of these proteins. Additionally, MARCH5 may also regulate the subcellular trafficking of Drp1 and/or division complex assembly/disassembly as the mitochondrial association of Drp1 is substantially increased when dominant negative forms of MARCH5 are expressed [73].
In addition to being ubiquitinated, Drp1 is a substrate for sumoylation. Increased sumoylation of Drp1, mediated by overexpression of SUMO1 or depletion of the SUMO protease SENP5, stabilizes Drp1, and this stabilization coincides with mitochondrial fragmentation [74, 75]. Thus, sumoylation may regulate division by modulating the cellular levels of Drp1. Interestingly, sumoylation, like ubiquitination, may also regulate the subcellular trafficking of Drp1. The increased and stable association of Drp1 with the mitochondrial membrane observed following stimulation of apoptosis coincides with increased sumoylation of the protein [22].
Phosphorylation of Drp1 by a number of different kinases also regulates its activity and cellular distribution. Two sites of phosphorylation have been identified in Drp1, Ser616 and Ser637 (amino acids corresponding to splice variant 1). Ser616 is phosphorylated in mitosis by Cdk1/cyclin B1 [76]. Phosphorylation at this site has been proposed to stimulate mitochondrial division in mitosis, which may serve as a possible mechanism to ensure equal transmission of mitochondria to daughter cells. Ser637 is phosphorylated by cAMP-dependent protein kinase (PKA) and calcium/calmodulin-dependent kinase I (CaMKI) and dephosphorylation is mediated by calcineurin [50, 77–79]. The effects of phosphorylation at this site are unclear as contradicting data has been reported. Three studies provide evidence that phosphorylation at this site inhibits mitochondrial division, possibly as a result of reduced Drp1 GTPase activity and/or inhibition of the translocation of Drp1 to mitochondria [77–79]. In contrast, another group reports that phosphorylation of Ser637 stimulates both the translocation of Drp1 to mitochondria and mitochondrial division [50]. Thus although the exact mechanism of regulation via phosphorylation is unclear, it is likely that phosphorylation of Drp1 can be used as a means to positively and negatively regulate its function in division.
Cellular roles of mitochondrial division
A fundamental role of mitochondrial division in all types of cells is to maintain the proper cellular distribution of mitochondria. In both yeast and higher eukaryotes, disruption of mitochondrial division leads to an extensively interconnected and collapsed mitochondrial network that leaves many areas of the cell devoid of the organelle. This altered distribution likely has some effects on cellular homeostasis in yeast. There is a modest defect in mitochondrial DNA inheritance in mitotic cells that lack Dnm1 [80]. In addition, in sporulating yeast cells that lack the essential mitochondrial division proteins there are defects in mtDNA inheritance as well as in the inheritance of the mitochondrial compartment itself [81]. In higher eukaryotes, more adverse effects are seen when mitochondrial distribution is disrupted as a result of attenuated mitochondrial division. In C. elegans loss of Drp1 activity results in embryonic lethality due in part to improper mitochondrial segregation [21]. Mitochondrial segregation defects are also observed during Drosophila spermatogenesis when Drp1 activity is disrupted [82]. Thus, maintenance of mitochondrial distribution is likely to be crucial for development in higher eukaryotes. Proper mitochondrial distribution is also critical for neuronal function. In neurons, mitochondria are distributed from the cell body to distant regions of the cell such as dendrites and the synaptic region of the axon, locations where mitochondrial functions such as ATP production and calcium buffering are in high demand [83]. Disruption of Drp1 function in neurons drastically reduces the numbers of synaptic and dendritic mitochondria, and as a result, sustained neurotransmission and dendritic morphogenesis, respectively, are disrupted [84, 85]. While the essentiality of the mitochondrial division proteins in mammals has not been directly assessed using mouse knock-out models, a lethal mutation in human Drp1 that causes early infant mortality has been reported, underscoring the essential role this process plays in humans [86].
Recently, it has been shown that disruption of mitochondrial division in mammalian cells results in general mitochondrial dysfunction: loss of membrane potential, increase in ROS, increase in oxidized proteins, and loss of mitochondrial DNA [87–90]. These sub-lethal stresses have been shown to induce senescence-associated phenotypic changes within cells [87, 88]. Thus mitochondrial division plays an important role in maintaining normal cellular function. These observations raise the possibility that mitochondrial division proteins are more directly involved in the regulation of mitochondrial function in cells. It has also been proposed that these phenotypes may be a consequence of the role division may play in protecting mitochondria from excessive damage [90]. Data suggests that mitochondrial division may be required to isolate damaged regions of the mitochondrial tubule and/or to create an appropriately sized substrate for autophagasome formation [89, 90]. Once severed from the mitochondrial network, damaged mitochondrial fragments are not able to refuse with the network due to their low membrane potential and thus remain isolated [89]. Evidence suggests that these damaged mitochondria are then targeted for autophagic degradation. Thus, mitochondrial division may function in quality control of the mitochondrial compartment [91].
Drp1 is less assembled at steady state likely in part because Drp1 self-assembly and/or assembly associated conformational changes have been harnessed in mammalian cells to integrate mitochondrial division with cellular signaling pathways. In contrast to yeast, where Dnm1 is mostly assembled at steady state, the assembly and targeting of Drp1 can be markedly stimulated under certain physiological conditions, resulting in an increase in Drp1-dependent mitochondrial division (Fig 1). For example, increases in cytosolic calcium levels, triggered by depolarization of mitochondria, changes in the cellular membrane potential, or release of ER calcium stores, promote the assembly and mitochondrial targeting of Drp1 and consequent mitochondrial fragmentation [50, 53, 77]. The functional significance of this increase in mitochondrial division as a response to calcium is unclear. However, it does suggest that regulation of Drp1 activity provides a means to regulate the shape and function of the mitochondrial network. In addition to calcium signaling, which is used in a variety of cellular processes, other intracellular signaling pathways may be used to regulate Drp1 activity, perhaps via post-translation modifications of Drp1.
Regulation of Drp1 activity is known to be functionally important in mitochondrial mediated or intrinsic apoptosis. A critical event in intrinsic apoptosis is mitochondrial outer membrane permeabilization (MOMP), which releases factors from the inner membrane space (IMS), such as cytochrome c, that mediate downstream cell death pathways [92–94]. Concurrent with MOMP, Drp1 self-assembly and mitochondrial targeting are increased resulting in increased mitochondrial division and fragmentation [22, 52, 53]. Inhibition of Drp1-dependent mitochondrial division delays and partially inhibits apoptosis [51, 52]. This raises the possibility that Drp1-mediated mitochondrial fragmentation may be required to regulate apoptosis. However, a recent study that makes use of a small molecular inhibitor of Drp1 assembly, mdivi-1, provides evidence that the role of Drp1 in apoptosis extends beyond its role in division [95]. Treatment of cells with mdivi-1 blocks the assembly and mitochondrial translocation of Drp1 and thus the mitochondrial fragmentation normally observed following the induction of apoptosis. Attenuation of cytochrome c release is also observed in these cells, consistent with a role for Drp1 activity in MOMP. Significantly, mdivi-1 also blocks the release of cytochrome c from isolated mitochondria in cell free MOMP assays where mitochondrial division does not occur. Thus, these data indicate that Drp1 plays a direct and critical role in MOMP that is independent of its role in division. Consistently, Drp1 colocalizes with Bax, an essential effector of MOMP, on the mitochondrial outer membrane suggesting that through its interaction with Bax, Drp1 regulates MOMP by an unknown mechanism [96]. Drp1 has also been shown to function in developmental programmed cell death. Disruption of Drp1 function in both C. elegans and Drosophila leads to the survival of cells normally slated for death [97, 98]. Thus the role of Drp1 in programmed cell death is well conserved.
hFis1 is also likely to play a role in the regulation of apoptosis that is distinct from its role in division. Depletion of hFis1 inhibits Bax translocation to the mitochondrial surface and delays the onset of apoptosis [51]. Interestingly a separation of function mutation has been identified in hFis1, hFis1K148R. In contrast to overexpression of WT hFis1, which induces both mitochondrial fragmentation and apoptosis, hFis1K148R overexpression only induces mitochondrial fragmentation, not apoptosis [58, 99]. Thus, the functions of hFis1 in division and apoptosis can be separated. While the mitochondrial division proteins likely have functions in apoptosis outside of their roles in division, it is conceivable that, while not required, mitochondrial fragmentation may facilitate MOMP and the progression of apoptosis.
Dnm1/Drp1 independent mitochondrial division
While Dnm1/Drp1-mediated mitochondrial division is the primary mechanism used to divide mitochondria, Dnm1/Drp1-independent mitochondrial division has been observed. In the absence of Dnm1, mitochondrial division can occur in yeast as a result of cytokinesis suggesting that external forces, akin to those generated by conformational changes of the Dnm1 helix, are sufficient to promote the scission of both mitochondrial membranes (J. Nunnari, unpublished data). In addition to cytokinetic mitochondrial division, other means of Dnm1/Drp1-independent mitochondrial division have also been observed. Mitochondria fragment in mature tetrads during sporulation in yeast in the absence of Dnm1 [81]. In mammalian cells, Drp1-independent mitochondrial fragmentation is observed in cells with decreased levels of LETM1, a mitochondrial inner membrane protein proposed to function in the regulation of mitochondrial morphology [100]. These observations suggest that novel mechanisms of mitochondrial division exist in yeast and higher eukaryotes.
Acknowledgments
We would like to thank Ann Cassidy-Stone for her comments on the manuscript. J.M.N. is supported by the NIH grant 5R01GM062942. L.L.L. is supported by the NIH postdoctoral fellowship 1F32GM078749.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- 2.Schmid SL. Clathrin-coated vesicle formation and protein sorting: an integrated process. Annu Rev Biochem. 1997;66:511–548. doi: 10.1146/annurev.biochem.66.1.511. [DOI] [PubMed] [Google Scholar]
- 3.Hinshaw JE. Dynamin and its role in membrane fission. Annu Rev Cell Dev Biol. 2000;16:483–519. doi: 10.1146/annurev.cellbio.16.1.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klockow B, Tichelaar W, Madden DR, Niemann HH, Akiba T, Hirose K, Manstein DJ. The dynamin A ring complex: molecular organization and nucleotide- dependent conformational changes. Embo J. 2002;21:240–250. doi: 10.1093/emboj/21.3.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Niemann HH, Knetsch ML, Scherer A, Manstein DJ, Kull FJ. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. Embo J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash B, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C. Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature. 2000;403:567–571. doi: 10.1038/35000617. [DOI] [PubMed] [Google Scholar]
- 7.Song BD, Schmid SL. A molecular motor or a regulator? Dynamin’s in a class of its own. Biochemistry. 2003;42:1369–1376. doi: 10.1021/bi027062h. [DOI] [PubMed] [Google Scholar]
- 8.Damke H, Binns DD, Ueda H, Schmid SL, Baba T. Dynamin GTPase domain mutants block endocytic vesicle formation at morphologically distinct stages. Mol Biol Cell. 2001;12:2578–2589. doi: 10.1091/mbc.12.9.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eccleston JF, Binns DD, Davis CT, Albanesi JP, Jameson DM. Oligomerization and kinetic mechanism of the dynamin GTPase. Eur Biophys J. 2002;31:275–282. doi: 10.1007/s00249-002-0226-2. [DOI] [PubMed] [Google Scholar]
- 10.van der Bliek AM. Functional diversity in the dynamin family. Trends Cell Biol. 1999;9:96–102. doi: 10.1016/s0962-8924(98)01490-1. [DOI] [PubMed] [Google Scholar]
- 11.Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM. A model for dynamin self-assembly based on binding between three different protein domains. J Biol Chem. 1999;274:14942–14947. doi: 10.1074/jbc.274.21.14942. [DOI] [PubMed] [Google Scholar]
- 12.Sever S, Muhlberg AB, Schmid SL. Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature. 1999;398:481–486. doi: 10.1038/19024. [DOI] [PubMed] [Google Scholar]
- 13.Low HH, Lowe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
- 15.Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otsuga D, Keegan BR, Brisch E, Thantcher JW, Hermann GJ, Bleazard W, Shaw J. The dynamin GTPase, Dnm1p, controls mitochondrial morphology in yeast. J Cell Biol. 1998;143:333–349. doi: 10.1083/jcb.143.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legesse-Miller A, Massol RH, Kirchhausen T. Constriction and dnm1p recruitment are distinct processes in mitochondrial fission. Mol Biol Cell. 2003;14:1953–1963. doi: 10.1091/mbc.E02-10-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naylor K, Ingerman E, Okreglak V, Marino M, Hinshaw JE, Nunnari J. Mdv1 interacts with assembled dnm1 to promote mitochondrial division. J Biol Chem. 2006;281:2177–2183. doi: 10.1074/jbc.M507943200. [DOI] [PubMed] [Google Scholar]
- 20.Smirnova E, Griparic L, Shurland DL, van Der Bliek AM. Dynamin-related protein drp1 is required for mitochondrial division in mammalian cells. Mol Biol Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labrousse AM, Zappaterra MD, Rube DA, van der Bliek AM. C. elegans dynamin-related protein DRP-1 controls severing of the mitochondrial outer membrane. Mol Cell. 1999;4:815–826. doi: 10.1016/s1097-2765(00)80391-3. [DOI] [PubMed] [Google Scholar]
- 22.Wasiak S, Zunino R, McBride HM. Bax/Bak promote sumoylation of DRP1 and its stable association with mitochondria during apoptotic cell death. J Cell Biol. 2007;177:439–450. doi: 10.1083/jcb.200610042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jensen RE, Hobbs AE, Cerveny KL, Sesaki H. Yeast mitochondrial dynamics: fusion, division, segregation, and shape. Microsc Res Tech. 2000;51:573–583. doi: 10.1002/1097-0029(20001215)51:6<573::AID-JEMT7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- 26.Yoon Y, Pitts KR, McNiven MA. Mammalian dynamin-like protein DLP1 tubulates membranes. Mol Biol Cell. 2001;12:2894–2905. doi: 10.1091/mbc.12.9.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bereiter-Hahn J, Voth M. Dynamics of mitochondria in living cells: shape changes, dislocations, fusion, and fission of mitochondria. Microsc Res Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 28.Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- 29.Danino D, Moon KH, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J Struct Biol. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 31.Mozdy AD, McCaffery JM, Shaw JM. Dnm1p GTPase-mediated mitochondrial fission is a multi-step process requiring the novel integral membrane component Fis1p. J Cell Biol. 2000;151:367–379. doi: 10.1083/jcb.151.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki M, Jeong SY, Karbowski M, Youle RJ, Tjandra N. The solution structure of human mitochondria fission protein Fis1 reveals a novel TPR-like helix bundle. J Mol Biol. 2003;334:445–458. doi: 10.1016/j.jmb.2003.09.064. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki M, Neutzner A, Tjandra N, Youle RJ. Novel structure of the N terminus in yeast Fis1 correlates with a specialized function in mitochondrial fission. J Biol Chem. 2005;280:21444–21452. doi: 10.1074/jbc.M414092200. [DOI] [PubMed] [Google Scholar]
- 34.Dohm JA, Lee SJ, Hardwick JM, Hill RB, Gittis AG. Cytosolic domain of the human mitochondrial fission protein fis1 adopts a TPR fold. Proteins. 2004;54:153–156. doi: 10.1002/prot.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tieu Q, Nunnari J. Mdv1p is a WD repeat protein that interacts with the dynamin-related GTPase, Dnm1p, to trigger mitochondrial division. J Cell Biol. 2000;151:353–365. doi: 10.1083/jcb.151.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cerveny KL, Jensen RE. The WD-repeats of Net2p interact with Dnm1p and Fis1p to regulate division of mitochondria. Mol Biol Cell. 2003;14:4126–4139. doi: 10.1091/mbc.E03-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griffin EE, Graumann J, Chan DC. The WD40 protein Caf4p is a component of the mitochondrial fission machinery and recruits Dnm1p to mitochondria. J Cell Biol. 2005;170:237–248. doi: 10.1083/jcb.200503148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karren MA, Coonrod EM, Anderson TK, Shaw JM. The role of Fis1p-Mdv1p interactions in mitochondrial fission complex assembly. J Cell Biol. 2005;171:291–301. doi: 10.1083/jcb.200506158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wells RC, Picton LK, Williams SC, Tan FJ, Hill RB. Direct binding of the dynamin-like GTPase, Dnm1, to mitochondrial dynamics protein Fis1 is negatively regulated by the Fis1 N-terminal arm. J Biol Chem. 2007;282:33769–33775. doi: 10.1074/jbc.M700807200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y, Chan DC. Structural basis for recruitment of mitochondrial fission complexes by Fis1. Proc Natl Acad Sci U S A. 2007;104:18526–18530. doi: 10.1073/pnas.0706441104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cerveny KL, McCaffery JM, Jensen RE. Division of mitochondria requires a novel DNM1-interacting protein. Net2p, Mol Biol Cell. 2001;12:309–321. doi: 10.1091/mbc.12.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tieu Q, Okreglak V, Naylor K, Nunnari J. The WD repeat protein, Mdv1p, functions as a molecular adaptor by interacting with Dnm1p and Fis1p during mitochondrial fission. J Cell Biol. 2002;158:445–452. doi: 10.1083/jcb.200205031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schauss AC, Bewersdorf J, Jakobs S. Fis1p and Caf4p, but not Mdv1p, determine the polar localization of Dnm1p clusters on the mitochondrial surface. J Cell Sci. 2006;119:3098–3106. doi: 10.1242/jcs.03026. [DOI] [PubMed] [Google Scholar]
- 44.Cerveny KL, Studer SL, Jensen RE, Sesaki H. Yeast mitochondrial division and distribution require the cortical num1 protein. Dev Cell. 2007;12:363–375. doi: 10.1016/j.devcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Kormanec J, Schaaff-Gerstenschlager I, Zimmermann FK, Perecko D, Kuntzel H. Nuclear migration in Saccharomyces cerevisiae is controlled by the highly repetitive 313 kDa NUM1 protein. Mol Gen Genet. 1991;230:277–287. doi: 10.1007/BF00290678. [DOI] [PubMed] [Google Scholar]
- 46.Farkasovsky M, Kuntzel H. Yeast Num1p associates with the mother cell cortex during S/G2 phase and affects microtubular functions. J Cell Biol. 1995;131:1003–1014. doi: 10.1083/jcb.131.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Farkasovsky M, Kuntzel H. Cortical Num1p interacts with the dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon Y, Krueger EW, Oswald BJ, McNiven MA. The mitochondrial protein hFis1 regulates mitochondrial fission in mammalian cells through an interaction with the dynamin-like protein DLP1. Mol Cell Biol. 2003;23:5409–4520. doi: 10.1128/MCB.23.15.5409-5420.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu T, Fox RJ, Burwell LS, Yoon Y. Regulation of mitochondrial fission and apoptosis by the mitochondrial outer membrane protein hFis1. J Cell Sci. 2005;118:4141–4151. doi: 10.1242/jcs.02537. [DOI] [PubMed] [Google Scholar]
- 50.Han XJ, Lu YF, Li SA, Kaitsuka T, Sato Y, Tomizawa K, Nairn AC, Takei K, Matsui H, Matsushita M. CaM kinase I alpha-induced phosphorylation of Drp1 regulates mitochondrial morphology. J Cell Biol. 2008;182:573–585. doi: 10.1083/jcb.200802164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee YJ, Jeong SY, Karbowski M, Smith CL, Youle RJ. Roles of the mammalian mitochondrial fission and fusion mediators Fis1, Drp1, and Opa1 in apoptosis. Mol Biol Cell. 2004;15:5001–5011. doi: 10.1091/mbc.E04-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank S, Gaume B, Bergmann-Leitner ES, Leitner W, Robert EG, Catez F, Smith CL, Youle RJ. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev Cell. 2001;1:515–525. doi: 10.1016/s1534-5807(01)00055-7. [DOI] [PubMed] [Google Scholar]
- 53.Breckenridge DG, Stojanovic M, Marcellus RC, Shore GC. Caspase cleavage product of BAP31 induces mitochondrial fission through endoplasmic reticulum calcium signals, enhancing cytochrome c release to the cytosol. J Cell Biol. 2003;160:1115–1127. doi: 10.1083/jcb.200212059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Germain M, Mathai JP, McBride HM, Shore GC. Endoplasmic reticulum BIK initiates DRP1-regulated remodelling of mitochondrial cristae during apoptosis. Embo J. 2005;24:1546–1556. doi: 10.1038/sj.emboj.7600592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Vos KJ, Allan VJ, Grierson AJ, Sheetz MP. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr Biol. 2005;15:678–683. doi: 10.1016/j.cub.2005.02.064. [DOI] [PubMed] [Google Scholar]
- 56.Varadi A, Johnson-Cadwell LI, Cirulli V, Yoon Y, Allan VJ, Rutter GA. Cytoplasmic dynein regulates the subcellular distribution of mitochondria by controlling the recruitment of the fission factor dynamin-related protein-1. J Cell Sci. 2004;117:4389–4400. doi: 10.1242/jcs.01299. [DOI] [PubMed] [Google Scholar]
- 57.Stojanovski D, Koutsopoulos OS, Okamoto K, Ryan MT. Levels of human Fis1 at the mitochondrial outer membrane regulate mitochondrial morphology. J Cell Sci. 2004;117:1201–1210. doi: 10.1242/jcs.01058. [DOI] [PubMed] [Google Scholar]
- 58.James DI, Parone PA, Mattenberger Y, Martinou JC. hFis1, a novel component of the mammalian mitochondrial fission machinery. J Biol Chem. 2003;278:36373–36379. doi: 10.1074/jbc.M303758200. [DOI] [PubMed] [Google Scholar]
- 59.Bhar D, Karren MA, Babst M, Shaw JM. Dimeric Dnm1-G385D interacts with Mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J Biol Chem. 2006;281:17312–17320. doi: 10.1074/jbc.M513530200. [DOI] [PubMed] [Google Scholar]
- 60.Karbowski M, Jeong SY, Youle RJ. Endophilin B1 is required for the maintenance of mitochondrial morphology. J Cell Biol. 2004;166:1027–1039. doi: 10.1083/jcb.200407046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cuddeback SM, Yamaguchi H, Komatsu K, Miyashita T, Yamada M, Wu C, Singh S, Wang HG. Molecular cloning and characterization of Bif-1. A novel Src homology 3 domain-containing protein that associates with Bax. J Biol Chem. 2001;276:20559–20565. doi: 10.1074/jbc.M101527200. [DOI] [PubMed] [Google Scholar]
- 62.Pierrat B, Simonen M, Cueto M, Mestan J, Ferrigno P, Heim J. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics. 2001;71:222–234. doi: 10.1006/geno.2000.6378. [DOI] [PubMed] [Google Scholar]
- 63.Ringstad N, Gad H, Low P, Di Paolo G, Brodin L, Shupliakov O, De Camilli P. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron. 1999;24:143–154. doi: 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 64.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. Mechanism of endophilin N-BAR domain-mediated membrane curvature. Embo J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 66.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niemann A, Ruegg M, La Padula V, Schenone A, Suter U. Ganglioside-induced differentiation associated protein 1 is a regulator of the mitochondrial network: new implications for Charcot-Marie-Tooth disease. J Cell Biol. 2005;170:1067–1078. doi: 10.1083/jcb.200507087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tondera D, Czauderna F, Paulick K, Schwarzer R, Kaufmann J, Santel A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J Cell Sci. 2005;118:3049–3059. doi: 10.1242/jcs.02415. [DOI] [PubMed] [Google Scholar]
- 70.Tondera D, Santel A, Schwarzer R, Dames S, Giese K, Klippel A, Kaufmann J. Knockdown of MTP18, a novel phosphatidylinositol 3-kinase-dependent protein, affects mitochondrial morphology and induces apoptosis. J Biol Chem. 2004;279:31544–31555. doi: 10.1074/jbc.M404704200. [DOI] [PubMed] [Google Scholar]
- 71.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–1022. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yonashiro R, Ishido S, Kyo S, Fukuda T, Goto E, Matsuki Y, Ohmura-Hoshino M, Sada K, Hotta H, Yamamura H, Inatome R, Yanagi S. A novel mitochondrial ubiquitin ligase plays a critical role in mitochondrial dynamics. Embo J. 2006;25:3618–3626. doi: 10.1038/sj.emboj.7601249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Harder Z, Zunino R, McBride H. Sumo1 conjugates mitochondrial substrates and participates in mitochondrial fission. Curr Biol. 2004;14:340–345. doi: 10.1016/j.cub.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 75.Zunino R, Schauss A, Rippstein P, Andrade-Navarro M, McBride HM. The SUMO protease SENP5 is required to maintain mitochondrial morphology and function. J Cell Sci. 2007;120:1178–1188. doi: 10.1242/jcs.03418. [DOI] [PubMed] [Google Scholar]
- 76.Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 77.Cereghetti GM, Stangherlin A, Martins de Brito O, Chang CR, Blackstone C, Bernardi P, Scorrano L. Dephosphorylation by calcineurin regulates translocation of Drp1 to mitochondria. Proc Natl Acad Sci U S A. 2008;105:15803–15808. doi: 10.1073/pnas.0808249105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chang CR, Blackstone C. Cyclic AMP-dependent protein kinase phosphorylation of Drp1 regulates its GTPase activity and mitochondrial morphology. J Biol Chem. 2007;282:21583–21587. doi: 10.1074/jbc.C700083200. [DOI] [PubMed] [Google Scholar]
- 79.Cribbs JT, Strack S. Reversible phosphorylation of Drp1 by cyclic AMP-dependent protein kinase and calcineurin regulates mitochondrial fission and cell death. EMBO Rep. 2007;8:939–944. doi: 10.1038/sj.embor.7401062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hanekamp T, Thorsness MK, Rebbapragada I, Fisher EM, Seebart C, Darland MR, Coxbill JA, Updike DL, Thorsness PE. Maintenance of mitochondrial morphology is linked to maintenance of the mitochondrial genome in Saccharomyces cerevisiae. Genetics. 2002;162:1147–1156. doi: 10.1093/genetics/162.3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gorsich SW, Shaw JM. Importance of mitochondrial dynamics during meiosis and sporulation. Mol Biol Cell. 2004;15:4369–4381. doi: 10.1091/mbc.E03-12-0875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aldridge AC, Benson LP, Siegenthaler MM, Whigham BT, Stowers RS, Hales KG. Roles for Drp1, a dynamin-related protein, and milton, a kinesin-associated protein, in mitochondrial segregation, unfurling and elongation during Drosophila spermatogenesis. Fly (Austin) 2007;1:38–46. doi: 10.4161/fly.3913. [DOI] [PubMed] [Google Scholar]
- 83.Hollenbeck PJ. Mitochondria and neurotransmission: evacuating the synapse. Neuron. 2005;47:331–333. doi: 10.1016/j.neuron.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 85.Verstreken P, Ly CV, Venken KJ, Koh TW, Zhou Y, Bellen HJ. Synaptic mitochondria are critical for mobilization of reserve pool vesicles at Drosophila neuromuscular junctions. Neuron. 2005;47:365–378. doi: 10.1016/j.neuron.2005.06.018. [DOI] [PubMed] [Google Scholar]
- 86.Waterham HR, Koster J, van Roermund CW, Mooyer PA, Wanders RJ, Leonard JV. A lethal defect of mitochondrial and peroxisomal fission. N Engl J Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 87.Lee S, Jeong SY, Lim WC, Kim S, Park YY, Sun X, Youle RJ, Cho H. Mitochondrial fission and fusion mediators, hFis1 and OPA1, modulate cellular senescence. J Biol Chem. 2007;282:22977–22983. doi: 10.1074/jbc.M700679200. [DOI] [PubMed] [Google Scholar]
- 88.Yoon YS, Yoon DS, Lim IK, Yoon SH, Chung HY, Rojo M, Malka F, Jou MJ, Martinou JC, Yoon G. Formation of elongated giant mitochondria in DFO-induced cellular senescence: involvement of enhanced fusion process through modulation of Fis1. J Cell Physiol. 2006;209:468–480. doi: 10.1002/jcp.20753. [DOI] [PubMed] [Google Scholar]
- 89.Twig G, Elorza A, Molina AJ, Mohamed H, Wikstrom JD, Walzer G, Stiles L, Haigh SE, Katz S, Las G, Alroy J, Wu M, Py BF, Yuan J, Deeney JT, Corkey BE, Shirihai OS. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. Embo J. 2008;27:433–446. doi: 10.1038/sj.emboj.7601963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Parone PA, Da Cruz S, Tondera D, Mattenberger Y, James DI, Maechler P, Barja F, Martinou JC. Preventing mitochondrial fission impairs mitochondrial function and leads to loss of mitochondrial DNA. PLoS ONE. 2008;3:e3257. doi: 10.1371/journal.pone.0003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Twig G, Hyde B, Shirihai OS. Mitochondrial fusion, fission and autophagy as a quality control axis: the bioenergetic view. Biochim Biophys Acta. 2008;1777:1092–1097. doi: 10.1016/j.bbabio.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell. 2003;112:481–490. doi: 10.1016/s0092-8674(03)00116-8. [DOI] [PubMed] [Google Scholar]
- 93.Antignani A, Youle RJ. How do Bax and Bak lead to permeabilization of the outer mitochondrial membrane? Curr Opin Cell Biol. 2006;18:685–689. doi: 10.1016/j.ceb.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 94.Chipuk JE, Bouchier-Hayes L, Green DR. Mitochondrial outer membrane permeabilization during apoptosis: the innocent bystander scenario. Cell Death Differ. 2006;13:1396–1402. doi: 10.1038/sj.cdd.4401963. [DOI] [PubMed] [Google Scholar]
- 95.Cassidy-Stone A, Chipuk JE, Ingerman E, Song C, Yoo C, Kuwana T, Kurth MJ, Shaw JT, Hinshaw JE, Green DR, Nunnari J. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev Cell. 2008;14:193–204. doi: 10.1016/j.devcel.2007.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karbowski M, Lee YJ, Gaume B, Jeong SY, Frank S, Nechushtan A, Santel A, Fuller M, Smith CL, Youle RJ. Spatial and temporal association of Bax with mitochondrial fission sites, Drp1, and Mfn2 during apoptosis. J Cell Biol. 2002;159:931–938. doi: 10.1083/jcb.200209124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jagasia R, Grote P, Westermann B, Conradt B. DRP-1-mediated mitochondrial fragmentation during EGL-1-induced cell death in C. elegans. Nature. 2005;433:754–760. doi: 10.1038/nature03316. [DOI] [PubMed] [Google Scholar]
- 98.Goyal G, Fell B, Sarin A, Youle RJ, Sriram V. Role of mitochondrial remodeling in programmed cell death in Drosophila melanogaster. Dev Cell. 2007;12:807–816. doi: 10.1016/j.devcel.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Alirol E, James D, Huber D, Marchetto A, Vergani L, Martinou JC, Scorrano L. The mitochondrial fission protein hFis1 requires the endoplasmic reticulum gateway to induce apoptosis. Mol Biol Cell. 2006;17:4593–4605. doi: 10.1091/mbc.E06-05-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dimmer KS, Navoni F, Casarin A, Trevisson E, Endele S, Winterpacht A, Salviati L, Scorrano L. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum Mol Genet. 2008;17:201–214. doi: 10.1093/hmg/ddm297. [DOI] [PubMed] [Google Scholar]


